Abstract
Background
SLC7A11 is a key regulator of ferroptosis, which mediates cysteine uptake for glutathione biosynthesis and maintains redox homeostasis. Emerging evidence has shown that SLC7A11 is upregulated in many human tumors. Nevertheless, the prognosis and posttranslational regulatory mechanism of SLC7A11 in renal cell carcinoma (RCC) remains obscure.
Methods
The Oncomine, Gene Expression Profiling Interactive Analysis (GEPIA), and The Cancer Genome Atlas (TCGA) databases were used to analyze the difference in SLC7A11 expression between malignant and normal tissues. Furthermore, the GEPIA, the University of ALabama at Birmingham CANcer data analysis Portal (UALCAN), and starBase databases were used to conduct the survival analyses. For correlation analysis, the UALCAN and starBase databases were employed. The Tumor Immune Estimation Resource (TIMER) database was used to approximate the abundance of immune infiltration.
Results
We confirmed that SLC7A11 was upregulated in most human cancers, including 3 types of RCC. SLC7A11 overexpression was linked to poor prognosis of individuals with kidney renal clear cell carcinoma (KIRC), kidney chromophobe cell carcinoma (KICH), and kidney renal papillary cell carcinoma (KIRP). SLC7A11 expression was also linked to immune cell infiltration levels. After performing a comprehensive analysis of the regulatory mechanisms of SLC7A11 expression, the results depicted a potential noncoding (ncRNA)–messenger RNA (mRNA) axis, incorporating SNHG6–miR-26a-5p–SLC7A11 networks in KICH, CASC19/CYTOR/LINC00997–miR-27b–3pSLC7A11 networks in KIRC, and CASC19/CYTOR/PVT1–miR27b-3p–SLC7A11 networks in KIRP as partially responsible for the functions of SLC7A11 in RCC. SLC7A11 expression was positively linked to infiltrated immune cells and their matching marker sets in 3 types of RCC, including CD8+ and myeloid dendritic cells.
Conclusions
Our research elucidated the crucial functions and the upstream long noncoding RNA (lncRNA)–microRNA (miRNA) regulatory network of SLC7A11 in RCC. Importantly, SLC7A11 can be used as a potential prognostic biomarker for 3 types of RCC and to determine the infiltration of immune cells in malignant tissues.
Keywords: Ferroptosis, renal cell carcinoma (RCC), ceRNA, immune infiltration, SLC7A11
Highlight box.
Key Findings
• The study elucidated the crucial functions and the upstream lncRNA–miRNA regulatory network of SLC7A11 in RCC. Importantly, SLC7A11 can be used as a potential prognostic biomarker for 3 types of RCC and to determine the infiltration of immune cells in malignant tissues.
What is known and what is new?
• The cystine/glutamate antiporter SLC7A11 is a key modulator of ferroptosis, which mediates cysteine uptake for glutathione biosynthesis and maintains redox homeostasis. SLC7A11 expression is upregulated in various cancers and has multiple effects on cancer growth and metastasis.
• Nevertheless, the prognosis and posttranslational regulatory mechanism of SLC7A11 in renal cell carcinoma (RCC) remains obscure. The study elucidated the crucial functions and the upstream lncRNA–miRNA regulatory network of SLC7A11 in RCC.
What is the implication, and what should change now?
• Importantly, SLC7A11 can be used as a potential prognostic biomarker for 3 types of RCC and to determine the infiltration of immune cells in malignant tissues.
Introduction
Renal cell carcinoma (RCC) is the sixth and ninth most prevalent malignancy among men and women, respectively, in the USA. Furthermore, a potential ~79,000 cases of RCC will be diagnosed in the USA in 2022 with at least 13,920 related mortalities (1). RCC has different pathological types according to histology and genetics. Among the various types of RCC, this study will focus on 3 of the most prevalent, including kidney chromophobe cell carcinoma (KICH), kidney renal papillary cell carcinoma (KIRP), and kidney renal clear cell carcinoma (KIRC) (2). With the continuous advancement and improvement in the diagnosis and management of RCC, the 5-year survival rate of RCC patients has risen to at least 74%, while 12% of patients develop advanced metastasis (3). In situations where the advanced kidney malignancy cannot be excised, the prognosis can be prolonged by application molecularly targeted therapies although this is only effective in 20–40% of cases (4). Therefore, investigations into the pathogenesis of RCC to find novel biomarkers for managing and evaluating the prognosis of RCC should be expedited.
The cystine/glutamate antiporter SLC7A11 (xCT) is a key modulator of ferroptosis, which mediates cysteine uptake for glutathione biosynthesis and maintains redox homeostasis (5). SLC7A11 expression is upregulated in various cancers and has multiple effects on cancer growth, invasion, and metastasis. Its overexpression has also been linked to the drug resistance of gemcitabine, cisplatin, and mitogen-activated protein kinase (MAPK) pathway inhibitors (6), resulting in poor prognosis (7). Furthermore, SLC7A11 expression and activity are exposed to tight modulation via various mechanisms, such as transcriptional modulation by transcription factors and epigenetic regulators as well as posttranscriptional modulation mechanisms to regulate its protein stability, messenger RNA (mRNA) levels, transporter activity, and subcellular localization (6). Long noncoding RNAs (lncRNAs) and microRNAs (miRNAs) are noncoding RNAs (ncRNAs) with important epigenetic regulation functions. They are involved in posttranscriptional regulation through mRNA degradation and translation inhibition and thus participate in the development of cancers (8). Therefore, this study comprehensively evaluated the prognostic mechanism of SLC7A11 in RCC and the upstream lncRNA–miRNA regulatory network.
Recently, 2 distinct studies shared similar findings, reporting that ferroptosis, which is stimulated by T cells in cancerous cells, can be a vital anticancer target, especially with programmed cell death protein 1/programmed death-ligand 1 (PD-1/PD-L1) antibody treatment. Moreover, among the ferroptosis-insensitive cancerous cells, the PD-L1 antibody only demonstrates a weak influence (9,10). In contrast, elevated ferroptosis levels have been shown to enhance the antitumor impact of immunotherapy, which highlights the positive feedback between immunotherapy and ferroptosis, which can synergistically destroy cancerous cells (9). Nevertheless, the participation of SLC7A11 in regulating the infiltration of immune cells and the progression processes in RCC has yet to be established.
In this study, we first evaluated the mRNA and protein levels of SLC7A11 in RCC. Second, we assessed the prognostic value of SLC7A11 in RCC. Finally, we investigated the fundamental upstream ncRNA-related mode of action and immunomodulatory impact of SLC7A11 in RCC. We identified a potential SLC7A11-related ncRNA–mRNA axis in RCC, which provides critical insight for creating potent therapeutic targets and potential biomarkers in RCC. We present the following article in accordance with the MDAR reporting checklist (available at https://tau.amegroups.com/article/view/10.21037/tau-22-663/rc).
