Abstract
Background and Aims
The Araceae are one of the most diverse monocot families with numerous morphological and ecological novelties. Plastid and mitochondrial genes have been used to investigate the phylogeny and to interpret shifts in the pollination biology and biogeography of the Araceae. In contrast, the role of whole-genome duplication (WGD) in the evolution of eight subfamilies remains unclear.
Methods
New transcriptomes or low-depth whole-genome sequences of 65 species were generated through Illumina sequencing. We reconstructed the phylogenetic relationships of Araceae using concatenated and species tree methods, and then estimated the age of major clades using TreePL. We inferred the WGD events by Ks and gene tree methods. We investigated the diversification patterns applying time-dependent and trait-dependent models. The expansions of gene families and functional enrichments were analysed using CAFE and InterProScan.
Key Results
Gymnostachydoideae was the earliest diverging lineage followed successively by Orontioideae, Lemnoideae and Lasioideae. In turn, they were followed by the clade of ‘bisexual climbers’ comprised of Pothoideae and Monsteroideae, which was resolved as the sister to the unisexual flowers clade of Zamioculcadoideae and Aroideae. A special WGD event ψ (psi) shared by the True-Araceae clade occurred in the Early Cretaceous. Net diversification rates first declined and then increased through time in the Araceae. The best diversification rate shift along the stem lineage of the True-Araceae clade was detected, and net diversification rates were enhanced following the ψ-WGD. Functional enrichment analyses revealed that some genes, such as those encoding heat shock proteins, glycosyl hydrolase and cytochrome P450, expanded within the True-Araceae clade.
Conclusions
Our results improve our understanding of aroid phylogeny using the large number of single-/low-copy nuclear genes. In contrast to the Proto-Araceae group and the lemnoid clade adaption to aquatic environments, our analyses of WGD, diversification and functional enrichment indicated that WGD may play a more important role in the evolution of adaptations to tropical, terrestrial environments in the True-Araceae clade. These insights provide us with new resources to interpret the evolution of the Araceae.
Keywords: Araceae, diversification, phylogenomics, WGDs, adaptation
INTRODUCTION
Molecular phylogenetic relationships are inferred, most frequently, by plastid and mitochondrial genome datasets. However, the exclusive use of uniparental organellar genes may result in biases and errors in some phylogenetic reconstructions (Davis et al., 2014). Nuclear genes with biparental inheritance often provide alternative evidence (Zeng et al., 2014; Kapli et al., 2020). In particular, recent advances in high-throughput sequencing technology have greatly facilitated the use of nuclear genes based on genome and transcriptome datasets. In ongoing research programmes on seed plants, they have been employed successfully to investigate phylogenetic relationships, whole-genome duplication (WGD) events, the history of diversification within lineages and the evolution of adaptations to extreme environments. Some recent studies (Ren et al., 2018; Leebens-Mack et al., 2019) on the Caryophyllales (N. Wang et al., 2019), Asteraceae (Huang et al., 2016; Mandel et al., 2019), Cucurbitaceae (Guo et al., 2020), Fabaceae (Koenen et al., 2021; Zhao et al., 2021) and Theaceae (Q. Zhang et al., 2022) have deepened our understanding of evolutionary pathways in angiosperm phylogeny.
Whole-genome duplication is a common but extreme mechanism of gene duplication (GD) in angiosperms, resulting in additional copies of the entire genome in the same cell (Panchy et al., 2016; Van De Peer et al., 2017). To date, at least three waves of WGD events have occurred in angiosperms, and been named tau (τ), sigma (σ) and rho (ρ), and gamma (γ), beta (β) and alpha (α) for monocots and eudicots, respectively (Jiao et al., 2011, 2012, 2014; Alix et al., 2017). WGDs have long been regarded as a major evolutionary force in plants providing raw genetic variation for natural selection, allowing generations to enhance adaptation and possibly drive speciation (Soltis et al., 2015; Mandakova and Lysak, 2018; Levin, 2019; Van de Peer et al., 2021). Wu et al. (2020) argued that ancient WGD events during the Cretaceous–Paleocene boundary [K–Pg, approx. 66 million years ago (Ma)] were associated with adaptations of angiosperms to cool and dark environments (Vanneste et al., 2014). Cai et al. (2019) suggested that past WGDs in the Malphigiales correlate with the enhancement of plant survival during climatic changes in the course of the Paleocene–Eocene transition (approx. 56–54 Ma) and the late Miocene (approx. 7 Ma) (Sessa, 2019). Additionally, GDs (including WGDs) in the Pooideae were associated with adaptation to lower temperatures and species diversification during the mid-Eocene (approx. 46 Ma) to late Oligocene (approx. 27 Ma) transition (L. Zhang et al., 2022).
Whole-genome duplications have been regarded as evolutionary dead-ends due to the negative effects they may produce. This includes severely abnormal meiosis and instable genomes (Stebbins, 1950; Arrigo and Barker, 2012). Some plant groups with comparatively recent trends in polyploidy have lower net diversification rates than their diploid relatives (Mayrose et al., 2011). Some recent studies failed to show a positive correlation between rates of WGDs and rates of diversification within lineages (Estep et al., 2014; Smith et al., 2018; N. Wang et al., 2019; Huang et al., 2020; Stull et al., 2021). In contrast, other studies suggested that WGDs are essential for speciation (Scarpino et al., 2014; Edger et al., 2015; Godden et al., 2019; Han et al., 2020; L. Zhang et al., 2022). Tank et al. (2015) showed that while increased diversification rates are rarely associated directly with WGD events, they usually follow them after a lag period, as explained by the WGD Radiation Lag-Time Model (Schranz et al., 2012). Younger WGDs are more likely to be followed by upshifts in diversification rates than older WGDs (Landis et al., 2018). However, the WGD events in these studies usually occurred after the K–Pg boundary. Although L. Zhang et al. (2020) reviewed ancient WGD events that occurred 100–120 Ma, the role of WGD events during two Ocean Anoxic Events (OAEs 1b and 1d) in the Early Cretaceous (approx. 100–110 Ma) has not received much investigation (Clark and Donoghue, 2018; Benton et al., 2022).
The Araceae family is defined currently as consisting of approx. 144 genera and 3645 species. Therefore, it is one of the largest monocot families (Croat, 2019). Previous studies have indicated that the ancestral habitats of ancestral aroids were aquatic and/or associated with wetlands. The complex programme of evolution within this family involved diversification of more aquatic life forms but also a greater trend towards colonization of terrestrial habits including geophytic, epiphytic and climbing plants (Cabrera et al., 2008; Cusimano et al., 2011a). Additionally, biogeographical analyses have also suggested that early lineages, including the Gymnostachydoideae and Orontioideae, were Laurasian and temperate in origin, making only a recent entry, but spectacular radiation into tropical biomes in Africa, South America, South-East Asia and Australasia (Nauheimer et al., 2012). Today, this family includes only a few temperate genera while 90 % of genera and 95 % of species are restricted to the tropics and sub-tropics (Mayo et al., 1997). To date, three groups representing eight subfamilies are recognized in the Araceae. Their current segregation into the Proto-Araceae group (Gymnostachydoideae and Orontioideae), lemnoids (Lemnoideae) and the True-Araceae clade (Lasioideae, Pothoideae, Monsteroideae, Zamioculcadoideae and Aroideae) are based on previous studies combining morphological traits and organellar genes (Mayo et al., 1997; Cusimano et al., 2011a; Henriquez et al., 2014). In Araceae, the Proto-Araceae group and lemnoids account for only 6 % of all genera and 1.3 % of all species. Almost all of their species are restricted to swampy-aquatic habitats (Cusimano et al., 2011a; Lee et al., 2019). In contrast, the True-Araceae clade accounts for 94 % of all genera and 98.7 % of all species. The majority take terrestrial-epiphytic habits with centres of diversity in tropical rain forests (Nauheimer et al., 2012). Currently, based on the genome and transcriptome sequences, some studies have detected WGD events from Colocasia esculenta (L.) Schott, Pistia stratiotes L., Typhonium blumei Nicolson & Sivad., Spirodela polyrhiza (L.) Schleid. and Lemna minuta Kunth in the True-Araceae clade, the Proto-Araceae group and the lemnoid clade (Cusimano et al., 2011b; Wang et al., 2014; Li and Barker, 2020; Yin et al., 2021). However, few studies have investigated the phylogeny, the roles of WGDs and the diversification history of the Araceae using large-scale nuclear gene datasets.
In this study, we generated 64 new transcriptome datasets and one new genome survey in the Araceae, and retrieved 22 transcriptome and three genome sequences from public databases to better understand evolution in this large family. Our goals are to (1) reconstruct phylogenetic relationships among eight subfamilies using large-scale nuclear genes; (2) identify additional WGDs; (3) infer the diversification history within the family; and (4) evaluate the potential roles of WGDs in the evolution of key traits and novelties.
