Abstract
Controlling the COVID-19 outbreak remains a challenge for Cameroon, as it is for many other countries worldwide. The number of confirmed cases reported by health authorities in Cameroon is based on observational data, which is not nationally representative. The actual extent of the outbreak from the time when the first case was reported in the country to now remains unclear. This study aimed to estimate and model the actual trend in the number of COVID -19 new infections in Cameroon from March 05, 2020 to May 31, 2021 based on an observed disaggregated dataset. We used a large disaggregated dataset, and multilevel regression and poststratification model was applied prospectively for COVID-19 cases trend estimation in Cameroon from March 05, 2020 to May 31, 2021. Subsequently, seasonal autoregressive integrated moving average (SARIMA) modeling was used for forecasting purposes. Based on the prospective MRP modeling findings, a total of about 7450935 (30%) of COVID-19 cases was estimated from March 05, 2020 to May 31, 2021 in Cameroon. Generally, the reported number of COVID-19 infection cases in Cameroon during this period underestimated the estimated actual number by about 94 times. The forecasting indicated a succession of two waves of the outbreak in the next two years following May 31, 2021. If no action is taken, there could be many waves of the outbreak in the future. To avoid such situations which could be a threat to global health, public health authorities should effectively monitor compliance with preventive measures in the population and implement strategies to increase vaccination coverage in the population.
Keywords: Cameroon, COVID-19, Forecasting, Observed, Post-stratification, Underestimated
Abbreviations: ACF, Autocorrelation Function; AIC, Akaike information criterion; COVID-19, Coronavirus Disease 2019; MAE, Mean Absolute Error; MAPE, Mean Absolute Percentage Error; MASE, Mean Absolute Scaled Error; ME, Mean Error; MPE, Mean Percentage Error; MRP, Multilevel Regression and Post-stratification; PACF, Partial Autocorrelation Function; PLACARD, Platform for Collecting, Analyzing and Reporting Data; SARIMA, Seasonal Autoregressive integrated moving average; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2
1. Introduction
The first case of COVID-19 was detected in Cameroon on March 5, 2020, and since then, the country has experienced several waves of the outbreak. At the initial stage, controlling the spreading of the virus in Cameroon has been very challenging for the public health authorities(Mbopi-Keou & Pondi, 2020). Testing is a key component of outbreak surveillance and control. In Cameroon, the first COVID-19 diagnostic laboratory was established in the central region and gradually decentralized to other regions of the country. Regional testing capacity was highly variable with some regions having six testing laboratories and others having only one. At the onset of the COVID-19 pandemic in Cameroon, only persons with symptoms, or persons in contact with a confirmed case, or travelers from high-risk countries were prioritized for testing. As the number of cases increased with community transmission and asymptomatic cases reported, testing was expanded to include every case. Despite this relaxation in prioritized testing, the majority of those tested were still in the priority group. It is well known that a large proportion of the population was asymptomatic (Aguilar et al., 2020; Chughtai & Malik, 2020; LiPei et al., 2020; Mizumoto et al., 2020; Yelin et al., 2020). In addition, regional variation in testing capacity was a limiting factor for individuals living in regions with low testing capacity to be tested for COVID-19. Therefore, the population getting tested for COVID-19 was not representative. In such cases, the number of confirmed cases reported would inevitably be skewed and would not reflect the true epidemiological situation. An important question that health authorities would like to answer is: what is the real magnitude of the pandemic in Cameroon?
Mathematical models such as in (Djaoue et al., 2020; Nabi et al., 2020; Nguemdjo et al., 2020) have attempted to answer this question using compartmental SIR models and other sophisticated mathematical models. These models are based on publicly available data reported by the Cameroonian Ministry of Public Health. Many other models have been developed for estimating the number of COVID-19 cases, including the Imperial College London (ICL) model and the Institute for Health Metrics and Evaluation (IHME) model (https://ourworldindata.org/covid-models). From these models, the estimated size of the outbreak in Cameroon from March 05, 2020 to October 30, 2020 was 915 647(3.67%) and 15 647 131(63%) from ICL and IHME models respectively. However, the total size of the outbreak in the same period based on reported data was 101 552(0.4%). Although these models are widely used, they are based on reported data that have numerous shortcomings (lack of overall tests number, demographic structure of the population, and many other relevant variables) which may still underestimate the size of the outbreak.
The main objective of our study was to estimate the actual trend of number of COVID-19 new infection in Cameroon based on an observed disaggregated dataset with demographic variables obtained from a routine laboratory information system. Then, to construct from this estimate a time series forecasting model for COVID-19 new infection number. Multilevel regression and poststratification models (MRP) were applied prospectively to emulate representative samples while accounting for the outbreak spreading. The target population was the total Cameroonian population distribution based on age, sex, and regions obtained from the Bureau of Census of Cameroon. Finally, Seasonal Autoregressive Integrated Moving Average (SARIMA) models were adopted to determine time series, forecasting models.
2. Methods and materials
2.1. Data
2.1.1. Data sources
Data were obtained from the following sources:
D1: Platform for Collecting, Analyzing and Reporting Data (PLACARD). That was the only data system that collected COVID-19 diagnostic data in a disaggregated form at the national level in Cameroon, a broad description of this platform is provided in (Eyangoh et al., 2021; Tchatchueng-Mbougua et al., 2022). The majority of individuals tested by PCR were reported in PLACARD. Each record contained the date of sample collection, demographic information of the patient and the PCR test results. From March 05, 2020 to May 31, 2021, about 330 000 samples tested by PCR were recorded in PLACARD of which 18 000 were PCR positive. Individuals reported in PLACARD were not from a sample survey but constituted a transversal observational sample.
D2: An auxiliary data set from the Central Bureau of Census and Population Studies (BUCREP, 2021). This data set contained details on the distribution of the total population of Cameroon based on sex, age, and region of residence, needed for applying the MRP model.