Methods
Gene expression analysis
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The difference in the expression level of SLC7A11 between cancer and adjoining normal tissues was estimated from the Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/), Oncomine (https://www.oncomine.org), and The Cancer Genome Atlas (TCGA; https://www.aclbi.com) databases. Variations in SLC7A11 expression by tumor stage, age, nodal metastasis, tumor grade, gender, and every subtype of RCC were determined from TCCA, UALCAN, and starBase (http://starbase.sysu.edu.cn/) databases. starBase was employed to analyze the differential expression of miRNAs and lncRNAs between the tumors and adjacent tissues.
Oncomine
Oncomine is not only the biggest cancer microarray database but also an integrated data-mining platform worldwide, which aims at mining cancer gene information. It incorporates RNA sequencing (RNA-seq) and DNA sequencing (DNA-seq) data from the TCGA and GEO databases, as well as the published study involving 715 gene expression datasets obtained from 86,733 samples (11).
starBase
starBase is an open-source platform and is freely available for public use. It is used for studying the ncRNA–RNA, miRNA–ncRNA, RNA–RNA, RBP–ncRNA, miRNA–mRNA, and RBP–mRNA interactions from degradome sequencing (seq), CLIP-seq, and RNA–RNA interactome data (12).
UALCAN
UALCAN is an online tool for evaluating cancer omics data [TCGA and the Clinical Proteomic Tumor Analysis Consortium (CPTAC)] that is comprehensive, convenient, and interactive. The Kaplan–Meier survival plot was generated for every gene in each TCGA cancer type, using “survival” package and “survminer” package. The survival curves of samples with high gene expression and low/medium gene expression were compared by log rank test (13).
Survival analysis
The survival and prognosis of patients with RCC were analyzed from the UALCAN, GEPIA, TCGA, and starBase databases.
LinkedOmics
LinkedOmics is a web tool to analyze the multiomics data based on TCGA database. LinkInterpreter evaluates functional enrichment against 26, 449 functional categories defined by Gene Ontology, pathways from the KEGG, Panther, Reactome and WikiPathways databases. Function and pathway enrichment analysis of SLC7A11 in the 3 RCCs was employed (14).
miRNA prediction
The miRSystem (http://mirsystem.cgm.ntu.edu.tw), an integrated database platform, was used to anticipate upstream miRNAs of SLC7A11 with the aid of 7- miRNA target gene prediction programs, which include TargetScan, miRanda, miRBridge, PITA, PicTar, DIANA, and rna22 (15). miRNAs that were present in more than 3 programs were screened and discussed in this paper.
LncRNA prediction
miRNet (http://www.mirnet.ca), an miRNA-centric network visual analytics platform, was used to determine upstream lncRNAs of miR-26a-5p or miR-27b-3p (16).
Correlation analysis
starBase was used to perform expression correlation analysis for both miRNA–lncRNA and lncRNA–mRNA pairs in 3 subtypes of RCC.
Tumor Immune Estimation Resource (TIMER) database analysis
