Abstract
Background
We found microRNA (miR)-1246 to be significantly differentially expressed between severe active alopecia areata (AA) patients and healthy individuals.
Objective
To explore the role and mechanism of miR-1246 in severe AA.
Methods
Expression of miR-1246, dual-specific tyrosine phosphorylation-regulated kinase 1A (DYRK1A), and nuclear factor of activated T cells 1c (NFATc1) in peripheral CD4+ T cells and in scalp tissues of patients were detected using RT-qPCR, Western blot, and immunohistochemistry assays. Peripheral CD4+ T cells from the AA patients were transfected with lentiviral vectors overexpressing miR-1246. RT-qPCR and Western blot analysis were used to measure mRNA or protein expression of retinoic-acid-receptor-related orphan nuclear receptor gamma (ROR-γt), interleukin (IL)-17, DYRK1A, NFATc1, and phosphorylated NFATc1. Flow cytometry was used to assay the CD4+IL-17+ cells proportion. ELISA was used to measure cytokine levels.
Results
miR-1246 levels decreased and DYRK1A and NFATc1 mRNA levels significantly increased in the peripheral CD4+ T cells and scalp tissues of severe active AA samples. NFATc1 protein expression was also significantly increased in the peripheral CD4+ T cells but not in the scalp tissues. NFATc1 positive cells were mainly distributed among infiltrating inflammatory cells around hair follicles. In peripheral CD4+ T cells of severe active AA, overexpression of miR-1246 resulted in significant downregulation of DYRK1A, NFATc1, ROR-γt, and IL-17 mRNA and phosphorylated NFATc1 protein, as well as a decrease in the CD4+IL-17+ cells proportion and the IL-17F level.
Conclusion
miR-1246 can inhibit NFAT signaling and Th17 cell activation, which may be beneficial in the severe AA treatment.
Keywords: Alopecia areata, MicroRNAs, Phosphorylation, Th17 cells, Transcription factors
INTRODUCTION
Alopecia areata (AA) is an organ-specific autoimmune disease caused by T cells that affects hair follicles. Due to the collapse of immune privilege dufring the anagen phase, activated T lymphocytes secrete inflammatory cytokines, resulting in hair follicles entering the catagen and telogen phases prematurely1. AA is characterized by its sudden onset, unpredictability of clinical outcome, ease of recurrence, and lack of preventability. Once the disease develops, it has significant impacts on mental health and quality of life2,3. Currently, there is no clinically effective treatment for AA4. While clinical trials of JAK inhibitors have demonstrated excellent efficacy and safety in recent years, it is still necessary to monitor, optimize, and resolve the challenges of long-term medication. As a result, additional research on the pathogenesis of AA is required for the development of a new theoretical foundation for clinical diagnosis and treatment.
MicroRNAs (miRNAs) are a well characterized class of noncoding RNA molecules that post-transcriptionally regulate gene expression by either targeting messenger RNAs for degradation or repressing their translation. Over the last few years, miRNAs have been identified as critical regulatory factors of the Th17/T regulatory (Treg) cell balance, which plays a role in the development and progression of autoimmune diseases5,6,7. Studies have demonstrated that miRNAs play an important role in the regulation of inflammatory skin diseases, such as atopic dermatitis and psoriasis, but little is known about the role of miRNAs in AA.
Several recent studies have suggested that Th17 cells play a role in the pathogenesis of AA. Lew et al.8 reported that a single nucleotide polymorphism in the interleukin (IL)-17 receptor A (IL17RA) gene was significantly different between AA patients and healthy individuals and was associated with an early onset of AA. Our previous study showed that the number of Th17 cells in the peripheral blood was significantly higher in AA patients than in healthy controls (HC), and the Th17 proportion was higher in patients in the active stage than those in the stable stage9. Immunofluorescence staining revealed a high number of Th17 cells in the upper dermis of scalp lesions in AA patients, particularly around the hair follicles10,11. In addition, serum IL-17 levels were elevated in the AA patients, which correlated with the severity of AA but not with its course12. Serum IL-17 levels were significantly higher in patients with early-onset AA and in patients under the age of 30 years13. After AA was improved by narrow-spectrum ultraviolet B treatment, peripheral IL-17 levels decreased significantly14. These findings suggest that Th17 cell subsets and their effector molecules may play a role in the pathological process of severe AA.
Ustekinumab, a human monoclonal immunoglobulin (Ig) G1 antibody that binds to and inhibits the function of the p40 subunit of IL-12 and IL-23, is approved by the Food and Drug Administration (FDA) for the treatment of psoriasis and psoriatic arthritis15. Ustekinumab has also been found to indirectly inhibit Th17 cell activation and IL-17 production. In one study, three extensive AA patients who received 90 mg ustekinumab subcutaneously experienced hair regrowth16. Another study of three children treated with ustekinumab for refractory AA reported similar results with only minor side effects17. However, a randomized controlled trial demonstrated that secukinumab, a human IgG1 monoclonal antibody that directly inhibits IL-17A, was ineffective in the treatment of severe AA18. These findings imply that modulating Th17 cell activation may be only one strategy for intervening in AA pathogenesis, and that combining this strategy with other approaches may be needed.
