Abstract
Understanding how organelles interact, exchange materials, assemble, disassemble, and evolve as a function of space, time, and environment is an exciting area at the very forefront of chemical and cell biology. Here, we bring attention to recent progress in the design and application of lipid-based tools to visualize and interrogate organelles in live cells, especially at super resolution. We highlight strategies that rely on modification of natural lipids or lipid-like small molecules ex cellula, where organelle specificity is provided by the structure of the chemically modified lipid, or in cellula using cellular machinery, where an enzyme labels the lipid in situ. We also describe recent improvements to the chemistry upon which lipid probes rely, many of which have already begun to broaden the scope of biological questions that can be addressed by imaging organelle membranes at the nanoscale.
Keywords: Super-resolution microscopy, Lipid, Organelle Interactome, Fluorogenic, Fluorophore, Bioorthogonal chemistry
Introduction
Organelles are how the cell segregates biochemistry. Alone and together, organelles drive eukaryotic cell biology. Many cellular functions are marked by dynamic changes in organelle structure and interaction, and organelle dysfunction underlies a plethora of metabolic and neurologic diseases. Understanding how organelles interact, exchange materials, assemble, disassemble, and evolve as a function of space, time, and environment is an exciting area at the very forefront of chemical and cell biology.
Historically, changes in organelle structure, function, and interaction have been visualized using fluorescence microscopy and organelle-specific proteins tagged with a fluorescent protein or small-molecule dye. Recently, lipids and lipid-like small molecules have been shown to offer distinct and unique advantages for imaging organelle structure, function, and interaction, especially at the nanoscale.
Besides demarking a more continuous organelle boundary, one advantage of labeling an organelle membrane with a lipid probe, as opposed to a membrane-embedded protein, is labeling density [1]. Excepting polymeric proteins such as actin and tubulin, the density of lipids in a membrane exceeds that of protein by more than 100-fold [2]. This increase in density facilitates super-resolution microscopy (SRM), as resolution depends integrally on the number of detectable molecules. The Nyquist-Shannon sampling criterion demands that the labeling density be greater than the desired resolution by at least a factor of two [3]. Additionally, the increase in labelling density also results in an almost equivalent increase in imaging time, even at super resolution. Moreover, as lipids and lipid-like small molecules (or their precursors) are often cell-permeant, they permit experimentation with primary or patient-derived cells as well as cultured cell lines that are notoriously difficult to genetically manipulate.
In this current opinion, we bring attention to recent progress in the design and application of tools that visualize organelles — predominantly in live cells — at super resolution using stimulated emission depletion (STED) [4,5] and single-molecule localization microscopy (SMLM) [6–9]. For those interested in tools suitable for predominantly confocal methods, the reader is referred to several excellent recent reviews: [10–12]. We focus broadly on three approaches: (1) modification of natural lipids ex cellula, where organelle specificity is provided by the structure of the chemically modified lipid (Figure 1a), (2) modification of lipids in cellula using cellular machinery, where an enzyme labels the lipid in situ (Figure 1b), and (3) the design of lipid-like small molecules that localize selectively within specific organelle membranes (Figure 1c). We also highlight improvements to the chemistry upon which lipid probes rely, including strategies to encode high fluorogenicity, and new, bright, versatile fluorophores and bioorthogonal reactions, many of which have already begun to broaden the scope of biological questions that can be addressed by imaging organelle membranes at the nanoscale.
Figure 1.
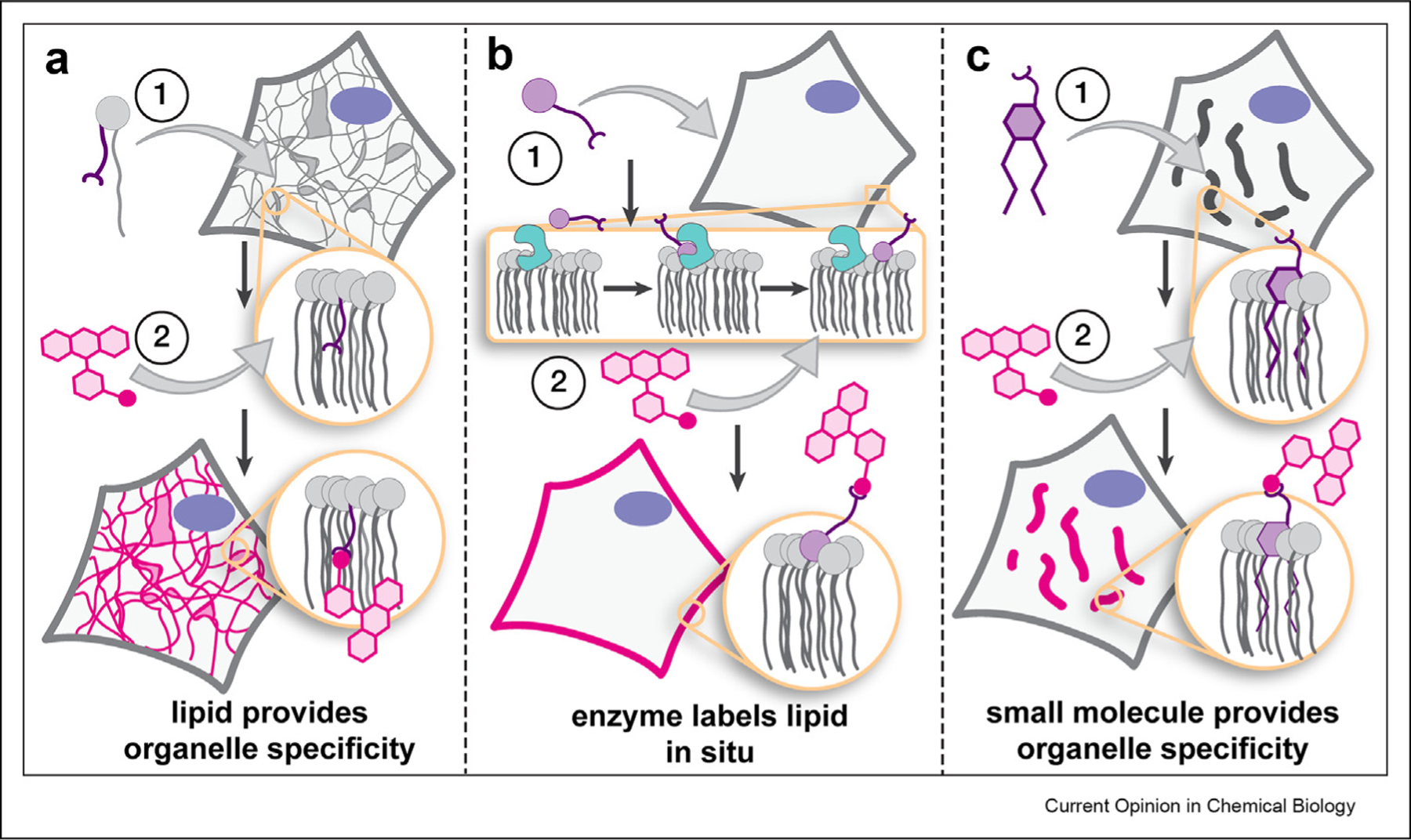
General strategies to image organelles and/or track lipids selectively, often at the nanoscale, which exploit (a) lipid structure; (b) organelle-restricted enzymes; and (c) organelle-targeting small molecules to limit fluorescence to the organelle of interest.
