Abstract
Background
Obesity is an established risk factor for colorectal cancer (CRC), but the evidence for the association is inconsistent across molecular subtypes of the disease.
Methods
We pooled data on body mass index (BMI), tumor microsatellite instability status, CpG island methylator phenotype status, BRAF and KRAS mutations, and Jass classification types for 11 872 CRC cases and 11 013 controls from 11 observational studies. We used multinomial logistic regression to estimate odds ratios (OR) and 95% confidence intervals (CI) adjusted for covariables.
Results
Higher BMI was associated with increased CRC risk (OR per 5 kg/m2 = 1.18, 95% CI = 1.15 to 1.22). The positive association was stronger for men than women but similar across tumor subtypes defined by individual molecular markers. In analyses by Jass type, higher BMI was associated with elevated CRC risk for types 1-4 cases but not for type 5 CRC cases (considered familial-like/Lynch syndrome microsatellite instability-H, CpG island methylator phenotype-low or negative, BRAF-wild type, KRAS-wild type, OR = 1.04, 95% CI = 0.90 to 1.20). This pattern of associations for BMI and Jass types was consistent by sex and design of contributing studies (cohort or case-control).
Conclusions
In contrast to previous reports with fewer study participants, we found limited evidence of heterogeneity for the association between BMI and CRC risk according to molecular subtype, suggesting that obesity influences nearly all major pathways involved in colorectal carcinogenesis. The null association observed for the Jass type 5 suggests that BMI is not a risk factor for the development of CRC for individuals with Lynch syndrome.
Colorectal cancer (CRC) is a heterogeneous disease that evolves through increasing genomic instability (1,2). These include microsatellite instability (MSI), which results from impaired DNA mismatch repair (MMR), and the CpG island methylator phenotype (CIMP), which results from extensive hypermethylation of promoter CpG island sites, causing inactivation of specific tumor suppressor genes. MSI status is commonly defined as MSI-high (MSI-H) or MSI-stable or low (MSS/MSI-L). MSI-H CRC is found in approximately 15% of cases (approximately 12% sporadic, approximately 3% familial-like/Lynch) (3,4). Additionally, somatic mutations in the BRAF and KRAS genes are major pathological features of CRC tumors with relevance to prognosis and prediction.
Overweight and obesity have been consistently associated with higher risk of CRC (5-7), but the evidence from studies that examined associations between body mass index (BMI) and molecular subtypes of CRC has been inconsistent. For MSI status, a recent meta-analysis of 6 studies reported positive associations of similar magnitudes for BMI with MSI-H vs MSS CRC, but high heterogeneity between studies was detected (8). For CIMP status, a recent analysis in the Darmkrebs: Chancen der Verhutung durch Screening Study (DACHS) case-control study reported a stronger positive association between BMI and CIMP-high CRC compared with CIMP-low or negative tumors for women only (9). However, no evidence of heterogeneity by CIMP status was found in a Netherlands Cohort Study analysis (10). For KRAS or BRAF tumor mutation status, the DACHS study found little evidence of heterogeneity with the association of BMI and CRC (9), whereas a Swedish cohort analysis reported differences in the BMI and CRC relationship according to KRAS or BRAF mutation status, with further heterogeneity found according to sex (11).
This inconsistent pattern of results reported across individual studies may be due to small study effects and chance, publication bias, between-study differences in tumor marker classifications, true population differences, and/or differential effects of confounding. The smaller sample sizes of previous studies precluded an examination of the associations between BMI and Jass subtypes [1-5; defined by the joint presentations of MSI, CIMP, BRAF, and KRAS status (2)]. A more detailed analysis according to Jass types would separate sporadic (Jass type 1) MSI-H tumors (also CIMP-high/BRAF-mutated) from familial-like/Lynch syndrome (Jass type 5) MSH-H tumors (also CIMP-low or negative). Such an approach is crucial because this could explain the heterogeneity in the BMI and MSI-H CRC associations reported by previous studies (8,9,12,13).
Here, we analyzed individual-level, harmonized data for 11 872 CRC cases and 11 013 controls from 11 observational studies within the Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO) and Colon Cancer Family Registry (CCFR) to examine the association between BMI and CRC overall and by tumor molecular subtypes.
Methods
Study participants
This study sample included CRC cases and controls within GECCO and CCFR with available tumor marker and BMI data (Table 1; Supplementary Table 1, available online). Additional information on contributing studies is included in the Supplementary Methods (available online). All participants provided written informed consent. Each study was approved by a research ethics committee or institutional review board.
Table 1.
Baseline characteristics of cases and controlsa
| Characteristics | Cases | Controls |
|---|---|---|
| Total No. | 11 872 | 11 013 |
| Age, mean (SD), y | 60.5 (12.2) | 63.2 (10.9) |
| Sex, No. (%) | ||
| Men | 6209 (52.3) | 5617 (51) |
| Women | 5663 (47.7) | 5396 (49) |
| Study, No. (%) | ||
| CCFR | 5073 (42.7) | 2180 (19.8) |
| CPSII | 790 (6.7) | 929 (8.4) |
| DACHS | 1966 (16.6) | 2744 (24.9) |
| DALS | 1083 (9.1) | 1148 (10.4) |
| EDRN | 188 (1.6) | 329 (3) |
| EPIC_Sweden | 145 (1.2) | 381 (3.5) |
| HPFS | 585 (4.9) | 591 (5.4) |
| MCCS | 490 (4.1) | 670 (6.1) |
| NFCCR | 489 (4.1) | 461 (4.2) |
| NHS | 764 (6.4) | 1197 (10.9) |
| NSHDS | 299 (2.5) | 383 (3.5) |
| Body mass index, No. (%) | ||
| 18.5 to <25 kg/m2 | 4154 (35) | 4510 (41) |
| 25 to <30 kg/m2 | 4991 (42) | 4650 (42.2) |
| ≥30 kg/m2 | 2727 (23) | 1853 (16.8) |
| Tobacco smoking, No. (%) | ||
| Never | 4947 (41.7) | 5156 (46.8) |
| Past or current | 6386 (53.8) | 5524 (50.2) |
| Unknown | 539 (4.5) | 333 (3) |
| Dietary intake | ||
| Red meat, mean (SD), servings/d | 0.8 (0.6) | 0.7 (0.6) |