A previous study used an Agilent microarray covering 2,549 human miRNAs to screen for and validate genes involved in AA. From that study, we identified microRNA 1246 (miR-1246) as being significantly differentially expressed between active AA patients and HCs19. miR-1246 has been extensively studied in recent years and may be used as a biological marker and interventional target for a variety of tumors20,21,22,23. Using bioinformatics analysis and dual-luciferase assays, previous studies have demonstrated that miR-1246 can down-regulate the expression of dual-specific tyrosine phosphorylation-regulated kinase 1A (DYRK1A) by targeting its 3'UTR mRNA sequence. MiR-1246 was also found to activate nuclear factor of activated T cells (NFAT) in non-neoplastic diseases, which is associated with the occurrence of Down syndrome24,25. Additionally, miR-1246 can protect the liver from ischemia/reperfusion injury by balancing the number of Treg and Th17 cells26. The miR-1246-DYRK1A-NFAT signaling axis has also been implicated in a variety of tumor diseases in recent years24,25,27. NFAT, which includes NFATc1-4 and NFAT5, is a type of transcription factor that is activated via the calcineurin (CaN) signaling pathway. NFATc1 and NFATc2 are most abundant in immune cells, and many studies have shown that NFATc1 can promote Th17 cell activation28,29. NFAT is also involved in the pathogenesis of a number of immune-related skin diseases, including psoriasis30,31, atopic dermatitis32, and lupus erythematosus33, but its role in the pathogenesis of AA has yet to be investigated. In the present study, we investigated whether miR-1246 affects NFAT signaling and explored its specific mechanisms in severe AA using patient specimens and in vitro cell models.
MATERIALS AND METHODS
Study subjects
Ten subjects with severe active AA for one to six months who were admitted to Huashan Hospital’s hair clinic between 2018 and 2020 were recruited in this study. Inclusion criteria were as follows: 1) age between 16 and 65 years; 2) conformity to the diagnosis of AA; 3) The Severity of Alopecia Tool (SALT) score greater than 50; and 4) positive for pull test. Exclusion criteria were as follows: 1) failure to meet the diagnostic criteria for AA; 2) co-existence of other inflammatory skin diseases such as psoriasis, atopic dermatitis, or vitiligo; 3) reported other infectious, neoplastic, or autoimmune diseases; or 4) received glucocorticoid or immunosuppressive therapy within the preceding six months. Ten age and gender matched HC were also included in this study. We collected peripheral venous blood samples from all enrolled patients and HC. Additionally, three AA patients underwent scalp biopsy to obtain tissue samples, and three HC patients provided normal scalp around the pigmented nevus obtained during nevus removal. The Huashan Hospital Ethics Committee approved the study protocol (Ethical approval No: 2020-157), and all study subjects signed the informed consent form.
Isolation, culture, and transduction of CD4+ T cells
A total of 107 peripheral blood mononuclear cells were separated using the Ficoll gradient density method (American Sigma Company), mixed and incubated with 80 µl MACS buffer and 20 µl MACS CD4 MicroBeads (Germany Miltenyi Company), and then sorted according to the manufacturer's instructions. The sorted CD4+ T cells were cultured in a 6-well plate with Gibco™CTS™OpTmizer™ T-Cell Expansion Serum Free Media (Thermo Fisher) at a concentration of 0.5×106/ml for 48 hours. The transduction was performed with the hsa-miR-1246 overexpression lentiviral vector (miR-1246 group) or the negative control vector (negative control group, PGMLV-6395; Shanghai Jiman Biotechnology Co., Ltd.) at the optimized moieties of infection of 50. The blank control was set as the blank control group. To increase transduction efficiency, cells were incubated with polybrene at a concentration of 10 µg/µl and centrifuged at a speed of 300 rpm for 2 hours at 24℃. The culture medium was changed 18 hours after transduction. After 48 hours of continued culture, the cells and supernatant were collected for further experiments.
Quantitative real-time PCR (RT-qPCR)
Total RNA was extracted from individual groups of peripheral CD4+ T cells or scalp tissues using TRIzol reagent and reverse transcribed into cDNA using the PrimeScript™RT Master Mix (TaKaRaBio) per the manufacturer’s protocol. The relative levels of miRNA and mRNA transcripts to the internal control U6 and GAPDH were quantified using qRT-PCR in duplicate with the Power SYBR Green PCR Master (Thermo Fisher) using the ABI750. The CT (cycle threshold) data were analyzed using the 2–ΔΔCt method. The specific primers are listed in Supplementary Table 1.
Immunohistochemical (IHC) staining
Briefly, paraffin-embedded scalp tissue samples were placed in an antigen repair solution, followed by microwave incubation. The sections were incubated with primary antibody against NFATc1 (1:500; Wanleibio) overnight at 4℃ after being blocked in goat serum. The slides were further washed and incubated with horseradish conjugated secondary antibody (Jackson) at 37℃ for 30 minutes. The stained tissues were developed in DAB colorant (Zsbio), and the slides were counterstained with hematoxylin. Five 400x magnification fields on each slide were randomly selected under the microscope, and the number of positively stained cells in each field was manually counted.