MATERIALS AND METHODS
Taxon sampling and transcriptome sequencing
We sampled 81 species representing 57 genera in the eight subfamilies of the Araceae, and nine additional species to represent outgroups (see Supplementary data Table S1). At least one representative from 43 of 44 clades identified by Cusimano et al. (2011a) were included. Three genome and 22 transcriptome datasets were downloaded from three public databases: Phytozome (http://phytozome.jgi.doe.gov), CoGe (https://genomevolution.org) and NCBI (http://www.ncbi.nlm.nih.gov/). In turn, we generated 64 new transcriptome datasets and one new genome survey. Following the collection of young leaves, we extracted total RNA using the SIGMA plant total RNA kit (Tiangen, Beijing, China). For Gymnostachys anceps R.Br., in the absence of fresh leaves, we used herbarium specimens (the Missouri Botanical Garden, Collection Number: 57203), extracting the total DNA using the CTAB (cetyltrimethylammonium bromide) method. For the library construction, Illumina HiSeq4000 sequencing and quality control was performed at Novogene Corporation, Beijing, China. We obtained published species numbers from the International Aroid Society (Croat, 2019). All clean reads in the study were deposited on the NCBI Short Reads Archive under the BioProject Accession Number PRJNA813353.
Orthologue identification, phylogeny inference and dating
We sequenced 6 Gb transcriptome sequences for each species except G. anceps. These transcriptome sequences were de novo assembled using Trinity-v2.0.6 (Haas et al., 2013). From each of the transcriptomes, CD-HIT v4.6 with the parameter: -c 0.99 -n 10 was carried out to obtain the non-redundant transcripts (Li and Godzik, 2006). TransDecoder-v5.1.0 was employed subsequently to predict amino acid sequences (Haas et al., 2013), but sequences <150 bp in length were discarded. For G. anceps, we sequenced 70 Gb genome survey datasets. De novo assembly was performed using platanus v1.2.4 with default parameters (Kajitani et al., 2014). RepeatMasker v4.1.0 was used to identify tandem repeats and transposable elements. Gene annotations were obtained using Augustus v3.3.3 and Exonerate v2.2.0, and then the gene set was integrated using EVidenceModeller v1.1.1 (Haas et al., 2008). The completeness of transcripts was assessed by comparison against the Benchmarking Universal Single-Copy Orthologs (BUSCO) datasets (Simao et al., 2015). Orthologous clusters were identified as described in Zhao et al. (2016) using 4180 core orthologues from ‘primer taxa’ [Arabidopsis thaliana (L.) Heynh., Glycine max (L.) Merr., Medicago truncatula Gaertn., Populus trichocarpa Torr. & A.Gray, Oryza sativa L., Sorghum bicolor (L.) Moench, Solanum lycopersicum L., Vitis vinifera L. and Zea mays L.] as seeds in HaMStR-v13 with strict parameters (-representative, -strict, -eval_limit = 0.00001 and -rbh) (Ebersberger et al., 2009). To reduce missing data, we retained only those clusters that included at least 60 % of the species, and then obtained 1081 clusters from 90 species.
Each cluster was aligned using MAFFT-v7.4 with default parameters (Katoh et al., 2005). Poorly aligned regions were trimmed by trimAlv1.4 with the parameter: -automated1 (Capella-Gutierrez et al., 2009). Final alignments were checked manually in MEGA-X (Kumar et al., 2018). Concatenation and coalescence methods were employed to infer phylogenetic relationships. For the concatenation method, all trimmed alignments were concatenated into a supermatrix using SCaFoS (Roure et al., 2007). Phylogenetic reconstructions were built using RAxML-8.2 with a PROTCATJTT model as suggested by ProtTestv3.4 (Stamatakis, 2014). For the coalescence method, each gene tree was inferred using RAxML8.2 with the PROTCATJTT model, 200 bootstrap replicates (-b) and a rapid bootstrapping algorithm (-r). Phylogenetic relationships were then constructed from the best-scoring maximum likelihood (ML) gene trees using ASTRALv5.5 (Mirarab and Warnow, 2015). Based on 922 rooted gene trees, we used PhyParts software (Smith et al., 2015) to evaluate the conflicts of gene trees.
Molecular clock analysis was performed to estimate divergence times by the penalized likelihood (PL) method implemented in TreePL, which can handle large-scale phylogenetic datasets (Smith and O’Meara, 2012). To determine the optimal smoothing value, the cross-validation tested 13 smoothing values separated by one order of magnitude, starting at 1 × 10–6. To estimate the confidence intervals of dating, we generated 1000 ML bootstrap trees with branch lengths using RAxML8.2. A maximum clade credibility (MCC) tree was calculated in TreeAnnotator in BEASTv2.5 (Bouckaert et al., 2019). We employed five fossil constraints in our dating estimation (Supplementary data Table S2) according to Nauheimer et al. (2012), Magallon et al. (2015), Iles et al. (2015) and H.T. Li et al. (2019). The maximum and minimum ages of the roots were assigned as 125 and 113 Ma, respectively.
Ks-based method for WGD inference from single species
To identify WGD events in 81 species in the Araceae, we first employed the Ks method (the number of synonymous substitutions per synonymous site). We performed wgd packages for the estimation of the Ks distribution from each species. The pipeline was a simple command-line tool for the analysis and visualization of WGD events (Zwaenepoel and Van de Peer, 2019). With the pipeline of wgd packages, we implemented all-to-all BLASTP with an e-value cut-off of 1e-5 and then carried out Markov cluster (MCL) algorithm to construct gene families using ‘wgd mcl’. We then plotted Ks distributions by Ks values of gene families using ‘wgd ksd’ as fitted by Gaussian mixture models (GMMs) using ‘wgd mix’ in the wgd packages. We excluded Ks values of <0.05 to avoid the effects of isoforms from transcriptomes (H.F. Wang et al., 2019; Zwaenepoel et al., 2019).
The gene tree-based method of WGD inference from multiple species
To locate the placement of WGDs in our datasets, we employed a gene tree sorting and counting algorithm, the multi-taxon paleopolyploidy search (MAPS) tool (Li et al., 2015). For each MAPS analysis, we followed the steps of Leebens-Mack et al. (2019). To minimize topology errors in gene trees, we collected eight species to infer shared WGD events, including two species representing the Proto-Araceae group (G. anceps and Lysichiton americanus Hulten & H.St.John), one species representing the lemnoid clade (Lemna minor L.), four species representing the True-Araceae clade [Dracontium dressleri Croat., Spathiphyllum floribundum (Linden & André) N.E.Br., Zamioculcas zamiifolia (Lodd.) Engl. and Arisaema franchetianum Engl.] and one outgroup species Acorus calamus L. Gene families were clustered from these OrthoFinder-2.3.3 results under the parameters: -S diamond and -M msa (Emms and Kelly, 2019). Using Perl script, we obtained the gene families that contained at least one copy from each taxon and discarded the remaining clusters. PASTA-1.8.5 was run until it reached three iterations without improving the likelihood score for the automatic alignments and the phylogenetic reconstruction of gene families (Mirarab et al., 2015). For each iteration, we constructed the alignments using MAFFT, merged these alignments using Muscle and inferred the gene tree using RAxML. The parameters of each software were defaulted in PASTA. All the best scoring gene trees were inputted into MAPS software to locate the shared WGD events.
Time-dependent diversifications
Based on the time-calibrated phylogeny, we inferred the diversification rates and the diversification shifts across eight subfamilies using BAMM v.2.5 (Title and Rabosky, 2019). BAMM employed a reversible jump Markov chain Monte Carlo (rjMCMC) to automatically detect a vast universe of models for lineage diversification. BAMM can automatically explore the clade’s rate shifts and diversification dynamics (Rabosky et al., 2017). We pruned the outgroups and estimated the priors. We set default values = 1.0 for poissonRatePrior because of small trees (<500 tips). Incomplete taxon sampling of eight subfamilies was considered by applying clade-specific sampling fractions. We ran BAMM with four rjMCMC chains, each for 50 000 000 generations, sampling every 10 000 generations. The first 10 % of sampled data were discarded as burn-in. The convergence was assessed by plotting the log-likelihood trace of the MCMC output and computing the effective sample size values (>200). The configuration of the diversification rate shifts was estimated by the posterior distribution, and Bayes factors were used to compare with alternative diversification models. We analysed the output file of BAMM and plotted the mean phylorate and the specific clade rate-through-time (RTT) curve by the R package BAMMtools v2.16 (Rabosky, 2014).
