Abstract
A number of proteins and nucleic acids have been explored as therapeutic targets. These targets are subjects of interest in different areas of biomedical and pharmaceutical research and in the development and evaluation of bioinformatics, molecular modeling, computer-aided drug design and analytical tools. A publicly accessible database that provides comprehensive information about these targets is therefore helpful to the relevant communities. The Therapeutic Target Database (TTD) is designed to provide information about the known therapeutic protein and nucleic acid targets described in the literature, the targeted disease conditions, the pathway information and the corresponding drugs/ligands directed at each of these targets. Cross-links to other databases are also introduced to facilitate the access of information about the sequence, 3D structure, function, nomenclature, drug/ligand binding properties, drug usage and effects, and related literature for each target. This database can be accessed at http://xin.cz3.nus.edu.sg/group/ttd/ttd.asp and it currently contains entries for 433 targets covering 125 disease conditions along with 809 drugs/ligands directed at each of these targets. Each entry can be retrieved through multiple methods including target name, disease name, drug/ligand name, drug/ligand function and drug therapeutic classification.
INTRODUCTION
Pharmaceutical agents generally exert their therapeutic effect by binding to a particular protein or nucleic acid target (1,2). So far, hundreds of proteins and nucleic acids have been explored as therapeutic targets (1). Rapid advances in genetic (3), structural (4) and functional (5) information of disease related genes and proteins not only raise strong interest in the search of new therapeutic targets, but also promote the study of various aspects of known targets including molecular mechanism of their binding agents and related adverse effects (6), and pharmacogenetic implications of sequence or proteomic variations (7), etc. The knowledge gained from such a study is important in facilitating the design of more potent, less toxic and personalized drugs. Development of advanced computational methods for bioinformatics (4), molecular modeling (8), drug design and pharmacokinetics analysis (9–11) increasingly uses known therapeutic targets to refine and test algorithms and parameters.
A publicly accessible database that provides comprehensive information about these targets is therefore helpful in catering for the need and interest of the relevant communities in general and those unfamiliar with a specific therapeutic target in particular. To the best of the authors’ knowledge, such a publicly accessible database is not yet available. In this work, we introduce a Therapeutic Target Database (TTD), which contains information about the known therapeutic protein and nucleic acid targets together with the targeted disease conditions, the pathway information and the corresponding drugs/ligands directed at each of these targets. Cross-links to other databases are introduced to facilitate the access of information regarding the function, sequence, 3D structure, nomenclature, drug/ligand binding properties and related literatures of each target.
The therapeutic targets collected in TTD are from a search of the available literature. It has been reported that, at present, approximately 500 therapeutic targets have been exploited in the currently available medical treatment (1). An effort has been made to collect as many of these known targets as possible. However, description of some of these targets in the literature was not specific enough to point to a particular protein or nucleic acid as the target. Hence these targets are not included in our database.
DATABASE STRUCTURE AND ACCESS
TTD has a web interface at http://xin.cz3.nus.edu.sg/Group/ttd/ttd.asp. The entries of this database are generated from a search of pharmacology textbooks (12,13), review articles (14–21) and a number of recent publications. Our database currently contains 433 entries of protein and nucleic acid targets found from the literature. These targets cover 125 different disease conditions, which are described in the database. Drugs and ligands directed at each of these targets are searched and included in the database. A total of 809 different drugs and ligands are listed in the database.
The TTD database web interface is shown in Figure 1. This database is searchable by target name or drug/ligand name. It can also be accessed by selection of disease name, drug/ligand function or drug therapeutic classification from the list provided in the corresponding selection field. Searches involving any combination of these five search or selection fields are also supported. The lists of disease names, drug/ligand functions and drug classifications are given in Tables 1, 2 and 3, respectively.
Figure 1.
The web interface of TTD. Five types of search mode are supported. This database is searchable by target name, disease name, drug/ligand name, drug/ligand function, drug classification or any combination of these.