Imaging organelles at the nanoscale with lipid probes
One way to image organelles selectively for nanoscopy is to fluorescently tag the molecules that comprise the membranes themselves — the lipids. This strategy exploits the fact that different organelles are often enriched in different lipids [13]. Pioneering work by Pagano [14,15] demonstrated over thirty years ago that ceramide-like lipids tagged with a boron dipyrromethene difluoride (BODIPY) or nitrobenzoxadiazole (NBD) fluorophore could visualize the Golgi apparatus and interrogate Golgi–plasma membrane trafficking. The utility of BODIPY-Cer and NBD-Cer catalyzed the development of commercially available ceramide-derived fluorescent probes to interrogate Golgi trafficking and sphingolipid transport/metabolism. Although the commercially available probes are used widely, their reliance on relatively nonphotostable NBD and BODIPY fluorophores limits their utility for super-resolution methods such as STED and SMLM.
One strategy [16,17] to expand the utility of ceramide-like probes for super-resolution imaging uses bespoke analogs that carry photostable fluorophores such as SiR [18] (useful for STED) and HMSiR [19] (useful for SMLM). These super-resolution lipid probes are assembled in cellula: a ceramide-derivative such as Cer-TCO (Figure 2a(i)) is added to cells, whereupon it localizes to the Golgi or the ER depending on temperature [20–22]. Next, a tetrazine-tagged fluorophore such as SiR-Tz or HMSiR-Tz is added, tetrazine ligation [23,24] ensues, and the organelle membrane is labeled selectively (Figure 2b). In this case, organelle selectivity is provided by the inherent trafficking/localization properties of the lipid (Figure 1a); subsequent reaction with the fluorophore enables three-dimensional (3D) live-cell nanoscopy [16,17]. In the case of the probe assembled from Cer-TCO and HMSiR-Tz, SMLM is achieved without toxic additives or laser-induced photoswitching.
Figure 2. Lipid-focused strategies to image organelles and/or study their trafficking.
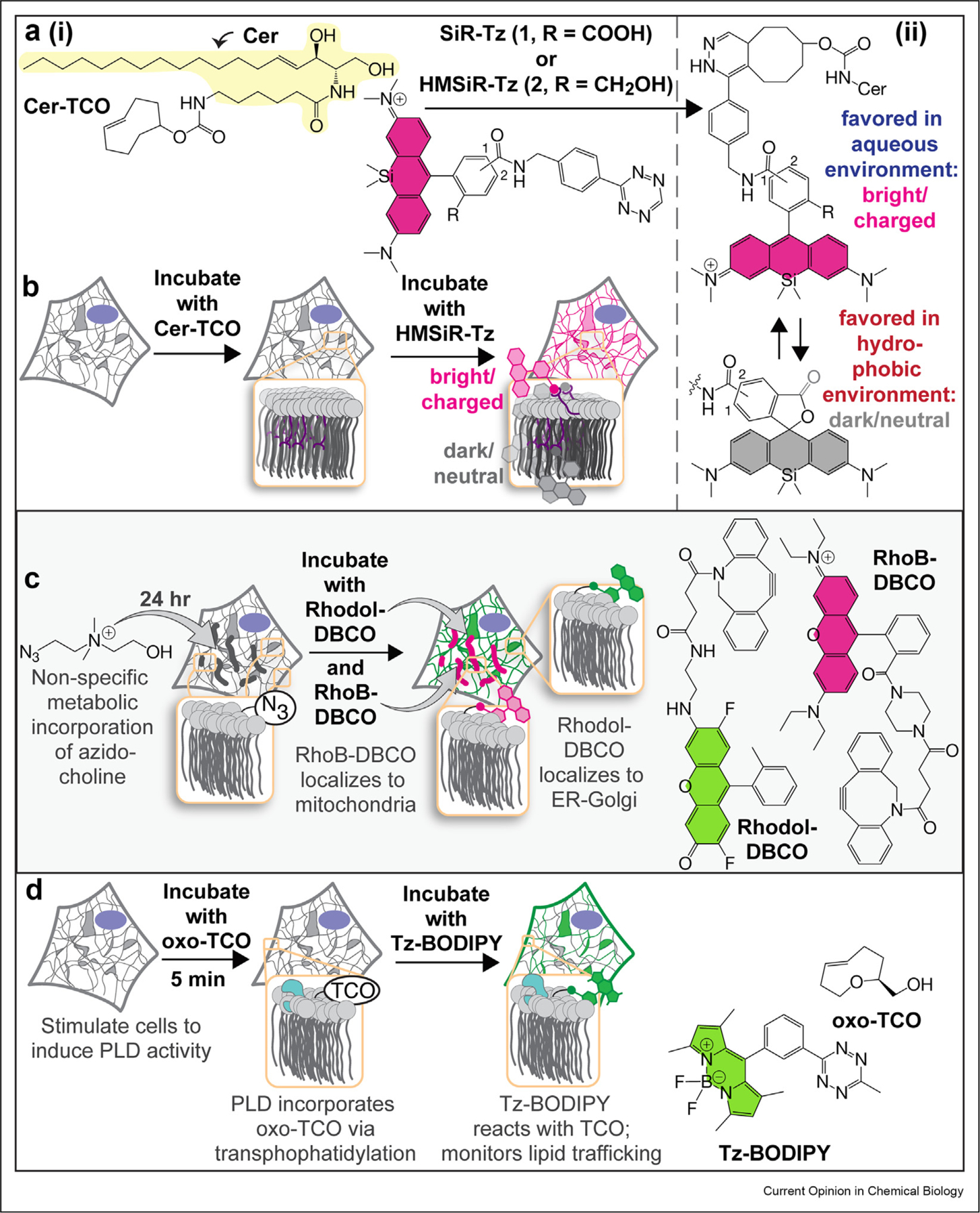
(a) (i) Cer-TCO, which localizes in a temperature-dependent way to the Golgi or ER and reacts and (ii) the environment-dependent spirocyclization equilibria of SiR and HMSiR that effectively extends imaging times by ‘hiding’ a reservoir of dark-state dyes in the membrane. (b) in cellula with tetrazine-containing fluorophores such as SiR and HMSiR to enable STED and SMLM, respectively. (c) Biosynthetic incorporation of azido-choline into phospholipids followed by coincubation with a click-compatible dye fused to an organelle-targeting small molecule results in selective localization of the labeled lipid to the organelle of interest. (d) Incorporation of oxo-TCO handle viaPLD-mediated transphosphatidylation, followed by fluorogenic Tz-BODIPY results in real-time visualization of PLD activity. BODIPY, boron dipyrromethene difluoride; ER, endoplasmic reticulum; PLD, phospholipase D; SMLM, single-molecule localization microscopy; STED, stimulated emission depletion; HMSiR, hydroxymethyl silicon rhodamine; SiR, silicon rhodamine; DBCO, dibenzocyclooctyne; TCO, trans-cyclooctyne.