2.1.2. Data flaws
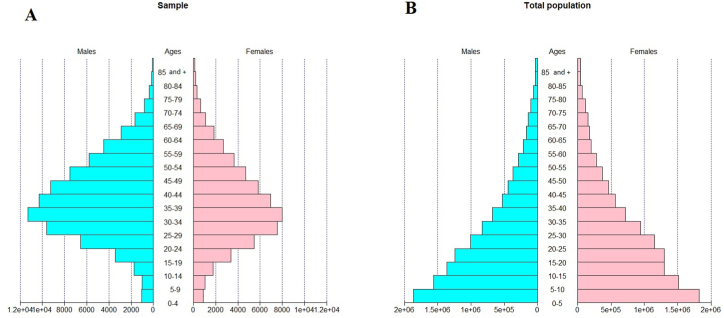
Similar to most sub-Saharan African countries, the age pyramid in Cameroon has a broad base and narrow top, with an almost equal number of males and females (see Fig. 1A). The shape of the pyramid reflects a predominantly younger population with more children and youths. The average age of the Cameroonian population is about 18 years and the male to female ratio is about 1.06. The pyramid structure of the participants in D1 was completely different from that of the overall Cameroonian population (see Fig. 1B). The D1 age pyramid had a narrow base and a distinct middle with an average age of 37 years and a sex ratio of 1.47. The D1 age pyramid was characterized by a population of predominantly adult males. As the D1 data structure differed from that of the overall population in terms of age and gender variables, it did not represent the general population of Cameroon (see Fig. 2).
Fig. 1.
Age pyramid of COVID-19 PCR samples (left) and the total population of Cameroon(right).
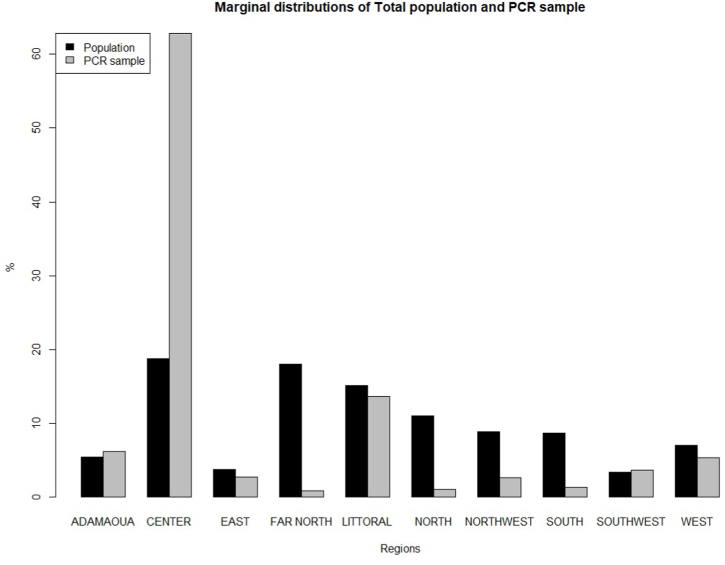
Fig. 2.
Marginal distribution of the total population of Cameroon and PCR sample respectively according to the region of residence.
Cameroon is divided into 10 regions, with the Central region, where the capital is located, being the most populated (18.75%), followed by the Far-North (18%) and the Littoral (15.12%) regions and the South-West region (3.37%) is the least populated. The distribution of testing data recorded in D1 differed from the overall population; more than 60% of the testing was done in the Center region, followed by the Littoral(17%) and Adamawa(6%) regions. As such, the regional distribution of the PCR observation testing data would not be truly representative to the entire Cameroon population.
2.2. Models
The aim is to determine from the observational data D1, the COVID-19 cases estimates over time that represent the total population of Cameroon. For that, the multilevel regression and post-stratification models (MRP) were fitted prospectively while accounting for the dynamics of outbreak spreading.
2.2.1. Basic MRP models
Downes et al.(Downes et al., 2018) provided a broad overview of the MRP models. MRP models are performed in two stages, the first stage consists of performing a multilevel logistic regression model to predict the individual-response in post-stratum based on a set of covariates. Then, the second stage is the post-stratification, using the prediction of the first stage to estimate the average number of responses in the different considered post-stratum using the distribution of the total population.
2.2.1.1. Multilevel regression
Let's consider we have j = 1, …, J post-stratum and denoting by j[i] the index of the individual i in the post-stratum j. The response variable will be denoted by Y and in the frame of this study, it would be a binary outcome defined by Yj[i] = 1 if the individual i in the stratum j is COVID-19 infected and Yj[i] = 0 otherwise. The multilevel regression consists of modeling the response Yj[i] as a linear function of covariates for post-stratum j, accounting for variations within the post-stratum. The multilevel regression modelling of the outcome Y in the post-stratum j is therefore specified as:
| (1) |
ηj = P(Yj = 1) represents the probability for any individuals of contracting COVID-19 infection in the post-stratum j (its estimate represents the proportion of COVID-19 infection in the post-stratum j), logit is the logistic function, β0 is the fixed intercept, t(β) is the fixed effect coefficient vector and is the varying coefficient effect of the category l of the variable in the post-stratum j. The parameters β0, β and are estimated based on Monte-Carlo-Markov-Chain(MCMC), with the prior assumed to be normally distributed with mean of zero and standard deviation estimated from the sample.
2.2.1.2. Poststratification
After the parameters are estimated from the multilevel regression, the model is fitted to census data to estimate the number of COVID-19 infection in the post-stratum j, which is estimated by:
| (2) |
Where nj and denote the actual number of COVID-19 infected cases and its estimate in the post-stratum j respectively, Nj denotes the total population of the post-stratum j.
2.2.2. Prospective MRP modeling for COVID-19
We applied the MRP models described above pointwise over the considered period to estimate the actual trend of COVID-19 infection, while accounting for the dynamic of the outbreak spread. Thus, let's assume that the dynamic of the outbreak spreading is such that after an initial infection, individuals are removed from the population of susceptible for some time to become again susceptible after the acquired natural immunity is lost.
Assuming that at the initial time, from t0 to tx, the virus had not yet spread throughout the country. Denoting by rj(t) the observed number of COVID-19 cases at time t, the observed number of infected estimates the actual number of cases during the initial period [t0; tx], that is , for any j in 1, …, x.
From tx+1, the population of susceptible individuals would increase, to a level that the whole population gets susceptible individuals. At time point tx+1, the population of susceptible would be Nj(tx+1) = δ(tx+1)(Nj(tx+1) − nj(tx)). Where δ(t) denotes the proportion of susceptible individuals at time t, in relation to the previous time.
This function would be asymptotic and convergent to 1, as COVID-19 spreads rapidly. For instance, one could choose δ(t) = 1 − exp τt, where τ is the increase rate per unit of time in the population of susceptible individuals. In the frame of this work, we assumed that this rate could be approximated by the weekly average percentage of change in COVID-19 transmission rate. From a repeated seroprevalence study carried out in Cameroon by Ndongo et al.(Ndongo et al., 2022), we estimated this rate at τ = 0.025.
