Abstract
Heterologous biosynthesis of natural products is meant to enable access to the vast array of valuable properties associated with these compounds. Often motivated by limitations inherent in native production hosts, the heterologous biosynthetic process begins with a candidate host regarded as technically advanced relative to original producing organisms. Given this requirement, E. coli has been a top choice for heterologous biosynthesis attempts as associated recombinant tools emerged and continue to develop. However, success requires overcoming challenges associated with natural product formation, including complex biosynthetic pathways and the need for metabolic support. These two challenges have been heavily featured in cellular engineering efforts completed to position E. coli as a viable surrogate host. This chapter outlines steps taken to engineer E. coli with an emphasis on genetic manipulations designed to support the heterologous production of polyketide, nonribosomal peptide, and similarly complex natural products.
Keywords: Heterologous biosynthesis, Cellular design, Natural products, Polyketide, Nonribosomal peptide, E. coli
1. Introduction
Heterologous natural product biosynthesis enables access to the beneficial properties of compounds that derive from their nonideal original hosts [1, 2]. The host selected for heterologous biosynthesis should provide a combination of innate and potential advantages relative to the native host in order to motivate the technical steps required for genetic content transfer and pathway reconstitution. The establishment of heterologous biosynthesis is the focal point of this chapter. Specifically, the latest alterations to the genetic background of an Escherichia coli host designed to support complex natural product biosynthesis are described.
E. coli holds an advantage over nearly every possible heterologous host because of its rapid growth kinetics. In turn, it is also the most thoroughly characterized bacterial host and features an impressive range of genetic manipulation protocols. These features provide both a motivation and an experimental basis for enacting heterologous biosynthesis. However, E. coli is not a native producer of complex natural products (such as polyketides and nonribosomal peptides). As such, the advantages offered by this potential surrogate host are offset by concerns regarding heterologous pathway transfer and reconstitution. Key issues include (1) enabling expression of foreign clusters that contain both numerous and individually large genes, (2) ensuring active protein products, and (3) providing the needed metabolic substrates required for complex natural product formation.
The cellular engineering steps outlined below describe the latest alterations to an E. coli strain, termed BAP1, capable of supporting both polyketide and nonribosomal peptide biosynthesis [3]. The BAP1 strain contains the following features designed to assist heterologous natural product biosynthesis: (1) a strong and processive T7 RNA polymerase [4-6] to aid in the expression of complex foreign natural product genetic pathways; (2) a promiscuous 4′-phosphopantetheinyl transferase [7, 8] to posttranslationally modify and, hence, activate polyketide synthases and nonribosomal peptide synthetases; and (3) a re-engineered prp operon that allows inducible upregulation of a propionyl-CoA synthetase gene (prpE) while deleting the remaining catabolic capabilities of the operon, in order to provide a key polyketide precursor, propionyl-CoA, while also eliminating a primary consumption pathway for this same metabolite. To this strain, additional genetic manipulations have been made to further support and advance heterologous biosynthesis by this host. The genotypes of all strains described in this chapter are presented in Table 1. As opposed to describing a single method used during the procedures to generate these strains, we more generally emphasize the goals associated with each (also outlined in Table 1) and the series of experimental steps completed during construction.
Table 1.
E. coli strains constructed in this study
| Strain | Description | Note |
|---|---|---|
| BAP1 | F– ompT hsdSB (rB–mB–) gal dcm (DE3) Δ prpRBCD::T7prom-sfp, T7prom-prpE | Parent strain; capable of polyketide substrate support and posttranslational modification |
| BT2 | BAP1 (araBADC::Tn10) | Disrupted arabinose degradation pathway for sustained induction |
| BT3 | BAP1 (araBADC::Tn10, lacZ::trfA-Kan) | Integration of trfA gene for plasmid copy-up |
| BTRA | BAP1 (araBADC::Tn10, lacZ::trfA-Kan, ΔrecA) | Deletion of recA to increase plasmid stability |
| BTRAP | BAP1 (araBADC::Tn10, lacZ::trfA-Kan, ΔrecA, pccAB<>codB) | Inclusion of PCC to enable enhanced substrate support |
2. Materials
2.1. Chromosomal Engineering
Ampicillin: 100 mg/L in water.
Kanamycin: 50 mg/L in water.
Chloramphenicol: 20 mg/L in ethanol.