A less obvious advantage of labeling an organelle membrane directly is a truly significant increase in imaging time [17]. In a particularly dramatic example, labeling the ER with the lipid probe generated by reaction of Cer-TCO and HMSiR-Tz generates SMLM images that last almost 30 min; when the ER is labeled with HMSiR attached to a HaloTag-Sec61β fusion protein, the images last for only a minute [17]. The extended imaging times also influence image quality for SMLM images, allowing a user to coalesce more blinking events over multiple frames for improved reconstruction. The increase in imaging time is due to the fact that dyes such as HMSiR and SiR exist in two states, only one of which is fluorescent (Figure 2a(ii)), and the nonfluorescent form is favored within the membrane environment. Thus, the membrane holds an excess reservoir of dye molecules that are ‘hiding’ in the lipid and can be used to replenish dyes that are bleached over time. This modular two-component approach using HIDE (HIgh Density Environment-Sensitive) probes demonstrates outstanding SRM imaging when compared with previous ceramide derivatives and protein-based labeling techniques.
Enzymatically modifying lipids for super-resolution imaging
A conceptually related approach to image organelles was reported recently by Hamachi [25**] (Figure 2c). In this case, azido-choline is enzymatically incorporated in cellula, to modify a choline-containing phospholipid. This modified phospholipid is then capable of a bioorthogonal click (SPAAC, Strain-Promoted Azide-Alkyne Cycloaddition) [26–28] reaction for subsequent fluorophore labelling.
Coincubation with a click-compatible dye fused to an organelle-targeting small molecule results in selective localization of the labeled lipid to the organelle of interest. This strategy is effectively the inverse of the HIDE strategy — rather than using the lipid to localize the dye, in this case, the dye is used to localize the lipid (Figure 1b). This strategy enabled structured illumination microscopy (SIM) of the mitochondria and confocal studies of interorganelle translocation of phosphatidyl-choline (PC).
Baskin et al. also recently reported a third creative way to selectively image organelle lipids, in this case by exploiting the promiscuity of phospholipase D (PLD) [29,30**]] (Figure 2d). PLD hydrolyzes phospholipids but can make use of alcohols as substrates in place of water — the result is a trans-phosphatidylation reaction, not hydrolysis. If the alcohol carries a trans-cyclooctene, the newly synthesized phospholipid can react with a fluorogenic tetrazine dye. Using this enzymatic tool, Baskin et al. achieved adequate temporal resolution to directly observe PLD activity and subsequent trafficking of its lipid substrates. Although this work did not report super-resolution experiments, the chemistry described could easily be adapted for this purpose. Lipid biosynthesis offers many potential avenues to selectively label lipids in cellula to observe their trafficking and metabolism [31–35].
Imaging organelle membranes at the nanoscale with lipid-like small molecules
Another versatile method for super-resolution imaging of organelle membranes exploits lipid-like small molecules that interact selectively with organelle membranes. Indeed, some of the earliest examples of live-cell super-resolution microscopy described by Zhuang exploited small-molecule fluorophores such as DiI, MitoTracker Red, ER-Tracker Red, and LysoTracker Red. These molecules could be induced to blink by strong laser excitation and subsequent reactivation by ultraviolet light to visualize the plasma membrane, mitochondrial inner membrane, endoplasmic reticulum, and lysosome, respectively, using Stochastic Optical Reconstruction Microscopy (STORM) [1].
One strategy to avoid the need for multiple lasers or additives to induce photoswitching makes use of the spontaneously blinking fluorophore HMSiR [19] paired with some of the small-molecule fluorophores described earlier, but tagged with trans-cyclooctene to enable in cellula tetrazine ligation [17,36**,37]. This strategy has been used to image multiple different organelles using SMLM methods, including the plasma membrane (HMSiR-DiI) (Figure 3b) and mitochondria (HMSiR-RhoB), both of which are assembled in cellula via a tetrazine ligation. Analogous experiments using DiI-C16-SiR, a lipid-like small molecule that localizes to endolysosomes, could detect both subtle and dramatic endosome motility defects in samples of patients with Niemann Pick disease using STED (Figure 3a). In all of these cases, because HMSiR and SiR exist in two states, imaging times are exceptionally long [36**,38**]. It should be noted that the membrane-targeting molecules in the aforementioned examples are intrinsically fluorescent and thereby occupy an imaging channel. For multicolor experiments targeting multiple organelles, this limits the number of channels that can be acquired simultaneously.
Figure 3. Membrane-targeting small-molecule probes for super-resolution microscopy of organelles.