Given the above assumptions and initial conditions, the estimated trend of COVID-19 new infections number would be determined as follows from k = x + 2:
-
1.
Determine the number of the susceptible individuals in the population at time tk, by
-
2.
Estimate the number of infected individuals at time tk using MRP on the observed population at the time tk, targeting the population Nj(tk),
-
3.
Repeat 1. and 2. Until tk − t0 = p, where p is the period over which natural immunity persists, for example p = 52 months(Dan et al., 2021).
-
4.
Then, the population of susceptible individuals would be:
.
-
5.
Repeat 1. to 4. until the end of the period.
We first used the variables sex, age groups, and region separately as poststratification variables, then, we combined all of them to evaluate the additive effect of the considered variables together. Table 1 displays the critical parameters adopted in the proposed prospective MRP for estimating the trend of COVID-19 new infection number.
Table 1.
List of model parameters and their interpretations.
| Parameter | Description |
|---|---|
| tx | Time point where the virus starts widespread in the general population |
| δ(t) | The fraction of susceptible at time t in the general population |
| τ | The weekly average increase in COVID-19 new infection number |
| p | The average duration in weeks of the natural immunity period after an initial infection |
2.2.3. SARIMA models
Once the estimated trend of confirmed cases is determined, one could determine a model that fits this trend for forecasting purposes. ARIMA is a type of time series analysis that has been widely applied in a variety of domains, including the COVID-19 outbreak, where it was utilized for forecasting and modelling(Al-qaness et al., 2020; Benvenuto et al., 2020; Ceylan, 2020; Chakraborty & Ghosh, 2020; Toga et al., 2021) – (Al-qaness et al., 2020; Benvenuto et al., 2020; Ceylan, 2020; Chakraborty & Ghosh, 2020; Toga et al., 2021). ARIMA model is defined by:
| (3) |
Where , and p, q and d model's parameters. The parameters p and q are usually determined based on the autocorrelation function (ACF) and the partial autocorrelation function (PACF), truncating the intercepts and tails. ARIMA is applied under the assumption that is stationary, otherwise the series d is differentiated times to be stationary, and . The parameters θi and φi are estimated using the maximum likelihood method (MLE).
Unlike other previous works that used ARIMA to model the COVID-19 trend, in this work we introduce seasonality into the model, called SARIMA. Seasonality would explain the occurrence of waves in the dynamics of the outbreak. SARIMA model is denoted by SARIMA(p, d, q)(P,D,Q)[s] and is defined by:
| (4) |
Where , and . With the additional parameters P, Q, D, Θi, and Φi are determined similarly to ARIMA, and s is the seasonal lag. In practice, different combinations of parameters (p, q, d) and (P, Q, D) are determined and the model in equation (4) is estimated accordingly. Then, based on a certain criterion (AIC and BIC), a model that is more suitable for predicting the trend is selected.
2.3. Materials
The R software(version 4.1.2) (R Core Team, 2021) was used in the statistical analysis. In particular, the R package rstanarm (Goodrich et al., 2020) was used to implement the prospective MRP for COVID-19 trend estimation. The statistical methods of the rstanarm package required a large amount of computation, so the r codes were run on a supercomputer with 48 cores and 64 Gigabytes(GB) of random access memory (RAM). The R codes files as well as datasets were shared as supplementary file.
3. Results
3.1. Reported trend versus estimated trend
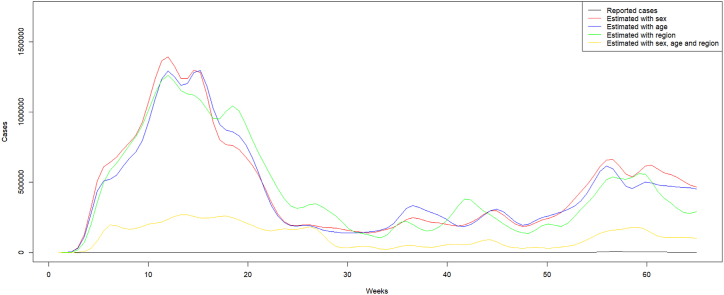
After cleaning the data and removing individuals with missing outcomes among the considered variables in the MRP models, the final dataset analyzed in this study had a total of 207 926 individuals recorded from March 05, 2020 to May 31, 2021. Fig. 3 shows the trend of estimated and reported numbers respectively. The estimated and reported trends were similar in terms of the numbers of waves as well as the starting point of each. However, the magnitude of waves from estimated and reported trends was discordant. In the trend based on reported data, the first wave was less intense compared to the second wave. While in the estimated trends, the first wave was more intense compared to the second wave. This result could be explained by the fact that the initial growth of COVID-19 cases in African countries including Cameroon was rapid as shown in (Musa et al., 2020).
Fig. 3.
Estimated versus reported trend of the number of weekly COVID-19 new infections in Cameroon from March 05, 2020 to May 31, 2021.
Overall, the trends of the estimated number of new cases were above the trend of reported number of cases. The estimated trends based on the variables sex, age, and region separately were similar. However, when considering these variables together in the model, the estimated trend, in this case, was below the trends estimated when the variables are considered separately. Though, we consider the estimates with all variables together as more reliable, as it has been found in many settings that these three variables were associated with COVID-19 positivity rates, which is particularly the case with the study by Tejiokem et al. (CyrilleT. M. J. B. S. M. SergeN. G. T. N. P. Alain et al., 2022) in Cameroon using the same data sources.
Table 2 displays the cumulative number of cases based on different estimates, and the ratio to compare the total estimated number of COVID-19 new cases to the total number of reported new cases. Generally, the total number of reported new cases was lower than the estimated total number of new cases. The level of underestimation depended on the variables considered for the MRP model. Considering the variables sex, age and region separately, the ratios of estimated versus reported are 379, 364, and 365 respectively which were larger than the ratio when the variables are considered together in the model (the ratio, in this case, was 94).
Table 2.