| Processed meat, mean (SD), servings/d | 0.5 (0.4) | 0.4 (0.4) |
| Fruits, mean (SD), servings/d | 1.7 (1.5) | 1.8 (1.6) |
| Vegetables, mean (SD), servings/d | 2.3 (1.9) | 2.4 (2.0) |
| Fiber, mean (SD), g/d | 23.3 (10.8) | 23.1 (10.3) |
| Total energy, mean (SD), kcal/d | 2133 (866) | 2015 (767) |
| Education level, No. (%) | ||
| Less than high school graduate | 2382 (20.1) | 1776 (16.1) |
| High school graduate | 2872 (24.2) | 2605 (23.7) |
| Vocational or technical school or some college/university | 2971 (25) | 2310 (21) |
| Undergraduate or graduate degree | 3394 (28.6) | 4063 (36.9) |
| Missing | 253 (2.1) | 259 (2.4) |
| First degree relative with colorectal cancer, No. (%) | ||
| No | 8777 (73.9) | 9323 (84.7) |
| Yes | 2712 (22.8) | 1134 (10.3) |
| Missing | 383 (3.2) | 556 (5) |
| Location of colorectal cancer, No. (%) | ||
| Proximal colon | 4510 (38) | — |
| Distal colon | 3525 (29.7) | — |
| Rectum (includes rectosigmoid junction) | 3331 (28.1) | — |
| Missing | 506 (4.3) | — |
| Colorectal cancer stage, No. (%) | ||
| Stage 1 or local | 2747 (23.1) | — |
| Stage 2/3 or regional | 6785 (57.2) | — |
| Stage 4 or distant | 1214 (10.2) | — |
| Missing | 1126 (9.5) | — |
| Microsatellite instability, No. (%) | ||
| MSI-H | 1809 (15.2) | — |
| MSS/MSI-L | 8967 (75.5) | — |
| Missing | 1096 (9.2) | — |
| CpG island methylator phenotype, No. (%) | ||
| High | 1386 (11.7) | — |
| Low/negative | 7160 (60.3) | — |
| Missing | 3326 (28.0) | — |
| BRAF, No. (%) | ||
| Mutated | 1297 (10.9) | — |
| Wild type | 9423 (79.4) | — |
| Missing | 1152 (9.7) | — |
| KRAS, No. (%) | ||
| Mutated | 2961 (24.9) | — |
| Wild type | 6011 (50.6) | — |
| Missing | 2900 (24.4) | — |
CCFR = Colon Cancer Family Registry; CPSII = Cancer Prevention Study II; DACHS = Darmkrebs: Chancen der Verhutung durch Screening Study; DALS = Diet Activity and Lifestyle Study; EDRN = Early Detection Research Network; EPIC = European Prospective Investigation into Cancer and Nutrition; HPFS = Health Professionals Follow-up Study; MCCS = Melbourne Collaborative Cohort Study; MSS = microsatellite stable; NFCCR = Newfoundland Familial Colorectal Cancer Study; NHS = Nurses’ Health Study; NSHDS = Northern Sweden Health and Disease Study.
Collection and harmonization of tumor marker data
Data collection and harmonization of GECCO and CCFR tumor marker data have been described elsewhere (14,15). Briefly, MSI testing was primarily conducted using polymerase chain reaction following accepted guidelines (CCFR, Cancer Prevention Study-II [CPS-II], Melbourne Collaborative Cohort Study [MCCS], Nurses’ Health Study [NHS]) (16) with 4 or more interpretable markers typically required to classify tumors (Supplementary Table 2, available online). DACHS used a mononucleotide panel of 3 markers. Tumors were classified as MSI-H if 30% or more of the markers showed instability. Other studies used immunohistochemistry for the correlated DNA MMR proteins (Northern Sweden Health and Disease Study [NSHDS], European Prospective Investigation into Cancer and Nutrition [EPIC]-Sweden, and subsets of CCFR and MCCS).
CIMP status was determined using methylation analyses as described in the Supplementary Materials (Supplementary Table 3, available online). Briefly, the CCFR, CPS-II, Health Professionals Follow-up Study, MCCS, NSHDS, EPIC-Sweden, and NHS used MethyLight to determine CIMP status. CPS-II, Health Professionals Follow-up Study, NSHDS, EPIC-Sweden, and NHS used an 8-gene panel; CCFR and MCCS used a 5-gene panel. DACHS determined CIMP status using a different 5-gene panel (17). CIMP categories for our analyses were CIMP-high and CIMP-low or negative.
Studies assessed BRAF and KRAS mutations using polymerase chain reaction, sequencing, and immunohistochemistry. Most studies evaluated BRAF via c.1799T>A (p.V600E) mutations in exon 15 and KRAS via mutations in codons 12 and 13, although any mutation identified by one of the studies in BRAF and KRAS genes was included. We further defined 5 combined colorectal tumor subtypes consistent with Jass classifications (2,18): type 1 (“sporadic”-MSI-H, CIMP-high, BRAF-mutated, KRAS-wild type), type 2 (MSS/MSI-L, CIMP-high, BRAF-mutated, KRAS-wild type), type 3 (MSS/MSI-L, CIMP-low or negative, BRAF-wild type, KRAS-mutated), type 4 (MSS/MSI-L, CIMP-low or negative, BRAF-wild type, KRAS-wild type), and type 5 (“Lynch syndrome”-MSI-H, CIMP-low or negative, BRAF-wild type, KRAS-wild type).
Exposure data
Data collection and harmonization are described elsewhere (14,15,19). Briefly, demographic and environmental risk factors were self-reported at in-person interviews or via questionnaires. Data were collected relevant to the time of study entry or recalled for a time period generally 1 to 2 years before study enrolment. The timing of height and body weight measurements for each contributing study ranged from 5 to 14 years before enrolment in the DACHS case-control study to baseline measurements in prospective cohort studies (Supplementary Table 1, available online). A multistep iterative data-harmonization procedure was applied, reconciling each study’s unique protocols and data collection instruments. Multiple quality-control checks were performed, and outlying values of variables were truncated to the minimum or maximum value of an established range for each variable. Variables were combined into a single dataset with common definitions, standardized coding, and standardized permissible values. BMI was calculated from self-reports or direct measures of body weight (kg) divided by height-squared (m2), with individuals with BMI less than 18.5 kg/m2 excluded from the analysis because of the observed nonlinear associations at the lower end of the BMI continuum in these data and elsewhere (20,21). Other variables included age, sex, education, smoking, physical activity, regular use of aspirin or nonsteroidal anti-inflammatory drugs, postmenopausal hormone therapy use (women), diabetes, first-degree family history of CRC, and history of endoscopy (colonoscopy or sigmoidoscopy). Dietary covariables were ascertained using food frequency questionnaires or diet history diaries or records. We defined age as age at CRC diagnosis for cases and age at enrolment for controls.