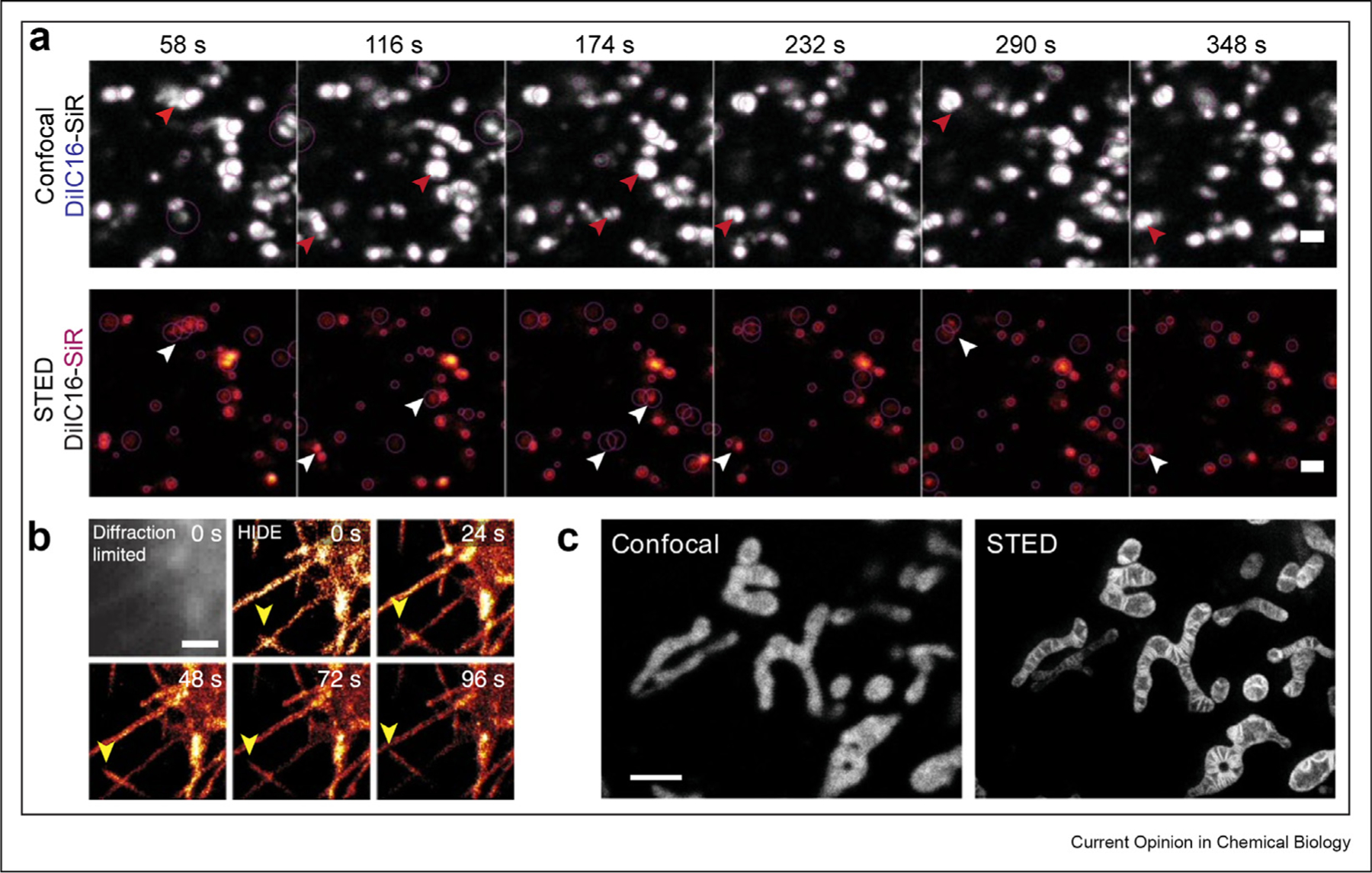
(a) Visualizing endosomal motility defects with DiI-C16-SiR in wild-type fibroblasts from patients with Niemann-Pick C disease; scale bar: 1 μm. Reprinted by permission from Springer Nature Customer Service Centre GmbH: Nature Chemical Biology, Endosome motility defects revealed at super resolution in live cells using HIDE probes, Gupta et al., 2020 (b) SMLM time-lapse images of plasma membrane filopodia acquired using DiI-HMSiR; scale bar: 1 μm. Reprinted by permission from Springer Nature Customer Service Centre GmbH: Nature Biotechnology, Long time-lapse nanoscopy with spontaneously blinking membrane probes, Takakura et al., 2017 (c) Confocal vs. STED images of the mitochondria in HeLa cells using MitoPB Yellow; scale bar: 2 μm. Reprinted from Wang C et al.,: A photostable fluorescent marker for the super-resolution live imaging of the dynamic structure of the mitochondrial cristae. Proc Natl Acad Sci U S A 2019, 116:15817–15822. SMLM, single-molecule localization microscopy; STED, stimulated emission depletion.
More recent examples of small molecules that enable plasma membrane–specific super resolution include multicolored, fluorogenic MemBright dyes [39*] (Figure 3c). These cyanine-derived molecules are amphiphilic and form non–fluorescent-soluble aggregates under cell culture conditions; the dyes light up when they distribute throughout the membrane. MemBright Cy3.5 supported the use of 3D STORM to dynamically image a synapse coiling around a dendrite, and in combination with immunostaining, multicolor STORM revealed the accumulation of glutamate receptors at axon–dendritic junctions in fixed cells. Styryl dyes were also recently improved for enhanced brightness and photostability and shown to support imaging of hippocampal neurons via STED [40*].
A unique and enabling attribute of certain fluorophores, such as Nile Red (Figure 3d), is solvatochromism, in which the emission spectrum varies as a function of solvent polarity. In early work, Xu et al. used Nile Red to image spatially resolved polarity changes across the plasma membrane and the membranes of nanoscale intracellular organelles in an unbiased manner [41]. More recently, Nile Red derivatives carrying lipid chains were shown to localize the dye to only the plasma membrane. Short-chain and long-chain derivatives with improved photophysical properties that display reversible and irreversible membrane binding have been designed, respectively [42*]. Irreversible binding is desirable for traditional confocal microscopy, and reversible labeling was utilized in Super Resolution-Points Accumulation for Imaging in Nanoscale Topography (SR-PAINT) microscopy to image nanoscale heterogeneity in lipid order within the plasma membrane. This kind of reversible binding shows promise for time-lapse STED microscopy [43], as new molecules are available to replace photobleached molecules over time.