State-dependent diversifications
We applied two state-dependent methods: BiSSE (Binary State Speciation and Extinction) (Maddison et al., 2007) and HiSSE (Hidden State Speciation and Extinction) to analyse diversification rates (Beaulieu and O’Meara, 2016). BiSSE allowed us to investigate the effect of the binary trait on diversification rates in the incompletely resolved phylogeny (FitzJohn et al., 2009). We conducted the BiSSE analyses in the R package Diversitree v0.9-11 based on the MCC tree (FitzJohn, 2012). Incomplete sampling was considered by clade-specific sampling fractions. For each trait, we used ML searches to compare four models: (1) full model allowed all variables to change independently; (2) λ0 ~ λ1 constrained speciation rates making them equal across states; (3) μ0 ~ μ1 allowed extinction rates to be equal across states; and (4) q01 ~ q10 constrained transition rates to be equal across states. To select the best-fitting model, we employed the lowest Akaike information criterion (AIC) values. We ran MCMC for 10 000 generations and sampled every 100 generations with an exponential prior at a rate of 1/(2r), where r was the diversification rate of the trait.
The HiSSE model extended the BiSSE models to account for unmeasured factors (i.e. hidden states) that could affect diversification rate estimations with the observed traits. Therefore, we also conducted an analysis of the diversification rate using HiSSE v1.9.5 (Herrera-Alsina et al., 2019). For HiSSE analyses, we implemented 25 models, comprising four BiSSE models, four trait-independent models and 17 HiSSE models following Beaulieu and O’Meara (2016). The best model was chosen according to the lowest ΔAIC values. We estimated the likeliest ancestral states for both internal nodes and tips of the phylogeny, and inferred the confidence interval of each parameter by sampling parameter values to ensure the difference of log-likelihood <2 in the ‘MarginRecon’ and ‘SupportRegion’ functions, respectively.
Gene family expansions and functional analyses
We employed CAFE v4.2.1 (Han et al., 2013) to infer the expansions from gene families that were clustered in eight species representing the Proto-Araceae group, the lemnoid clade and the True-Araceae clade using OrthoFinder-2.3.3 (Emms and Kelly, 2019). We performed InterProScan v5.41 to obtain functional annotation of the InterPro signatures and the Gene Ontology (GO) terms (Jones et al., 2014). The functional enrichment of GO terms was analysed by comparing the foreground sample (expanded gene families) with all annotated gene families based on Fisher’s exact test and Bonferroni correction (Ashburner et al., 2000).
RESULTS
Phylogenetic backbone of the Araceae
A total of 81 Araceae species were included in this study to represent all eight subfamilies, 57 out of 144 genera and 43 out of 44 clades. Among the 81 Araceae datasets, 65 were newly generated for this study. In addition, nine species from other monocot families were used as outgroup taxa for phylogenetic reconstruction. The non-redundant unigenes ranged in size from 10 407 bp to 72 972 bp. BUSCO recovery in assembled unigenes averaged 70 ± 11 % for the combination of the complete and fragmented orthologues (Supplementary data Table S1).
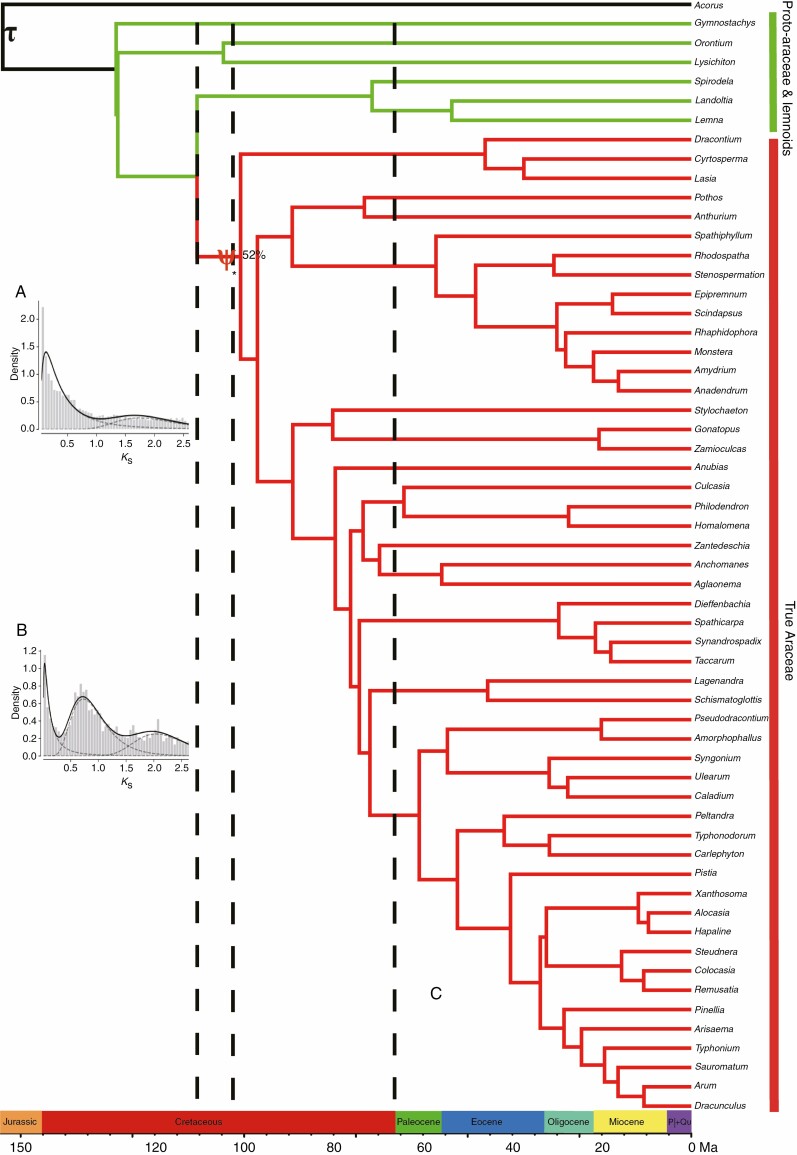
A total of 1081 orthologous groups were identified from 90 species for phylogenetic tree reconstructions. Sequence alignments of orthologous groups were concatenated, and were composed of 490 218 amino acid sites. The concatenated ML tree was estimated, and gained a highly supported phylogeny with bootstrap values of 100 for all nodes, excluding the Orontioideae node with a 58 % bootstrap value (Supplementary data Fig. S1). The maximum quartet support species tree was also inferred by using ASTRAL-II, and gained a similar phylogeny with high support, except for two nodes of the Orontioideae and Lasioideae with 31 % and 26 % bootstrap values, respectively (Supplementary data Fig. S2). It was worth noting that the concatenated ML tree and the maximum quartet support species tree were congruent with respect to the phylogenetic relationships of eight subfamilies (Fig. 1). In our results, the Gymnostachydoideae was the earliest diverging lineage within the family, successively followed by the Orontioideae, Lemnoideae and Lasioideae. In turn, they were followed by the ‘bisexual climbers’ clade comprised of the Pothoideae and Monsteroideae. These last two subfamilies were resolved as sisters to the clade with unisexual flowers, as comprised of the Zamioculcadoideae and Aroideae. The Lasioideae was the sister of a clade formed by the clades of the ‘bisexual climbers’ and the ‘unisexual flowers’ (Fig. 1). In addition, our results offered strong support for adding the genus Stylochaeton Lep. to the Zamioculcadoideae (Henriquez et al., 2014).
Fig. 1.
Phylogenetic tree of the Araceae using RAxML and ASTRAL methods based on 1081 orthologous groups from 60 representative species. Nodes with no values have 100 % bootstrap support. ‘-’ indicates that the two placements reconstructed by ASTRAL are inconsistent with the results inferred by RAxML.
Estimation of divergence time
The ages of the stem and crown groups for the Araceae in our estimation were approx. 131.6 and 128.8 Ma, with a 95 % highest probability density (HPD): 129.1–132.7 and 125.2–130.6 Ma, respectively. Our calculation was older than previous analyses based on chloroplast genes (Janssen and Bremer 2004; Nauheimer et al., 2012) (Supplementary data Figs S3 and S4). The Proto-Araceae group, the lemnoid clade and the True-Araceae clade diverged at 100.7–110.5 Ma from the early Cretaceous (Fig. 2). The stem age of the Gymnostachydoideae was estimated at 128.8 Ma (95 % HPD 125.2–130.6 Ma). The Orontioideae was estimated at 128.3 Ma (95 % HPD 123.9–129.8 Ma), the Lemnoideae at 110.5 Ma (95 % HPD 68.9.3–81.1 Ma), the Lasioideae at 110.5 Ma (95 % HPD 97.7–107.5 Ma), the Pothoideae at 89.2 Ma (95 % HPD 86.4–97.0 Ma), the Monsteroideae at 89.2 Ma (95 % HPD 86.4–97.0 Ma), the Zamioculcadoideae at 89.1 Ma (95 % HPD 86.5–97.0 Ma) and the Aroideae at 89.1Ma (95 % HPD 86.5–97.0 Ma; Supplementary data Figs S3 and S4). Our results suggested that the Araceae originated in the early Cretaceous (Hauterivian), and all eight subfamilies diverged before the K–Pg boundary.