Statistical analyses
We used logistical and multinomial models to estimate odds ratios (OR) and 95% confidence intervals (CIs) for the association between BMI and CRC overall and with CRC subtypes defined by tumor markers: MSI-H vs MSS/MSI-L, CIMP high vs low or negative, and BRAF or KRAS mutated vs nonmutated. Analyses were conducted for sexes combined and separately. Heterogeneity by sex was assessed by calculating χ2 statistics. BMI was defined continuously (per 5 kg/m2; primary analyses) and categorically (18.5 to <25 [reference group], 25 to <30, and ≥30 kg/m2). For the Jass type analyses, type 4 was used as the reference group in the case-only analysis, whereas in the multinomial analysis, controls were used as the reference group. Both analyses used multinomial logistic regression to compare odds of exposure in each molecular pathological subtype with the reference group while accounting for covariables. The multivariable models included a set of a priori–determined CRC risk factors: age, sex, smoking status, education, and red meat intake as covariates. Additional adjustment for family history of CRC; history of endoscopy; aspirin or nonsteroidal anti-inflammatory drug use; energy intake; and consumption of alcohol, processed meat, fruits, and vegetables resulted in virtually unchanged odds ratio estimates. Analyses between BMI and individual molecular subtypes of CRC according to study were also undertaken. For Jass type–defined pathways of CRC, we conducted analyses stratified by case-control and cohort studies. We used Bonferroni corrected P values (<.05/12 = .004; 4 subtypes being tested in the sexes combined, and men and women separately) to assess statistical significance for the case-only analyses of the primary subtypes (MSI, CIMP-status BRAF, and KRAS). For secondary analyses, we considered a 2-sided P value less than .05 as statistically significant. In a sensitivity analysis using multivariable logistic regression models, we calculated study-specific odds ratios for the associations between BMI and CRC tumor molecular subtypes and then pooled using random effects meta-analysis models. Analyses were performed using R v4.0.0 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Compared with controls, CRC cases were younger; were more likely to be men, heavier (≥30 kg/m2), past or current smokers, and to have a first-degree relative with CRC; and were less likely to have attained an undergraduate or graduate degree (Table 1). Among cases, 15.2% were MSI-H (n = 1809), 11.7% were CIMP-high (n = 1386), 10.9% were BRAF-mutated (n = 1297), and 24.9% were KRAS-mutated (n = 2961). Baseline characteristics according to contributing study are presented in Supplementary Table 4 (available online).
Higher BMI was associated with elevated CRC risk in multivariable models (OR per 5 kg/m2 = 1.18, 95% CI = 1.15 to 1.22; OR for ≥30 vs 18.5 to <25 kg/m2 = 1.47, 95% CI = 1.36 to 1.59; Ptrend < .001), with a stronger positive association observed for men than women (Pheterogeneity = .02) (Table 2). BMI was positively associated with CRC risk for all individual molecular subtypes, with minimal evidence of heterogeneity observed. For women, a stronger positive association was observed for CIMP-high cases (OR per 5 kg/m2 = 1.23, 95% CI = 1.15 to 1.33) compared with CIMP-low or negative cases (OR per 5 kg/m2 = 1.14, 95% CI = 1.08 to 1.19). This heterogeneity was higher than the Bonferroni-corrected P value threshold (Pdifference = .008) and largely driven by results from the DACHS study (Pdifference with DACHS excluded = .22). Results for the association between BMI and individual molecular subtypes of CRC according to study are presented in Supplementary Table 5 (available online). Similar associations between BMI and individual molecular subtypes of CRC were found when individual study odds ratios were calculated and then pooled in a meta-analysis (Supplementary Table 6, available online).
Table 2.
| Microsatellite instability |
CpG island methylator phenotype |
BRAF
|
KRAS
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CRC | MSS/MSI-L | MSI-H | CIMP-low/negative | CIMP-high | BRAF-wild type | BRAF-mutated | KRAS-wild type | KRAS-mutated | |
| Exposure | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) | OR (95% CI) |
| Both sexes | |||||||||
| No. cases | 11 872 | 8967 | 1809 | 7160 | 1386 | 9423 | 1297 | 6011 | 2961 |
| 18.5 to <25 kg/m2 | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| 25 to <30 kg/m2 | 1.18 (1.11 to 1.25) | 1.18 (1.11 to 1.26) | 1.20 (1.06 to 1.35) | 1.20 (1.11 to 1.29) | 1.20 (1.05 to 1.37) | 1.20 (1.12 to 1.28) | 1.17 (1.02 to 1.34) | 1.24 (1.15 to 1.34) | 1.13 (1.03 to 1.24) |
| ≥30 kg/m2 | 1.47 (1.36 to 1.59) | 1.47 (1.35 to 1.59) | 1.55 (1.34 to 1.78) | 1.39 (1.27 to 1.52) | 1.67 (1.43 to 1.96) | 1.47 (1.36 to 1.60) | 1.54 (1.31 to 1.81) | 1.54 (1.40 to 1.69) | 1.40 (1.25 to 1.57) |
| Ptrend | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 |
| Per 5 kg/m2 | 1.18 (1.15 to 1.22) | 1.19 (1.15 to 1.23) | 1.18 (1.12 to 1.24) | 1.17 (1.12 to 1.21) | 1.23 (1.16 to 1.31) | 1.18 (1.15 to 1.23) | 1.20 (1.13 to 1.28) | 1.20 (1.16 to 1.25) | 1.17 (1.12 to 1.22) |
| Pdifferencec | .91 | .04 | .35 | .22 | |||||