We used an open platform known as TIMER, which contains data on 32 types of cancers and includes 10,987 TCGA samples in order to assess the immune inner infiltrate abundance (http://cistrome.org/TIMER/) (17). The TIMER database was employed to examine the link between SLC7A11 expression and the abundance of 6 kinds of infiltrating immune cells including macrophages, CD4+ T cells, B cells, dendritic cells (DCs), CD8+ T cells, and neutrophils among patients with RCC. The link between the SLC7A11 expression and tumor purity was also clarified, while that between SLC7A11 expression and immune infiltration as well as markers of varied subsets of immune cells was evaluated using the TIMER database. Statistically significant differences were set at a P value <0.05.
Statistics analysis
The survival curves of samples with high gene expression and low/medium gene expression were compared by log rank test. Data are displayed as mean ± SD, t-test was used to assess the differences. Statistically significant differences were set at a P value <0.05.
Results
Expression and prognostic values of SLC7A11 in RCC
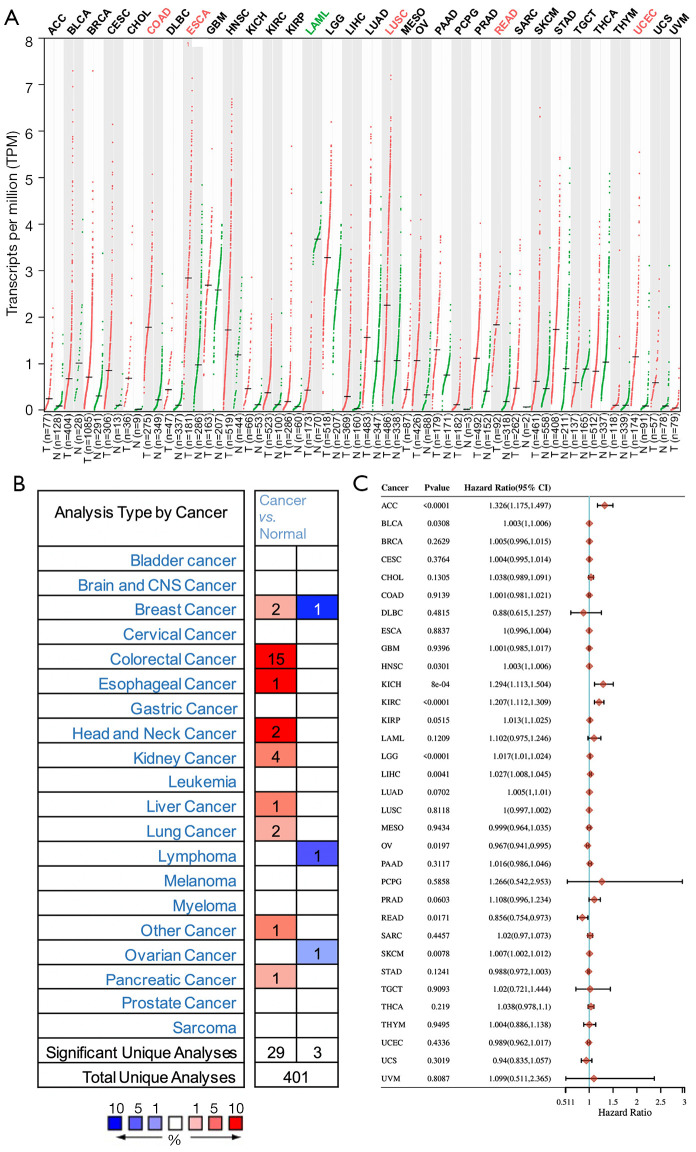
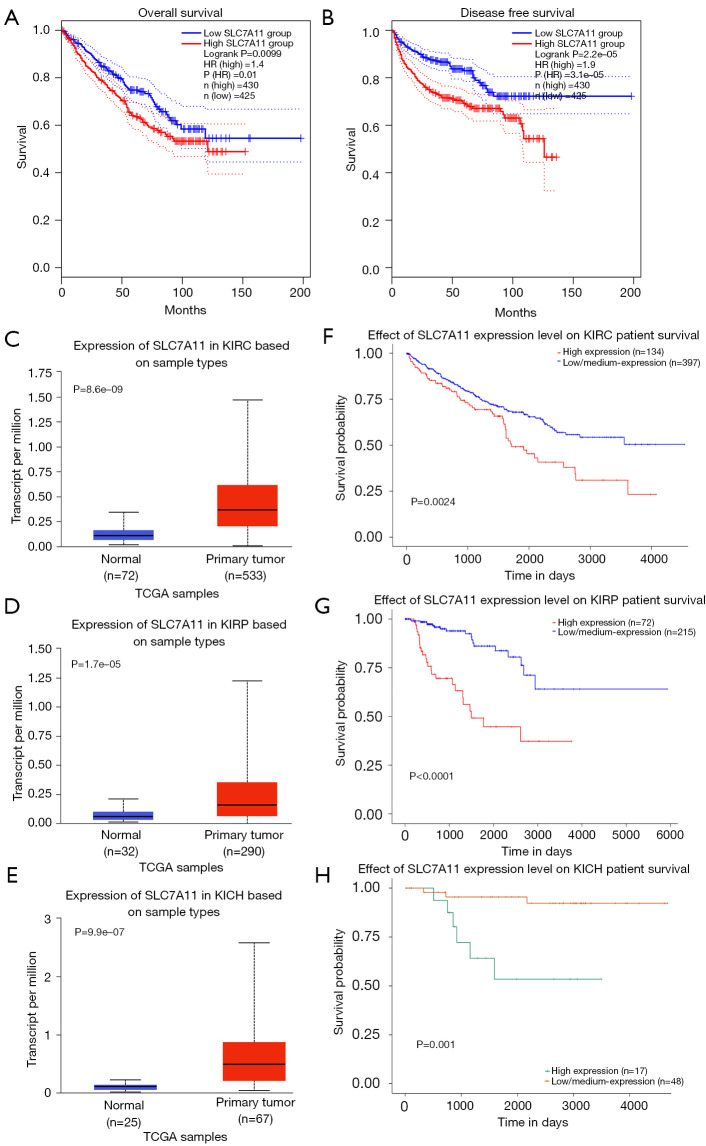
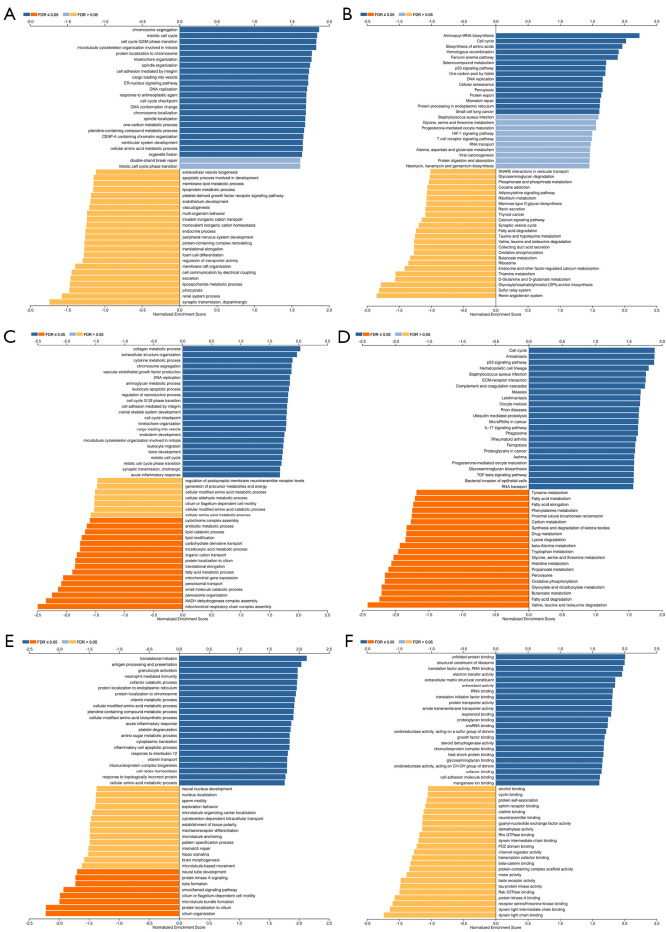
We used the GEPIA database to examine the data on RNA-seq expression status based on several samples recruited from both the genotype-tissue expression (GTEx) and TCGA projects to estimate the mRNA level of SLC7A11 in malignant and normal tissues in multiple cancers. The results are displayed in Figure 1A. The SLC7A11 expression was increased in cancers compared with adjacent normal samples, including KICH, KIRC, and KIRP. The differences in the gene expression profile of SLC7A11 across cancer samples and paired normal tissues were also assessed using the Oncomine database. This database contains a total of 456 distinct analyses for SLC7A11. SLC7A11 expression in tumors was upregulated in 29 distinct analyses and downregulated in 3 analyses. Specifically, SLC7A11 expression was elevated in kidney, liver, colorectal, esophageal, and head and neck malignancies compared to the normal samples (Figure 1B). Furthermore, TCGA data set was used to analyze the prognostic values of SLC7A11 in 3 types of RCC. The findings showed that high expression of SLC7A11 in KIRP, KICH, and KIRC was associated with a worse prognosis (Figure 1C). A Kaplan-Meier plot was used to evaluate the prognostic functions of SLC7A11 in all patients with RCC and for each of the 3 subtypes of RCC. The findings in the GEPIA database revealed that a high expression of SLC7A11 was significantly linked to worse overall survival [OS; P=0.01; hazard ratio (HR) =1.4] and disease-free survival (DFS; P<0.001; HR =1.9) in RCC (Figure 2A,2B). Lastly, the UALCAN and starBase databases were used for survival analysis of the 3 subtypes of RCC. The findings indicated that SLC7A11 expression in KICH, KIRP, and KIRC was significantly increased compared to normal tissues (Figure 2C-2E), whereas high expression of SLC7A11 was also linked to shorter OS duration (Figure 2F-2H). The LinkedOmics database was employed to analyze the enrichment of functions in 3 different types of RCC (Figure 3). In KICH, SLC7A11 was mainly enriched in the biological process of chromosome segregation and aminoacyl–transfer RNA (tRNA) biosynthesis pathway. In KIRC, SLC7A11 was mainly enriched in the collagen metabolic process and cell cycle pathway. In KIRP, SLC7A11 was mainly enriched in the translational initiation and unfolded protein binding pathway. Taken together, these findings imply that SLC7A11 may act as a potential diagnostic and prognostic biomarker in RCC.