A naphthophosphole fluorophore demonstrating remarkable photostability (MitoPB Yellow) was developed with a mitochondria-targeting triphenylphosphine moiety and a protein-cross-linking epoxide, facilitating STED imaging (Figure 3c) even after membrane potential loss due to fixation [44*]. Cyanine- and coumarin-based derivatives have also been reported for imaging lipid droplets with two-photon excitation microscopy and STED [45,46], and 3D STED has enabled the quantification of lipid droplet count and size in cells. All the probes that have been discussed earlier have been categorized and listed in a supplementary table (Fig. S1).
Looking to the future: enabling chemistry for more sophisticated experimentation
There has also been excellent recent progress in technologies that increase the sophistication of experiments possible with lipid and lipid-like small-molecule probes. One big issue is labeling specificity, which can be improved with dyes whose emission intensity increases only when properly localized or assembled. Several groups have reported fluorogenic tetrazine and azide probes whose fluorescence is quenched until bound by its target [47–52]. A particularly creative approach to improved fluorogenicity was reported by Wombacher et al., yielding a spontaneously blinking HMSiR analog (f-HM-SiR) useful for live-cell SMLM whose fluorescence increases 10-fold upon reaction with bicyclo [6.1.0]non-4-yne (BCN). f-HM-SiR allows for no-wash imaging and reduces unwanted background signal to improve localization precision [53**] (Figure 4a).
Figure 4. Supporting technologies for super-resolution imaging with lipid-based probes.
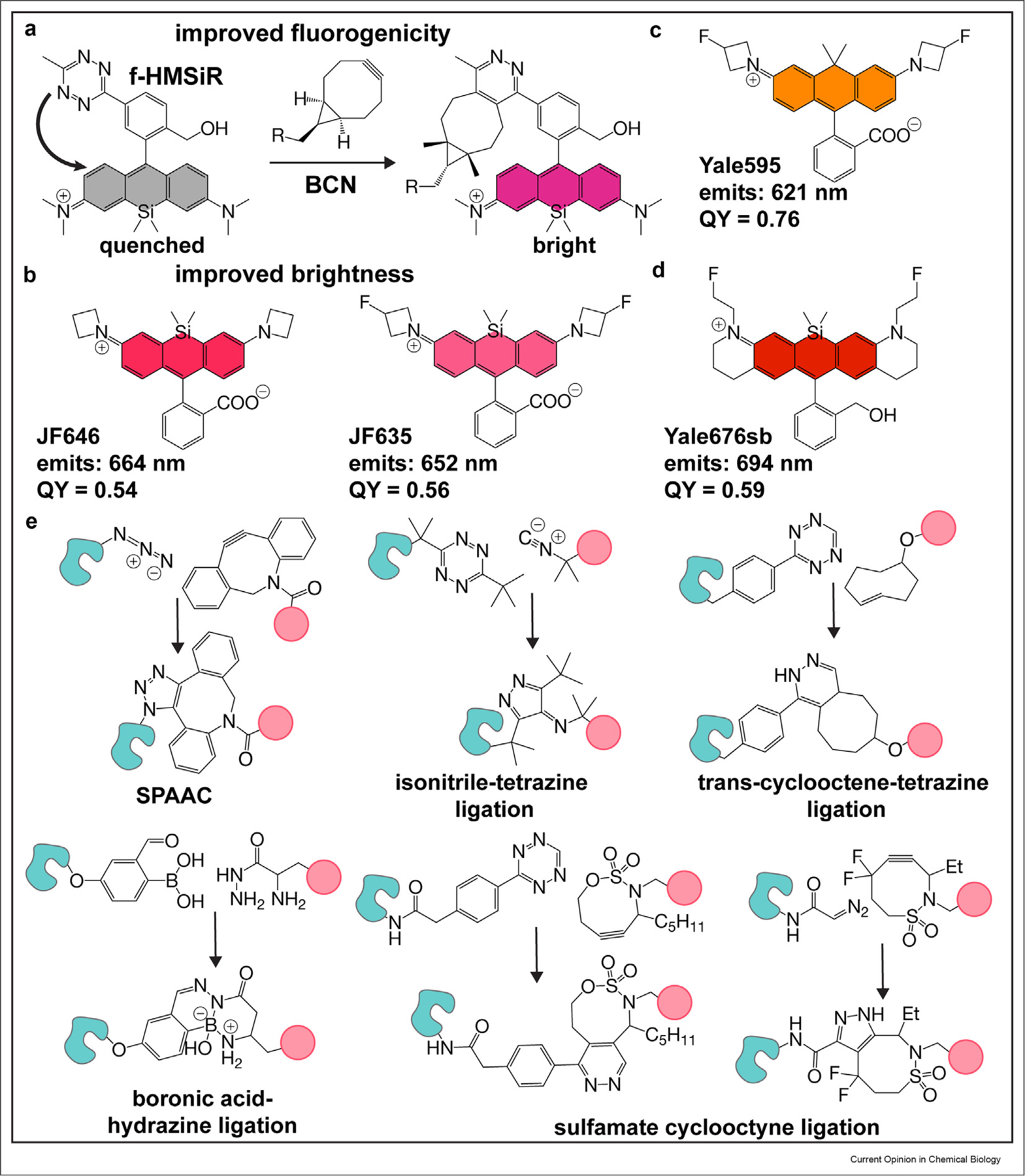
(a) Fluorogenic spontaneously blinking fluorophores reported by Wombacher et al. Upon reacting with bicyclo [6.1.0]non-4-yne (BCN), the fluorescence intensity of the HM-SiR analog increases, thereby enabling no-wash, live-cell SRM. (b) Brighter fluorophores. Improvements to SiR by Lavis et al. resulted in near-infrared fluorophores that absorb at similar wavelengths to SiR, but with increased brightness and better photophysical properties. (c) Tuning dyes for hydrophobic environments. Yale595 was developed by Chu et al. to high brightness in hydrophobic environments. (d) Development of a near-infrared, spontaneously blinking fluorophore. Yale676sb was developed as a bright, spectrally separable fluorophore from HMSiR. The development of this fluorophore has enabled two-color ratiometric SMLM, using NIR dyes and a single laser. (e) Mutually orthogonal bioorthogonal ligations. Improvements to the isonitrile-tetrazine ligation by Franzini et al., and development of the sulfamate-cyclooctyne ligation by Schomaker et al. have led to the development of triply orthogonal biorthogonal reaction pairs. SRM, super-resolution microscopy; SMLM, single-molecule localization microscopy; NIR, near-infrared.