| Men | |||||||||
| No. cases | 6209 | 4923 | 764 | 3954 | 528 | 5118 | 474 | 3144 | 1563 |
| 18.5 to <25 kg/m2 | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| 25 to <30 kg/m2 | 1.19 (1.09 to 1.31) | 1.22 (1.11 to 1.34) | 1.06 (0.88 to 1.28) | 1.20 (1.09 to 1.33) | 1.16 (0.93 to 1.43) | 1.19 (1.09 to 1.31) | 1.25 (1.00 to 1.57) | 1.28 (1.15 to 1.42) | 1.11 (0.97 to 1.27) |
| ≥30 kg/m2 | 1.51 (1.35 to 1.70) | 1.53 (1.35 to 1.72) | 1.43 (1.14 to 1.79) | 1.42 (1.25 to 1.62) | 1.56 (1.19 to 2.04) | 1.52 (1.35 to 1.72) | 1.44 (1.08 to 1.91) | 1.52 (1.33 to 1.74) | 1.44 (1.22 to 1.71) |
| Ptrend | <.001 | <.001 | .004 | <.001 | .002 | <.001 | .01 | <.001 | <.001 |
| Per 5 kg/m2 | 1.24 (1.18 to 1.30) | 1.25 (1.18 to 1.31) | 1.16 (1.05 to 1.28) | 1.22 (1.15 to 1.29) | 1.22 (1.09 to 1.38) | 1.24 (1.17 to 1.31) | 1.20 (1.06 to 1.35) | 1.24 (1.16 to 1.31) | 1.21 (1.13 to 1.31) |
| Pdifferencec | .19 | .89 | .6 | .54 | |||||
| Women | |||||||||
| No. cases | 5663 | 4044 | 1045 | 3206 | 858 | 4305 | 823 | 2867 | 1398 |
| 18.5 to <25 kg/m2 | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) | 1 (referent) |
| 25-<30 kg/m2 | 1.17 (1.07 to 1.28) | 1.16 (1.05 to 1.28) | 1.27 (1.08 to 1.49) | 1.20 (1.08 to 1.33) | 1.23 (1.03 to 1.46) | 1.22 (1.11 to 1.35) | 1.10 (0.93 to 1.32) | 1.21 (1.09 to 1.35) | 1.17 (1.02 to 1.34) |
| ≥30 kg/m2 | 1.44 (1.29 to 1.60) | 1.43 (1.27 to 1.60) | 1.57 (1.31 to 1.89) | 1.36 (1.20 to 1.55) | 1.74 (1.43 to 2.11) | 1.44 (1.28 to 1.62) | 1.60 (1.32 to 1.95) | 1.57 (1.39 to 1.78) | 1.37 (1.17 to 1.61) |
| Ptrend | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | <.001 | .001 |
| Per 5 kg/m2 | 1.15 (1.11 to 1.20) | 1.16 (1.11 to 1.21) | 1.17 (1.10 to 1.25) | 1.14 (1.08 to 1.19) | 1.23 (1.15 to 1.33) | 1.16 (1.11 to 1.21) | 1.19 (1.11 to 1.28) | 1.19 (1.13 to 1.24) | 1.14 (1.08 to 1.21) |
| Pdifferencec | .41 | .008 | .14 | .18 | |||||
Controls are used as reference for all odds ratios. CI = confidence interval; CIMP = CpG island methylator phenotype; CRC = colorectal cancer; MSI = microsatellite instability; MSS = microsatellite stable; OR = odds ratio.
Odds ratios are adjusted for study, age, sex, smoking status, education, and red meat intake.
Case-only analyses used to calculate Pdifference.
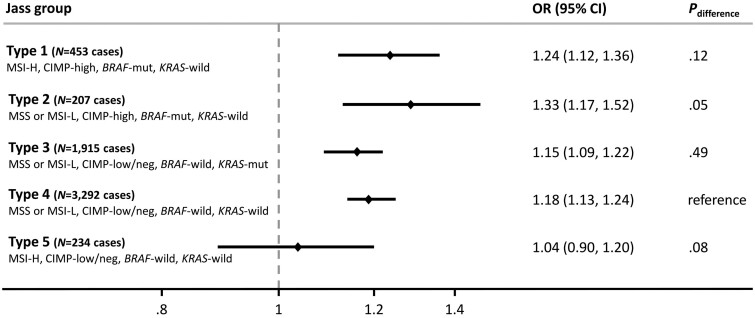
Analyses for Jass types of CRC presenting with MSI-H found positive associations for type 1 cases (OR per 5 kg/m2 = 1.24, 95% CI = 1.12 to 1.36) but not for type 5 cases (OR per 5 kg/m2 = 1.04, 95% CI = 0.90 to 1.20) (Figure 1; Supplementary Table 7, available online). Null associations for Jass type 5 CRC were also observed when case-control (OR per 5 kg/m2 = 1.02, 95% CI = 0.87 to 1.21) and cohort (OR per 5 kg/m2 = 1.08, 95% CI = 0.79 to 1.47) studies were analyzed separately (Supplementary Table 8, available online). Higher BMI was associated with elevated risk of Jass types 2, 3, and 4–defined CRC (Figure 1; Supplementary Table 7, available online). A similar pattern of associations was observed between BMI and Jass-classified CRC according to sex (Supplementary Table 7, available online) and when individual study odds ratios were calculated and pooled in a meta-analysis (Supplementary Table 9, available online).
Figure 1.
Association between body mass index and Jass classified types of colorectal cancer. Controls were used as reference for all odds ratios. Odds ratios were adjusted for study, age, sex, smoking status, education, and red meat intake. Multinomial logistic regression was used to compare each type with the reference group (Type 4; Pdifference). CI = confidence interval; CIMP = CpG island methylator phenotype; MSI = microsatellite instability; MSS = microsatellite stable; mut = mutated; OR = odds ratio; wild = wild type.
Discussion
In our analysis of pooled individual-level data from 11 872 CRC cases and 11 013 controls, higher BMI was associated with increased CRC risk, with little evidence of heterogeneity across molecular subtypes observed. Higher BMI was consistently associated with elevated risks of Jass types 1-4 CRC, suggesting that obesity influences all major pathways. The null association found for the Jass type 5 indicates that high BMI may not be a risk factor for the development of CRC for individuals with Lynch syndrome.
Inconsistent results were previously reported regarding the associations between BMI and CRC by CIMP status. While a case-control analysis reported a positive association between BMI with CIMP-low or negative, but not CIMP-high, colon cancers (22), a Netherlands Cohort Study prospective analysis reported relatively consistent associations between BMI and CRC according to CIMP status (10). These earlier investigations were not stratified by sex. Our sex-specific analyses, similarly to the DACHS study (9), found a stronger positive association for BMI with CIMP-high compared with CIMP-low or negative CRC for women only. Importantly, this heterogeneity of association by CIMP status was mainly driven by our inclusion of the DACHS study. When we excluded DACHS, there was little evidence of divergent associations between BMI and CRC by CIMP status, suggesting the differential association was specific to only the DACHS study.
