Abstract
Genew, the Human Gene Nomenclature Database, is the only resource that provides data for all human genes which have approved symbols. It is managed by the HUGO Gene Nomenclature Committee (HGNC) as a confidential database, containing over 16 000 records, 80% of which are represented on the Web by searchable text files. The data in Genew are highly curated by HGNC editors and gene records can be searched on the Web by symbol or name to directly retrieve information on gene symbol, gene name, cytogenetic location, OMIM number and PubMed ID. Data are integrated with other human gene databases, e.g. GDB, LocusLink and SWISS-PROT, and approved gene symbols are carefully co-ordinated with the Mouse Genome Database (MGD). Approved gene symbols are available for querying and browsing at http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/searchgenes.pl.
INTRODUCTION
The Human Gene Nomenclature Database (Genew) was developed in the 1980s by the HUGO Gene Nomenclature Committee (HGNC) as a private (off-line) database of information relevant to gene symbol assignment. The HGNC, formed in the 1970s, is the authority responsible for assigning consistent, standardized human gene nomenclature (1), which is vital for the interoperability of databases. Nomenclature committees were established for both the human and mouse genes, and guidelines were published in conjunction with the reports from the Human Genome Mapping meetings. The first public database containing gene nomenclature was EDIT10 (2). However, subsequently, ‘approved’ gene nomenclature was only available online via The Genome Database (GDB) (3), managed as part of their curation system.
The Genew database is not directly searchable on the Web, as it contains confidential and administrative information in addition to the gene symbol, name, chromosomal localization and other data. Therefore, in 1997 we made data from seven key fields within Genew available as a searchable list via the Web (http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/searchgenes.pl). This online version of the database is updated weekly and now receives over 25 000 hits per month. It is possible to search it using any symbol or name and retrieve displayed information on the approved symbol, name, cytogenetic location, PubMed ID (PMID) and Online Mendelian Inheritance in Man (OMIM) number for each gene where this information is known. Further information is found by following the direct links to the PubMed (4) and OMIM (4) database web sites or by the indirect links to GDB (3), GenAtlas (5), GeneCards (6) and LocusLink (7). It is also possible to retrieve all the aliases or synonyms recorded for each gene from Genew using the list aliases search.
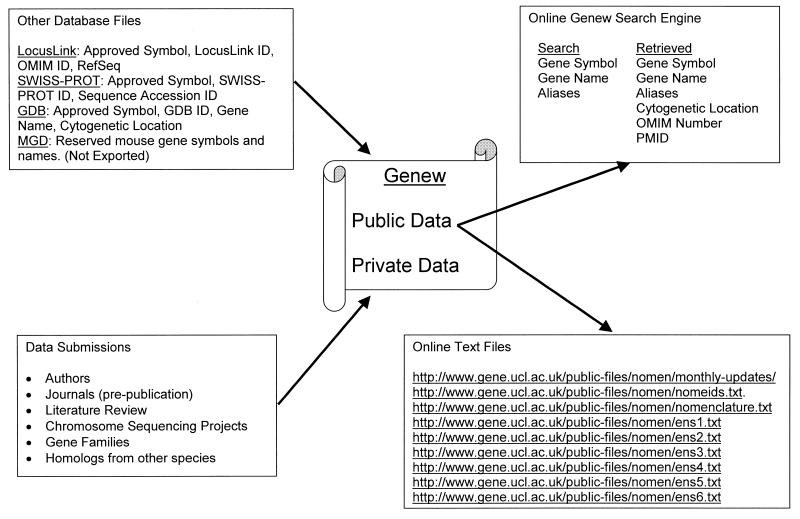
Gene data are collated and integrated from a variety of sources (Fig. 1) including author submissions, gene family specific databases, human gene databases, the scientific literature and other organism databases, especially The Mouse Genome Database (MGD) (8). Data are directly imported and mapped from MGD, SWISS-PROT (9), GDB (3) and LocusLink (7) to our gene symbols.