Figure 1.
Differential expression of SLC7A11 in cancers. (A) The mRNA level of SLC7A11 in malignant and normal tissues in multiple cancers according to the GEPIA database, red dots: tumor samples, green dots: normal samples; (B) the differences in the gene expression profile of SLC7A11 across cancer samples and paired normal tissues according to the Oncomine database; (C) high expression of SLC7A11 in KIRP, KICH, and KIRC was associated with a worse prognosis according to TCGA database. GEPIA, Gene Expression Profiling Interactive Analysis; KIRP, kidney renal papillary cell carcinoma; KICH, kidney chromophobe cell carcinoma; KIRC, kidney renal clear cell carcinoma; TCGA, The Cancer Genome Atlas.
Figure 2.
The survival analysis of the patients with high and low SLC7A11 expression in RCC. (A) The overall survival curves of RCC; (B) the disease-free survival curves of RCC; (C) the expression of SLC7A11 in KIRC and normal tissues; (D) the expression of SLC7A11 in KIRP and normal tissues; (E) the expression of SLC7A11 in KICH and normal tissues; (F) the overall survival curves of KIRC; (G) the overall survival curves of KIRP; (H) the overall survival curves of KICH. The survival analysis were performed used the GEPIA database in (A) and (B) and the UALCAN database in (C-H). RCC, renal cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; KICH, kidney chromophobe cell carcinoma; GEPIA, Gene Expression Profiling Interactive Analysis.
Figure 3.
Function and pathway enrichment analysis of different types of RCC. (A,B) The BP and KEGG pathway of SLC7A11 in KICH were analyzed; (C,D) the BP and KEGG pathway of SLC7A11 in KIRC were analyzed; (E,F) the BP and KEGG of SLC7A11 in KIRP were analyzed. RCC, renal cell carcinoma; BP, biological processes; KEGG, Kyoto Encyclopedia of Genes and Genome; KICH, kidney chromophobe cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; FDR, false discovery rate.
Analysis of upstream miRNAs of SLC7A11 in RCC
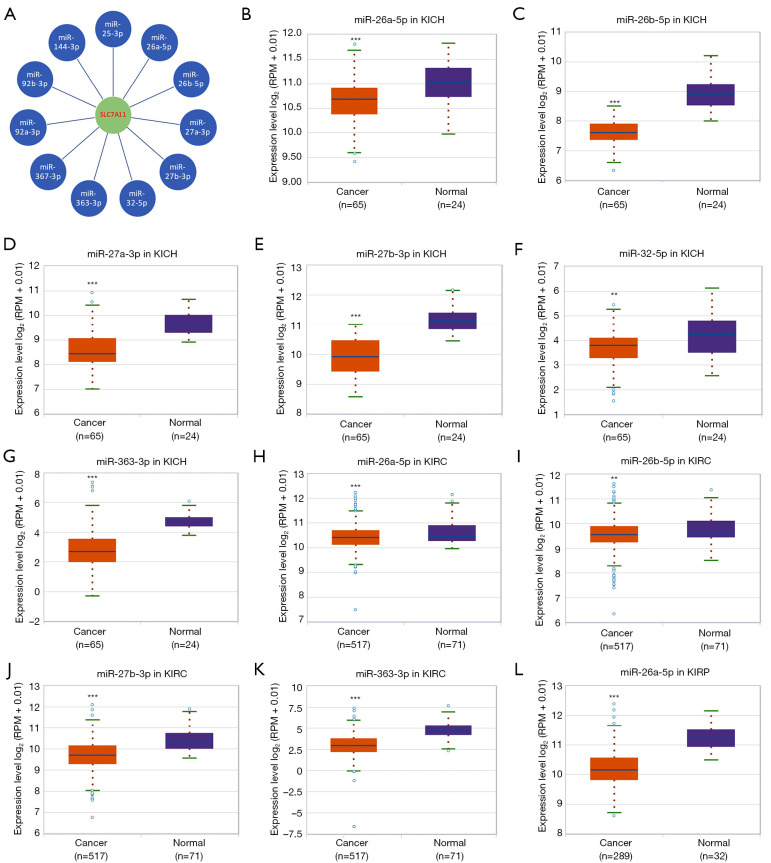
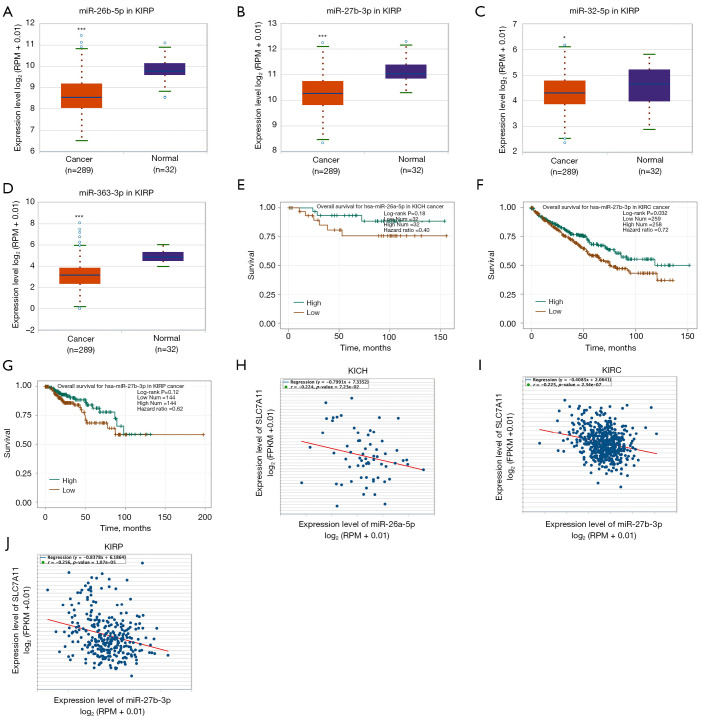
To predict the upstream miRNAs of SLC7A11, the miRSystem platform was used with 7 miRNA target gene prediction programs, including miRanda, PicTar, PITA, miRBridge, rna22, DIANA, and TargetScan. A total of 11 miRNAs were identified in more than 3 programs and subsequently used for further analysis (Figure 4A). The candidate miRNAs play a suppressive role in RCC by binding to SLC7A11. The findings in the starBase database revealed that the expression levels of miR-26b-5p, miR-32-5p, miR-27b-3p, miR-26a-5p, miR-363-3p, and miR-27a-3p in both KICH and KIRC were significantly reduced compared to those in normal tissues (Figure 4B-4K). Additionally, the expression levels of miR-27b-3p, miR-26b-5p, miR-27a-3p, miR-32-5p, miR-26a-5p, and miR-363-3p in KIRP were significantly lowered compared to those normal tissues (Figures 4L,5A-5D). The remaining miRNAs were not statistically differentially expressed between RCC and normal tissues. Furthermore, we found that a low miR-26a-5p expression was linked to poor prognosis and negatively associated with SLC7A11 in KICH (Figure 5E, Figure S1A-S1E). Low miR-27b-3p expression was linked to poor outcomes and negatively correlated with SLC7A11 in KIRC and KIRP (Figure 5F-5G, Figure S1F-S1N). Correspondingly, a low miR-26a-5p expression was negatively associated with SLC7A11 in KICH (Figure 5H, Figure S1M-S1N). Low miR-27b-3p expression was negatively correlated with SLC7A11 in KIRC and KIRP (Figure 5I,5J, Figure S1O). Altogether, these results show that miR-26a-5p and miR-27b-3p may serve as 2 potential upstream miRNAs of SLC7A11 in RCC.