Western blot analysis
Total protein in each sample was extracted using RIPA lysis buffer (Beyotime Biotechnology). The protein concentration was determined using the BCA protein concentration quantification kit (Beyotime Biotechnology). Denatured proteins were separated using 5× SDS-PAGE and transferred to PVDF membranes followed by blocking with 5% nonfat dry milk in Tris-buffered saline with 0.05% Tween 20 and probed with primary antibodies overnight at 4℃. Following washing, membranes were incubated with goat peroxidase-conjugated secondary antibodies at 37℃ for 2 hours. The reactions were detected using the Pierce ECL Western Blotting Substrate (Thermo Fisher). The bands were visualized via exposure to an x-ray beam in a dark room and semi-quantified analyzed using Tanon Image software. The ratio of the densitometric values of NFATc1, phosphorylated NFATc1, or DYRK1A bands to those of β-actin bands were used to determine relative expression. The primary and secondary antibodies included: mouse anti-β-actin (43 kDa, 1:1,000 dilution; Santa Cruz), mouse anti-NFATc1 (110,140 kDa, 1:500; Wanleibi), rabbit anti-NFATc1-P (phospho S237, 110, 140 kDa, 1:1,000; Abcam), rabbit anti-DYRK1A (46, 60, 86 kDa, 1:1,000; Abcam), Peroxidase Affini Pure Donkey Anti-Mouse IgG (H+L, 1:5,000; Jackson), and Peroxidase Affini Pure Goat Anti-Rabbit IgG (H+L, 1:10,000; Jackson).
Flow cytometry
Peripheral CD4+ T cells sorted with magnetic beads were adjusted to a final concentration of 2×106/ml with pre-cooled stain buffer, and 106 cell suspension was incubated for 20 minutes at room temperature in the dark with 10 µl allophycocyamin (APC) conjugated Anti-human CD4 (eBioscience). After being washed twice with stain buffer, the cells were resuspended with 500 µl of stain to detect the purity of CD4+ T cells using flow cytometry. To determine the phenotype of Th17 cells, CD4-APC stained cells were added to fixative Medium A 100l and incubated for 15 minutes at room temperature in the dark. After resuspension in phosphate buffered saline (PBS), the cells were incubated at room temperature for 20 minutes in the dark with permeabilization buffer Medium B and PerCP-Cy5.5 conjugated antihuman IL-17A (eBioscience), followed by a PBS wash and resuspension for flow cytometry analysis of the CD4+IL-17+ cell ratio.
ELISA
Concentrations of IL-17A/IL-17F/IL-21/IL-22/TNF-α in the cell culture supernatant were determined using ELISA kits (Shanghai Xitang Biological Co., Ltd.) according to the manufacturer's instructions. The absorbance values were used to create a standard curve, which was then used to calculate the sample concentrations (pg/ml).
Statistical analysis
SPSS 23.0 (IBM Corp.) and GraphPad Prism 8.0 (GraphPad Software Inc.) were used to analyze the data presented as mean±standard deviation (SD). For measurement data, results are presented as the mean±SD. For pairwise comparison, the student’s t-test was used for normally distributed data and the rank sum test was used for non-normally distributed data. One-way ANOVA was used to compare multiple groups and the Bonferroni test was used for multiple comparisons between multiple samples. All experiments were repeated at least three times and p<0.05 was set to indicate statistical significance. Each experiment was repeated for three times.
Ethics statement
The Huashan Hospital Ethics Committee approved the study protocol (Ethical approval No: 2020-157). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
RESULTS
Expression of the miR-1246/DYRK1A/NFATc1 axis in CD4+ T cells in the peripheral blood
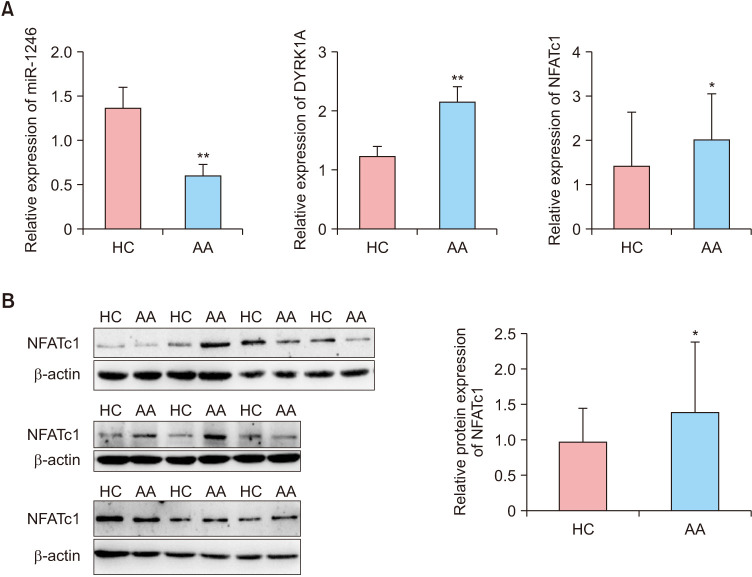
In patients with severe active AA, the level of miR-1246 in the peripheral CD4+ T cells was significantly decreased compared to HC. In contrast, DYRK1A and NFATc1 mRNA levels were significantly increased (p<0.01 and p<0.05, respectively; Fig. 1A). Western blot analysis confirmed the increase in NFATc1 protein expression (p<0.05; Fig. 1B).
Fig. 1. Expression of miR-1246/DYRK1A/NFATc1 in peripheral CD4+ T cells. (A) mRNA levels of miR-1246, DYRK1A, and NFATc1 in severe active AA and HC. (B) Protein expression of NFATc1 in severe active AA and HC. AA: alopecia areata, HC: healthy controls, miR-1246: microRNA-1246, DYRK1A: dual-specific tyrosine phosphorylation-regulated kinase 1A, NFAT: nuclear factor of activated T cells. *p<0.05, **p<0.01.