Tetracycline: 10 mg/L in ethanol.
l-arabinose: 2 mg/L in water for plasmid copy-up studies.
IPTG: isopropyl β-d-1-thiogalactopyranoside, 100 μM in water.
Lysogeny broth (LB) medium: 10 g Bacto-tryptone, 5 g yeast extract, and 10 g NaCl per liter.
LB agar plates: 10 g Bacto-tryptone, 5 g yeast extract, 10 g NaCl, and 1 % agarose per liter.
M9 medium: 12.8 g Na2HPO4·7H2O, 3 g KH2PO4, 0.5 g NaCl, 1.0 g NH4Cl, 2 mM MgSO4, and 0.1 mM CaCl2 per liter.
P1 phage lysate prepared using chloroform and stored at 4 °C.
Plasmids pKD46, pCP20, and pKD3 used for chromosomal DNA manipulation as described by Datsenko et al. [9].
Phusion DNA polymerase obtained from New England Biolabs (Ipswich, MA).
Restriction enzymes.
Qiaprep Spin miniprep kit for plasmid preparation, Qiagen (Valencia, CA).
QIAquick gel extraction kit for PCR product purification, Qiagen (Valencia, CA).
GeneJET gel extraction kit, Thermo Scientific (Pittsburgh, PA).
MicroPulser electroporator from Bio-Rad (Hercules, CA).
Thermo Scientific Barnstead Micropure Water Purification System used to provide water for reagents and reactions.
2.2. Phenotypic Assays
l-arabinose: 1 g/L in water.
pCC1FOS™ fosmid vector and the accompanying EPI3000 strain, Epicenter (Madison, WI).
UVP GelDoc-It TS™ Imaging System Transilluminator (Upland, CA).
API 3000 Triple Quad LC-MS with a Turbo Ion Spray source (PE Sciex) coupled with a Shimadzu Prominence LC system.
3. Methods
3.1. ara Operon Deletion
The first step for the desired strain construction is the removal of the araBADC operon from the E. coli BAP1 chromosome in order to prevent arabinose degradation and to thus enable a constant concentration of arabinose inducer during later gene expression or plasmid copy-up steps in the context of heterologous biosynthesis.
For this step, transfer the araBADC::Tn10 cassette from E. coli LMG194 (Life Sciences, Grand Island, NY) to BAP1 by P1 transduction. Grow E. coli LMG194 overnight in LB medium containing tetracycline at 37 °C with shaking. Dilute the overnight culture at a volume ratio of 1:100 in LB medium containing 5 mM CaCl2. Incubate the diluted culture at 37 °C for 1 h.
Add approximately 107 phage from a previous lysate to 1 mL of the bacterial subculture in step 1 and incubate at 37 °C for 1 h until the culture becomes clear, indicating cell lysis. Add chloroform (50 μL) to the cell lysate (see Note 1), followed by vortexing for 1 min. Clear cell debris after lysis by centrifugation; save the supernatant in a fresh tube at 4 °C as the P1 phage lysate solution.
Collect 1 mL overnight LB culture of recipient BAP1 cells by centrifugation and resuspend in 200 μL fresh LB medium containing 5 mM CaCl2 and 100 mM MgSO4. Mix this culture (100 μL) with 100 μL of the P1 phage lysate solution prepared in step 2. After a 30 min incubation at 37 °C, add 300 μL of 0.1 M sodium citrate and 1 mL LB medium to the reaction and incubate the mixture at 37 °C for 1 h to allow sufficient time for expression of the tetracycline resistance marker.
Harvest the transduced cells after P1 transduction by centrifugation and resuspend in 0.1 M sodium citrate solution. Plate the resulting suspension on LB agar containing tetracycline for isolation of individual colonies.
After overnight incubation, re-streak several colonies from the plate onto a fresh LB agar plate containing 0.1 M sodium citrate and tetracycline. Individual colonies are then picked and tested for the desired gene knockout by PCR and phenotypic analysis.