Table 1. Disease names listed in TTD (synonyms of disease names are also included to facilitate searching).
| Acute lymphoblastic leukemia | Erectile dysfunction | Neuropathic |
| Addiction | Fever | Obesity |
| Advanced pancreatic tumor | Fungal infection | Obstructive pulmonary disease |
| Affective disorder | Gastric tumor | Ocular hypertension/glaucoma |
| AIDS | Glaucoma | Oral |
| Allergic rhinitis | Gout | Osteoporosis |
| Allergy | Heart disease | Ovarian |
| Alzheimer’s | Heart failure | Pain |
| Analgesic | Helminth infection | Parkinson’s |
| Anesthesia | Hepatitis C | Peptic ulcer |
| ANF degradation | Herpes | Phaeochromocytoma |
| Angiogenesis | High blood glucose level | Platelet adhesion |
| Anxiety | High blood sugar level | Platelet disease |
| Arthritis | High cholesterol | Posterior pituitary disorder |
| Asthma | Hirsutism | Postsurgical |
| Autoimmune disease | Hormone-dependent tumors | Prostate adenocarcinoma |
| B cell | Human African trypanosomiasis | Prostate tumor |
| Bacterial infection | Hypertension | Prostatic hyperplasia |
| Baldness | Hyperthyroidism | Psychiatric illness |
| Blood coagulation | Hypocalcaemia | Psychomotor |
| Bone Loss | Immune response | Reproduction |
| Brain ischaemia | Immunodeficiency | Respiration |
| Breast | In transplantation, etc. | Rheumatoid |
| Calcium deficiency | Inflammation | Riboflavin deficiency |
| Cancer | Influenza A and B | Schizophrenia |
| Carcinoid syndrome | Insomnia | Seizure |
| Cardiac failure | Irritable bowel syndrome | Smoking |
| Cardiovascular disease | Kidney failure | Smooth muscle |
| Chronic myelogenous leukemia | Leukemia | Solid tumor |
| Cognitive dysfunction | Liposarcoma | Thiamine deficiency |
| Colon | Liver | Tuberculosis |
| Common cold | Local anesthetic | Urinary tract infection |
| Common roundworm | Lung | Urticaria |
| Congestive heart failure | Lupus | Uterus contraction |
| Cystic fibrosis | Malaria | Vascular disease |
| Dementia | Malignant pain | Viral infection |
| Depression | Melanoma | Visceral |
| Diabetes | Metastasis | Vitamin A deficiency |
| Diabetic retinopathy | Migraine | Vitamin B12 deficiency |
| Diarrhea | Morning sickness | Vitamin B6 deficiency |
| Drug dependence | Motion sickness | Vitamin C deficiency |
| Drug induced | Motor disorder | Vitamin D deficiency |
| Dry eye | Movement disorder | Vomiting |
| Dysrhythmic | Nasal congestion | Zollinger-Ellison syndrome |
| Emphysema | Neurodegeneration | |
| Epilepsy | Neurological symptom |
Table 2. Drug functions listed in TTD (synonyms of drug functions are also included to facilitate searching).
| Activator | Cofactor |
| Agonist | Immunotoxin |
| Alkylator | Inactivator |
| Antagonist | Inhibitor |
| Antibody | Intercalator |
| Antisense | Opener |
| Blocker | Stimulator |
| Chain breaker | Substrate |
| Coenzyme | Vaccine |
Table 3. Drug classifications listed in TTD (synonyms of drug classifications are also included to facilitate searching).