Fig. 2.
The time-calibrated phylogenetic tree and evidence for WGD events in the Araceae. (A) Histograms of Ks distribution in the Proto-Araceae group and the lemnoid clade. (B) Histograms of Ks distribution of the True-Araceae clade. (C) The τ-WGD event was shared by the Araceae and other monocot families. The ψ-WGD event was shared by the True-Araceae clade. Fifty-two per cent of gene sub-trees were consistent with the species tree, and possibly supported a shared WGD by the True-Araceae clade using MAPS analyses. The three dotted lines indicate the OAE 1b, OAE 1d and K–Pg event from left to right.
Inferences of WGDs
Ks frequency plots suggested the presence of multiple WGD events for many aroids, in which peaks of Ks ranged from 0.60 to 2.05. From Ks frequency plots, we found that Araceae shared one round of ancient WGD events (τ) with other monocot families, consistent with results in previous studies (Jiao et al., 2014; Abramson et al., 2022). Interestingly, the True-Araceae clade may have shared the special WGD event (namely ψ), with Ks ranging between 0.60 and 0.95. However, the ψ-WGD event was not shared with the Proto-Araceae group and the lemnoid clade (Fig. 2), which have their own WGD events with peak Ks values of 0.60 for O. aquaticum and 1.23 for S. polyrhiza (Supplementary data Table S3 and Fig. S5).
We also used a gene tree mapping approach, MAPS, to test whether the True-Araceae clade shared the ψ-WGD event according to the methods of Leebens-Mack et al. (2019). Gene family clustering across eight species obtained 2309 clusters with at least one copy from each sampled species. At the node of the True-Araceae clade, >52 % of gene sub-trees were consistent with the species tree, supporting the ψ-WGD event in the True-Araceae clade across the phylogeny. The shared ψ-WGD event at this node was consistent with the observed result from single species Ks plots in the True-Araceae clade (Supplementary data Figs S5 and S6).
Diversification rates through time
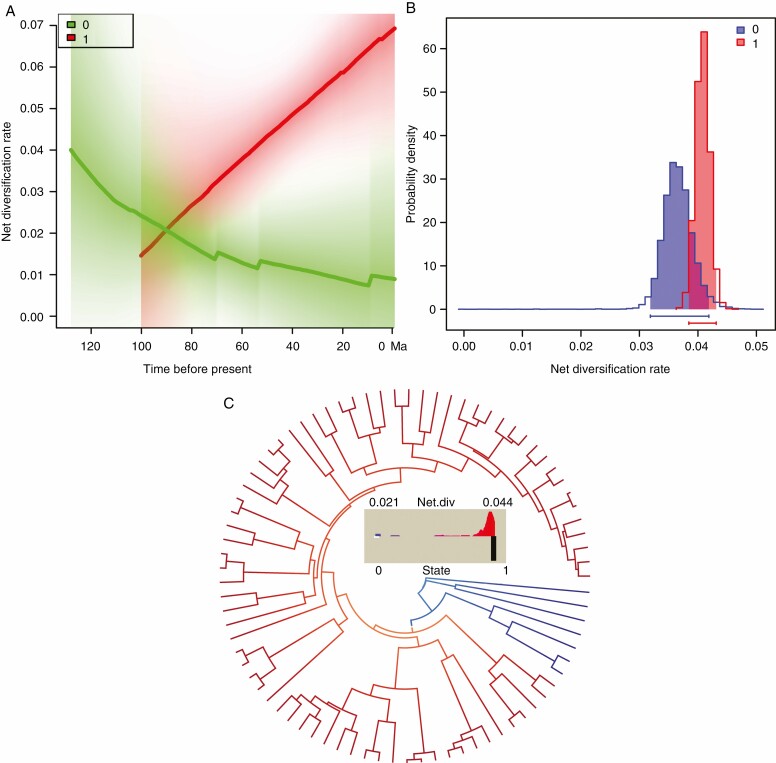
We estimated that the mean speciation rate, extinction rate and net diversification rate for the Araceae was 0.2991, 0.2527 and 0.0464 lineages per million years, respectively. Rate-through-time analysis suggested that the diversification of the Araceae began in the early Cretaceous, followed by three stages (Fig. 3A and see Supplementary data Fig. S7). The net diversification rate declined from the Barremian to the Cenomanian. However, the net diversification rate rose slowly from the Cenomanian to the Maastrichtian. Finally, the net diversification rate increased rapidly after the K–Pg boundary. The post burn-in posterior distribution indicated that the best shift occurred with a posterior probability (PP) of 0.42 (Supplementary data Table S4 and Fig. S8). The 95 % credible set of shift configurations and marginal shift probabilities favoured the model that included the best shift along branches with the True-Araceae clade (Supplementary data Figs S9 and S10), consistent with the previous study by Givnish et al. (2018).
Fig. 3.
Macroevolutionary dynamics of Araceae estimated by BAMM. (A) The pattern of net diversification rate across time. The rate-through-time plot indicates an increase in the diversification rate after the ψ-WGD event. (B) The placement of the best shift for the diversification of the Araceae. Branch colours represent the relative net diversification rates on the left side of the tree. Number 1 indicates the placement of the ψ-WGD event.
Diversification rates in the Proto-Araceae, lemnoids and True-Araceae
We compared net diversification rates between the Proto-Araceae group, the lemnoid clade and the True-Araceae clade. Using BAMM, we calculated the mean speciation rate, extinction rate and net diversification rate for the Proto-Araceae group and the lemnoid clade to be 0.1043, 0.0895 and 0.0148 lineages per million years, respectively (Supplementary data Fig. S11). The mean speciation rate, extinction rate and net diversification rate for the True-Araceae clade were 0.3481, 0.2937 and 0.0544 lineages per million years, respectively (Supplementary data Fig. S12). The BAMM analyses indicated that the net diversification rate was enhanced 3.67 times from the Proto-Araceae group and the lemnoid clade to the True-Araceae clade (Fig. 4A). We further corroborated the results by using BiSSE and HiSSE packages, respectively. In BiSSE analyses, the best-fitting BiSSE model was the full model with different speciation, extinction and transition rates (Table 1). In the best-fit model, both speciation and extinction rates were high for the True-Araceae clade (Supplementary data Figs S13 and S14), and the net diversification rate was approx. 2-fold higher than in the Proto-Araceae group and the lemnoid clade (Fig. 4B). In HiSSE analyses, the full HiSSE model with ‘τ0A = τ1A = τ0B, ε0A = ε1A = ε0B, q0B1B = q1B0B = 0 and all other equals’ was the best-fit model (Table 2). The circle-plot clearly showed a 2-fold higher mean net diversification rate in the True-Araceae clade compared with the Proto-Araceae group and the lemnoid clade (Fig. 4C).
Fig. 4.
Differences in net diversification rates for the Proto-Araceae group, the lemnoid clade and the True-Araceae clade using three methods. Number 0 indicates the Proto-Araceae group and the lemnoid clade. Number 1 indicates the True-Araceae clade. (A) Diversification rate-through-time plots estimated by BAMM across time. (B) Results of BiSSE analyses; net diversification rates under the best-fit model are shown. (C) Results of HiSSE analyses with net diversification rates under the best-fit model. Net diversification rates are indicated as colour shadings along branch edges (from blue to red).
Table 1.
Comparative fit of different BiSSE models for the Proto-Araceae group, the lemnoid clade and the True-Araceae clade using Diversitree packages*
| Model | LogL | AIC | ΔAIC |
|---|---|---|---|
| Full | –20.220 | 52.441 | 0 |
| Equal λ | –24.081 | 58.160 | 5.719 |
| Equal μ | –23.819 | 57.639 | 5.198 |
| Equal q | –23.681 | 57.362 | 4.921 |
*Bold represents the best model. LogL, the loglikelihood of the model; AIC, the Akaike information criterion.
Table 2.