3.2. trfA Chromosomal Integration
The second step is to equip E. coli with the ability to increase the copy number of specific copy-up plasmids, enabling the production of varying metabolic levels of newly introduced natural product pathways. For this step, integrate the trfA gene into the chromosome of the E. coli strain produced according to Subheading 3.1 above. The trfA gene product initiates plasmid replication from oriV and thus increases copy number by 10–50-fold [10-13]. Based upon a method described previously [14], replace the lacZ gene in the chromosome of BAP1 (araBADC::Tn10) with a cassette containing the trfA gene and a kanamycin resistance gene. Specifically, transfer plasmid pJW410, a derivative of pBRINT-TsKm (ampicillin resistant), containing the araC-para-trfA-Kan cassette flanked by lacZ homology sequences into BAP1 (araBADC::Tn10) by electroporation. Grow the transformants on an LB plate containing ampicillin and kanamycin at 30 °C.
Pick single colonies and re-streak onto a fresh LB plate containing kanamycin only. Grow re-streaked single colonies in LB medium containing kanamycin at 30 °C for 3 h. Dilute the culture with fresh LB medium and plate on LB agar plates containing kanamycin, followed by an overnight incubation at 42 °C.
Pick individual ampicillin-sensitive colonies (indicating pJW410 plasmid loss) and test for the desired trfA gene integration by PCR and the copy-up phenotype assay.
3.3. recA Deletion
Delete the recA gene from the chromosome of BAP1 (araBADC::Tn10, lacZ::trfA-Kan) to improve the stability of extrachromosomal plasmid DNA [15]. This step will potentially limit loss or rearrangements of large foreign natural product gene clusters localized to plasmids during reconstitution efforts.
PCR amplify a DNA fragment containing a chloramphenicol-resistance gene (cat) from plasmid pIKD3 [9] using primers recA1 and recA2.
Gel-purify the resulting PCR product using a QIAquick gel extraction kit.
The PCR product is transformed via electroporation (see Note 2) into E. coli K-12 strain BW25113 (obtained from the E. coli Genetic Stock Center, Yale University) (see Note 3) harboring pKD46.
Once recombination is completed and confirmed, prepare genomic DNA from the recA- BW25113 strain for a second PCR by centrifuging 50 μL of an overnight 2 mL LB culture, washing the pelleted cells once, and resuspending in 50 μL water prior to incubation at 95 °C for 10 min using a heating block. The resulting supernatant can be used as a PCR template. Amplify the recA::cat cassette using primers recA3 and recA4 and insert into BAP1 (araBADC::Tn10, lacZ::trfA-Kan) by another round of lambda Red recombination (see Note 4).
Remove the cat resistance marker by FRT-site-mediated recombination with pCP20, a FLP synthesis plasmid [9]. Specifically, the pCP20 plasmid is transformed into BAP1 (araBADC::Tn10, lacZ::trfA-Kan, recA::cat) to express the desired flippase and remove the cat resistance gene. The resulting transformant is incubated at 42 °C to allow the loss of the temperature-sensitive pCP20 plasmid. The resulting strain E. coli BAP1 (araBADC::Tn10, lacZ::trfA-Kan, ΔrecA) is referred to as BTRA.
Confirm the deletion of the recA gene by PCR and a UV exposure assay.
3.4. Propionyl-CoA Carboxylase (PCC) Chromosomal Integration
The final and most recent step in augmenting the genetic and metabolic background of E. coli in support of complex natural product biosynthesis is the replacement of the nonessential chromosomal gene codB with a pccAB-cat cassette. The encoded PCC enables the conversion of propionyl-CoA to (2S)-methylmalonyl-CoA, which is an important metabolic substrate for polyketide biosynthesis [16, 17]. The steps in the integration process are outlined below:
Amplify the pccAB genes by PCR from pBP144 [3] using the primers specified in Table 2. The primers are designed to provide flanking NdeI (5′) and BamHI (3′) sites to the resulting PCR product.
Isolate the PCR fragment using the GeneJET gel extraction kit, digest with the flanking restriction sites, and ligate into plasmid pET21c.
PCR amplify the cat gene, flanked by FRT sites, from pKD3 using primers indicated in Table 2. This set of primers introduces SacI (5′) and XhoI (3′) flanking restriction sites to the PCR product.
Digest the cat PCR product using the flanking restriction sites and insert into the plasmid containing the pccAB genes to generate the pccAB-cat cassette.
PCR amplify the pccAB-cat cassette (Table 2), to be used for homologous recombination by means of the lambda Red procedure.
Complete homologous recombination into strain BW25113 using the protocol for lambda Red recombination described previously (see Note 5).
Confirm integration of the pccAB-cat cassette into strain BW25113 by PCR amplification and sequencing of the inserted cassette. Use the genomic DNA of this confirmed strain for a second round of lambda Red recombination.