| Anesthetic | Antimalarial | Lipid-lowering |
| Anti-allergic | Antimotility | Local anesthetic |
| Anti-allergy | Anti-neurodegenerative | Lupus |
| Anti-androgen | Anti-obesity | Nasal decongestion |
| Anti-angiogenic | Antiplatelet | Neurological |
| Anti-asthmatic | Antipsychotic | Opioid overdose |
| Antibacterial | Antipyretic | Osteoporosis |
| Anticancer | Antirheumatoid | Ovulation induction |
| Anti-cholesterol | Antiseptics | Pain-killer |
| Anticoagulant | Antiviral | Parkinson’s |
| Anticonvulsant | Anxiolytic | Platelet |
| Antidepressant | Anxiotic | Procoagulant |
| Antidiabetic | Arthritis | Psychomotor stimulant |
| Antidiarrheal | Bronchodilator | Psychostimulant |
| Antidiuretic | Cardiotonic | Psychotomimetic |
| Antidysrhythmic | Contraceptive | Respiratory stimulant |
| Anti-emetic | Convulsant | Sedative |
| Anti-emetics | Depressant | Supplement |
| Antiepileptic | Diuretics | Uterine contractant |
| Antifungal | Drug dependence | Uterine relaxant |
| Anti-gastric secretion | Erectile dysfunction | Vasodilator |
| Antihelminthic | Glaucoma treatment | Vitamin |
The search is case insensitive. In a query, a user can specify full name or any part of the name in a text field, or choose one item from a selection field. Wild characters of ‘%’ and ‘_’ are supported in text field. Here, ‘_’ represents any one character and ‘%’ represents a string of characters of any length. For example, input of ‘phosphatase’ in the target name field finds entries containing ‘phosphatase’ in their name, such as Cdc25A phosphatase or tyrosine phosphatase. On the other hand, input of ‘Cdc25_ phosphatase’ finds entries with names like Cdc25A phosphatase, Cdc25B phosphatase and Cdc25C phosphatase. Likewise, input of Cdc% phosphatase finds the same entries as above. In this case, ‘%’ represents ‘25A’, ‘25B’, ‘25C’, respectively.
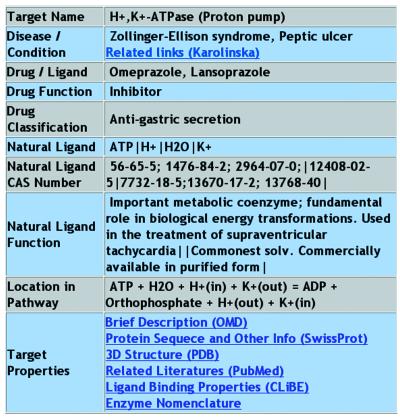
The result of a typical search is illustrated in Figure 2. In this interface, all the therapeutic targets that satisfy the search criteria are listed along with the disease conditions to be treated, drugs or ligands directed at the target, and the drug class. More detailed information of a target can be obtained by clicking the corresponding target name. The result is displayed in an interface shown in Figure 3. From this interface, one finds target name, corresponding disease condition and cross-link to Karolinska disease database (http://www.kib.ki.se/), target function in pathway and corresponding natural ligand, known drugs or ligands directed at the target, drug function (such as inhibitor, antagonist and blocker, etc.), drug therapeutic classification, and additional cross-links to other databases that provide useful information about the target.
Figure 2.
The interface of a search result on TTD. All the targets that satisfy the specified search criteria are listed along with disease, drug/ligand name and drug classification.
Figure 3.
Interface of the detailed information of target in TTD. Information related to disease, drug/ligand, pathway and some of the cross-database shortcuts are provided. In the case of one target having multi ligands, the ligands are separated with ‘│’, as well as their functions and CAS numbers.
The functional properties of an identified target can be obtained through cross-linking to the On-line Medical Dictionary (OMD) database (http://www.graylab.ac.uk/omd/) and the SWISS-PROT database (22). The target sequence can be retrieved from cross-link to the SWISS-PROT database. The available 3D structure of this target can be accessed through cross-linking to the Protein Data Bank (PDB) database (23). For an enzymatic target, its nomenclature can be obtained from cross-link to the Enzyme Data Bank (24). Ligand-binding properties may be obtained from cross-link to the Computed Ligand Binding Energy database (CliBE) (http://xin.cz3.nus.edu.sg/group/CLiBE.asp). The related literature can be accessed from cross-link to the relevant entries in the PubMed database (25).
As the research in proteomics (26) and pathways (27) progresses, the relevant information can be incorporated or the corresponding databases can be cross-linked to TTD to provide more comprehensive information about the drug targets and their relationship to other biomolecules and cellular processes.