Expression of the miR-1246/DYRK1A/NFATc1 axis in scalp tissues
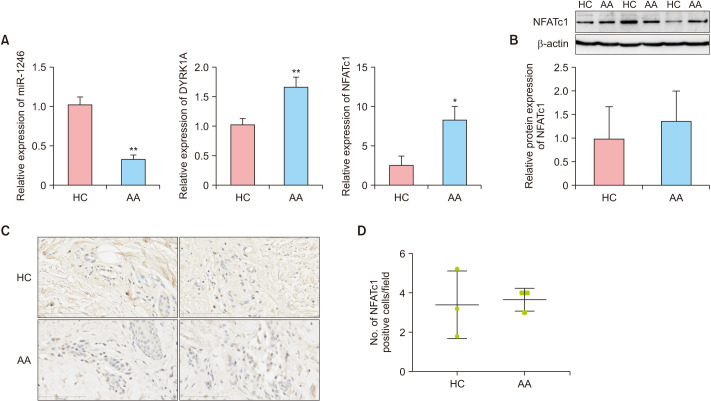
In patients with severe active AA, the level of miR-1246 in scalp tissues was significantly decreased compared to HC. In contrast, DYRK1A and NFATc1 mRNA levels were significantly increased (p<0.01 and p<0.05, respectively; Fig. 2A). Western blot analysis confirmed the increase in NFATc1 protein expression, but it was not statistically significant (p>0.05, Fig. 2B). IHC showed that NFATc1 positive cells were mainly distributed among the infiltrating inflammatory cells around the hair follicles (Fig. 2C), but there was no significant difference between the AA group and the HC group (p>0.05; Fig. 2D).
Fig. 2. Expression of miR-1246/DYRK1A/NFATc1 in scalp tissues. (A) mRNA levels of miR-1246, DYRK1A, and NFATc1 in severe active AA and HC. (B) Protein expression of NFATc1 in severe active AA and HC. (C) Immunohistochemistry of NFATc1 in scalp tissues (×400). (D) Number of NFATc1 positive cells under high power field (mean±standard deviation). AA: alopecia areata, HC: healthy controls, miR-1246: microRNA-1246, DYRK1A: dual-specific tyrosine phosphorylation-regulated kinase 1A, NFAT: nuclear factor of activated T cells. *p<0.05, **p<0.01.
Establishment of CD4+ T cells overexpressing miR-1246
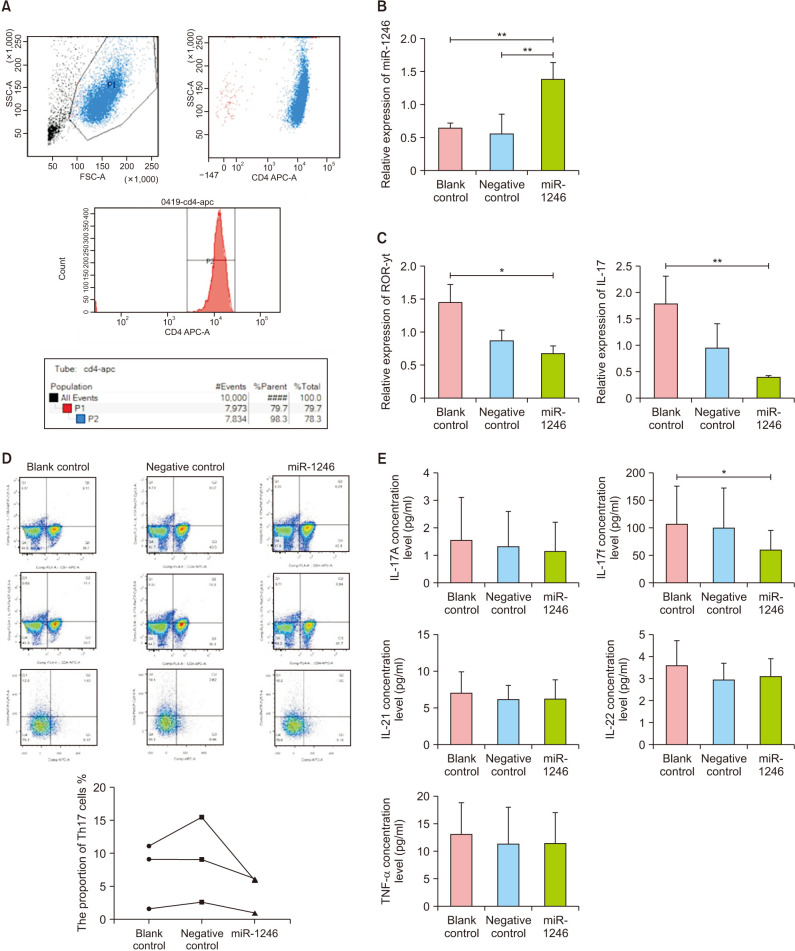
Three patients with severe active AA each had 60 ml of peripheral venous blood drawn for CD4+ T cell sorting. As illustrated in Fig. 3A, flow cytometry analysis indicated that the purity of CD4+ T cells isolated using magnetic bead sorting was greater than 98%. RT-qPCR analysis revealed that the miR-1246 expression level was significantly increased in the miR-1246 group (Fig. 3B), indicating that the cell model established here could be used for further experimentation.
Fig. 3. Effect of miR-1246 overexpression on Th17 subsets in peripheral CD4+ T cells. (A) Flow cytometry assay of the purity of peripheral CD4+ T cells sorted by magnetic beads, including the miR-1246 lentiviral vector transduced cells (miR-1246 group), the negative control vector transduced cells (negative control group), and the blank control group. (B) miR-1246 expression mediated by transduction. (C) ROR-γt and IL-17 mRNA levels of transduced cells. (D) Proportion of Th17 cells in the transduced cells. (E) Concentration of cytokines in the supernatant of the transduced cell culture. miR-1246: microRNA-1246, ROR-γt: retineic-acid-receptor-related orphan nuclear receptor gamma, Th17: T helper 17, IL: interleukin, TNF: tumor necrosis factor. *p<0.05, **p<0.01.