The relatively few previous studies that examined associations between BMI and CRC risk according to KRAS and BRAF tumor mutation status have reported discordant results. For example, a Malmö Diet and Cancer cohort analysis (n = 494 CRC cases) reported a stronger positive association between BMI and CRC for mutated KRAS than for wild-type tumors for men but not women (11). In contrast, the DACHS study (n = 2217 CRC cases) reported no evidence of heterogeneity according to KRAS mutation status for men yet for women found that BMI was positively associated with wild-type KRAS but not KRAS mutated tumors (9). For BRAF, a Malmö Diet and Cancer cohort analysis reported that BMI was positively associated with wild-type BRAF CRC for men and women (11). The DACHS study reported a stronger positive association for BRAF mutated tumors compared with wild-type tumors, but this heterogeneity was not statistically significant after correction for multiple comparisons (9). Our current pooled analysis, which included more than 8900 and more than 10 700 CRC cases with data on KRAS and BRAF tumor mutations, respectively, found consistent positive associations according to mutation status of these 2 oncogenes, suggesting high BMI is positively associated with CRC with and without KRAS or BRAF mutations.
For MSI status, like our findings, a meta-analysis of 4 previous studies reported similar positive associations between BMI and CRC for MSI-H and MSS/MSI-L tumors (8). Across individual studies included and not included in this meta-analysis, heterogeneous findings for the BMI and MSI-H CRC associations have been reported (8,9,12,23,24). In particular, divergent results were reported by 2 of the larger case-control studies, with higher BMI associated with MSI-H CRC in the population-based DACHS study (9,13) but a null association found by the CCFR, a large case-control study of CRC families likely enriched for individuals with Lynch syndrome (12,25).
Part of the discordance in the association of BMI by MSI status might be explained by the predominant source of MSI-H cases in each study. In our analyses according to Jass types, for the first time to our knowledge, the BMI and CRC association was investigated after further separating sporadic MSI-H tumors from the less common familial-like/Lynch MSI-H tumors. Consistent with the earlier results from the CCFR study (12), we observed a null association between BMI and risk of familial-like/Lynch syndrome (type 5) MSI-H CRC, with similar null associations found according to sex and for case-control and cohort (which excluded all CCFR participants by design) studies when analyzed separately. The current findings for increasing BMI and higher risk of Jass type 1 tumors, which are largely considered sporadic MSI-H, were also consistent with the earlier DACHS findings, which would have been largely comprised of sporadic MSI-H tumors. That is, from these studies, it seems that high BMI is a risk factor for sporadic MSI-H tumors but perhaps less relevant to risk of tumors developing on a background of Lynch syndrome.
Interpretation of the null association between BMI and Jass type 5/Lynch syndrome MSI-H tumors in this study is further complicated by previous studies of apparent Lynch syndrome families, defined by germline mutations in a MMR gene or by being a member of Amsterdam criteria I or revised Bethesda guidelines families (26-28). In those few previous studies, including a recent meta-analysis of 4 studies in Lynch syndrome patients, a positive association was reported between obesity (compared with the nonobese group) and CRC/adenoma for men (relative risk = 2.09, 95% CI = 1.23 to 3.55) but a weaker association for women (relative risk = 1.41, 95% CI = 0.46 to 4.27) (28). More than 89% of the cases in this previous meta-analysis were included in 2 earlier CCFR publications (26,27); those studies defined Lynch syndrome according to clinical family history criteria (26) or by MMR carrier status (27). Notably, these studies had several design features that may have contributed to the apparently discordant results with this study. First, the study by Win and colleagues (27) used a retrospective cohort, time-to-event study design with noncases drawn from Lynch family members who did not have a CRC diagnosis. Those study participants may be leaner and otherwise more health conscious, owing to their genetic predisposition to cancer, compared with controls in this study who would generally not have a strong family history of CRC. Second, the larger study by Campbell and colleagues (26) used Amsterdam and Bethesda criteria to identify potential Lynch syndrome families, and thus the interpretation of those results is not specific to Lynch syndrome. Additionally, neither study considered tumor MSI status as a separate outcome. Collectively, these results suggest that high BMI is not associated with MSH-H tumors consistent with Lynch syndrome, but, given the many disease outcomes and cancers associated with obesity (29), persons predisposed to Lynch syndrome should still be advised to maintain a healthy body weight.
Beyond Lynch syndrome, our results suggest that obesity can affect all other major pathways of CRC tumorigenesis, including the traditional adenoma to cancer pathway and the serrated pathway. However, our analysis used broad categories to define molecular subtypes. Next-generation sequencing of tumor samples will allow a deeper classification of tumor subtypes by identifying somatically mutated genes. Examining associations between BMI and newly identified CRC subtypes could provide important insights into the molecular mechanisms underlying the BMI and CRC positive association.
Ours was the largest study yet to investigate associations between BMI and molecular subtypes of CRC. The large sample size obtained by pooling individual-level data from 11 studies, harmonized data, and standardized statistical analytical approach meant our analyses are less prone to between-study heterogeneity and publication biases. Crucially, the large sample size allowed an examination of the association between BMI and all 5 Jass types, providing insights into how excess adiposity is associated with different pathways of tumorigenesis. Limitations of our analysis include that 5 of the 11 studies in our analysis used a case-control design, which may be vulnerable to reverse causality; however, the positive association we found between BMI and CRC in the current analysis for case-control studies was of similar strength to that from prospective studies (6,30), indicating that any bias from reverse causation is likely small. A further limitation was that data on central adiposity measurements (eg, waist circumference) were not available to be included in our study. Finally, as is common for studies including tumor samples, tissue samples were not available for all CRC cases within contributing studies and may have been related with stage and/or size of tumor; however, prior investigations in some of the contributing studies reported that lifestyle and demographic characteristics differed little between CRC cases for whom tumor samples could or could not be obtained (31-33).
In conclusion, we found little evidence of heterogeneity for the positive association between BMI and CRC according to individual molecular tumor subtypes. Analyses by Jass type suggest that obesity influences all major pathways involved in colorectal carcinogenesis. The lack of association found for Jass type 5 tumors suggests that high BMI may not be a risk factor for the development of CRC for individuals with an inherited predisposition to Lynch syndrome. Further studies are needed to confirm this finding and increase understanding of the role of obesity in Lynch syndrome cancers.
Supplementary Material
Contributor Information
Neil Murphy, Nutrition and Metabolism Branch, International Agency for Research on Cancer, Lyon, France.
Christina C Newton, Population Science Department, American Cancer Society (ACS), Atlanta, GA, USA.
Mingyang Song, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA; Department of Nutrition, Harvard T.H. Chan School of Public Health, Boston, MA, USA; Division of Gastroenterology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
Nikos Papadimitriou, Nutrition and Metabolism Branch, International Agency for Research on Cancer, Lyon, France.