Comparative fit of alternative models for the Proto-Araceae group, lemnoids and the True-Araceae clade using HiSSE packages*
| Model | LogL | AIC | ΔAIC |
|---|---|---|---|
| BiSSE model: all free | –370.707 | 753.413 | 29.862 |
| BiSSE model: ε0 = ε1 | –357.702 | 725.404 | 1.853 |
| BiSSE model: qs equal | –371.311 | 752.623 | 29.072 |
| BiSSE model: ε0 = ε1, qs equal | –361.871 | 731.741 | 8.19 |
| CID-2: qs equal | –360.573 | 731.145 | 7.594 |
| CID-2: εs, qs equal | –364.393 | 736.786 | 13.235 |
| CID-4: εs equal, qs equal | –361.436 | 741.874 | 18.323 |
| CID-4: qs equal | –361.735 | 734.470 | 10.919 |
| HiSSE full model | –355.575 | 743.150 | 19.599 |
| HiSSE: τ0A = τ1A, εs equal, qs equal | –360.583 | 733.165 | 9.614 |
| HiSSE: τ0A=τ1A, ε0A = ε1A, q0B1B = 0, q1B0B=0, all other qs equal | –367.084 | 744.168 | 20.617 |
| HiSSE: εs equal, q0B1B = 0, q1B0B = 0, all other qs equal | –369.235 | 746.471 | 22.92 |
| HiSSE: q0B1B = 0, q1B0B = 0, all other qs equal | –360.669 | 735.339 | 11.788 |
| HiSSE: τ0A = τ1A, εs equal, q0B1B = 0, q1B0B = 0, all other qs equal | –361.742 | 733.485 | 9.934 |
| HiSSE: τ0A = τ0B, ε0A = ε0B, q0B1B = 0, q1B0B = 0, all other q’s equal | –356.678 | 727.356 | 3.805 |
| HiSSE: τ0A = τ0B, εs equal, q0B1B = 0, q1B0B = 0, all other qs equal | –362.644 | 735.288 | 11.737 |
| HiSSE: εs equal, qs equal | –356.567 | 731.134 | 7.583 |
| HiSSE: qs equal | –358.793 | 729.586 | 6.035 |
| HiSSE: τ0A = τ0B, ε0A = ε0B, qs equal | –358.641 | 725.284 | 1.733 |
| HiSSE: τ0A = τ1A, ε0A = ε1A, qs equal | –356.621 | 727.242 | 3.691 |
| HiSSE: τ0A = τ1A = τ0B, εs equal, qs equal | –357.263 | 724.526 | 0.975 |
| HiSSE: τ0A = τ0B, εs equal, qs equal | –356.798 | 727.597 | 4.046 |
| HiSSE: τ0A = τ1A = τ0B, ε0A = ε1A= ε1B, qs equal | –358.835 | 727.671 | 4.12 |
| HiSSE: τ0A = τ1A = τ0B, ε0A = ε1A = ε0B, q0B1B = q1B0B = 0, all other qs equal | –356.775 | 723.551 | 0 |
| HiSSE: τ0A = τ1A = τ0B, εs equal, q0B1B = 0, q1B0B =0, all other qs equal | –356.691 | 731.382 | 7.831 |
*Bold represents the best model. LogL, the loglikelihood of the model; AIC, the Akaike information criterion.
Enrichment of gene functions in the True-Araceae clade
We identified >500 expanded gene families by CAFE in the True-Araceae clade (Supplementary data Table S5). With the annotation of InterPro, the functions of expanded gene families were mostly related to development, stress responses, metabolic pathways and signal transduction, including genes encoding cytochrome P450, glycosyl hydrolase, RING fingers, heat shock proteins, pectin acetylesterase, cellulose synthase and ABC transporters. For example, cytochrome P450 represented one of the largest enzyme families consisting of multiple subfamilies, which are involved in various processes related to plant development and metabolism. This includes root hair development (Pan et al., 2018), lignification (Lui et al., 2020), defence against herbivores (Liu et al., 2019) and drought stress response (J. Zhang et al., 2020). In particular, Renault et al. (2017) found that the membrane topology of cytochrome P450, which was associated with the metabolism of the plant phenolic pathway, was altered by gene duplications in plants (J. Zhang et al., 2020). Other gene families have also expanded in the True-Araceae including glycosyl hydrolases and RING fingers. The glycosyl hydrolase superfamily forms complex and diverse families that combine with glycosyltransferases for the formation and hydrolysis of glycosidic bonds. We detected the expansions of glycosyl hydrolase families, such as 1, 3, 14, 38 and 63, which played important roles in response to biotic and abiotic stresses (Okrent and Wildermuth, 2011; Cao et al., 2017) and cell wall formation (Wang et al., 2020) in plants.
We performed functional GO term enrichment analysis and identified 49 GO terms for molecular functions, biological processes and cellular components that were significantly (P-value ≥0.05) over-represented in the True-Araceae clade (Table 3). Over-represented functions were mostly related to binding, receptor and channel activity in the molecular functions terms, including heat shock protein binding, ion binding, transmembrane signalling receptor activity, ionotropic glutamate receptor activity and extracellular ligand-gated ion channel activity, among others. At the level of biological processes, many metabolic functions, such as DNA repair and cellular stress response, were involved, suggesting that some types of reception were more developed and possessed a wide range of metabolic pathways in the True-Araceae clade. Overall, these results indicated that these expansions may contribute to adaptations encouraging radiations of ancestors of the True-Araceae into tropical and terrestrial biomes.
Table 3.
List of GO terms enriched in the expanded families of the clade representing the True-Araceae.
| GO term | GO category | Description | P-value |
|---|---|---|---|
| GO:0005488 | Molecular function | Binding | 6.40E-05 |
| GO:0031072 | Molecular function | Heat shock protein binding | 6.86E-05 |
| GO:0043167 | Molecular function | Ion binding | 0.0001 |
| GO:0097159 | Molecular function | Organic cyclic compound binding | 0.0006 |
| GO:1901363 | Molecular function | Heterocyclic compound binding | 0.0005 |
| GO:0043168 | Molecular function | Anion binding | 0.0006 |
| GO:0032559 | Molecular function | Adenyl ribonucleotide binding | 0.0018 |
| GO:0032553 | Molecular function | Ribonucleotide binding | 0.0019 |
| GO:0004888 | Molecular function | Transmembrane signalling receptor activity | 0.0019 |
| GO:0004970 | Molecular function | Ionotropic glutamate receptor activity | 0.0019 |
| GO:0005230 | Molecular function | Extracellular ligand-gated ion channel activity | 0.0019 |
| GO:0005524 | Molecular function | ATP binding | 0.0021 |
| GO:0030554 | Molecular function | Adenyl nucleotide binding | 0.0022 |
| GO:0097367 | Molecular function | Carbohydrate derivative binding | 0.0023 |
| GO:0001883 | Molecular function | Purine nucleoside binding | 0.0025 |
| GO:0032549 | Molecular function | Ribonucleoside binding | 0.0025 |
| GO:0005216 | Molecular function | Ion channel activity | 0.0029 |
| GO:0017076 | Molecular function | Purine nucleotide binding | 0.0031 |
| GO:0036094 | Molecular function | Small molecule binding | 0.0042 |
| GO:0000166 | Molecular function | Nucleotide binding | 0.0055 |
| GO:1901265 | Molecular function | Nucleoside phosphate binding | 0.0049 |
| GO:0003993 | Molecular function | Acid phosphatase activity | 0.0091 |
| GO:0015267 | Molecular function | Channel activity | 0.0103 |
| GO:0022803 | Molecular function | Passive transmembrane transporter activity | 0.0103 |
| GO:0015276 | Molecular function | Ligand-gated ion channel activity | 0.0171 |
| GO:0030151 | Molecular function | Molybdenum ion binding | 0.0171 |
| GO:0099600 | Molecular function | Transmembrane receptor activity | 0.0171 |
| GO:0016160 | Molecular function | Amylase activity | 0.0215 |
| GO:0003972 | Molecular function | RNA ligase (ATP) activity | 0.0261 |
| GO:0004097 | Molecular function | Catechol oxidase activity | 0.0261 |
| GO:0004556 | Molecular function | α-Amylase activity | 0.0261 |
| GO:0008452 | Molecular function | RNA ligase activity | 0.0261 |
| GO:0000287 | Molecular function | Magnesium ion binding | 0.0301 |
| GO:0006259 | Biological process | DNA metabolic process | 0.0021 |
| GO:0008033 | Biological process | tRNA processing | 0.0068 |
| GO:0006281 | Biological process | DNA repair | 0.0087 |
| GO:0006974 | Biological process | Cellular response to DNA damage stimulus | 0.0121 |
| GO:0033554 | Biological process | Cellular response to stress | 0.0121 |
| GO:0090304 | Biological process | Nucleic acid metabolic process | 0.0132 |
| GO:0043170 | Biological process | Macromolecule metabolic process | 0.0172 |
| GO:0006468 | Biological process | Protein phosphorylation | 0.0221 |
| GO:0046653 | Biological process | Tetrahydrofolate metabolic process | 0.0241 |
| GO:0046654 | Biological process | Tetrahydrofolate biosynthetic process | 0.0241 |
| GO:0044260 | Biological process | Cellular macromolecule metabolic process | 0.0252 |
| GO:0034470 | Biological process | ncRNA processing | 0.0281 |
| GO:0016310 | Biological process | Phosphorylation | 0.0301 |
| GO:0006260 | Biological process | DNA replication | 0.0383 |
| GO:0006259 | Biological process | DNA metabolic process | 0.0021 |
| GO:0005666 | Cellular component | DNA-directed RNA polymerase III complex | 0.04 |
DISCUSSION
Phylogenetic relationships of eight subfamilies in Araceae
In previous studies, the phylogeny of the Araceae was inferred by chloroplast gene sequences (Cusimano et al., 2011a; Henriquez et al., 2014). However, in plants, these genes are generally inherited from only one parent and they are much affected by rampant horizontal gene transfer (HGT). This may lead to biases when inferring phylogenetic relationships. In our study, our phylogenetic analyses yielded the same relationships for eight subfamilies using concatenation and coalescence methods based on 1081 orthologous nuclear genes. Nevertheless, for the Gymnostachydoideae and Lasioideae, the phylogenetic relationships of eight subfamilies based on nuclear genes did not agree with conclusions inferred from earlier chloroplast sequences (Supplementary data Fig. S15). In our phylogenetic analysis, the Gymnostachydoideae was recovered as a sister group to all other Araceae, with 58 % and 31 % bootstrap values based on the concatenation and coalescence estimations, respectively (Supplementary data Figs S1 and S2). The Lasioideae was the sister of the ‘bisexual climbers’ clade and the unisexual flowers clade, with only 26 % bootstrap values based on coalescence estimation (Supplementary data Figs S1 and S2). To assess the reasons for low support and conflicts at the nodes, we used PhyParts software to map 922 rooted single-gene trees to species trees. Only 37.5 % of single gene trees (346) supported the placement of the Gymnostachydoideae, and 78.1 % of single gene trees (720) supported the sister relationship of the ‘bisexual climbers’ clade and the ‘unisexual flowers’ clade. For the two inconsistent placements reconstructed by ASTRAL and RAxML (Fig. 1), 94.1 % of single gene trees (868) and 85.7 % of single gene trees (790) supported the phylogenetic relationships reconstructed by RAxML. The results indicated that the incongruent phylogenies of the genes may be one of the main factors for the low support and the conflicts of some nodes.