Overexpression of miR-1246 inhibits Th17 cell activation in AA patients
The RT-qPCR assay revealed that the miR-1246 group had significantly lower levels of retineic-acid-receptor-related orphan nuclear receptor gamma (ROR-γt) and IL-17 mRNA compared to the blank control group, but there was no statistical difference between the negative control and blank control groups (Fig. 3C). Flow cytometry analysis revealed that the miR-1246 group had a significantly lower proportion of CD4+IL-17+ cells compared to the blank control group (6.21% vs. 9.11%; 5.94% vs. 11.1%; 1.03% vs. 1.63%; Fig. 3D). ELISA analysis revealed that the miR-1246 group had significantly less IL-17F than the blank control group (57.37±25.80 pg/ml vs. 128±63.23 pg/ml; p<0.05), whereas the levels of IL-17A, IL-21, IL-22, and TNF-α were not significantly different between the three groups (Fig. 3E, Table 1).
Table 1. Cytokine concentrations in transduced CD4+ T cell culture supernatant (pg/ml).
| Cytokine | Blank control group (n=9) | Negative control group (n=9) | miR-1246 group (n=9) | p-value |
|---|---|---|---|---|
| IL-17A | 1.58±1.53 | 1.35±1.25 | 1.18±1.02 | 0.809 |
| IL-17F | 128±63.23 | 101.70±70.64 | 57.37±25.80 | 0.042 |
| IL-21 | 7.14±2.80 | 6.30±1.84 | 6.30±2.62 | 0.706 |
| IL-22 | 3.61±1.09 | 2.96±0.73 | 3.12±0.77 | 0.280 |
| TNF-α | 13.09±5.64 | 11.46±6.52 | 11.52±5.46 | 0.804 |
Values are presented as mean±standard deviation. IL: interleukin, miR-1246: microRNA-1246, TNF: tumor necrosis factor.
miR-1246 inhibits NFATc1 phosphorylation
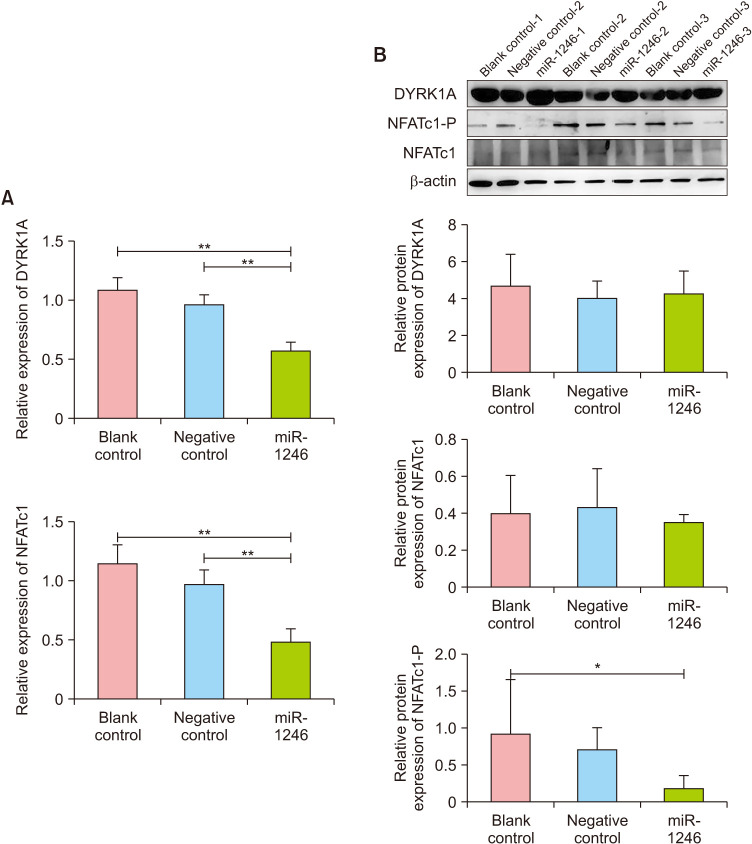
The qPCR test revealed that CD4+ T cells had significantly lower levels of DYRK1A and NFATc1 mRNA in the miR-1246 group compared to the blank control group (p<0.01), whereas there was no significant difference between the negative control and blank control groups (p>0.05, Fig. 4A). Western blot analysis revealed that while DYRK1A and NFATc1 protein expression were not significantly different between the three groups (p>0.05, Fig. 4B), the level of phosphorylated NFATc1 (NFATc1-P) was significantly decreased in the miR-1246 group (p<0.05, Fig. 4B). This result suggests that overexpression of miR-1246 may inhibit NFATc1's nucleocytoplasmic shuttle by lowering the level of cellular NFATc1-P protein, thereby inhibiting the NFAT signaling pathway.
Fig. 4. Effect of miR-1246 overexpression on DYRK1A and NFATc1 in peripheral CD4+ T cells. (A) DYRK1A and NFATc1 mRNA levels in transduced cells. (B) Protein expression of DYRK1A, total NFATc1, and phosphorylated NFATc1 in transduced cells. DYRK1A: dual-specific tyrosine phosphorylation-regulated kinase 1A, miR-1246: microRNA-1246, NFAT: nuclear factor of activated T cells.*p<0.05, **p<0.01.