Michael Hoffmeister, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Amanda I Phipps, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Tabitha A Harrison, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Polly A Newcomb, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Elom K Aglago, Department of Epidemiology and Biostatistics, Imperial College London, School of Public Health, London, UK.
Sonja I Berndt, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Hermann Brenner, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Daniel D Buchanan, Colorectal Oncogenomics Group, Department of Clinical Pathology, The University of Melbourne, Parkville, VIC, Australia; University of Melbourne Centre for Cancer Research, Victorian Comprehensive Cancer Centre, Melbourne, VIC, Australia; Genomic Medicine and Family Cancer Clinic, Royal Melbourne Hospital, Parkville, VIC, Australia.
Yin Cao, Division of Public Health Sciences, Department of Surgery, Washington University School of Medicine, St Louis, MO, USA; Division of Gastroenterology, Department of Medicine, Washington University School of Medicine, St Louis, MO, USA; Alvin J. Siteman Cancer Center, St Louis, MO, USA.
Andrew T Chan, Division of Gastroenterology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA; Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA, USA.
Xuechen Chen, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany; Medical Faculty Heidelberg, Heidelberg University, Heidelberg, Germany.
Iona Cheng, Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, CA, USA.
Jenny Chang-Claude, Division of Cancer Epidemiology, German Cancer Research Center (DKFZ), Heidelberg, Germany; Cancer Epidemiology Group, University Cancer Center Hamburg (UCCH), University Medical Center Hamburg-Eppendorf (UKE), Hamburg, Germany.
Niki Dimou, Nutrition and Metabolism Branch, International Agency for Research on Cancer, Lyon, France.
David Drew, Division of Gastroenterology, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital and Harvard Medical School, Boston, MA, USA.
Alton B Farris, Department of Pathology, Emory University, Atlanta, GA, USA.
Amy J French, Division of Laboratory Genetics, Department of Pathology and Laboratory Medicine, Mayo Clinic, Rochester, MN, USA.
Steven Gallinger, Lunenfeld Tanenbaum Research Institute, Mount Sinai Hospital, University of Toronto, Toronto, ON, Canada.
Peter Georgeson, Colorectal Oncogenomics Group, Department of Clinical Pathology, The University of Melbourne, Parkville, VIC, Australia; University of Melbourne Centre for Cancer Research, Victorian Comprehensive Cancer Centre, Melbourne, VIC, Australia.
Marios Giannakis, Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA; Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Graham G Giles, Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, VIC, Australia; Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, VIC, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, VIC, Australia.
Stephen B Gruber, Department of Medical Oncology & Therapeutics Research, City of Hope National Medical Center, Duarte, CA, USA.
Sophia Harlid, Department of Radiation Sciences, Oncology Unit, Umeå University, Umeå, Sweden.
Li Hsu, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA; Department of Biostatistics, University of Washington, Seattle, WA, USA.
Wen-Yi Huang, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD, USA.
Mark A Jenkins, Centre for Epidemiology and Biostatistics, Melbourne School of Population and Global Health, The University of Melbourne, Melbourne, VIC, Australia.
Ruhina S Laskar, Nutrition and Metabolism Branch, International Agency for Research on Cancer, Lyon, France.
Loic Le Marchand, University of Hawaii Cancer Center, Epidemiology Program, Honolulu, HI, USA.
Paul Limburg, Mayo Clinic, Rochester, MN, USA.
Yi Lin, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Marko Mandic, Division of Clinical Epidemiology and Aging Research, German Cancer Research Center (DKFZ), Heidelberg, Germany.
Johnathan A Nowak, Program in Molecular Pathological Epidemiology, Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
Mereia Obón-Santacana, Unit of Biomarkers and Susceptibility (UBS), Oncology Data Analytics Program (ODAP), Catalan Institute of Oncology (ICO), L’Hospitalet del Llobregat, Barcelona, Spain; ONCOBELL Program, Bellvitge Biomedical Research Institute (IDIBELL), L'Hospitalet de Llobregat, Barcelona, Spain; Consortium for Biomedical Research in Epidemiology and Public Health (CIBERESP), Madrid, Spain.
Shuji Ogino, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA; Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA, USA; Program in Molecular Pathological Epidemiology, Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA; Department of Oncologic Pathology, Dana-Farber Cancer Institute, Boston, MA, USA.
Conghui Qu, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Lori C Sakoda, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA; Division of Research, Kaiser Permanente Northern California, Oakland, CA, USA.
Robert E Schoen, Department of Medicine and Epidemiology, University of Pittsburgh Medical Center, Pittsburgh, PA, USA.
Melissa C Southey, Department of Clinical Pathology, The University of Melbourne, Melbourne, VIC, Australia; Cancer Epidemiology Division, Cancer Council Victoria, Melbourne, VIC, Australia; Precision Medicine, School of Clinical Sciences at Monash Health, Monash University, Clayton, VIC, Australia.
Zsofia K Stadler, Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Robert S Steinfelder, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Wei Sun, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Stephen N Thibodeau, Mayo Clinic, Rochester, MN, USA.
Amanda E Toland, Departments of Cancer Biology and Genetics and Internal Medicine, Comprehensive Cancer Center, The Ohio State University, Columbus, OH, USA.
Quang M Trinh, Ontario Institute for Cancer Research, Toronto, ON, Canada.
Kostas K Tsilidis, Department of Epidemiology and Biostatistics, Imperial College London, School of Public Health, London, UK; Department of Hygiene and Epidemiology, University of Ioannina School of Medicine, Ioannina, Greece.
Tomotaka Ugai, Department of Epidemiology, Harvard T.H. Chan School of Public Health, Harvard University, Boston, MA, USA; Program in Molecular Pathological Epidemiology, Department of Pathology, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, USA.
Bethany Van Guelpen, Department of Radiation Sciences, Oncology Unit, Umeå University, Umeå, Sweden; Wallenberg Centre for Molecular Medicine, Umeå University, Umeå, Sweden.
Xiaoliang Wang, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA.
Michael O Woods, Memorial University of Newfoundland, Discipline of Genetics, St. John's, NL, Canada.
Syed H Zaidi, Ontario Institute for Cancer Research, Toronto, ON, Canada.
Marc J Gunter, Nutrition and Metabolism Branch, International Agency for Research on Cancer, Lyon, France.
Ulrike Peters, Public Health Sciences Division, Fred Hutchinson Cancer Center, Seattle, WA, USA; Department of Epidemiology, University of Washington, Seattle, WA, USA.