In addition to a robust phylogeny based on nuclear genes, we employed five fossil constraints to determine divergence times of Araceae by PL analysis. However, the age of the crown group was approx. 10 Ma older than that of Janssen and Bremer (2004) based exclusively on chloroplast sequences. Overall, this study provided the first well-supported phylogeny based on nuclear genes for elucidating the evolutionary history of Araceae.
WGD contributed to Araceae adaptation to different environments
In this study, we identified a new WGD event (ψ, approx. 100.7–110.5 Ma) that occurred possibly during the OAEs 1b and 1d (Fig. 2) when the occurrence of extensive submarine volcanic activity resulted in the release of excess greenhouse gases which led to a warming climate (Heimhofer et al., 2005; Jenkyns, 2010; Rodriguez-Cuicas et al., 2019). It is interesting that this WGD event was shared by the True-Araceae clade, but not by the Proto-Araceae group and the lemnoid clade, despite O. aquaticum with a separate WGD event in the Proto-Araceae group, and S. polyrhiza with one special WGD event in the lemnoid clade (Wang et al., 2014). It seems that WGD could be derived from allopolyploidization, as introgression was detected in the early history of the Araceae (Chen et al., 2022). In fact, the ψ-WGD event was followed by upshifts in the rate of diversification in the True-Araceae clade, the opposite of the Pro-Araceae group and the lemnoid clade. Some studies have shown that anthocyanin biosynthesis is related to R2R3-MYB and bHLH gene families mediated by WGD/GD, resulting in the colour changes of spathes (C. Li et al., 2019; Wu et al., 2022). Spathes of the True-Araceae clade are generally showy, attractive and probably adaptive. In contrast, spathes are simple or absent in the Proto-Araceae group and the lemnoid clade (Cusimano et al., 2011a). It was suggested that upshifts in the rate of diversification in the True-Araceae clade may be association with the development of spathes as key innovations (Soltis and Soltis, 2016; Clark and Donoghue, 2018). The best shift along the stem lineage of the True-Araceae clade in diversification rates was identified. This suggested that the shift may have been associated with the change in habit from aquatic to terrestrial environments. These findings indicated that WGD may play other roles, such as a macroevolutionary increase in adaptive and key traits to niches in new environments, rather than a direct increase in speciation rates in the Araceae. In previous studies, comparative genomics between S. polyrhiza and other terrestrial monocots found that some gene families in S. polyrhiza were lost, and this meant a loss of functions in certain biosynthetic and secondary metabolic processes including water transport by aquaporins, lignin biosynthesis, cell wall organization and terpenoid production (Wang et al., 2014; An et al., 2019). Furthermore, the genome of Wolffia australiana (Benth.) Hartog & Plas lost several hundred genes, including those associated with the cell wall, flavonoid biosynthesis, protein phosphorylation, immune response and terpene biosynthesis (Michael et al., 2021). These results suggested that for Spirodela and Wolffia, once adapted to aquatic environments, complex metabolites were no longer needed to cope with variable temperature, light and drought regimes indicative of terrestrial environments. In contrast, our analyses showed expansions of gene families and functional enrichments. The functions of these expanded families were related to heat shock protein, ABC transporter, glycosyl hydrolase, RING finger, pectin acetylesterase, cytochrome P450 and oxidative stress-responsive kinase. Such results indicated that the True-Araceae clade, in particular, showed a successful and long-term trend expanding into tropical regions (Van de Peer et al., 2021). Specifically, the past WGDs in this clade facilitated the macroevolution of adaptations to exploit terrestrial niches, leading to what we now see if the successful colonization and diversification of the Araceae into novel niches.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following materials. Table S1: summary of sampled taxa and dataset statistics. Table S2: description and placement of five fossil calibrations used in this study. Table S3: list of Ks values of WGD events in 81 species of the Araceae and outgroup, Acorus calamus. Table S4: summary of diversification sites in BAMM. Table S5: family name, taxa number, gene number and Pfam function in each cluster for the expanded gene families. Figure S1: phylogenetic tree of Araceae using the RAxML method based on 1081 orthologous groups derived from 90 species. Figure S2: the phylogenetic tree of the Araceae using the ASTRAL method based on 1081 orthologous groups from 90 species. Figure S3: the time-calibrated phylogenetic tree of the Araceae estimated by TreePL based on five fossil constraints. Figure S4: a molecular time scale of the Araceae inferred by TreePL based on 1000 ML bootstrap trees and five fossil calibrations. Figure S5: histograms of Ks distribution in 81 species of the Araceae and Acorus calamus based on wgd tools packages. Figure S6: species trees with gene duplication at each node as inferred by MAPS. Figure S7: speciation and extinction rates of the Araceae estimated by BAMM. Figure S8: the post burn-in posterior distribution for shifts estimated by BAMM. Figure S9: the 95 % credible set of shift configurations estimated by BAMM. Figure S10: the marginal shift probabilities estimated by BAMM. Figure S11: speciation and extinction rates of the Proto-Araceae group and the lemnoid lineages. Figure S12: speciation and extinction rates of the True-Araceae clade after the ψ-WGD event estimated by BAMM. Figure S13: speciation rates of the Proto-Araceae group, the lemnoid clade and the True-Araceae clade estimated by BiSSE. Lambda0 indicates the Proto-Araceae group and the lemnoid clade. Figure S14: extinction rates of the Proto-Araceae group, the lemnoid clade and the True-Araceae clade estimated by BiSSE. Figure S15: a comparative figure for the phylogenetic relationships of eight subfamilies based on nuclear and chloroplast genes.
ACKNOWLEDGEMENTS
We are grateful to Drs Jin-Mei Lu, Jie Cai, Ting Zhang and other staff of Germplasm Bank of Wild Species in South-west China for various assistance. We are grateful to the curators and staff of the Kunming Botanical Garden and the Missouri Botanical Garden for sample collection. We are also grateful to Drs Jacob B. Landis, Bin-E Xue, Hang-Hui Kong, Zong-Xin Ren and Chao-Nan Fu for sharing scripts and discussion. Professor Peter Bernhardt (the Missouri Botanical Garden) critically revised the manuscript and polished the English.
Contributor Information
Lei Zhao, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China; Kunming College of Life Sciences, University of Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Ying-Ying Yang, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China; Kunming College of Life Sciences, University of Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Xiao-Jian Qu, Shandong Provincial Key Laboratory of Plant Stress Research, College of Life Sciences, Shandong Normal University, Ji’nan, Shandong 250014, China.