Prepare BW25113 (pccAB-cat) genomic DNA for a second round of PCR and lambda Red recombination: wash 500 μL of a 3 mL overnight LB culture (incubated at 37 °C with shaking) once and resuspend in 200 μL water. Boil the sample at 95 °C for 5 min in a heating block; use the resulting supernatant for PCR.
Complete PCR using the BW25113 (pccAB-cat) genomic DNA template with primers (Table 2) that will yield an amplified product with a minimum of 200 base pairs on either side of the pccAB-cat cassette (see Note 6).
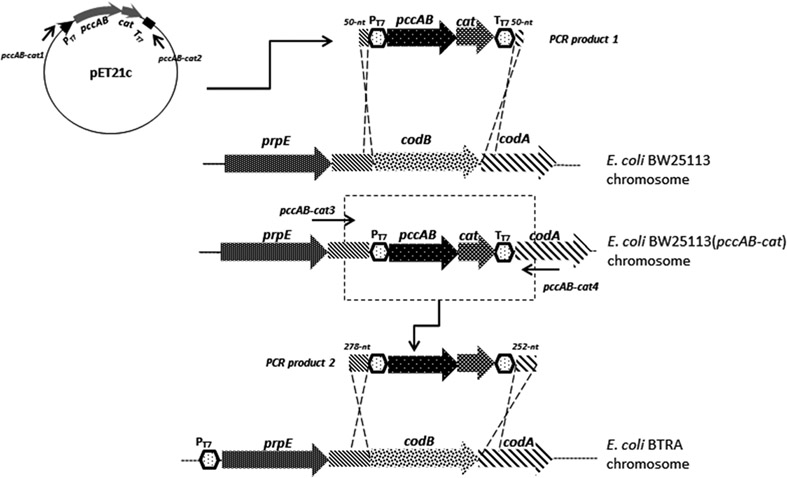
Repeat the lambda Red procedure within BTRA using a PCR product which has a 278-nt left homology arm and a 252-nt right homology arm (Fig. 1).
Transform plasmid pCP20 into BTRA (pccAB-cat<>codB) to eliminate the cat chloramphenicol resistance gene. The resulting E. coli strain, named BTRAP, is confirmed by diagnostic PCR and tested for (2S)-methylmalonyl-CoA metabolism through the phenotypic assay described below.
Table 2.
Primers used in the engineering of E. coli
| Name | Sequence |
|---|---|
| recA1 | Forward: 5′-ATGCGACCCTTGTGTATCAAACAAGACGATTAAAAATCTTCGTTAGTTTCGTGTAGGCTGGAGCTGCTTC-3′ |
| recA2 | Reverse: 5′-CAGAACATATTGACTATCCGGTATTACCCGGCATGACAGGAGTAAAAATGATGGGAATTAGCCATGGTCC-3′ |
| recA3 | Forward: 5′-CTTGTCGAGCCCAAGGAACA-3′ |
| recA4 | Reverse: 5′-GAACCCGTCGTGGTGGAAAT-3′ |
| pccAB1 | Forward: 5′-GATCATATGGCTGCTCCGGCTTCTG-3′ |
| pccAB2 | Reverse: 5′-GTAGGATCCTTACAGCGGGATG-3′ |
| cat1 | Forward: 5′-GCGGAGCTCGTGTAGGCTGGAGCTGCTTC-3′ |
| cat2 | Reverse: 5′-GCGCTCGAGCATATGAATATCCTCCTTGG-3′ |
| pccAB-cat1 | Forward: 5′-TAGAATGCGGCGGATTTTTTGGGTTTCAAACAGCAAAAAGGGGGAATTTCAGATCTCGATCCCGCG-3′ |
| pccAB-cat2 | Reverse: 5′-ACCGGGCGTTAATAATTGTTTGTAAAGCGTTATTCGACACTGTTAGCCTCCAAAAAACCCCTCAAG-3′ |
| pccAB-cat3 | Forward: 5′-TCCTGCGTCGTTGGATCAGA-3′ |
| pccAB-cat4 | Reverse: 5′-GTGCCGGACTGATTCCAGTT-3′ |
Fig. 1.