Peter T Campbell, Department of Epidemiology and Population Health, Albert Einstein College of Medicine, Bronx, NY, USA.
Funding
This study was supported by Cancer Research UK (C18281/A29019) and Cancer Research UK (PPRCPJT\100005; Dr Tsilidis). Dr Obón-Santacana received a post-doctoral fellowship from the Spanish Association Against Cancer Scientific Foundation (AECC; POSTD037OBÓN).
Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO): National Cancer Institute, National Institutes of Health, U.S. Department of Health and Human Services (U01 CA137088, R01 CA059045, R01CA201407). Genotyping/Sequencing services were provided by the Center for Inherited Disease Research (CIDR) contract number HHSN268201700006I and HHSN268201200008I. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA015704. Scientific Computing Infrastructure at Fred Hutch funded by Office of Research Infrastructure Program (ORIP) Grant S10OD028685.
The Colon Cancer Family Registry (CCFR, www.coloncfr.org) is supported in part by funding from the National Cancer Institute (NCI), National Institutes of Health (NIH) (award U01 CA167551). Support for case ascertainment was provided in part from the Surveillance, Epidemiology and End Results (SEER) Program and the following U.S. state cancer registries: AZ, CO, MN, NC, NH; and by the Victoria Cancer Registry (Australia) and Ontario Cancer Registry (Canada). The CCFR Set-1 (Illumina 1M/1M-Duo) and Set-2 (Illumina Omni1-Quad) scans were supported by NIH awards U01 CA122839 and R01 CA143247 (to GC). The content of this article does not necessarily reflect the views or policies of the NCI, NIH or any of the collaborating centers in the Colon Cancer Family Registry (CCFR), nor does mention of trade names, commercial products or organizations imply endorsement by the US Government, any cancer registry or the CCFR.
CPS-II: The American Cancer Society funds the creation, maintenance and updating of the Cancer Prevention Study-II (CPS-II) cohort. Our study was conducted with Institutional Review Board approval.
DACHS: This work was supported by the German Research Council (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4, CH 117/1-1, HO 5117/2-1, HE 5998/2-1, KL 2354/3-1, RO 2270/8-1 and BR 1704/17-1), the Interdisciplinary Research Program of the National Center for Tumor Diseases (NCT), Germany and the German Federal Ministry of Education and Research (01KH0404, 01ER0814, 01ER0815, 01ER1505A and 01ER1505B).
DALS: National Institutes of Health (R01 CA48998 to M. L. Slattery).
EDRN: This work is funded and supported by the NCI, EDRN Grant (U01 CA 84968-06).
EPIC_Sweden: Swedish Cancer Society, Swedish Research Council and County Councils of Skåne and Västerbotten (Sweden).
Harvard cohorts: HPFS is supported by the National Institutes of Health (P01 CA055075, UM1 CA167552, U01 CA167552, R01 CA137178, R01 CA151993, and R35 CA197735), NHS by the National Institutes of Health (P01 CA087969, UM1 CA186107, R01 CA137178, R01 CA151993, and R35 CA197735).
MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 509348, 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database.
NFCCR: This work was supported by an Interdisciplinary Health Research Team award from the Canadian Institutes of Health Research (CRT 43821); the National Institutes of Health, U.S. Department of Health and Human Services (U01 CA74783); and National Cancer Institute of Canada grants (18223 and 18226). The authors wish to acknowledge the contribution of Alexandre Belisle and the genotyping team of the McGill University and Génome Québec Innovation Centre, Montréal, Canada, for genotyping the Sequenom panel in the NFCCR samples. Funding was provided to Michael O. Woods by the Canadian Cancer Society Research Institute.
NSHDS: The research was supported by Biobank Sweden through funding from the Swedish Research Council (VR 2017-00650, VR 2017-01737), the Swedish Cancer Society (CAN 2017/581), Region Västerbotten (VLL-841671, VLL-833291), Knut and Alice Wallenberg Foundation (VLL-765961) and the Lion's Cancer Research Foundation (several grants) and Insamlingsstiftelsen, both at Umeå University.
Notes
Role of the funder: The funders had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: Marios Giannakis reports potential financial conflicts of interest through research funding from Bristol-Myers Squibb, Merck, Servier and Janssen unrelated to our study. Paul Limburg serves as Chief Medical Officer for Screening at Exact Sciences through a contracted services agreement with Mayo Clinic. Paul Limburg and Mayo Clinic have contractual rights to receive royalties through this agreement. Jonathan Nowak reports prior research funding (for pilot projects) from NanoString and Illumina and ongoing research funding (for technology development) from Akoya Biosciences, unrelated to our study. All other authors declare no conflicts of interest.
MS, a JNCI Associate Editor and co-author on this article, was not involved in the editorial review or decision to publish this manuscript.
Author contributions: Conceptualization: NM, UP, PTC. Data Curation: NM, UP, PTC Formal Analysis: CCN, NM, PTC. Investigation: all authors. Methodology: NM, CCN, UP, PTC. Writing—original draft: NM, PTC. Writing—review & editing: all authors.
Acknowledgements: CCFR: The Colon CFR graciously thanks the generous contributions of their study participants, dedication of study staff and the financial support from the US National Cancer Institute, without which this important registry would not exist. The authors would like to thank the study participants and staff of the Seattle Colon Cancer Family Registry and the Hormones and Colon Cancer study (CORE Studies). CPS-II: The authors thank the CPS-II participants and Study Management Group for their invaluable contributions to this research. The authors would also like to acknowledge the contribution to our study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program. DACHS: We thank all participants and cooperating clinicians, and everyone who provided excellent technical assistance. Harvard cohorts (HPFS, NHS): The study protocol was approved by the institutional review boards of the Brigham and Women's Hospital and Harvard T.H. Chan School of Public Health, and those of participating registries as required. We would like to thank the participants and staff of the HPFS and NHS for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data. NSHDS investigators thank the Västerbotten Intervention Programme, the Northern Sweden MONICA study, the Biobank Research Unit at Umeå Universitet and Biobanken Norr at Region Västerbotten for providing data and samples and acknowledge the contribution from Biobank Sweden, supported by the Swedish Research Council.
Disclaimer: Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Data availability
Tumor marker and epidemiologic data is available upon request and permission. Please contact gecco@fredhutch.org to request the standardized proposal form. The principal investigators of each contributing study will evaluate and approve the proposal, and data access will be managed centrally.