Hong Ma, Department of Biology, Huck Institutes of the Life Sciences, Pennsylvania State University, University Park, PA 16802, USA.
Yi Hu, Department of Biology, Huck Institutes of the Life Sciences, Pennsylvania State University, University Park, PA 16802, USA.
Hong-Tao Li, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
Ting-Shuang Yi, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
De-Zhu Li, Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, Yunnan 650201, China.
FUNDING
This study is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, China (grant no. XDB31000000), the Program of Science and Technology Talents Training of Yunnan Province, China (grant no. 2017HA014), the National Natural Science Foundation of China key international (regional) cooperative research project (grant no. 31720103903); the Science and Technology Basic Resources Investigation Program of China (grant no. 2019FY100900); the Large-scale Scientific Facilities of the Chinese Academy of Sciences (grant no. 2017-LSF-GBOWS-02); the National Natural Science Foundation of China (grant nos 31270274 and 31570333), the Key R and D program of Yunnan Province, China (grant no. 202103AC100003), the Key Basic Research program of Yunnan Province, China (grant no. 202101BC070003) and the open research project of the ‘Cross-Cooperative Team’ of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences.
CONFLICT OF INTEREST
The authors declare no competing interests.
LITERATURE CITED
- Abramson BW, Novotny M, Hartwick NT, et al. 2022. The genome and preliminary single-nuclei transcriptome of Lemna minuta reveals mechanisms of invasiveness. Plant Physiology 188: 879–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix K, Gerard PR, Schwarzacher T, et al. 2017. Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Annals of Botany 120: 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Zhou Y, Li CS, et al. 2019. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proceedings of the National Academy of Sciences, USA 116: 18893–18899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo N, Barker MS. 2012. Rarely successful polyploids and their legacy in plant genomes. Current Opinion in Plant Biology 15: 140–146. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. 2000. Gene Ontology: tool for the unification of biology. Nature Genetics 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, O’Meara BC. 2016. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Systematic Biology 65: 583–601. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Wilf P, Sauquet H. 2022. The Angiosperm Terrestrial Revolution and the origins of modern biodiversity. New Phytologist 233: 2017–2035. [DOI] [PubMed] [Google Scholar]
- Bouckaert R, Vaughan TG, Barido-Sottani J, et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Computational Biology 15: e1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera LI, Salazar GA, Chase MW, et al. 2008. Phylogenetic relationships of aroids and duckweeds (Araceae) inferred from coding and noncoding plastid DNA. American Journal of Botany 95: 1153–1165. [DOI] [PubMed] [Google Scholar]
- Cai L, Xi Z, Amorim AM, et al. 2019. Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytologist 221: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao YY, Yang JF, Liu TY, et al. 2017. A phylogenetically informed comparison of GH1 hydrolases between Arabidopsis and rice response to stressors. Frontiers in Plant Science 8: 350. doi: 10.3389/fpls.2017.00350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Lu B, Morales-Briones DF, et al. 2022. Phylogenomic analyses of Alismatales shed light into adaptations to aquatic environments. Molecular Biology and Evolution 39: msac079, 10.1093/molbev/msac079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JW, Donoghue PCJ. 2018. Whole-genome duplication and plant macroevolution. Trends in Plant Science 23: 933–945. [DOI] [PubMed] [Google Scholar]
- Croat TB. 2019. Araceae, a family with great potential. Annals of the Missouri Botanical Garden 104: 3–9. [Google Scholar]
- Cusimano N, Bogner J, Mayo SJ, et al. 2011a. Relationships within the Araceae: comparison of morphological patterns with molecular phylogenies. American Journal of Botany 98: 654–668. [DOI] [PubMed] [Google Scholar]
- Cusimano N, Sousa A, Renner SS. 2011b. Maximum likelihood inference implies a high, not a low, ancestral haploid chromosome number in Araceae, with a critique of the bias introduced by ‘x’. Annals of Botany 109: 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CC, Xi Z, Mathews S. 2014. Plastid phylogenomics and green plant phylogeny: almost full circle but not quite there. BMC Biology 12: 11. doi: 10.1186/1741-7007-12-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersberger I, Strauss S, von Haeseler A. 2009. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evolutionary Biology 9: 157. doi: 10.1186/1471-2148-9-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Heidel-Fischer HM, Bekaert M, et al. 2015. The butterfly plant arms-race escalated by gene and genome duplications. Proceedings of the National Academy of Sciences, USA 112: 8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biology 20: 238. doi: 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep MC, McKain MR, Diaz DV, et al. 2014. Allopolyploidy, diversification, and the Miocene grassland expansion. Proceedings of the National Academy of Sciences, USA 111: 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzJohn RG. 2012. Diversitree: comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution 3: 1084–1092. [Google Scholar]
- FitzJohn RG, Maddison WP, Otto SP. 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Systematic Biology 58: 595–611. [DOI] [PubMed] [Google Scholar]
- Givnish TJ, Zuluaga A, Spalink D, et al. 2018. Monocot plastid phylogenomics, timeline, net rates of species diversification, the power of multi-gene analyses, and a functional model for the origin of monocots. American Journal of Botany 105: 1888–1910. [DOI] [PubMed] [Google Scholar]
- Godden GT, Kinser TJ, Soltis PS, Soltis DE. 2019. Phylotranscriptomic analyses reveal asymmetrical gene duplication dynamics and signatures of ancient polyploidy in mints. Genome Biology Evolution 11: 3393–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Xu W, Hu Y, et al. 2020. Phylotranscriptomics in Cucurbitaceae reveal multiple whole-genome duplications and key morphological and molecular innovations. Molecular Plant 13: 1117–1133. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, et al. 2008. Automated eukaryotic gene structure annotation using EVidenceModeller and the program to assemble spliced alignments. Genome Biology 9: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MV, Thomas GW, Lugo-Martinez J, Hahn MW. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Molecular Biology and Evolution 30: 1987–1997. [DOI] [PubMed] [Google Scholar]
- Han TS, Zheng QJ, Onstein RE, et al. 2020. Polyploidy promotes species diversification of Allium through ecological shifts. New Phytologist 225: 571–583. [DOI] [PubMed] [Google Scholar]
- Heimhofer U, Hochuli PA, Burla S, et al. 2005. Timing of Early Cretaceous angiosperm diversification and possible links to major paleoenvironmental change. Geology 33: 141–144. [Google Scholar]
- Henriquez CL, Arias T, Pires JC, et al. 2014. Phylogenomics of the plant family Araceae. Molecular Phylogenetics and Evolution 75: 91–102. [DOI] [PubMed] [Google Scholar]
- Herrera-Alsina L, van Els P, Etienne RS. 2019. Detecting the dependence of diversification on multiple traits from phylogenetic trees and trait data. Systematic Biology 68: 317–328. [DOI] [PubMed] [Google Scholar]
- Huang CH, Zhang CF, Liu M, et al. 2016. Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Molecular Biology and Evolution 33: 2820–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XC, German DA, Koch MA. 2020. Temporal patterns of diversification in Brassicaceae demonstrate decoupling of rate shifts and mesopolyploidization events. Annals of Botany 125: 29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles WJD, Smith SY, Gandolfo MA, Graham SW. 2015. Monocot fossils suitable for molecular dating analyses. Botanical Journal of the Linnean Society 178: 346–374. [Google Scholar]
- Janssen T, Bremer K. 2004. The age of major monocot groups inferred from 800+ rbcL sequences. Botanical Journal of the Linnean Society 146: 385–398. [Google Scholar]
- Jenkyns HC. 2010. Geochemistry of oceanic anoxic events. Geochemistry Geophysics Geosystems 11: Q03004, doi: 10.1029/2009GC002788 [DOI] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Jiao YN, Leebens-Mack J, Ayyampalayam S, et al. 2012. A genome triplication associated with early diversification of the core eudicots. Genome Biology 13: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Li J, Tang H, Paterson AH. 2014. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. The Plant Cell 26: 2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R, Toshimoto K, Noguchi H, et al. 2014. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Research 24: 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapli P, Yang ZH, Telford MJ. 2020. Phylogenetic tree building in the genomic age. Nature Reviews. Genetics 21: 428–444. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen EJM, Ojeda DI, Bakker FT, et al. 2021. The origin of the legumes is a complex paleopolyploid phylogenomic tangle closely associated with the Cretaceous–Paleogene (K–Pg) mass extinction event. Systematic Biology 70: 508–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, et al. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JB, Soltis DE, Li Z, et al. 2018. Impact of whole-genome duplication events on diversification rates in angiosperms. American Journal of Botany 105: 348–363. [DOI] [PubMed] [Google Scholar]