Figure 1.
Genew data flow diagram. Data are imported from four databases along with updates from sources such as submitting authors and reviewed literature. Public data only are exported to the main text files; those that are used by the online search engine and those generated from users’ requests.
IMPLEMENTATION
Genew is implemented in the Microsoft Access 97 (version 8.0) relational database management system. The database consists of 12 tables containing over 150 fields and 16 000 gene records.
The Genew search engine (http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/searchgenes.pl) is based on Perl and CGI scripts and uses text files exported from the off-line database.
OTHER INFORMATION
Electronic data submission
We enter data into Genew both manually and using automatic methods (Fig. 1). Only scientific editors from the HGNC are able to curate Genew and we carefully check each gene record to ensure high quality. We would like to encourage authors to submit their request for a new gene symbol via the Web-based submission form (http://www.gene.ucl.ac.uk/nomenclature/submit.html) or to contact us directly by email (nome@galton.ucl.ac.uk) if they have large data sets. The submission form involves entering contact details along with any other relevant information, including a suggested gene name and short-form gene symbol. We also require sequence data for each new gene submitted. The HGNC editors validate every record that is entered in this way by further analyses and database searching. We also rely on assistance from a number of specialist advisors, http://www.gene.ucl.ac.uk/nomenclature/advisors.html, and communication with the scientific community in general via our gene family specific web pages, http://www.gene.ucl.ac.uk/nomenclature/genefamily.shtml, to maintain accuracy.
Each gene record is given a unique HGNC ID and status as it is entered into Genew. Confidential records (not available online) include those genes in progress, without approved symbols which have a ‘Pending’ status and those that have approved symbols but have a ‘Reserved’ status and are not released for a period of up to 6 months. All other gene records are accessible via the search engine and have one of three different states: ‘Approved’ gene symbols, ‘Symbol Withdrawn’ for genes which now have different approved symbols and ‘Entry Withdrawn’ for records that refer to a gene that has since been shown not to exist.
LBlast
We have developed an in-house BLAST (10) analysis tool termed ‘LBlast’ to validate new gene submissions and avoid duplication of symbols. LBlast searches all the databases generated from the variety of sources kept by HGNC, e.g. chromosome specific sequence, sequence from Genew and sequence from archived text files. This is very useful and has been enhanced with the addition of the downloadable RefSeq sequences.
Confidentiality
We ensure that release of any data stored in Genew is agreed with the submitting authors via completion of our confidentiality form: http://www.gene.ucl.ac.uk/nomenclature/information/confidentiality.shtml. However, we have recently agreed where possible to release a sequence accession ID for each gene. To ensure that no confidential data are released by mistake, without having to check each entry individually, we have only exported those IDs which matched against gene symbols in both Genew and LocusLink (7) or Genew and SWISS-PROT (9).
Accessibility
The Genew data available via the online search engine are also downloadable as a tab-delimited text file in compressed (GNU zip), http://www.gene.ucl.ac.uk/public-files/nomen/nomenclature.gz, or uncompressed format, http://www.gene.ucl.ac.uk/public-files/nomen/nomenclature.txt
Integration
We import limited data sets from four other databases: LocusLink (7), SWISS-PROT (9), GDB (3) and MGD (8). These data are stored in separate tables and are not directly curated within Genew. However, our strong collaboration with these databases ensures that the data are accurate. We use some of this mapped information to assist in our validation of Genew and to export as part of our output file, http://www.gene.ucl.ac.uk/public-files/nomen/nomeids.txt.
User support
We have received requests in the past for specific data subsets which we are willing to fulfill (as long as we can ensure that confidentiality is not compromised). We make these available as tab-delimited text files: http://www.gene.ucl.ac.uk/public-files/nomen/ens1.txt containing HGNC ID, SWISS-PROT ID, RefSeq ID; http://www.gene.ucl.ac.uk/public-files/nomen/ens2.txt containing HGNC ID, Gene Approved Symbol; http://www.gene.ucl.ac.uk/public-files/nomen/ens4.txt containing HGNC ID, Approved Symbol, Aliases, Previous Symbol(s); http://www.gene.ucl.ac.uk/public-files/nomen/ens5.txt containing HGNC ID, Approved Symbols, Gene Name, EC Number, SWISS-PROT ID; http://www.gene.ucl.ac.uk/public-files/nomen/ens6.txt containing HGNC ID, Symbol, Accession ID.