Schematic for chromosomal engineering using lambda Red recombination and a sequential process of genetic transfer between K and B strains of E. coli. Table 2 primers are indicated in association with first and second PCR steps. PT7: T7 promoter; TT7: T7 terminator
3.5. Genetic and Phenotypic Analysis of Strain Construction
Assays to confirm the above genetic manipulations can be divided into genetic and phenotypic categories. At the genetic level, PCR amplification and fragment sequencing allow confirmation of gene insertion or deletion (Fig. 2). Complementary assays, to be described below, use a phenotype associated with the desired genetic change. Eventually, the phenotypic assays should support the larger objective of complex natural product heterologous biosynthesis.
Fig. 2.
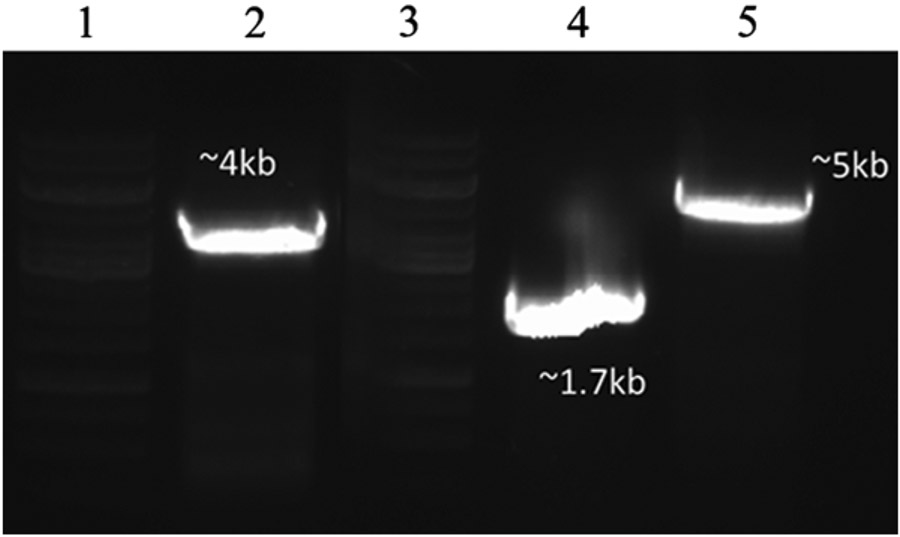
PCR verification of chromosomal insertion of pccAB. Lanes 1 and 3: marker (top band, 10 kb); Lane 2: PCR of final BTRAP genomic DNA containing pccAB (post-cat removal); Lane 4: negative control: PCR of starting BTRA genomic DNA insertion region; Lane 5: PCR of BTRAP genomic DNA containing pccAB-cat
The simplest phenotypic assay associated with the various genetic construction steps is the correct antibiotic resistance patterns associated with each step. Though simple, this is the first and essential signature needed for strain verification.
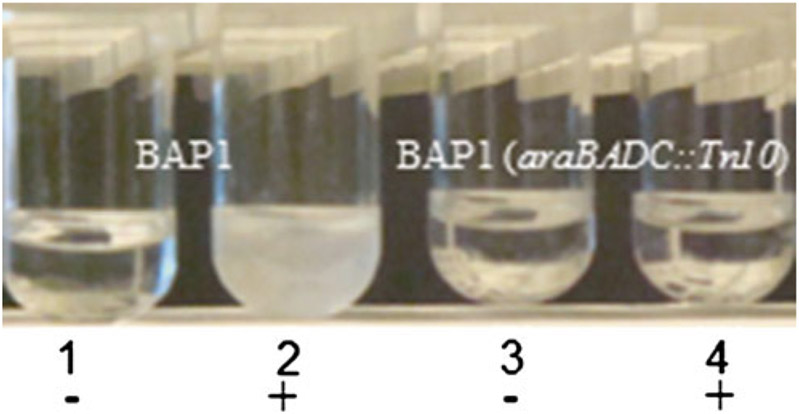
As an example, strain BAP1 (araBADC::Tn10) or BT2 is resistant to the antibiotic tetracycline. The deletion of the araBADC cassette is also confirmed by a phenotype assay in which the desired inability of the strain to grow in M9 medium with arabinose as the sole carbon source is tested (Fig. 3), indicating that the arabinose utilization pathway is successfully disrupted (see Note 7).
Fig. 3.