References
- 1. Ogino S, Chan AT, Fuchs CS, Giovannucci E.. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50(1):113-130. [DOI] [PubMed] [Google Scholar]
- 3. Gatalica Z, Vranic S, Xiu J, Swensen J, Reddy S.. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Familial Cancer. 2016;15(3):405-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guastadisegni C, Colafranceschi M, Ottini L, Dogliotti E.. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer. 2010;46(15):2788-2798. [DOI] [PubMed] [Google Scholar]
- 5. WCRF-AICR. World Cancer Research Fund. Diet, Nutrition, Physical Activity and Colorectal Cancer. Continuous Update Project. 2017. https://www.wcrf.org/wp-content/uploads/2021/02/Colorectal-cancer-report.pdf. Accessed February 10, 2022.
- 6. Murphy N, Jenab M, Gunter MJ.. Adiposity and gastrointestinal cancers: epidemiology, mechanisms and future directions. Nat Rev Gastroenterol Hepatol. 2018;15(11):659-670. [DOI] [PubMed] [Google Scholar]
- 7. Bull CJ, Bell JA, Murphy N, et al. Adiposity, metabolites, and colorectal cancer risk: Mendelian randomization study. BMC Med. 2020;18(1):396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carr PR, Alwers E, Bienert S, et al. Lifestyle factors and risk of sporadic colorectal cancer by microsatellite instability status: a systematic review and meta-analyses. Ann Oncol. 2018;29(4):825-834. [DOI] [PubMed] [Google Scholar]
- 9. Carr PR, Amitay EL, Jansen L, et al. Association of BMI and major molecular pathological markers of colorectal cancer in men and women. Am J Clin Nutr. 2020;111(3):562-569. [DOI] [PubMed] [Google Scholar]
- 10. Hughes LAE, Simons CCJM, van den Brandt PA, et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP). PLoS One. 2011;6(4):e18571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brändstedt J, Wangefjord S, Nodin B, et al. Associations of anthropometric factors with KRAS and BRAF mutation status of primary colorectal cancer in men and women: a cohort study. PLoS One. 2014;9(2):e98964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campbell PT, Jacobs ET, Ulrich CM, et al. ; Colon Cancer Family Registry. Case–control study of overweight, obesity, and colorectal cancer risk, overall and by tumor microsatellite instability status. J Natl Cancer Inst. 2010;102(6):391-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoffmeister M, Bläker H, Kloor M, et al. Body mass index and microsatellite instability in colorectal cancer: a population-based study. Cancer Epidemiol Biomarkers Prev. 2013;22(12):2303-2311. [DOI] [PubMed] [Google Scholar]
- 14. Hidaka A, Harrison TA, Cao Y, et al. Intake of dietary fruit, vegetables, and fiber and risk of colorectal cancer according to molecular subtypes: a pooled analysis of 9 studies. Cancer Res. 2020;80(20):4578-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Labadie JD, Harrison TA, Banbury B, et al. Postmenopausal hormone therapy and colorectal cancer risk by molecularly defined subtypes and tumor location. JNCI Cancer Spectr. 2020;4(5):pkaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248-5257. [PubMed] [Google Scholar]
- 17. Warth A, Kloor M, Schirmacher P, Bläker H.. Genetics and epigenetics of small bowel adenocarcinoma: the interactions of CIN, MSI, and CIMP. Mod Pathol. 2011;24(4):564-570. [DOI] [PubMed] [Google Scholar]
- 18. Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148(1):77-87.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zaidi SH, Harrison TA, Phipps AI, et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun. 2020;11(1):3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng W, McLerran DF, Rolland B, et al. Association between body-mass index and risk of death in more than 1 million Asians. N Engl J Med. 2011;364(8):719-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Angelantonio E, Bhupathiraju SN, Wormser D, et al. Body-mass index and all-cause mortality: individual-participant-data meta-analysis of 239 prospective studies in four continents. Lancet. 2016;388(10046):776-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120(3):656-663. [DOI] [PubMed] [Google Scholar]
- 23. Brändstedt J, Wangefjord S, Borgquist S, et al. Influence of anthropometric factors on tumour biological characteristics of colorectal cancer in men and women: a cohort study. J Transl Med. 2013;11(1):293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanyuda A, Ogino S, Qian ZR, et al. Body mass index and risk of colorectal cancer according to tumor lymphocytic infiltrate. Int J Cancer. 2016;139(4):854-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Newcomb PA, Baron J, Cotterchio M, et al. ; Colon Cancer Family Registry. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331-2343. [DOI] [PubMed] [Google Scholar]
- 26. Campbell PT, Cotterchio M, Dicks E, Parfrey P, Gallinger S, McLaughlin JR.. Excess body weight and colorectal cancer risk in Canada: associations in subgroups of clinically defined familial risk of cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(9):1735-1744. [DOI] [PubMed] [Google Scholar]
- 27. Win AK, Dowty JG, English DR, et al. Body mass index in early adulthood and colorectal cancer risk for carriers and non-carriers of germline mutations in DNA mismatch repair genes. Br J Cancer. 2011;105(1):162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lazzeroni M, Bellerba F, Calvello M, et al. A meta-analysis of obesity and risk of colorectal cancer in patients with lynch syndrome: the impact of sex and genetics. Nutrients. 2021;13(5):1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Campbell PT. Obesity: a certain and avoidable cause of cancer. Lancet. 2014;384(9945):727-728. [DOI] [PubMed] [Google Scholar]
- 30. Murphy N, Ward HA, Jenab M, et al. Heterogeneity of colorectal cancer risk factors by anatomical subsite in 10 European countries: a multinational cohort study. Clin Gastroenterol Hepatol. 2019;17(7):1323-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chan AT, Ogino S, Fuchs CS.. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med. 2007;356(21):2131-2142. [DOI] [PubMed] [Google Scholar]
- 32. Slattery ML, Edwards SL, Palmer L, et al. Use of archival tissue in epidemiologic studies: collection procedures and assessment of potential sources of bias. Mutat Res. 2000;432(1-2):7-14. [DOI] [PubMed] [Google Scholar]
- 33. Campbell PT, Deka A, Briggs P, et al. Establishment of the cancer prevention study II nutrition cohort colorectal tissue repository. Cancer Epidemiol Biomarkers Prev. 2014;23(12):2694-2702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Tumor marker and epidemiologic data is available upon request and permission. Please contact gecco@fredhutch.org to request the standardized proposal form. The principal investigators of each contributing study will evaluate and approve the proposal, and data access will be managed centrally.