Distribution
Genew is available from our Web site and is mirrored as part of EBI’s SRS package (11). However, all public data are also added to LocusXref (a privately edited database, hosted by NCBI) which feeds data directly into LocusLink (7) within 24 h.
FUTURE DEVELOPMENTS
We are intending to migrate the data in Genew to a MySQL database management system in the next year. This will enable us to have more up-to-date data release (within 24 h of input) and online editorial capacity, as the public database views will be presented online dynamically.
AVAILABILITY
The databases and associated information files are freely available to users for non-commercial purposes only. If there is a financial return users are requested to contact the corresponding author for further information.
CITATION
Authors are requested to cite this article and the database in the following format: Genew, HUGO Gene Nomenclature Committee (HGNC), Department of Biology, University College London, Wolfson House, 4 Stephenson Way, London NW1 2HE, UK (http://www.gene.ucl.ac.uk/cgi-bin/nomenclature/searchgenes.pl). Include month and year when you retrieved the data cited.
SUPPLEMENTARY MATERIAL
Supplementary Material (history of Genew, nomenclature resources and useful URLs) is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The HGNC is supported by NIH contract N01-LM-9-3533 and by the UK Medical Research Council. Many thanks to John Attwood (Sanger Centre), Julia White (Open University) and Gary Walsh for their past contributions to the maintenance and development of Genew. Thanks are also due to the HGNC editors Drs Elspeth Bruford, Ruth Lovering and Mathew Wright, whose accurate curation and attention to detail ensure the validity of the gene records.
REFERENCES
- 1.Shows T.B., Alper,C.A., Bootsma,D., Dorf,M., Douglas,T., Huisman,T., Kit,S., Klinger,H.P., Kozak,C., Lalley,P.A. et al. (1979) International system for human gene nomenclature (1979) ISGN. Cytogenet. Cell Genet., 25, 96–116. [DOI] [PubMed] [Google Scholar]
- 2.Miller R.L., Chan,H.S., Cavanaugh,M.L., Alles,W.P., Mador,M.L. and Kidd,K,K. (1989) EDIT10: a database interface for HGM10. Cytogenet. Cell Genet., 51, 9–11. [DOI] [PubMed] [Google Scholar]
- 3.Talbot C.C. Jr and Cuticchia,A.J. (1999) Human Mapping Databases, Current Protocols in Human Genetics. John Wiley & Sons, New York, NY, 1.13.1–1.13.12. [DOI] [PubMed]
- 4.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. Updated article in this issue: Nucleic Acids Res. (2002), 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frezal J. (1998) Genatlas database, genes and development defects. C. R. Acad. Sci. III, 321, 805–817. [DOI] [PubMed] [Google Scholar]
- 6.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lancet,D. (1998) GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics, 14, 656–664. [DOI] [PubMed] [Google Scholar]
- 7.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake J.A., Eppig,J.T., Richardson,J.E., Bult,C.J. and Kadin,J.A. (2001) The Mouse Genome Database (MGD): integration nexus for the laboratory mouse. Nucleic Acids Res., 29, 91–94. Updated article in this issue: Nucleic Acids Res. (2002), 30, 113–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bairoch A. and Apweiler R. (2000) SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 11.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Lombard,V., Lopez,R., Parkinson,H., Redaschi,N., Sterk,P., Stoehr,P. and Tuli,M.A. (2001) The EMBL nucleotide sequence database. Nucleic Acids Res., 29, 17–21. Updated article in this issue: Nucleic Acids Res. (2002), 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.