Cumulated estimated number of COVID-19 cases in Cameroon.
| Poststratification variables | Cumulative cases | Estimated/Reported |
|---|---|---|
| Sex | 29933808 | 379.2 |
| Age | 28756513 | 364.3 |
| Region | 28845325 | 365.4 |
| Sex + Age + Region | 7450935 | 94.4 |
3.2. Time series analysis
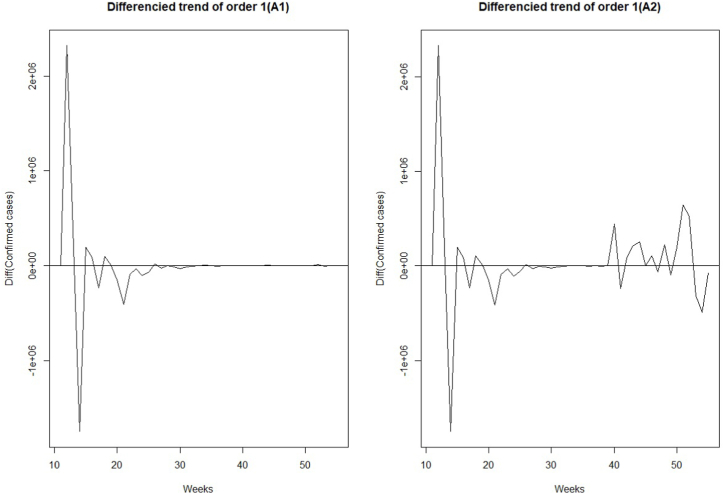
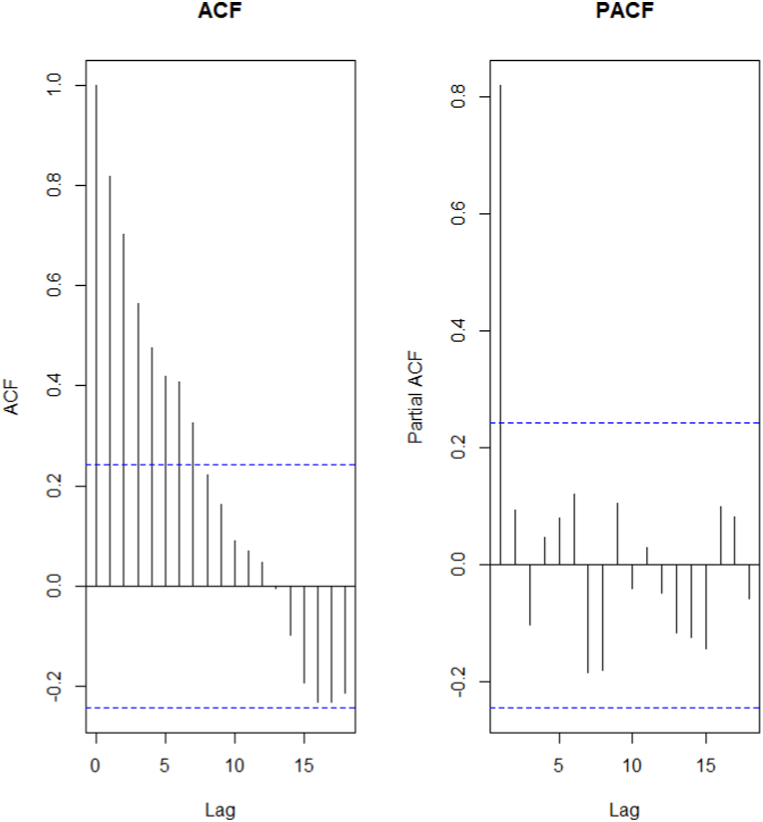
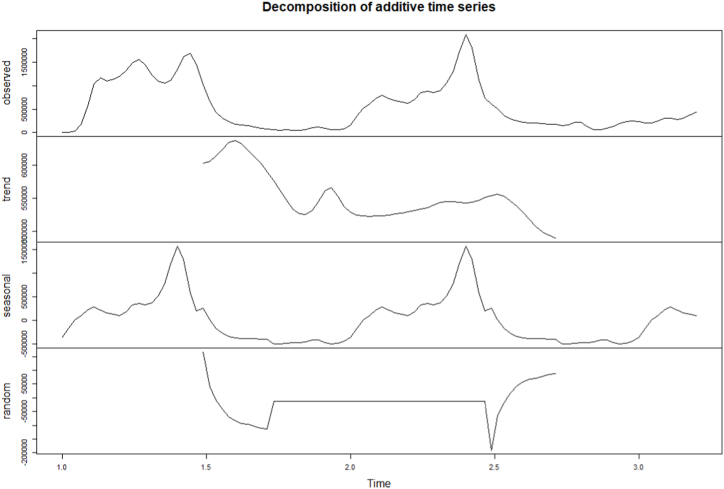
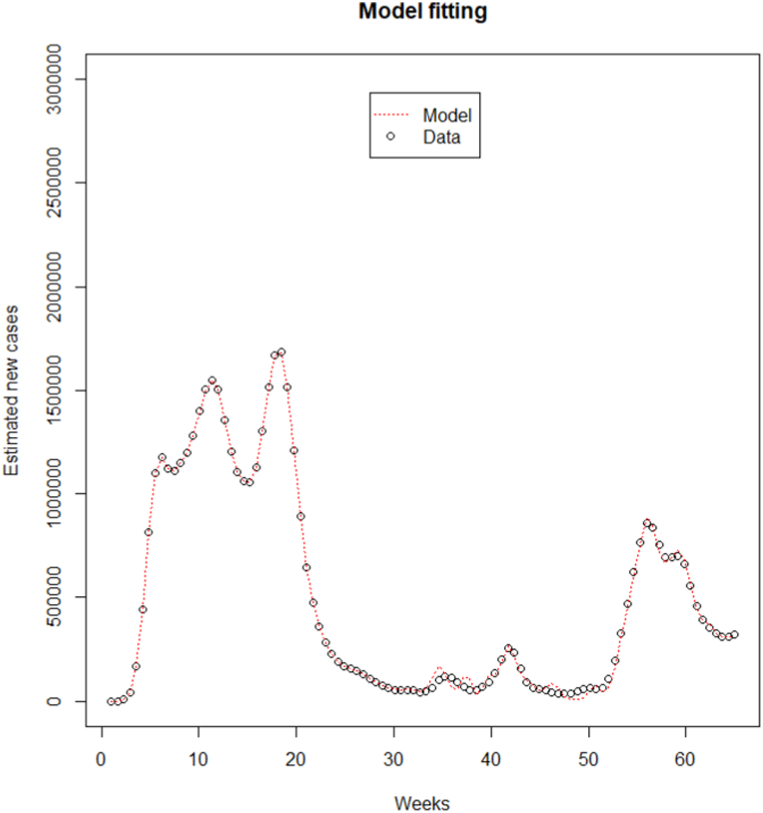
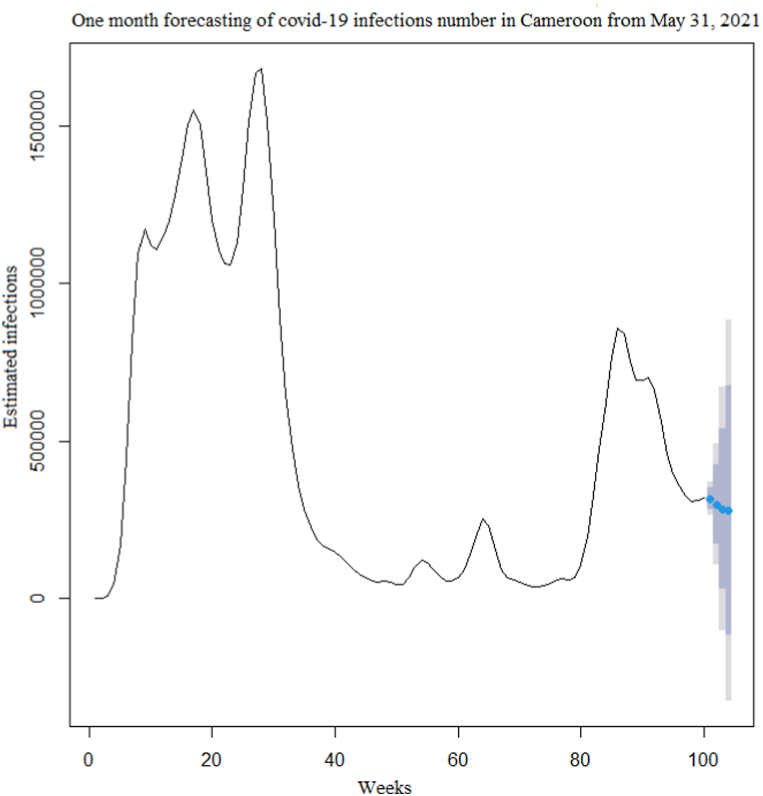
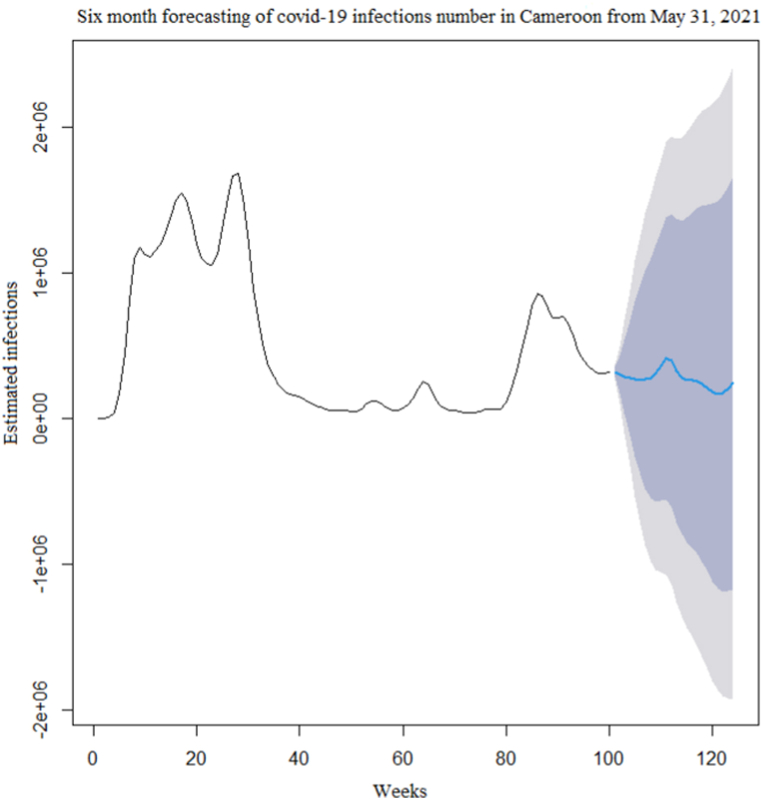
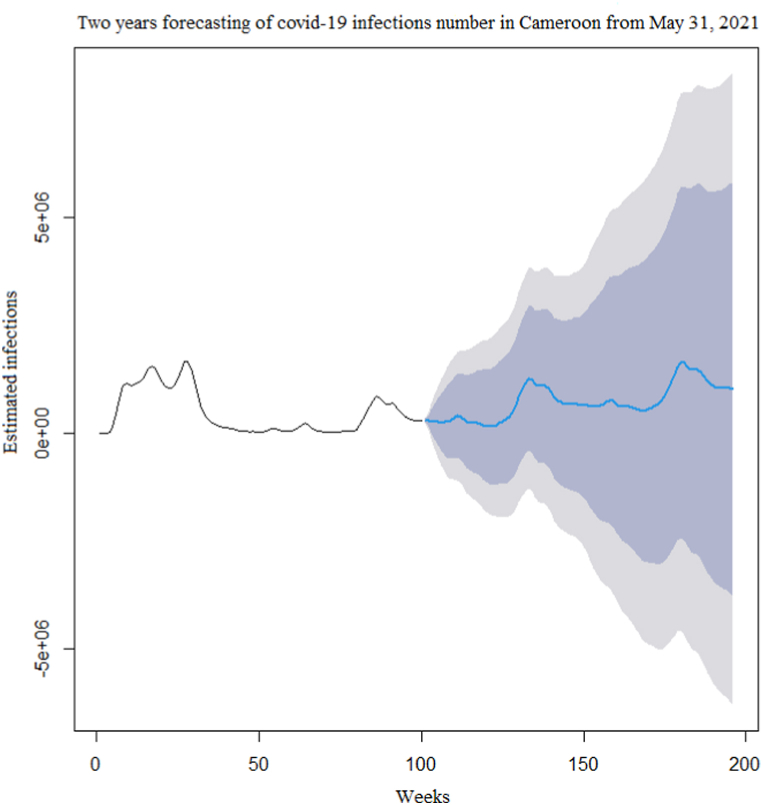
The time series of the estimated number of COVID-19 new cases in Cameroon as shown in Fig. 3 was not stationary. The Augmented Dickey-Fuller Test with p − value = 0.24 confirmed this observation. After a differentiation (d = 1), as shown in Fig. 4, the series became stationary (p − value = 0.01). Based on acf and pacf cuts off and tails off displayed in Fig. 5, the parameters p and q of the ARIMA model belongs to1, 2. After decomposition (Fig. 6), a clear seasonal component was observed that corresponded to the occurrence of the two observed waves. The model was selected manually by combining different plausible combinations of the parameters. The tentative model that best fit the data was identified such that the Akaike information criterion (AIC) was minimized, the SARIMA(2, 1, 1)(1,1,1)[47] model was identified to fit the data. Fig. 7 displays the fitting of the identified model on data and Table 3 presents the different error measures of this model. This identified model was used for forecasting; the short, medium and long-term forecasting are displayed in Fig. 8, Fig. 9, Fig. 10 respectively. Periods of one month, six months and two years were considered for short-, medium- and long-term forecasting respectively (see Fig. 6).