Another potentially useful approach utilizes polarity-dependent photoactivation, which leads to a fluorescent product only within apolar media. A probe based on this molecular logic gate was demonstrated to label lipid droplets with very high specificity [54].
All of the experiments described earlier would be impossible without photostable fluorophores whose physical properties — absorption and emission maxima, quantum yield, spirocyclization equilibria (pKcycle), dielectric constant, fluorogenicity — are appropriate for the super-resolution modality (STED, SMLM, SIM, etc.), cell environment (pH), and optical setup of the commercial or custom instrument used. Lavis et al. developed a number of azetidine-containing Janelia dyes with improved brightness, photostability, and fluorogenicity relative to classical tetramethyl-rhodamines such as the commonly used SiR and tetramethyl rhodamine (TMR) [55,56]. In particular, JF646 and JF635 (Figure 4b) exhibit higher quantum yields (0.54 and 0.56, respectively) [55] than SiR (0.41) but emit at approximately the same wavelength [56].
Despite these advances, real challenges remain, most notably the lack of fluorophores that satisfy all the criteria cited earlier but also emit at a wavelength that does not overlap with widely used silicon-rhodamine dyes such as SiR-COOH (for STED/SIM), JF646 (for STED) or HMSiR (for SMLM). Such dyes are essential for multicolor experiments needed to reveal organelle interactions in both space and time. Notable progress includes the recent report of Yale595, an azetidine dye whose electron-withdrawing substituents have been tuned to promote high brightness even within a nonpolar lipid environment [38**]. Yale595 (Figure 4c) was used alongside SiR to enable two-color super resolution (STED) and 3D confocal imaging of the plasma membrane and either the ER or mitochondria. In more recent progress, Tyson et al. judiciously balanced the pKcycle and emission wavelength of a spontaneously blinking silicon-rhodamine fluorophore to develop Yale676sb, which was paired with HMSiR to enable two-color SMLM of the ER and mitochondria using a single far-red laser [57**]. Yale676sb (Figure 4d) is distinguished not only by its emission wavelength (694 nm) and pKcycle but also by an exceptional quantum yield (0.59).
Several improvements have also been made to expand the bioorthogonal ligation toolkit to include new, mutually orthogonal chemistries. Franzini et al. described an isonitrile-tetrazine ligation [58**] (Figure 4e) that is promoted by dispersion forces. In addition to favorable reaction kinetics, this new reaction is mutually orthogonal to both SPAAC [26–28] and TCO-tetrazine ligations [23,24]. Similarly, Schomaker et al. demonstrated mutual orthogonality between sulfamate-containing cyclooctynes by tuning the electronics of the reaction pairs [59]. Using two newly developed reaction partners, and a boronic acid/hydrazine ligation, they too demonstrated simultaneous triple labelling (Figure 4e). These mutually orthogonal chemistries enable the modular delivery of multiple fluorophore-probe pairs for multicolor imaging. With the development of additional mutually orthogonal chemistries, in tandem with other technologies described earlier [25,29,30], one could easily envision experiments that combine different lipid reporters to interrogate time-dependent lipid trafficking between multiple organelles, in addition to observing individual organelles at subdiffraction resolutions.
Conclusions
The last few years have witnessed great progress in the design and application of lipids and lipid-like small molecules for imaging organelle structure, function, and interaction, especially at the nanoscale. Multiple approaches have been described for both labeling and localizing lipids selectively, and in certain cases, these events have led to new biological knowledge that would be impossible to obtain without the lipid probe. There has also been much progress in chemistry upon which lipid probes rely, including strategies to encode high fluorogenicity, and new, bright, versatile fluorophores and bioorthogonal reactions. Many of these advances have already begun to broaden the scope of biological questions that can be addressed by imaging organelle membranes at the nanoscale.
Supplementary Material
Acknowledgements
This work was supported by the NIH (1R01GM131372 and 1R35GM134963 to A.S.).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cbpa.2021.09.003.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Shim S-H, Xia C, Zhong G, Babcock HP, Vaughan JC, Huang B, et al. : Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc Natl Acad Sci U S A 2012, 109:13978–13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinn P, Griffiths G, Warren G: Density of newly synthesized plasma membrane proteins in intracellular membranes II. Biochemical studies. J Cell Biol 1984, 98:2142–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernández-Suárez M, Ting AY: Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol 2008, 9: 929–943. [DOI] [PubMed] [Google Scholar]
- 4.Klar TA, Jakobs S, Dyba M, Egner A, Hell SW: Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci Unit States Am 2000, 97:8206–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hell SW, Kroug M: Ground-state-depletion fluorscence microscopy: a concept for breaking the diffraction resolution limit. Appl Phys B 1995, 60:495–497. [Google Scholar]
- 6.Rust MJ, Bates M, Zhuang X: Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Methods 2006, 3:793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. : Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313:1642–1645. [DOI] [PubMed] [Google Scholar]
- 8.Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, et al. : Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed 2008, 47:6172–6176. [DOI] [PubMed] [Google Scholar]
- 9.Hess ST, Girirajan TPK, Mason MD: Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J 2006, 91:4258–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores J, White BM, Brea RJ, Baskin JM, Devaraj NK: Lipids: chemical tools for their synthesis, modification, and analysis. Chem Soc Rev 2020, 10.1039/d0cs00154f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neef AB, Schultz C: Selective fluorescence labeling of lipids in living cells. Angew Chem Int Ed 2009, 48:1498–1500. [DOI] [PubMed] [Google Scholar]
- 12.Bumpus TW, Baskin JM: Greasing the wheels of lipid biology with chemical tools. Trends Biochem Sci 2018, 43:970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Meer G, Voelker DR, Feigenson GW: Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008, 9:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagano RE, Martin OC, Kang HC, Haugland RP: A novel fluorescent ceramide analogue for studying membrane traffic in animal cells: accumulation at the Golgi apparatus results in altered spectral properties of the sphingolipid precursor. J Cell Biol 1991, 113:1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipsky N, Pagano R: A vital stain for the Golgi apparatus. Science 1985, 228:745–747. [DOI] [PubMed] [Google Scholar]