DISCUSSION
In the present study, we discovered that miR-1246 expression is significantly decreased in peripheral CD4+ T cells of patients with severe active AA, whereas DYRK1A and NFATc1 mRNA and NFATc1 protein expression are significantly increased, implying that the miR-1246-DYRK1A-NFAT axis is involved in the pathogenesis of severe AA. Furthermore, the expression of miR-1246 decreased significantly in scalp tissue samples of severe AA patients, whereas the mRNA levels of DYRK1A and NFATc1 increased significantly; however, the protein expression of NFATc1 remained unchanged. IHC revealed that NFATc1-positive cells predominantly infiltrated hair follicles, but the number of cells was small and there was no significant difference between the AA and control groups. This result could be explained by the small sample size and low total protein content of our scalp samples. Therefore, while our findings indicate that NFATc1 signaling plays a role in the pathogenesis of severe AA in the peripheral blood immune system, its role in the scalp microenvironment needs to be further determined. Additionally, we discovered that overexpression of miR-1246 in circulating CD4+ T cells from severe active AA patients down-regulated Th17-related transcription factors ROR-γt, IL-17, and IL-17F, as well as reduced the proportion of Th17 cells. Thus, our data indicate that up-regulation of miR-1246 in peripheral CD4+ T cells in severe active AA can inhibit the number and function of peripheral blood Th17 cells, which may be beneficial for treating severe AA.
The miR-1246-DYRK1A-NFAT signaling axis has been implicated in a variety of tumor diseases and Down syndrome in recent years24,25,27. The dual-luciferase reporter gene system was used previously to confirm that miR-1246 targets DYRK1A, indicating that miR-1246 can inhibit DYRK1A expression and its downstream NFAT activity24,25. As a result, miR-1246 has the potential to be used as an interventional target in a wide variety of diseases. For instance, in the liver ischemia/reperfusion injury model, up-regulation of CD4+ T cell miR-1246 can reduce inflammation by adjusting the Th17/Treg ratio26. In patients with lupus erythematosus, up-regulation of B cell miR-1246 can reduce the expression of cell surface costimulatory molecules CD40, CD80, and CD86 and reduce B cell activity34. Additionally, NFATc1 has been shown to promote Th17 cell activation35, resulting in improvements in disease symptoms in a mouse model31,36. On the other hand, emerging evidence indicates that dysregulated miRNAs play a role in the development of autoimmune diseases by disrupting the Th17/Treg balance37. For example, Li et al.38 found miR-1246 was down-regulated in the circulating mononuclear cells of Chinese children with Henoch–Schonlein purpura (HSP) and was associated with Th17 activation during HSP development. In another study, researchers discovered that miR-1246 was highly expressed in the peripheral blood of rheumatoid arthritis patients regardless of their naive or memory phenotype, indicating that miR-1246 is involved in the maintenance of a specific Treg phenotype39.
Our results indicate that overexpression of miR-1246 can specifically target DYRK1A and inhibit its transcription, but has no effect on DYRK1A translation, which contradicts the findings of Fujita et al40. They reported that when miR-1246 was transfected into the hepatocellular carcinoma Li-7 cell line, DYRK1A was down-regulated and caspase-9 was up-regulated, which correlated with an increase in apoptosis. Zhang et al.25 also demonstrated that overexpression of miR-1246 decreased DYRK1A protein expression and induced apoptosis. In the future, we can increase the number of transduced CD4+ T cells and optimize lentiviral transduction efficiency to determine the extent to which miR-1246 influences DYRK1A protein expression and whether there are differences between cell lines. However, we found that the overexpression of miR-1246 in CD4+ T cells in peripheral blood of severe active AA significantly reduced the phosphorylation level of NFATc1, which is consistent with the results of Zhang et al25. According to previous research, activation of NFAT can be divided into several steps: intracellular calcium mobilization, calmodulin activation, CaN activation, CaN dephosphorylation of NFAT, nuclear translocation of NFAT, binding to target DNA, synergistic effect with other nuclear proteins, and re-phosphorylated NFAT exiting the nucleus28,29. Obstructing any of these steps could impair NFAT signaling pathway activation and may become a target for new inhibitors. Therefore, the use of miR-1246 to inhibit the NFAT signaling pathway in severe active AA warrants further investigation.
To the best of our knowledge, our report is the first study to examine the role of the miR-1246-DYRK1A-NFAT signaling axis in patients with AA. However, this study has some limitations that should be noted. Our study included only acute AA cases, not chronic cases. We will investigate the role of the miR-1246-DYRK1A-NFAT signaling axis in all types of AA in the future. Second, our study only included a small sample size and the results of the peripheral blood CD4+ T cell study have not been confirmed in scalp tissues, which we will address in future studies.
In summary, we propose that the miR-1246/DYRK1A/NFATc1 axis plays a role in the development of severe AA. Upregulation of miR-1246 may inhibit DYRK1A transcription and NFATc1 protein phosphorylation, as well as affect NFATc1 nuclear export, thereby impairing NFATc1 signaling and the activation of Th17 cells, which may be beneficial in the treatment of severe AA.
Footnotes
CONFLICTS OF INTEREST: The authors have nothing to disclose.
FUNDING SOURCE: This work was supported by the National Natural Science Foundation of China (Program No. 82103760).
DATA SHARING STATEMENT
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.
SUPPLEMENTARY MATERIALS
Supplementary data can be found via http://anndermatol.org/src/sm/ad-22-126-s001.pdf.