Figure 4.
Analysis of upstream miRNAs of SLC7A11 in RCC. (A) The miRSystem platform was used to predict the upstream miRNAs of SLC7A11; (B) the expression of miR-26a-5p in KICH and normal tissues; (C) the expression of miR-26b-5p in KICH and normal tissues; (D) the expression of miR-27a-3p in KICH and normal tissues; (E) the expression of miR-27b-3p in KICH and normal tissues; (F) the expression of miR-32-5p in KICH and normal tissues; (G) the expression of miR-363-3p in KICH and normal tissues; (H) the expression of miR-26a-5p in KIRC and normal tissues; (I) the expression of miR-26b-5p in KIRC and normal tissues; (J) the expression of miR-27b-3p in KIRC and normal tissues; (K) the expression of miR-363-3p in KIRC and normal tissues; (L) the expression of miR-26a-5p in KIRP and normal tissues. Differential expressions from the starBase database in (B-L). **P<0.01; ***P<0.001. RCC, renal cell carcinoma; KIRP, kidney renal papillary cell carcinoma; KICH, kidney chromophobe cell carcinoma; KIRC, kidney renal clear cell carcinoma.
Figure 5.
Two potential upstream miRNAs of SLC7A11 in RCC from the starBase database. (A) The expression of miR-26b-5p in KIRP and normal tissues; (B) the expression of miR-27b-3p in KIRP and normal tissues; (C) the expression of miR-32-5p in KIRP and normal tissues; (D) the expression of miR-363-3p in KIRP and normal tissues; (E) the overall survival for miR-26a-5p in KICH; (F) the overall survival for miR-27b-3p in KIRC; (G) the overall survival for miR-27b-3p in KIRP; (H) the correlation of miR-26a-5p with SLC7A11 expression in KICH; (I) the correlation of miR-27b-3p with SLC7A11 expression in KIRC; (J) the correlation of miR-27b-3p with SLC7A11 expression in KIRP. *P<0.05; ***P<0.001. RCC, renal cell carcinoma; KIRP, kidney renal papillary cell carcinoma; KICH, kidney chromophobe cell carcinoma; KIRC, kidney renal clear cell carcinoma.
Analysis of the upstream competing endogenous RNA network of lncRNA/miRNA/SLC7A11 in RCC
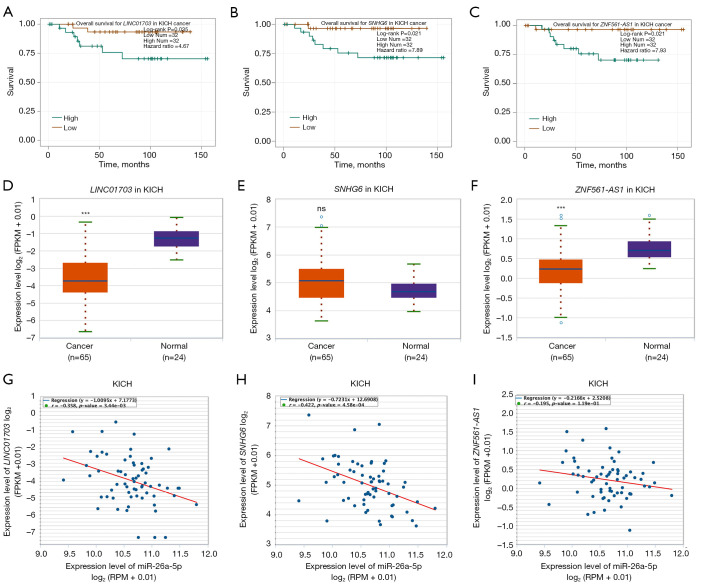
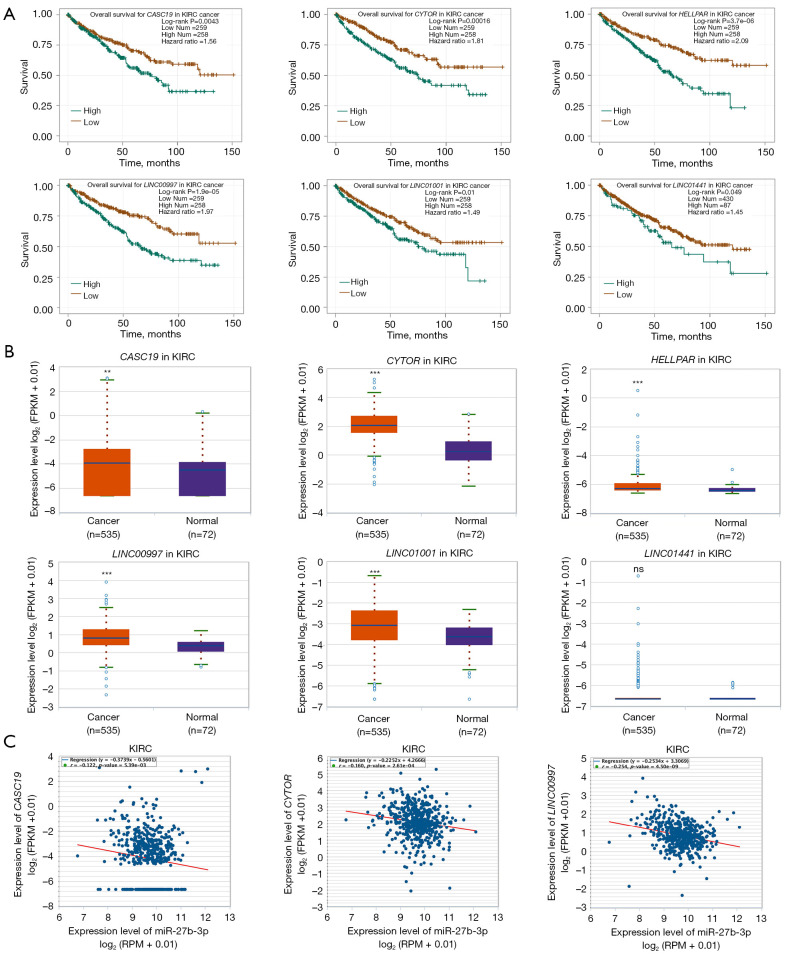
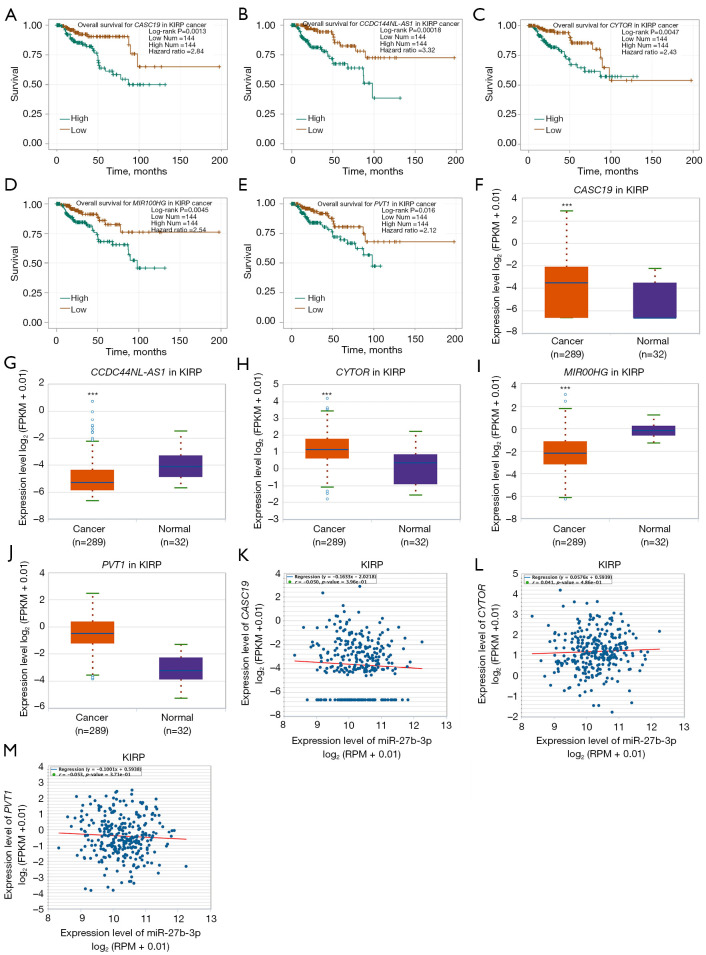