- 16.Erdmann RS, Toomre D, Schepartz A: STED imaging of Golgi dynamics with cer-SiR: a two-component, photostable, high-density lipid probe for live cells. In Super-resolution microscopy: methods and protocols Edited by Erfle H; 2017:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takakura H, Zhang Y, Erdmann RS, Thompson AD, Lin Y, McNellis B, et al. : Long time-lapse nanoscopy with spontaneously blinking membrane probes. Nat Biotechnol 2017, 35: 773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukinavičius G, Umezawa K, Olivier N, Honigmann A, Yang G, Plass T, et al. : A near-infrared fluorophore for live-cell super-resolution microscopy of cellular proteins. Nat Chem 2013, 5: 132–139. [DOI] [PubMed] [Google Scholar]
- 19.Uno S, Kamiya M, Yoshihara T, Sugawara K, Okabe K, Tarhan MC, et al. : A spontaneously blinking fluorophore based on intramolecular spirocyclization for live-cell super-resolution imaging. Nat Chem 2014, 6:681–689. [DOI] [PubMed] [Google Scholar]
- 20.Kuliawat R, Arvan P: Protein targeting via the “constitutive-like” secretory pathway in isolated pancreatic islets: passive sorting in the immature granule compartment. J Cell Biol 1992, 118:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths G, Pfeiffer S, Simons K, Matlin K: Exit of newly synthesized membrane proteins from the trans cisterna of the Golgi complex to the plasma membrane. J Cell Biol 1985, 101: 949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon JP, Ivanov IE, Adesnik M, Sabatini DD: The production of post-Golgi vesicles requires a protein kinase C-like molecule, but not its phosphorylating activity. J Cell Biol 1996, 135: 355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackman ML, Royzen M, Fox JM: Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels−Alder reactivity. J Am Chem Soc 2008, 130:13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devaraj NK, Weissleder R, Hilderbrand SA: Tetrazine-based cycloadditions: application to pretargeted live cell imaging. Bioconjugate Chem 2008, 19:2297–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.**. Tamura T, Fujisawa A, Tsuchiya M, Shen Y, Nagao K, Kawano S, et al. : Organelle membrane-specific chemical labeling and dynamic imaging in living cells. Nat Chem Biol 2020, 16: 1361–1367. This work labeled phospholipids of the entire cell with azido-containing choline via metabolic incorporation. Imaging of ER, mitochondria and lipid trafficking was subsequently enabled by bioorthogonal labeling with organelle specific dyes.
- 26.Agard NJ, Prescher JA, Bertozzi CR: A strain-promoted [3 + 2] Azide−Alkyne cycloaddition for covalent modification of biomolecules in living systems. J Am Chem Soc 2004, 126: 15046–15047. [DOI] [PubMed] [Google Scholar]
- 27.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, et al. : Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci Unit States Am 2007, 104: 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sletten EM, Bertozzi CR: From mechanism to mouse: a tale of two bioorthogonal reactions. Acc Chem Res 2011, 44:666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bumpus TW, Baskin JM: Clickable substrate mimics enable imaging of phospholipase D activity. ACS Cent Sci 2017, 3: 1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.**. Liang D, Wu K, Tei R, Bumpus TW, Ye J, Baskin JM: A real-time, click chemistry imaging approach reveals stimulus-specific subcellular locations of phospholipase D activity. Proc Natl Acad Sci Unit States Am 2019, 116:15453–15462. This work demonstrated precise subcellular locations of PLD activity. The successful utilization of TCO-containing alcohol for PLD catalyzed transphosphatidylation enabled a real-time observation.
- 31.Thiele C, Papan C, Hoelper D, Kusserow K, Gaebler A, Schoene M, et al. : Tracing fatty acid metabolism by click chemistry. ACS Chem Biol 2012, 7:2004–2011. [DOI] [PubMed] [Google Scholar]
- 32.Pérez AJ, Bode HB: ω-Azido fatty acids as probes to detect fatty acid biosynthesis, degradation, and modification. J Lipid Res 2014, 55:1897–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng T, Hang HC: Bifunctional fatty acid chemical reporter for analyzing S-palmitoylated membrane protein-protein interactions in mammalian cells. J Am Chem Soc 2015, 137: 556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won SJ, Martin BR: Temporal profiling establishes a dynamic S-palmitoylation cycle. ACS Chem Biol 2018, 13:1560–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thinon E, Hang HC: Chemical reporters for exploring protein acylation. Biochem Soc Trans 2015, 43:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.**. Gupta A, Rivera-Molina F, Xi Z, Toomre D, Schepartz A: Endosome motility defects revealed at super-resolution in live cells using HIDE probes. Nat Chem Biol 2020, 16:408–414. This publication reports a bioorthogonally reactive DiIC16 endosome probe that can intracellularly attach to a STED-compatible fluorophore.
- 37.Thompson AD, Omar MH, Rivera-Molina F, Xi Z, Koleske AJ, Toomre DK, et al. : Long-term live-cell STED nanoscopy of primary and cultured cells with the plasma membrane HIDE probe DiI-SiR. Angew Chem Int Ed 2017, 56:10408–10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.**. Chu L, Tyson J, Shaw JE, Rivera-Molina F, Koleske AJ, Schepartz A, et al. : Two-color nanoscopy of organelles for extended times with HIDE probes. Nat Commun 2020, 11:4271. A azetidine-based carborhodamine, Yale595, is reported that enables two-color STED imaging of organelles for extended time frames. This probe is the first to describe two-color imaging with HIDE probes by simultaneously employing Tz-ligation and SPAAC. The authors successfully image PM and mitochondria and PM and ER simultaneously in live cells over >5 min under continuous illumination.
- 39.*. Collot M, Ashokkumar P, Anton H, Boutant E, Faklaris O, Galli T, et al. : MemBright: a family of fluorescent membrane probes for advanced cellular imaging and neuroscience. Cell Chem Biol 2019, 26:600 [−+]. A series of six cyanine membrane dyes were reported that selectively label the plasma membrane, and they were used in multicolor 3D STORM experiments to visualize clustering of glutamate receptors in dendritic spines.
- 40.*. Collot M, Boutant E, Fam KT, Danglot L, Klymchenko AS: Molecular tuning of styryl dyes leads to versatile and efficient plasma membrane probes for cell and tissue imaging. Bioconjugate Chem 2020, 31:875–883. The authors improved the photophysical properties of styrl plasma membrane probe FM1–43 to achieve STED imaging of neurons.