Fig. 4.
Differentiated series of the estimated trend of weekly COVID-19 new infections number in Cameroon from March 05, 2020 to May 31, 2021.
Fig. 5.
ACF and PACF of the series of the estimated trend of weekly COVID-19 new infections number in Cameroon from March 05, 2020 to May 31, 2021.
Fig. 6.
Decomposition of the series of the estimated trend of weekly COVID-19 new infections number in Cameroon from March 05, 2020 to May 31, 2021.
Fig. 7.
SARIMA model for fitting the series of the estimated trend of weekly COVID-19 new infections number in Cameroon from March 05, 2020 to May 31, 2021.
Table 3.
Error measures of the selected model for COVID-19 forecasting in Cameroon.
| ME | RMSE | MAE | MPE | MAPE | MASE | ACF1 |
|---|---|---|---|---|---|---|
| 350.36 | 18824.93 | 11156.77 | −0.99 | 12.35 | 0.17 | 0.39 |
Fig. 8.
Short-term(one month) forecasting of the COVID-19 new infections number in Cameroon after May 31, 2021.
Fig. 9.
Medium-term(6 months) forecasting of the COVID-19 new infections number in Cameroon after May 31, 2021.
Fig. 10.
Long-term(Two years) forecasting of the COVID-19 new infections number in Cameroon after May 31, 2021.
During the month following May 31, 2021(i.e June 2021), the second wave would still be declining. In the medium-term forecasting, six months following May 31, 2021, the country would have fully experienced the second wave, with an additional 6 539 204 estimated number of infection cases. In the long-term forecasting, the next two years from May 31, 2021, the country would have experienced two additional waves of the outbreak. Overall, there will be successive waves of the outbreak in the country with a periodicity of about 47 weeks, and, during each wave, almost the entire population will become infected.
However, the width of confidence intervals of the predicted number of COVID-19 infections in the medium and long-term forecasting was very large. These would suggest low accuracy of medium and long-term forecasting. While, in short-term forecasting, the confidence interval width was relatively narrow, indicating that our model is more appropriate for short-term forecasting.
4. Discussion
Since the onset of the outbreak of COVID-19 in Wuhan in December 2019, there have been numerous scientific works aimed at modelling, estimating, and forecasting the number of COVID-19 cases, deaths, recoveries, contacts and attack rates in various settings(Al-qaness et al., 2020; Benvenuto et al., 2020; Ceylan, 2020; Chakraborty & Ghosh, 2020; Dobreva et al., 2022; Musa et al., 2022; Toga et al., 2021) – (Al-qaness et al., 2020; Benvenuto et al., 2020; Ceylan, 2020; Chakraborty & Ghosh, 2020; Dobreva et al., 2022; Musa et al., 2022; Toga et al., 2021). Most of these works were based on publicly available data, generally reported by public health authorities in an aggregated form. This is one of the few studies that have used disaggregated data at an individual level with demographic variables for estimating, modelling, and forecasting purposes.
Serosurvey studies have been implemented in many settings to estimate the actual size of the outbreak. In Cameroon, a study by (Nwosu et al., 2021) in an urban district estimate the seroprevalence at 29.2% by November 26, 2020. This number was far less compared to the estimated size of the outbreak found in this study by the same date. This discrepancy could be partly explained by the period considered, from the onset up to November 26, 2020 which corresponds to more than 9 months. Antibodies produced by memory B cells especially IgG antibodies begin to decline by week seven after the onset, and by three months almost none is detectable (Stephens & McElrath, 2020; Yamayoshi et al., 2021). Therefore, as pointed out as a limitation in (Nwosu et al., 2021), many individuals who have been infected had lost their immunity at the time of serosurvey which could have resulted to them not detected. The delay in seroreconversion among some previously infected individuals could be another reason for the observed discrepancy between the estimated seroprevalence and our findings. Even though serosurveys are relevant to estimate the actual size of the outbreak, they present many limitations that might underestimate the actual size of the outbreak(Long et al., 2020; Rostami et al., 2021).
It has been found that there was a large variation in COVID-19 trend estimates when considering the set of most common representative models for estimating the trend of COVID-19(https://ourworldindata.org/covid-models): ICL and IHME models, one would observe that there are large discrepancies between the estimates. These observed discrepancies between trend estimates of different methods would reflect the challenges of making assumptions on the dynamic of the outbreak on one hand and the difficulties of correcting the representation bias in reported data. Even though our trend estimate presented a similar shape as the one of IHME model, our estimates were larger than those obtained from both methods. Comparatively to other methods, our methods were based on a disaggregated dataset with demographic variables which allowed us to mimic the true population of Cameroon. Therefore, our trend estimate could be closer to the actual trend of the outbreak compared to other estimates.
In Cameroon, the period of the lockdown and the monitoring of preventive measures was very short (only during the period March 2020 to April 2020). Compliance to preventive measures has also gradually declined(Fodjo et al., 2021). To date, due to the perception of low personal risk to infection in the majority of the population, only very few individuals are practicing social distancing and other preventive measures. In such conditions, added to the very low rate of vaccination (less than 1%), only naturally acquired immunity from a previous infection would protect individuals from infections. Unfortunately, this natural immunity only lasts for few a months (3–13 months). Therefore, the obtained estimates in this study seem more realistic in the Cameroonian setting and would be probably the same in countries where there is no monitoring and observing of preventive measures. Fortunately, with the age structure of the Cameroonian population as in other Sub-saharan countries, the outbreak is less severe(less fatal and severe cases) as children, adolescents and young adults are mostly asymptomatic (Aguilar et al., 2020; Chughtai & Malik, 2020; de Lusignan et al., 2020; LiPei et al., 2020; Mizumoto et al., 2020; Yelin et al., 2020).