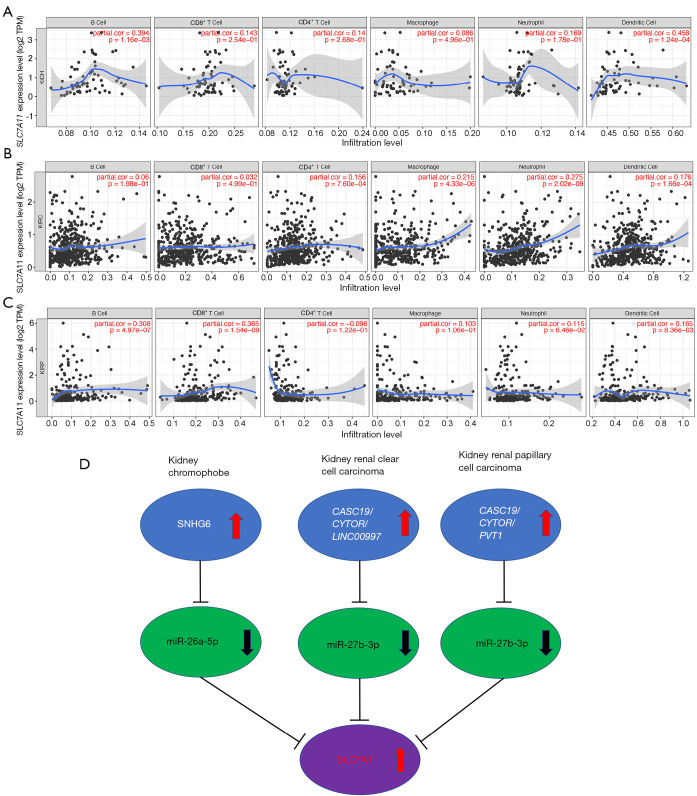
To ascertain the upstream molecular mechanism of the miR-26a-5p–miR-27b-3p–SLC7A11 axis in RCC, the miRNet and starBase databases were used to identify those lncRNAs that may bind to miR-26a-5p or miR-27b-3p. As indicated online (https://cdn.amegroups.cn/static/public/10.21037tau-22-663-1.xlsx and https://cdn.amegroups.cn/static/public/10.21037tau-22-663-2.xls), 43 and 59 lncRNAs appeared frequently in the dual prediction lncRNA sets, specifically miR-26a-5p and miR-27b-3p, respectively. Based on the competing endogenous RNA (ceRNA) hypothesis, the promising upstream lncRNAs of miR-26a-5p/miR-27b-3p-SLC7A11 in RCC should serve as tumor oncogenic lncRNAs in RCC. First, the levels of prognostic lncRNAs of miR-26a-5p or miR-27b-3p were evaluated using the starBase database. Second, a survival analysis for the upregulated lncRNAs was performed. As shown in Figure 6A-6C, high expression of LINC01703, SNHG6, and ZNF561-AS1 was linked to poor prognosis in patients with KICH. Subsequently, TCGA database showed that only SNHG6 was highly expressed in KICH (fold change 1.4; P=0.064; Figure 6D-6F). As displayed in Figure 6G-6I, there was 1 lncRNA–miRNA pair (SNHG6–miR-26a-5p) with a negative expression correlation in KICH. As demonstrated in Figure 7A, high expression of CASC19, CYTOR, HELLPAR, LINC00997, LINC01001, and LINC01441 had a poor prognosis in patients with KIRC. Subsequently, TCGA database showed that CASC19, CYTOR, and LINC00997 had a significantly higher expression in KIRC (Figure 7B). As demonstrated in Figure 7C, there were 3 negatively correlated lncRNA–miRNA pairs (CASC19, CYTOR, LINC00997/miR-27b-3p) in KIRC. As demonstrated in Figure 8A-8E, a high expression of CASC19, CCDC144NL-AS1, CYTOR, MIR100HG, and PVT1 was associated with poor prognosis in patients with KIRP. Subsequently, TCGA database showed that CASC19, CYTOR, and PVT1 had a significantly higher expression in KIRP (Figure 8F-8J). As shown in Figure 8K-8M, 3 lncRNA–miRNA pairs (CASC19, CYTOR, PVT1/miR-27b-3p) had a negative expression correlation in KIRP.