- 41.Moon S, Yan R, Kenny SJ, Shyu Y, Xiang L, Li W, et al. : Spectrally resolved, functional super-resolution microscopy reveals nanoscale compositional heterogeneity in live-cell membranes. J Am Chem Soc 2017, 139:10944–10947. [DOI] [PubMed] [Google Scholar]
- 42.*. Danylchuk D, Moon S, Xu K, Klymchenko AS: Switchable solvatochromic probes for live-cell super-resolution imaging of plasma membrane organization. Angew Chem Int Ed 2019, 58:14920–14924. Two switchable polarity-sensitive membrane probes are reported, one of which was shown to be suitable for SR-PAINT and was used to achieve super-resolution imaging of plasma membrane lipid order in human cells.
- 43.Spahn C, Grimm JB, Lavis LD, Lampe M, Heilemann M: Whole-cell, 3D, and multicolor STED imaging with exchangeable fluorophores. Nano Lett 2018, 19:500–505. [DOI] [PubMed] [Google Scholar]
- 44.*. Wang C, Taki M, Sato Y, Tamura Y, Yaginuma H, Okada Y, et al. : A photostable fluorescent marker for the superre-solution live imaging of the dynamic structure of the mitochondrial cristae. Proc Natl Acad Sci U S A 2019, 116:15817–15822. The authors report MitoPB Yellow, a super-photostable mitochondrial inner-membrane probe that enabled STED imaging of mitochondrial cristae.
- 45.Collot M, Fam TK, Ashokkumar P, Faklaris O, Galli T, Danglot L, et al. : Ultrabright and fluorogenic probes for multicolor imaging and tracking of lipid droplets in cells and tissues. J Am Chem Soc 2018, 140:5401–5411. [DOI] [PubMed] [Google Scholar]
- 46.Xu H, Zhang H, Liu G, Kong L, Zhu X, Tian X, et al. : Coumarin-based fluorescent probes for super-resolution and dynamic tracking of lipid droplets. Anal Chem 2019, 91:977–982. [DOI] [PubMed] [Google Scholar]
- 47.Wieczorek A, Werther P, Euchner J, Wombacher R: Green- to far-red-emitting fluorogenic tetrazine probes – synthetic access and no-wash protein imaging inside living cells. Chem Sci 2017, 8:1506–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J, Šečkute_ J, Cole CM, Devaraj NK: Live-cell imaging of cyclopropene tags with fluorogenic tetrazine cycloadditions. Angew Chem Int Ed 2012, 51:7476–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galeta J, Dzijak R, Obořil J, Dračínský M, Vrabel M: A systematic study of coumarin–tetrazine light-up probes for bioorthogonal fluorescence imaging. Chem Eur J 2020, 26:9945–9953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kozma E, Girona GE, Paci G, Lemke EA, Kele P: Bioorthogonal double-fluorogenic siliconrhodamine probes for intracellular super-resolution microscopy. Chem Commun 2017, 53: 6696–6699. [DOI] [PubMed] [Google Scholar]
- 51.Shieh P, Hangauer MJ, Bertozzi CR: Fluorogenic azido-fluoresceins for biological imaging. J Am Chem Soc 2012, 134:17428–17431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wieczorek A, Buckup T, Wombacher R: Rigid tetrazine fluorophore conjugates with fluorogenic properties in the inverse electron demand Diels–Alder reaction. Org Biomol Chem 2014, 12:4177–4185. [DOI] [PubMed] [Google Scholar]
- 53.**. Werther P, Yserentant K, Braun F, Kaltwasser N, Popp C, Baalmann M, et al. : Live-cell localization microscopy with a fluorogenic and self-blinking tetrazine probe. Angew Chem Int Ed 2020, 59:804–810. This work reports a fluorogenic HM-SiR analog for live-cell SMLM. Given that localization precision in SMLM can be affected by background signal, the development of fluorogenic spontaneously blinking fluorophores allows for live-cell super-resolution microscopy without the need for additional wash procedures or additives. This increases biocompatibility and complements the microscopy technique.
- 54.Eördögh Á, Paganini C, Pinotsi D, Arosio P, Rivera-Fuentes P: A molecular logic gate enables single-molecule imaging and tracking of lipids in intracellular domains. ACS Chem Biol 2020, 15:2597–2604. [DOI] [PubMed] [Google Scholar]
- 55.Grimm JB, Muthusamy AK, Liang Y, Brown TA, Lemon WC, Patel R, et al. : A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods 2017, 14:e04236. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, et al. : A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 2015, 12:244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.**. Tyson J, Hu K, Zheng S, Kidd P, Dadina N, Chu L, et al. : Extremely bright, near-IR emitting spontaneously blinking fluorophores enable ratiometric multicolor nanoscopy in live cells. ACS Cent Sci 2021, 7:1419–1426. This report introduces the spontaneously blinking fluorophore Yale676sb-the highest quantum yield (0.59), nanoscopy-compatible Si-Rhodamine fluorophore reported to date. This fluorophores utilizes a novel design strategy that combines both electron-withdrawing and rotationally restricted diakylamino groups in order to simultaneously increase excitation/emission maximum and quantum yield, while maintaining pKcycle in a range appropriate for live cell SMLM without chemical additives. The authors also demonstrate that this fluorophore can be combined with HMSiR to enable 2-color live cell nanoscopy with a single 642 nm excitation laser.
- 58.**. Tu J, Svatunek D, Parvez S, Liu AC, Levandowski BJ, Eckvahl HJ, et al. : Stable, reactive, and orthogonal tetrazines: dispersion forces promote the cycloaddition with isonitriles. Angew Chem Int Ed 2019, 58:9043–9048. The authors report a mutually orthogonal triad of bioorthogonal reactions. They demonstrate that the sterically hindered isonitrile-tetrazine reaction is orthogonal to the tetrazine TCO ligation and SPAAC.
- 59.**. Hu Y, Roberts JM, Kilgore HR, Lani ASM, Raines RT, Schomaker JM: Triple, mutually orthogonal bioorthogonal pairs through the design of electronically activated sulfamate-containing cycloalkynes. J Am Chem Soc 2020, 142:18826–18835. Similar to work by Tu et al., Schomaker and coworkers develop a bioorthognal triad and demonstrate its orthogonality in live-cell microscopy experiments.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