A study by (Nkwayep et al., 2020) in May 2020 in a short-term forecasting study, concluded that COVID-19 would persit in the country in the absence of control measures. These conclusions are consistent with our findings. Moreover, the persistence of the outbreak will be in the form of waves. This should alert public health authorities to implement effective preventive and control measures within the population to avoid long-term consequences of the spreading of the virus such as the development of new variants of the virus.
Even though COVID-19 mortality was generally very low in Sub-Saharan countries, including Cameroon, not considering COVID-19 mortality in our model could be viewed as a limitation of our study. However, our data system, well described in (Tchatchueng-Mbougua et al., 2022) did not collect data on COVID-19 mortality, and we are not aware of any data system in Cameroon which has collected such data.
5. Conclusion
Reported data on COVID-19 in Cameroon as well as in many Sub-Saharan countries are particularly affected by representative flaws which underestimate the actual trend of the outbreak. Serosurveys are important to estimate the actual size of the outbreak. However, the cross-sectional nature of such surveys will still underestimate the size of the COVID-19 outbreak, as the memories of SARS-CoV-2 antibodies last only a few months. This study has provided estimates which are closer to the actual trend as it was based on the emulation of representative samples at the national level.
The estimates and the forecasting from this study shows that the size of the outbreak is far larger than what has been reported. To avoid the development of new variants of the virus resulting from the persistence of the spreading of the virus, public health authorities should implement effective strategies for monitoring preventive and control measures against the spread of the virus. This particularly includes the strategies to improve the vaccination rate which is currently very low in Cameroon (less than 10%).
Ethics
Approval was obtained from the Cameroon National Ethics Committee for Research in Human Health (N°2020/05/1221/CE/CNERSH/SP).
Funding
This study was funded by the French Ministry for Europe and Foreign Affairs via the project “REPAIR COVID-19-Africa” coordinated by the Pasteur International Network association.
Author contributions
Conceptualization: Arsène Brunelle Sandie.
Data curation: Arsène Brunelle Sandie.
Formal analysis: Arsène Brunelle Sandie.
Investigation: Arsène Brunelle Sandie, Mathurin Cyrille Tejiokem, Jules Brice Tchatchueng-Mbougua.
Methodology: Arsène Brunelle Sandie and Jules Brice Tchatchueng-Mbougua.
Writing – original draft: Arsène Brunelle Sandie.
Writing – review and editing: Arsène Brunelle Sandie, Jules Brice Tchatchueng-Mbougua, Mathurin Cyrille Tejiokem, Cheikh Mbacké Faye, Achta Hamadou, Serge Sadeuh Mbah, Paul Alain Tagnouokam-Ngoupo, Richard Njouom, Maurice Tchuente, Ngu Karl Abanda, Sara Eyangoh, Mathurin Cyrille Tejiokem, Maryam Diarra, Slimane Ben Miled, REPAIR consortium.
All authors reviewed the manuscript and approved the final version.
Availability of data and materials
The data that support the findings of this study are available from the Pasteur Center of Cameroon but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Pasteur Center of Cameroon.
Declaration of competing interest
The authors declare that they have no known competing or conflicting financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
REPAIR consortium: Slimane Ben Miled, Maryam Diarra, Cheikh Loucoubar,Lulla Opatowski, Mohamed Hamidouche, Amira Kebir, Dorra Louati, Ram-atoulaye Hamidou Lazoumar, Cheikh Talla, Walid Ben Aribi, Nesrine Be Yahia, Ahmed Nasri, Bechir Naffeti.
Handling Editor: Dr He Daihai
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2023.02.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Aguilar J.B., Faust J.S., Westafer L.M., Gutierrez J.B. Investigating the impact of asymptomatic carriers on covid-19. medXiv. 2020;19(1—12):888–896. doi: 10.1101/2020.03.18.20037994. [DOI] [Google Scholar]
- Al-qaness M.A.A., Ewees A.A., Fan H., Aziz M.A.E. Optimization method for forecasting confirmed cases of covid-19 in China. Journal of Clinical Medicine. 2020 doi: 10.3390/jcm9030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Giovanetti M., Vassallo L., Angeletti S., Ciccozzi M. Application of the arima model on the covid-2019 epidemic dataset. Data in Brief. 2020 doi: 10.1016/j.dib.2020.105340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUCREP . 2021. 3ème rgph : Projections démographiques. [Google Scholar]
- Ceylan Z. Estimation of covid-19 prevalence in Italy, Spain, and France. The Science of the Total Environment. 2020 doi: 10.1016/j.scitotenv.2020.138817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Ghosh I. Real-time forecasts and risk assessment of novel coronavirus (covid-19) cases: A data-driven analysis. Chaos, Solitons & Fractals. 2020 doi: 10.1016/j.chaos.2020.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai A., Malik A.A. Is coronavirus disease (covid-19) case fatality ratio underestimated? Global Biosecurity. 2020;2 doi: 10.31646/gbio.56. [DOI] [Google Scholar]
- Cyrille T.M., Serge S.-M., Brice T.M.J., Alain T.N.P., Grace N., Joseph F., Achta H., Gisèle N., Julius N., Marcel T., Melissa S., Lucy N., Ronald P., Claire O.A.M., Walter P.Y.E., Alain E.M.G., Richard N., Sara E. Clinical presentation of covid-19 at the time of testing and factors associated with pre-symptomatic cases in Cameroon. IJID Reg. 2022 doi: 10.1016/j.ijregi.2022.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Stephen, Peters R.B., Krammer F., Simon V., Saphire E.O., Smith D.M., Weiskopf D., Sette A., Crotty S. Immunological memory to sars-cov-2 assessed for up to 8 months after infection. Science. 2021 doi: 10.1126/science.abf4063. https://doi.org/10.1101/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djaoue S., Kolaye G.G., Abboubakar H., Ari A.A.A., Damakoa I. Mathematical modeling analysis and numerical simulation of the covid-19 transmission with mitigation of control strategies used in Cameroon. Chaos, Solitons & Fractals. 2020 doi: 10.1016/j.chaos.2020.110281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva Z., Gimma A., Rohan H., Djoudalbaye B., Tshangela A., Jarvis C.I., van Zandvoort K., Quaife M. Characterising social contacts under covid-19 control measures in africa. BMC Medicine. 2022 doi: 10.1186/s12916-022-02543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes M., Gurrin L.C., English D.R., Pirkis J., Currier D., Spittal M.J., Carlin J.B. Multilevel regression and poststratification: A modeling approach to estimating population quantities from highly selected survey samples. American Journal of Epidemiology. 2018 doi: 10.1093/aje/kwy070. [DOI] [PubMed] [Google Scholar]