Figure 6.
Analysis of the upstream ceRNA network of lncRNAs/miRNAs/SLC7A11 in KICH according to the starBase database. (A) The overall survival for LINC01703 in KICH; (B) the overall survival for SNHG6 in KICH; (C) the overall survival for ZNF561-AS1 in KICH; (D) the expression of LINC01703 in KICH and normal tissues; (E) the expression of SNHG6 in KICH and normal tissues; (F) the expression of ZNF561-AS1 in KICH and normal tissues; (G) the correlation of LINC01703 with SLC7A11 expression in KICH; (H) the correlation of SNHG6 with SLC7A11 expression in KICH; (I) the correlation of ZNF561-AS1 with SLC7A11 expression in KICH. ***P<0.001. KICH, kidney chromophobe cell carcinoma; ns, no significance.
Figure 7.
Analysis of the upstream ceRNA network of lncRNAs/miRNAs/SLC7A11 in KIRC using the starBase database. (A) High expression of CASC19, CYTOR, HELLPAR, LINC00997, LINC01001, and LINC01441 had a poor prognosis in patients with KIRC; (B) CASC19, CYTOR, and LINC00997 had a significantly high expression in KIRC; (C) three negatively correlated lncRNA–miRNA pairs (CASC19, CYTOR, LINC00997/miR-27b-3p). **P<0.01; ***P<0.001. KIRC, kidney renal clear cell carcinoma; ns, no significance.
Figure 8.
Analysis of the upstream ceRNA network of lncRNAs/miRNAs/SLC7A11 in KIRP using the starBase database. (A-E) High expression of CASC19, CCDC144NL-AS1, CYTOR, MIR100HG, and PVT1 was associated with poor prognosis in patients with KIRP; (F-J) CASC19, CYTOR, and PVT1 had a significantly high expression in KIRP; (K-M) Three lncRNA–miRNA pairs (CASC19, CYTOR, PVT1/miR-27b-3p) whose expression was negatively correlated with KIRP. ***P<0.001. KIRP, kidney renal papillary cell carcinoma.
Correlation analysis between SLC7A11 expression and infiltrating immune cells
Tumor-infiltrating lymphocytes have an impact on the survival of individuals with different types of tumors. We used 6 types of cells including CD4+ T cells, macrophages, B cells, CD8+ T cells, neutrophils, and DCs, and correlated them with tumor purity to determine SLC7A11 expression. The results showed that the SLC7A11 expression had a positive link to infiltrating levels of B cells (r=0.394; P=1.16e-03) and DCs (r=0.458; P=1.24e-04) in KICH, but no link to CD8+ T cells, macrophages, CD4+ T cells, or neutrophils (Figure 9A). SLC7A11 expression in KIRC had a positive association with infiltrating levels of CD4+ T cells (r=0.156; P=7.60e-04), macrophages (r=0.215; P=4.33e-06), neutrophils (r=0.275; P=2.02e-09), and DCs (r=0.176; P=1.66e-04), but no link to B cells or CD8+ T cells (Figure 9B). SLC7A11 expression in KIRP had a positive association with infiltrating levels of B cells (r=0.308; P=4.97e-07), CD8+ T cells (r=0.365; P=1.54e-09), and DCs (r=0.165; P=8.36e-03), but no link to neutrophils, CD4+ T cells, or macrophages (Figure 9C).
Figure 9.
The correlation analysis of SLC7A11 with immune cells infiltration in KICH (A), KIRC (B), and KIRP (C). (D) A schematic model for the potential ceRNA network of the lncRNAs/miRNAs/SLC7A11 in 3 types of RCC. KIRP, kidney renal papillary cell carcinoma; KICH, kidney chromophobe cell carcinoma; KIRC, kidney renal clear cell carcinoma; RCC, renal cell carcinoma.
To comprehensively investigate the possible function of SLC7A11 in the infiltration of different immune cells in RCC, we employed the TIMER database to probe into the link between SLC7A11 expression and various immune marker sets (immunocytes) including natural killer cells, CD8+ T cells, T cells (general), M1/M2 macrophages, tumor-associated macrophages (TAMs), neutrophils, monocytes, B cells, and DCs in KICH, KIRC, and KIRP. In addition, different functional T cells, including exhausted T, T helper (TH) 1, Th2, Th9, Th17, Th22, regulator T (Treg), and T follicular helper (Tfh) cells were also examined in our experiment. For KICH, the levels of the majority of immune sets marking different T cells, TAMs, B cells, Th17, Th22, T cells, Tregs, and macrophages were linked to SLC7A11 expression (Table 1). For KIRC, the levels of the immune marker set TAMs, T cells, macrophages, M1/M2 macrophages, DCs, and neutrophils were linked to SLC7A11 expression (Table 2). For KIRP, the levels of the immune marker set TAMs, B cells, different T cells, natural killer cells, and M2 macrophages were linked to SLC7A11 expression (Table 3). According to these findings, SLC7A11 may exert a special function in RCC immune invasion.