Primer sequences
References
- 1.Gilhar A. Collapse of immune privilege in alopecia areata: coincidental or substantial? J Invest Dermatol. 2010;130:2535–2537. doi: 10.1038/jid.2010.260. [DOI] [PubMed] [Google Scholar]
- 2.Tan H, Lan XM, Yu NL, Yang XC. Reliability and validity assessment of the revised Symptom Checklist 90 for alopecia areata patients in China. J Dermatol. 2015;42:975–980. doi: 10.1111/1346-8138.12976. [DOI] [PubMed] [Google Scholar]
- 3.Qi S, Xu F, Sheng Y, Yang Q. Assessing quality of life in Alopecia areata patients in China. Psychol Health Med. 2015;20:97–102. doi: 10.1080/13548506.2014.894641. [DOI] [PubMed] [Google Scholar]
- 4.Messenger AG, McKillop J, Farrant P, McDonagh AJ, Sladden M. British Association of Dermatologists' guidelines for the management of alopecia areata 2012. Br J Dermatol. 2012;166:916–926. doi: 10.1111/j.1365-2133.2012.10955.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhu S, Pan W, Qian Y. MicroRNA in immunity and autoimmunity. J Mol Med (Berl) 2013;91:1039–1050. doi: 10.1007/s00109-013-1043-z. [DOI] [PubMed] [Google Scholar]
- 6.Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett. 2019;454:134–145. doi: 10.1016/j.canlet.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Park EJ, Shimaoka M, Kiyono H. MicroRNA-mediated dynamic control of mucosal immunity. Int Immunol. 2017;29:157–163. doi: 10.1093/intimm/dxx019. [DOI] [PubMed] [Google Scholar]
- 8.Lew BL, Cho HR, Haw S, Kim HJ, Chung JH, Sim WY. Association between IL17A/IL17RA gene polymorphisms and susceptibility to alopecia areata in the Korean population. Ann Dermatol. 2012;24:61–65. doi: 10.5021/ad.2012.24.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han YM, Sheng YY, Xu F, Qi SS, Liu XJ, Hu RM, et al. Imbalance of T-helper 17 and regulatory T cells in patients with alopecia areata. J Dermatol. 2015;42:981–988. doi: 10.1111/1346-8138.12978. [DOI] [PubMed] [Google Scholar]
- 10.Tanemura A, Oiso N, Nakano M, Itoi S, Kawada A, Katayama I. Alopecia areata: infiltration of Th17 cells in the dermis, particularly around hair follicles. Dermatology. 2013;226:333–336. doi: 10.1159/000350933. [DOI] [PubMed] [Google Scholar]
- 11.Loh SH, Moon HN, Lew BL, Sim WY. Role of T helper 17 cells and T regulatory cells in alopecia areata: comparison of lesion and serum cytokine between controls and patients. J Eur Acad Dermatol Venereol. 2018;32:1028–1033. doi: 10.1111/jdv.14775. [DOI] [PubMed] [Google Scholar]
- 12.Atwa MA, Youssef N, Bayoumy NM. T-helper 17 cytokines (interleukins 17, 21, 22, and 6, and tumor necrosis factor-α) in patients with alopecia areata: association with clinical type and severity. Int J Dermatol. 2016;55:666–672. doi: 10.1111/ijd.12808. [DOI] [PubMed] [Google Scholar]
- 13.El-Morsy EH, Eid AA, Ghoneim H, Al-Tameemi KA. Serum level of interleukin-17A in patients with alopecia areata and its relationship to age. Int J Dermatol. 2016;55:869–874. doi: 10.1111/ijd.12994. [DOI] [PubMed] [Google Scholar]
- 14.Morsy H, Maher R, Negm D. Correlation between serum IL-17A level and SALT score in patients with alopecia areata before and after NB-UVB therapy. J Cosmet Dermatol. 2018;17:533–537. doi: 10.1111/jocd.12664. [DOI] [PubMed] [Google Scholar]
- 15.Langley RG, Lebwohl M, Krueger GG, Szapary PO, Wasfi Y, Chan D, et al. Long-term efficacy and safety of ustekinumab, with and without dosing adjustment, in patients with moderate-to-severe psoriasis: results from the PHOENIX 2 study through 5 years of follow-up. Br J Dermatol. 2015;172:1371–1383. doi: 10.1111/bjd.13469. [DOI] [PubMed] [Google Scholar]
- 16.Guttman-Yassky E, Ungar B, Noda S, Suprun M, Shroff A, Dutt R, et al. Extensive alopecia areata is reversed by IL-12/IL-23p40 cytokine antagonism. J Allergy Clin Immunol. 2016;137:301–304. doi: 10.1016/j.jaci.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 17.Aleisa A, Lim Y, Gordon S, Her MJ, Zancanaro P, Abudu M, et al. Response to ustekinumab in three pediatric patients with alopecia areata. Pediatr Dermatol. 2019;36:e44–e45. doi: 10.1111/pde.13699. [DOI] [PubMed] [Google Scholar]
- 18.Guttman-Yassky E, Nia JK, Hashim PW, Mansouri Y, Alia E, Taliercio M, et al. Efficacy and safety of secukinumab treatment in adults with extensive alopecia areata. Arch Dermatol Res. 2018;310:607–614. doi: 10.1007/s00403-018-1853-5. [DOI] [PubMed] [Google Scholar]
- 19.Sheng Y, Qi S, Hu R, Zhao J, Rui W, Miao Y, et al. Identification of blood microRNA alterations in patients with severe active alopecia areata. J Cell Biochem. 2019;120:14421–14430. doi: 10.1002/jcb.28700. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, et al. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol. 2020;25:89–99. doi: 10.1007/s10147-019-01532-9. [DOI] [PubMed] [Google Scholar]