- Lee JS, Kim SH, Lee S, et al. 2019. New insights into the phylogeny and biogeography of subfamily Orontioideae (Araceae). Journal of Systematics and Evolution 57: 616–632. [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, et al. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA. 2019. Plant speciation in the age of climate change. Annals of Botany 124: 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qiu J, Huang S, et al. 2019. AaMYB3 interacts with AabHLH1 to regulate proanthocyanidin accumulation in Anthurium andraeanum (Hort.)—another strategy to modulate pigmentation. Horticulture Research 6: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HT, Yi TS, Gao LM, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nature Plants 5: 461–470. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659. [DOI] [PubMed] [Google Scholar]
- Li Z, Barker MS. 2020. Inferring putative ancient whole-genome duplications in the 1000 Plants (1KP) initiative: access to gene family phylogenies and age distributions. Gigascience 9: giaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Baniaga AE, Sessa EB, et al. 2015. Early genome duplications in conifers and other seed plants. Science Advances 1: e1501084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Khakimov B, Cardenas PD, et al. 2019. The cytochrome P450 CYP72A552 is key to production of hederagenin-based saponins that mediate plant defense against herbivores. New Phytologist 222: 1599–1609. [DOI] [PubMed] [Google Scholar]
- Lui ACW, Lam PY, Chan KH, et al. 2020. Convergent recruitment of 5ʹ-hydroxylase activities by CYP75B flavonoid B-ring hydroxylases for tricin biosynthesis in Medicago legumes. New Phytologist 228: 269–284. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character’s effect on speciation and extinction. Systematic Biology 56: 701–710. [DOI] [PubMed] [Google Scholar]
- Magallon S, Gomez-Acevedo S, Sanchez-Reyes LL, Hernandez-Hernandez T. 2015. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytologist 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Mandakova T, Lysak MA. 2018. Postpolyploid diploidization and diversification through dysploid changes. Current Opinion in Plant Biology 42: 55–65. [DOI] [PubMed] [Google Scholar]
- Mandel JR, Dikow RB, Siniscalchi CM, et al. 2019. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proceedings of the National Academy of Sciences, USA 116: 14083–14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo SJ, Bogner J, Boyce PC. 1997. The genera of Araceae. Kew: Royal Botanic Gardens. [Google Scholar]
- Mayrose I, Zhan SH, Rothfels CJ, et al. 2011. Recently, formed polyploid plants diversify at lower rates. Science 333: 1257–1257. [DOI] [PubMed] [Google Scholar]
- Michael TP, Ernst E, Hartwick N, et al. 2021. Genome and time-of-day transcriptome of Wolffia australiana link morphological minimization with gene loss and less growth control. Genome Research 31: 225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Warnow T. 2015. ASTRAL-II: coalescent-based species tree estimation with many hundreds of taxa and thousands of genes. Bioinformatics 31: i44–i52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirarab S, Nguyen N, Guo S, et al. 2015. PASTA: ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. Journal of Computational Biology 22: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauheimer L, Metzler D, Renner SS. 2012. Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytologist 195: 938–950. [DOI] [PubMed] [Google Scholar]
- Okrent RA, Wildermuth MC. 2011. Evolutionary history of the GH3 family of acyl adenylases in rosids. Plant Molecular Biology 76: 489–505. [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Baerson SR, Wang M, et al. 2018. A cytochrome P450 CYP71 enzyme expressed in Sorghum bicolor root hair cells participates in the biosynthesis of the benzoquinone allelochemical sorgoleone. New Phytologist 218: 616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171: 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9: e89543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabosky DL, Mitchell JS, Chang J. 2017. Is BAMM flawed? Theoretical and practical concerns in the analysis of multi-rate diversification models. Systematic Biology 66: 477–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, Wang H, Guo C, et al. 2018. Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Molecular Plant 11: 414–428. [DOI] [PubMed] [Google Scholar]
- Renault H, De Marothy M, Jonasson G, et al. 2017. Gene duplication leads to altered membrane topology of a cytochrome P450 enzyme in seed plants. Molecular Biology and Evolution 34: 2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cuicas ME, Montero-Serrano JC, Garban G. 2019. Paleoenvironmental changes during the late Albian oceanic anoxic event 1d: an example from the Capacho Formation, southwestern Venezuela. Palaeogeography Palaeoclimatology Palaeoecology 521: 10–29. [Google Scholar]
- Roure B, Rodriguez-Ezpeleta N, Philippe H. 2007. SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evolutionary Biology 7: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpino SV, Levin DA, Meyers LA. 2014. Polyploid formation shapes flowering plant diversity. American Naturalist 184: 456–465. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP. 2012. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Current Opinion in Plant Biology 15: 147–153. [DOI] [PubMed] [Google Scholar]
- Sessa EB. 2019. Polyploidy as a mechanism for surviving global change. New Phytologist 221: 5–6. [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologues. Bioinformatic 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Smith SA, O’Meara BC. 2012. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics 28: 2689–2690. [DOI] [PubMed] [Google Scholar]
- Smith SA, Moore MJ, Brown JW, Yang Y. 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evolutionary Biology 15: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Brown JW, Yang Y, et al. 2018. Disparity, diversity, and duplications in the Caryophyllales. New Phytologist 217: 836–854. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Current Opinion in Plant Biology 30: 159–165. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE. 2015. Polyploidy and genome evolution in plants. Current Opinion in Genetics and Development 35: 119–125. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and postanalysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Stull GW, Qu XJ, Parins-Fukuchi C, et al. 2021. Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nature Plants 7: 1015–1025. [DOI] [PubMed] [Google Scholar]
- Tank DC, Eastman JM, Pennell MW, et al. 2015. Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytologist 207: 454–467. [DOI] [PubMed] [Google Scholar]
- Title PO, Rabosky DL. 2019. Tip rates, phylogenies and diversification: what are we estimating, and how good are the estimates? Methods in Ecology and Evolution 10: 821–834. [Google Scholar]
- Van De Peer Y, Mizrachi E, Marchal K. 2017. The evolutionary significance of polyploidy. Nature Reviews Genetics 18: 411–424. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Ashman TL, Soltis PS, Soltis DE. 2021. Polyploidy: an evolutionary and ecological force in stressful times. The Plant Cell 33: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K, Maere S, Van de Peer Y. 2014. Tangled up in two: a burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Philosophical Transactions of the Royal Society B: Biological Sciences 369: 20130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, Guo CC, Ma H, Qi J. 2019. Reply to Zwaenepoel et al.: meeting the challenges of detecting polyploidy events from transcriptomic data. Molecular Plant 12: 137–140. [DOI] [PubMed] [Google Scholar]
- Wang N, Yang Y, Moore MJ, et al. 2019. Evolution of Portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Molecular Biology and Evolution 36: 112–126. [DOI] [PubMed] [Google Scholar]
- Wang SB, Li LZ, Li HY, et al. 2020. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nature Plants 6: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Haberer G, Gundlach H, et al. 2014. The Spirodela polyrhiza genome reveals insights into its neotenous reduction fast growth and aquatic lifestyle. Nature Communication 5: 3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Han B, Jiao Y. 2020. Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Molecular Plant 13: 59–71. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wen J, Xia Y, et al. 2022. Evolution and functional diversification of R2R3-MYB transcription factors in plants. Horticulture Research 9: uhac058. 10.1093/hr/uhac058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JM, Jiang L, Wang L, et al. 2021. A high-quality genome of taro (Colocasia esculenta (L.) Schott), one of the world’s oldest crops. Molecular Ecology Resources 21: 68–77. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Sun R, et al. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nature Communation 5: 4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fu XX, Li RQ, et al. 2020. The hornwort genome and early land plant evolution. Nature Plants 6: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wu S, Chang X, et al. 2020. The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant, Cell & Environment 43: 2847–2856. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhu X, Zhao Y, et al. 2022. Phylotranscriptomics resolves the phylogeny of Pooideae and uncovers factors for their adaptive evolution. Molecular Biology and Evolution 39: msac026. 10.1093/molbev/msac026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhao L, Folk RA, et al. 2022. Phylotranscriptomics of Theaceae: generic level relationships, reticulation and whole-genome duplication. Annals of Botany 129: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Li X, Zhang N, et al. 2016. Phylogenomic analyses of large-scale nuclear genes provide new insights into the evolutionary relationships within the rosids. Molecular Phylogenetics and Evolution 105: 166–176. [DOI] [PubMed] [Google Scholar]
- Zhao YY, Zhang R, Jiang KW, et al. 2021. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Molecular Plant 14: 748–773. [DOI] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. 2019. Wgd-simple command line tools for the analysis of ancient whole-genome duplications. Bioinformatics 35: 2153–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A, Li Z, Lohaus R, Van de Peer Y. 2019. Finding evidence for whole genome duplications: a reappraisal. Molecular Plant 12: 133–136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.