Engineered araBADC disruption and assessment of arabinose catabolism. BAP1 [1 and 2] and BAP1 (araBADC::Tn10) [3 and 4] cultured in M9 minimal medium without [−] and with [+] l-arabinose
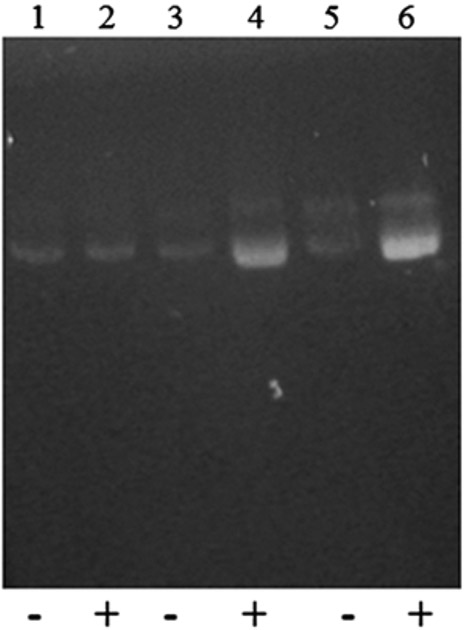
Likewise, strain BAP1 (araBADC::Tn10, lacZ:: trfA-Kan) or BT3 can be selected for resistance to tetracycline and kanamycin. In addition, a copy-up assay can be performed to confirm the induced copy-up capability of a fosmid with an oriV origin of replication (Fig. 4). In this assay, E. coli BAP1 (negative control) and EPI300 (positive control) are transformed with the pCC1FOS™ fosmid and grown in LB medium with and without the inducer arabinose (see Note 8). Strains can be grown overnight, adjusted to the same optical density, and fosmid DNA can be isolated from each strain using a Qiaprep Spin miniprep kit (see Note 9). As indicated, the original BAP1 is unable to increase the copy number of the pCC1FOS™ fosmid vector; whereas, strains EPI300 and BAP1 (araBADC::Tn10, lacZ::trfA-Kan) containing the inducible trfA gene show copy-up capability (see Note 10).
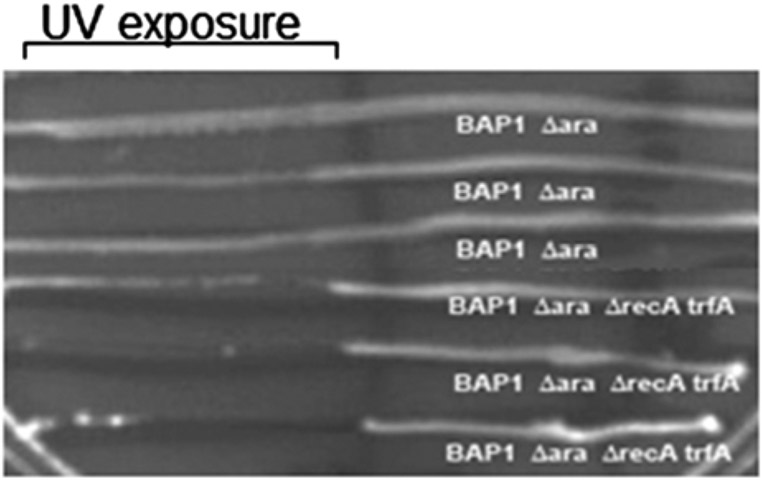
To confirm the reeA deletion in BTRA, assay for sensitivity to UV light, as recA strains are more sensitive than recA+ strains. Grow BAP1 (araBADC::Tn10) and BTRA overnight at 37 °C with shaking in LB medium and then streak strains horizontally across an LB agar plate. Expose one half of the plate to UV light (UVLMS-38 8-W lamp (UVP, Upland, CA)) at 254 nm wavelength and at a distance of 22 cm for 15 s; shield the other half of the plate from UV exposure. Compare the growth of UV-exposed and non-UV-exposed cells after overnight incubation at 37 °C (Fig. 5).