- Eyangoh S., Tchatchueng J., Njouom R., Okomo M.C., Mballa A.E. Scaling up laboratory testing for covid-19 in Cameroon: Challenges, lessons learnt and perspectives. Lab Culture. 2021 [Google Scholar]
- Fodjo J.N.S., Ngarkaand L., Njamnshi W.Y., Nfor L.N., Mengnjo M.K., Mendo E.L., Angwafor S.A., Basseguin J.G.A., Nkouonlack C., Njit E.N., et al. Covid-19 preventive behaviours in Cameroon: A six-month online national survey. International Journal of Environmental Research and Public Health. 2021 doi: 10.3390/ijerph18052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich B., Gabry J., Ali I., Brilleman S. 2020. rstanarm: Bayesian applied regression modeling via Stan.https://mc-stan.org/rstanarm r package version 2.21.1. [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Wan Y., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (sars-cov2) Science. 2020 doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Tang X.J., Shi Q.L., et al. Clinical and immunological assessment of asymptomatic sars-cov-2 infections. Nature Medicine. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- de Lusignan S., Dorward J., Correa A., et al. Risk factors for sars-cov-2 among patients in the oxford royal college of general practitioners research and surveillance centre primary care network: A cross-sectional study. The Lancet Infectious Diseases. 2020 doi: 10.1016/S1473-3099(20)30371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbopi-Keou F.X., Pondi M.A.S.J.E. Covid-19 in Cameroon: A crucial equation to resolve. The Lancet Infectious Diseases. 2020 doi: 10.1016/S1473-3099(20)30373-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (covid-19) cases on board the diamond princess cruise ship, yokohama, Japan, 2020. European Communicable Disease Bulletin. 2020 doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa S.S., Wang X., Zhao S., Li S., Hussaini N., Wang W., He D. The heterogeneous severity of covid-19 in african countries: A modeling approach. Bulletin of Mathematical Biology. 2022 doi: 10.1007/s11538-022-00992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa S.S., Zhao S., Wang M.H., Habib A.G., Mustapha U.T., He D. Estimation of exponential growth rate and basic reproduction number of the coronavirus disease 2019 (covid-19) in africa. Infect Dis Poverty. 2020 doi: 10.1186/s40249-020-00718-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabi K.N., Abboubakar H., Kumar P. Forecasting of covid-19 pandemic: From integer derivatives to fractional derivatives. Chaos, Solitons & Fractals. 2020 doi: 10.1016/j.chaos.2020.110283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndongo F.A., Guichet E., Mimbé E.D., Ndié J., Pelloquin R., Varloteaux M., Esemu L., Mpoudi-Etame M., Lamare N., Edoul G., Wouambo R.K., Djomsi D.M., Tongo M., Tabala F.N., Dongmo R.K., Diallo M.S.K., Bouillin J., Thaurignac G., Ayouba A.…Mpoudi-Ngolé E. Rapid increase of community sars-cov-2 seroprevalence during second wave of covid-19, yaoundé, Cameroon. Emerging Infectious Diseases. 2022 doi: 10.3201/eid2806.212580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguemdjo U., Meno F., Dongfack A., Bruno V. Simulating the progression of the covid-19 disease in Cameroon using sir models. PLoS One. 2020 doi: 10.1371/journal.pone.0237832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkwayep C.H., Bowong S., Tewa J.J., Kurths J. Short-term forecasts of the covid-19 pandemic: A study case of Cameroon. Chaos, Solitons & Fractals. 2020 doi: 10.1016/j.chaos.2020.110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu K., Fokam J., Wanda F., Orel L.M.E., Ray N., Mekeand J., Tassegning A., Takou D., Mimbe E., Stoll B., Guillebert J., Comte E., Keiser O., Ciaffi L. Sars-cov-2 antibody seroprevalence and associated risk factors in an urban district in Cameroon. Nature Communications. 2021 doi: 10.1038/s41467-021-25946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2021. R: A language and environment for statistical computing.https://www.R-project.org/ URL. [Google Scholar]
- Rostami A., Sepidarkish M., Leeflang M.M.G., Riahi S.M., Shiadehand M.N., Esfandyari S., Mokdad A.H., Hotez P.J., Gasser R.B. Sars-cov-2 seroprevalence worldwide: A systematic review and meta-analysis. Clinical Microbiology and Infections. 2021 doi: 10.1016/j.cmi.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens D.S., McElrath M.J. Covid-19 and the path to immunity. JAMA. 2020 doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchatchueng-Mbougua J.B., Essengue L., Yuya F.J.S., Kamtchogom V., Hamadou A., Sadeuh-Mbah S.A., Tagnouokam-Ngoupo P.A., Tchuente M., Njouom R., Eyangoh S., Tejiokem M.C. Improving the management and security of covid 19 diagnostic test data with a digital platform in resource-limited settings: The case of placard in Cameroon. Plos Digital Health. 2022 doi: 10.1371/journal.pdig.0000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga G., Atalay B., Toksari M. Forecasting using autoregressive integrated moving average (arima) and artificial neural networks (ann): Case of Turkey. Journal of Infection and Public Health. 2021 doi: 10.1016/j.jiph.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S., et al. Antibody titers against sars-cov-2 decline, but do not disappear for several months. EClinicalMedicine. 2021 doi: 10.1016/j.eclinm.2021.100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelin I., Aharony N., Shaer-Tamar E., Argoetti A., Messer E., Berenbaum D., Shafran E., Kuzli A., Gandali N., Hashimshony T., Mandel-Gutfreund Y., Halberthal M., Geffen Y., SzwarcwortCohen M., Kishony R. Evaluation of covid-19 rt-qpcr test in multisample pools. medRxiv. 2020 doi: 10.1093/cid/ciaa531. https://doi.org/10.1101/ [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the Pasteur Center of Cameroon but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Pasteur Center of Cameroon.