- 21.Wei J, Yang L, Wu YN, Xu J. Serum miR-1290 and miR-1246 as potential diagnostic biomarkers of human pancreatic cancer. J Cancer. 2020;11:1325–1333. doi: 10.7150/jca.38048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CM, Liao YW, Hsieh PL, Yu CH, Chueh PJ, Lin T, et al. miR-1246 as a therapeutic target in oral submucosa fibrosis pathogenesis. J Formos Med Assoc. 2019;118:1093–1098. doi: 10.1016/j.jfma.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Yang F, Xiong H, Duan L, Li Q, Li X, Zhou Y. MiR-1246 promotes metastasis and invasion of A549 cells by targeting GSK-3β-Mediated Wnt/β-catenin pathway. Cancer Res Treat. 2019;51:1420–1429. doi: 10.4143/crt.2018.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao JM, Zhou X, Zhang Y, Lu H. MiR-1246: a new link of the p53 family with cancer and Down syndrome. Cell Cycle. 2012;11:2624–2630. doi: 10.4161/cc.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Liao JM, Zeng SX, Lu H. p53 downregulates Down syndrome-associated DYRK1A through miR-1246. EMBO Rep. 2011;12:811–817. doi: 10.1038/embor.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie K, Liu L, Chen J, Liu F. Exosomal miR-1246 derived from human umbilical cord blood mesenchymal stem cells attenuates hepatic ischemia reperfusion injury by modulating T helper 17/regulatory T balance. IUBMB Life. 2019;71:2020–2030. doi: 10.1002/iub.2147. [DOI] [PubMed] [Google Scholar]
- 27.Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu XK, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem. 2004;279:52762–52771. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- 29.Reppert S, Zinser E, Holzinger C, Sandrock L, Koch S, Finotto S. NFATc1 deficiency in T cells protects mice from experimental autoimmune encephalomyelitis. Eur J Immunol. 2015;45:1426–1440. doi: 10.1002/eji.201445150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Daraji WI, Malak TT, Prescott RJ, Abdellaoui A, Ali MM, Dabash T, et al. Expression, localisation and functional activation of NFAT-2 in normal human skin, psoriasis, and cultured keratocytes. Int J Clin Exp Med. 2009;2:176–192. [PMC free article] [PubMed] [Google Scholar]
- 31.Alrefai H, Muhammad K, Rudolf R, Pham DA, Klein-Hessling S, Patra AK, et al. NFATc1 supports imiquimod-induced skin inflammation by suppressing IL-10 synthesis in B cells. Nat Commun. 2016;7:11724. doi: 10.1038/ncomms11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson SR, Thé L, Batia LM, Beattie K, Katibah GE, McClain SP, et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013;155:285–295. doi: 10.1016/j.cell.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujii Y, Fujii K, Iwata S, Suzuki K, Azuma T, Saito K, et al. Abnormal intracellular distribution of NFAT1 in T lymphocytes from patients with systemic lupus erythematosus and characteristic clinical features. Clin Immunol. 2006;119:297–306. doi: 10.1016/j.clim.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Luo S, Liu Y, Liang G, Zhao M, Wu H, Liang Y, et al. The role of microRNA-1246 in the regulation of B cell activation and the pathogenesis of systemic lupus erythematosus. Clin Epigenetics. 2015;7:24. doi: 10.1186/s13148-015-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Rodriguez J, Sahu N, Handon R, Davidson TS, Anderson SM, Kirby MR, et al. Differential expression of interleukin-17A and-17F is coupled to T cell receptor signaling via inducible T cell kinase. Immunity. 2009;31:587–597. doi: 10.1016/j.immuni.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch S, Reppert S, Finotto S. NFATc1 deletion in T lymphocytes inhibits the allergic trait in a murine model of asthma. Clin Exp Allergy. 2015;45:1356–1366. doi: 10.1111/cea.12493. [DOI] [PubMed] [Google Scholar]
- 37.Liu C, Yang H, Shi W, Wang T, Ruan Q. MicroRNA-mediated regulation of T helper type 17/regulatory T-cell balance in autoimmune disease. Immunology. 2018;155:427–434. doi: 10.1111/imm.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Chen M, Wang J, Lu L, Li X, Le Y. MicroRNA profiling in Chinese children with Henoch-Schonlein purpura and association between selected microRNAs and inflammatory biomarkers. Acta Paediatr. 2021;110:2221–2229. doi: 10.1111/apa.15789. [DOI] [PubMed] [Google Scholar]
- 39.Smigielska-Czepiel K, van den Berg A, Jellema P, van der Lei RJ, Bijzet J, Kluiver J, et al. Comprehensive analysis of miRNA expression in T-cell subsets of rheumatoid arthritis patients reveals defined signatures of naive and memory Tregs. Genes Immun. 2014;15:115–125. doi: 10.1038/gene.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujita K, Iwama H, Sakamoto T, Okura R, Kobayashi K, Takano J, et al. Galectin-9 suppresses the growth of hepatocellular carcinoma via apoptosis in vitro and in vivo. Int J Oncol. 2015;46:2419–2430. doi: 10.3892/ijo.2015.2941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences
Data Availability Statement
The datasets generated and analyzed during the present study are available from the corresponding author on reasonable request.