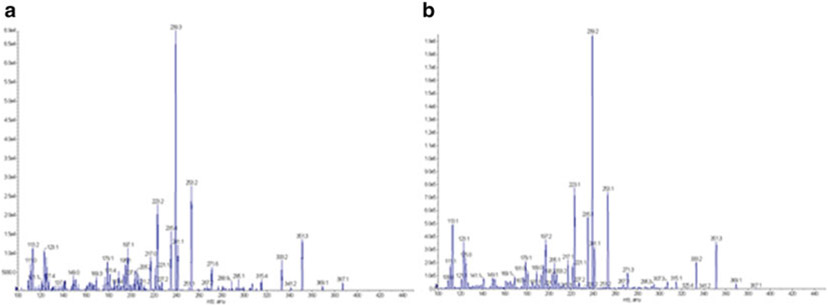
In order to test functionality of the pccAB gene products in BTRAP, obtain the DEBS1 gene from pBP144 [3] via digestion with NdeI and EcoRI and introduce into pET21c to generate plasmid pLF04. The polyketide product 6-deoxyerythronolide B (6dEB), which requires propionyl- and (2S)-methylmalonyl-CoA, is produced by the DEBS1, 2, and 3 enzymes encoded on plasmids pBPJW130 [18] and pLF04. As such, 6dEB production is tested by culturing BTRAP (pLF04/pBPJW130) in 2 mL of LB medium containing selection antibiotics and 20 mM sodium propionate for 5 days at 22 °C with shaking prior to extraction (1:1 by volume) with ethyl acetate, evaporating the solvent under vacuum, and resuspending the extract in methanol for LC-MS analysis (Fig. 6); successful production then confirms the functionality of the introduced pccAB genes (see Note 11).
All MS analyses should be conducted in positive ion mode and chromatography performed on a Waters X Terra C18 column (5 μm, 2.1 mm × 250 mm). After an injection of 10 μL of extract or 6dEB standard, a linear gradient of 30 % buffer A (95 % water–5 % acetonitrile–0.1 % formic acid) to 100 % buffer B (5 % water–95 % acetonitrile–0.1 % formic acid) should be used at a flow rate of 0.2 mL/min.
Fig. 4.
Copy-up test of the constructed E. coli BAP1 (araBADC::Tn10, lacZ::trfA-Kan) by agarose gel electrophoresis. Lanes 1 and 2: negative control BAP1 containing pCCIFOS™ fosmid vector with (+) and without (−) induction (no DNA yield improvement was observed); Lanes 3 and 4: positive control EPI300 containing the pCC1FOS™ fosmid vector with and without induction (DNA yield was improved after induction); Lanes 5 and 6: BAP1 (araBADC::Tn10, lacZ::trfA-Kan) containing the pCC1FOS™ fosmid vector with and without induction (DNA yield was improved after induction)
Fig. 5.
UV test for recA gene deletion. Comparison of BAP1 (araBADC::Tn10) and BAP1 (araBADC::Tn10, lacZ::trfA-Kan, ΔrecA) after UV exposure. The removal of recA results in UV sensitivity
Fig. 6.
Mass spectrum of 6dEB produced from BTRAP (pLF04/pBPJW144) (a) when compared to 6dEB standard (b)
4. Notes
The chloroform used in the P1 transduction procedure may lyse recipient cells, reducing the transduction efficiency. As such, it is recommended that chloroform be evaporated from the P1 phage lysate by air drying before adding P1 to recipient cells.
It is important to give the lambda Red recombination system ample time to function. Therefore, let electroporated cells incubate at 37 °C for a minimum of 3 h; this extends the recovery period and also provides more time for chromosomal integrates to produce sufficient amounts of the selection antibiotic resistance protein.
The BW25113 strain was chosen based upon its common usage and higher efficiency in the lambda Red recombination protocols outlined by Datsenko et al. [9].
Our group has observed a reduced efficiency for the lambda Red procedure when using B strains of E. coli (such as BL21(DE3)) when compared to K strains. Thus, we have adopted an approach in which we first complete the procedure in a K strain of E. coli and then transfer the chromosomal modification to the desired B strain by a second round of lambda Red recombination or P1 transduction.
This is another example of a means of transferring an engineered cassette from a K to a B strain of E. coli.
In our experience, 50 bp homology arms, though efficient when using K E. coli strains, are considered too short for chromosomal engineering in a B strain. We therefore use PCR fragments with a minimum of 200 bp homology for both flanking regions. As a second example, the deletion of recA outlined in Subheading 3.3 featured flanking PCR fragment homology regions of 340 and 850 bp.
For the phenotypic assay to test growth on arabinose, it is important that the inoculum prepared from the LB starter culture is thoroughly washed before inoculating into M9 medium containing arabinose. Residual amounts of nutrients in the LB medium may cause false negative results if not completely removed in the wash step.