REFERENCES
- 1.Drews J. (2000) Drug discovery: a historical perspective. Science, 287, 1960–1964. [DOI] [PubMed] [Google Scholar]
- 2.Ohlstein E.H., Ruffolo,R.R.,Jr and Ellroff,J.D. (2000) Drug discovery in the next millennium. Annu. Rev. Pharmacol. Toxicol., 40, 177–191. [DOI] [PubMed] [Google Scholar]
- 3.Peltonen L. and Mckusick,V.A. (2001) Genomics and medicine: dissecting human disease in the postgenomics era. Science, 291, 1224–1232. [DOI] [PubMed] [Google Scholar]
- 4.Sali A. (1998) 100,000 protein structures for biologist. Nature Struct. Biol., 5, 1029–1032. [DOI] [PubMed] [Google Scholar]
- 5.Koonin E.V., Tatusov,R.L. and Michael,Y.G. (1998) Beyond complete genomes: from sequence to structure and function. Curr. Opin. Struct. Biol., 8, 355–363. [DOI] [PubMed] [Google Scholar]
- 6.Wallace K.B. and Starkov,A.A. (2000) Mitochondrial targets of drug toxicity. Annu. Rev. Pharmacol. Toxicol., 40, 353–388. [DOI] [PubMed] [Google Scholar]
- 7.Vesell E.S. (2000) Advances in pharmacogenetics and pharmacogenomics. J. Clin. Pharmacol., 40, 930–938. [DOI] [PubMed] [Google Scholar]
- 8.Cornell W.D., Cieplak,P., Bayly,C.I., Gould,I.R., Mer,K.M.,Jr, Ferguson,D.M., Spellmeyer,D.C., Fox,T., Caldwell,J.W. and Kollman,P.A. (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc., 117, 5179–5197. [Google Scholar]
- 9.Blundell T.L. (1996) Structure-based drug design. Nature, 384 (Suppl.), 23–26. [DOI] [PubMed] [Google Scholar]
- 10.Podlogar B.L. and Terguson,D.M. (2000) QSAR and CoMFA: a perspective on the practical application to drug discovery. Drug Des. Discov., 17, 4–12. [PubMed] [Google Scholar]
- 11.Chen Y.Z. and Zhi,D.G. (2001) Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins, 43, 217–226. [DOI] [PubMed] [Google Scholar]
- 12.Rang H.P., Dale,M.M. and Ritter,J.M. (1999) Pharmacology, 4th Edn. Churchill Livingstone, New York, NY.
- 13.Katznug B.G. (1998) Basic and Clinical Pharmacology, 7th Edn. Appleton & Lange, New Jersey, NJ.
- 14.Navia M.A. and Murcko,M.A. (1992) Use of structural information in drug design. Curr. Opin. Struct. Biol., 2, 202–210. [Google Scholar]
- 15.Gibbs J.B. (2000) Mechanism-based target identification and drug discovery in cancer research. Science, 287, 1969–1973. [DOI] [PubMed] [Google Scholar]
- 16.Rao R.N. (1996) Targets for cancer therapy in the cell cycle pathway. Curr. Opin. Oncol., 8, 516–524. [DOI] [PubMed] [Google Scholar]
- 17.Brower V. (1999). Tumor angiogenesis – new drugs on the block. Nat. Biotechnol., 17, 963–968. [DOI] [PubMed] [Google Scholar]
- 18.Moir D.J., Shaw,K.J., Hare,R.S. and Vovis,G.F. (1999) Genomics and antimicrobial drug discovery. Antimicrob. Agents Chemother., 43, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olliaro P.L. and Yuthavong,Y. (1999) An overview of chemotherapeutic targets for antimalarial drug discovery. Pharmacol. Ther., 81, 91–110. [DOI] [PubMed] [Google Scholar]
- 20.Brower V. (2000) New paths to pain relief. Nat. Biotechnol., 18, 387–391. [DOI] [PubMed] [Google Scholar]
- 21.Persidis A. (2000) Industry trends supplement. Nat. Biotechnol., 18, IT3–IT29.11055590 [Google Scholar]
- 22.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2002), 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bairoch A. (2000) The ENZYME database in 2000. Nucleic Acids Res., 28, 304–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEntyre J. and Lipman,D. (2001) PubMed: bridging the information gap. Can. Med. Assoc. J., 164, 317–1319. [PMC free article] [PubMed] [Google Scholar]
- 26.Dove A. (1999) Proteomics: translating genomics into products? Nat. Biotechnol., 17, 233–236. [DOI] [PubMed] [Google Scholar]
- 27.Scharpe S. and De Meester,I. (2001) Peptide truncation by dipeptidyl peptidase IV: a new pathway for drug discovery? Verh. K. Acad. Geneeskd. Belg., 63, 5–32. [PubMed] [Google Scholar]