Although a high concentration of arabinose may improve the signal associated with the copy-up assay, the deletion of the arabinose operon ensures that even a low concentration of arabinose (2 mg/L) is sufficient for copy number amplification.
For the copy-up comparison, cultures capable of arabinose induction displayed a lower cell density, as the increased copy number of the fosmid presumably imposed intracellular metabolic burden. As such, it is essential that the cell density of the cultures being compared be normalized before isolation and assessment of the fosmid.
In addition to the copy-up test, the replacement of the lacZ gene with the trfA-Kan cassette could also be confirmed by a blue-white screening test. The parent strain with an intact lacZ gene would generate blue colonies on an X-gal plate; whereas, the BT3 strain would only generate white colonies.
Intracellular generation of the (2S)-methylmalonyl-CoA metabolite could also be tested directly by LC-MS analysis, as described previously [19].
Acknowledgement
This work was supported by grants from the NIH (AI074224) and NSF (0712019 and 0924699) in collaboration with EarthGenes Pharmaceuticals. Guidance in strain design and assessment was provided by Drs. Asuncion Martinez, David Rothstein, and Lance Davidow of EarthGenes.
References
- 1.Zhang H, Boghigian BA, Armando J et al. (2011) Methods and options for the heterologous production of complex natural products. Nat Prod Rep 28:125–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ongley SE, Bian X, Neilan BA et al. (2013) Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep 30:1121–1138 [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer BA, Admiraal SJ, Gramajo H et al. (2001) Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790–1792 [DOI] [PubMed] [Google Scholar]
- 4.Golomb M, Chamberlin M (1974) Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem 249:2858–2863 [PubMed] [Google Scholar]
- 5.McAllister WT, Morris C, Rosenberg AH et al. (1981) Utilization of bacteriophage T7 late promoters in recombinant plasmids during infection. J Mol Biol 153:527–544 [DOI] [PubMed] [Google Scholar]
- 6.Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130 [DOI] [PubMed] [Google Scholar]
- 7.Lambalot RH, Gehring AM, Flugel RS et al. (1996) A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol 3:923–936 [DOI] [PubMed] [Google Scholar]
- 8.Quadri LE, Weinreb PH, Lei M et al. (1998) Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585–1595 [DOI] [PubMed] [Google Scholar]
- 9.Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hradecna Z, Wild J, Szybalski W (1998) Conditionally amplifiable inserts in pBAC vectors. Microb Comp Genomics 3:58 [Google Scholar]
- 11.Wild J, Hradecna Z, Szybalski W (2002) Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res 12:1434–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wild J, Szybalski W (2004) Copy-control pBAC/oriV vectors for genomic cloning. Methods Mol Biol 267:145–154 [DOI] [PubMed] [Google Scholar]
- 13.Wild J, Hradecna Z, Szybalski W (2001) Single-copy/high-copy (SC/HC) pBAC/oriV novel vectors for genomics and gene expression. Plasmid 45:142 [Google Scholar]
- 14.Le Borgne S, Palmeros B, Valle F et al. (1998) pBRINT-Ts: a plasmid family with a temperature-sensitive replicon, designed for chromosomal integration into the lacZ gene of Escherichia coli. Gene 223:213–219 [DOI] [PubMed] [Google Scholar]
- 15.Phue JN, Lee SJ, Trinh L et al. (2008) Modified Escherichia coli B (BL21), a superior producer of plasmid DNA compared with Escherichia coli K (DH5alpha). Biotechnol Bioeng 101:831–836 [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez E, Gramajo H (1999) Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolor A3 (2). Microbiology 145:3109–3119 [DOI] [PubMed] [Google Scholar]
- 17.Chan YA, Podevels AM, Kevany BM et al. (2009) Biosynthesis of polyketide synthase extender units. Nat Prod Rep 26:90–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang H, Wang Y, Wu J, Skalina K et al. (2010) Complete biosynthesis of erythromycin A and designed analogs using E. coli as a heterologous host. Chem Biol 17:1232–1240 [DOI] [PubMed] [Google Scholar]
- 19.Armando JW, Boghigian BA, Pfeifer BA (2012) LC-MS/MS quantification of short-chain acyl-CoA’s in Escherichia coli demonstrates versatile propionyl-CoA synthetase substrate specificity. Lett Appl Microbiol 54:140–148 [DOI] [PubMed] [Google Scholar]