Table 1. Correlation analysis between SLC7A11 and markers of immune cells for KICH in TIMER.
| Description | Gene markers | SLC7A11 | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| r | P | r | P | |||
| B cell | CD27 | 0.237 | 5.52e-02 | 0.352 | 3.99e-03 | |
| CD19 | 0.179 | 1.5e-01 | 0.225 | 7.18e-02 | ||
| CD38 | 0.282 | 2.18e-02 | 0.392 | 1.25e-03 | ||
| CD8+ T cell | CD8A | 0.151 | 2.26e-01 | 0.291 | 1.86e-02 | |
| CD8B | 0.157 | 2.08e-01 | 0.268 | 3.06e-02 | ||
| T cell (general) | CD3D | 0.194 | 1.18e-01 | 0.313 | 1.12e-02 | |
| CD3E | 0.223 | 7.24e-02 | 0.340 | 5.56e-03 | ||
| CD2 | 0.245 | 4.78e-02 | 0.391 | 1.26e-03 | ||
| Monocyte | CD14 | 0.119 | 3.40e-01 | 0.225 | 7.13e-02 | |
| CD115 | 0.183 | 1.40e-01 | 0.338 | 5.91e-03 | ||
| Tfh | BCL6 | 0.295 | 1.64e-02 | 0.279 | 2.42e-02 | |
| ICOS | 0.282 | 2.18e-02 | 0.381 | 1.75e-03 | ||
| CXCR5 | 0.100 | 4.25e-01 | 0.185 | 1.40e-01 | ||
| Th1 | T-bet (TBX21) | 0.088 | 4.80e-01 | 0.170 | 1.77e-01 | |
| STAT1 | 0.323 | 8.38e-03 | 0.355 | 3.66e-03 | ||
| STAT4 | 0.142 | 2.55e-01 | 0.271 | 2.92e-02 | ||
| IL12RB2 | 0.095 | 4.49e-01 | 0.110 | 3.81e-01 | ||
| WSX1(IL27RA) | 0.057 | 6.47e-01 | 0.171 | 1.73e-01 | ||
| IFN-γ (IFNG) | 0.094 | 4.51e-01 | 0.183 | 1.44e-01 | ||
| TNF-α (TNF) | 0.186 | 1.35e-01 | 0.268 | 3.11e-02 | ||
| Th2 | STAT6 | 0.022 | 8.58e-01 | 0.012 | 9.22e-01 | |
| GATA3 | −0.069 | 5.83e-01 | −0.065 | 6.08e-01 | ||
| STAT5A | 0.149 | 2.33e-01 | 0.225 | 7.14e-02 | ||
| CCR3 | 0.350 | 3.92e-03 | 0.383 | 1.65e-03 | ||
| Th9 | TGFBR2 | −0.081 | 5.18e-01 | −0.057 | 6.53e-01 | |
| PU.1(SPI1) | 0.143 | 2.51e-01 | 0.246 | 4.78e-02 | ||
| IRF4 | 0.186 | 1.34e-01 | 0.297 | 1.65e-02 | ||
| Th17 | IL-17A(IL17A) | NA | NA | NA | NA | |
| IL-21R(IL21R) | 0.237 | 5.55e-02 | 0.365 | 2.75e-03 | ||
| IL-23R(IL23R) | 0.109 | 3.83e-01 | 0.134 | 2.87e-01 | ||
| STAT3 | 0.332 | 6.67e-03 | 0.390 | 1.32e-03 | ||
| Th22 | AHR | 0.313 | 1.09e-02 | 0.329 | 7.41e-03 | |
| CCR10 | −0.177 | 1.55e-01 | −0.163 | 1.94e-01 | ||
| Treg | CD25(IL2RA) | 0.267 | 2.99e-02 | 0.369 | 2.47e-03 | |
| FOXP3 | 0.185 | 1.36e-01 | 0.238 | 5.67e-02 | ||
| CCR8 | 0.390 | 1.19e-03 | 0.441 | 2.37e-04 | ||
| T cell exhaustion | CTLA4 | 0.307 | 1.23e-02 | 0.416 | 5.59e-04 | |
| PD-1 (PDCD1) | 0.187 | 1.32e-01 | 0.288 | 2.00e-02 | ||
| TIM-3 (HAVCR2) | 0.096 | 4.40e-01 | 0.177 | 1.57e-01 | ||
| LAG3 | 0.236 | 5.70e-02 | 0.299 | 1.57e-02 | ||
| TAM | CD80 | 0.242 | 5.01e-02 | 0.306 | 1.32e-02 | |
| CD86 | 0.242 | 5.08e-02 | 0.387 | 1.46e-03 | ||
| CCL2 | 0.004 | 9.77e-01 | 0.068 | 5.90e-01 | ||
| CCR5 | 0.287 | 1.96e-02 | 0.420 | 5.03e-04 | ||
| Macrophage | CD68 | 0.251 | 4.23e-02 | 0.403 | 8.67e-04 | |
| CD11b (ITGAM) | 0.166 | 1.83e-01 | 0.317 | 1.01e-02 | ||
| M1 | IRF5 | 0.252 | 4.11e-02 | 0.353 | 3.88e-03 | |
| COX2 (PTGS2) | 0.158 | 2.04e-01 | 0.154 | 2.20e-01 | ||
| INOS (NOS2) | −0.074 | 5.53e-01 | −0.026 | 8.36e-01 | ||
| M2 | ARG1 | −0.037 | 7.69e-01 | −0.016 | 9.01e-01 | |
| CD16 (FCGR3B) | 0.169 | 1.75e-01 | 0.255 | 4.08e-02 | ||
| MRC1 | 0.094 | 4.52e-01 | 0.165 | 1.90e-01 | ||
| MS4A4A | 0.215 | 8.33e-02 | 0.371 | 2.34e-03 | ||
| Neutrophil | CD11b (ITGAM) | 0.166 | 1.83e-01 | 0.317 | 1.01e-02 | |
| CD15 (FUT4) | 0.305 | 1.32e-02 | 0.340 | 5.61e-03 | ||
| CD66b (CEACAM8) | −0.023 | 8.55e-01 | −0.023 | 8.56e-01 | ||
| Natural killer cell | CD7 | 0.137 | 2.73e-01 | 0.199 | 1.12e-01 | |
| XCL1 | 0.139 | 2.67e-01 | 0.196 | 1.17e-01 | ||
| KIR3DL1 | 0.039 | 7.58e-01 | 0.062 | 6.22e-01 | ||
| Dendritic cell | CD141(THBD) | −0.020 | 8.72e-01 | 0.028 | 8.27e-01 | |
| CD11c (ITGAX) | 0.210 | 9.10e-02 | 0.320 | 9.25e-03 | ||
| CD1C | 0.071 | 5.71e-01 | 0.168 | 1.81e-01 | ||
| Monocyte | CD14 | 0.119 | 3.40e-01 | 0.225 | 7.13e-02 | |
| CD16 (FCGR3B) | 0.169 | 1.75e-01 | 0.255 | 4.08e-02 | ||
| CD115 | 0.183 | 1.40e-01 | 0.338 | 5.91e-03 | ||
KICH, kidney chromophobe cell carcinoma; TIMER, Tumor Immune Estimation Resource.
Table 2. Correlation analysis between SLC7A11 and markers of immune cells for KIRC in TIMER.
| Description | Gene markers | SLC7A11 | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| r | P | r | P | |||
| B cell | CD27 | −0.024 | 5.8e-01 | −0.080 | 8.65e-02 | |
| CD19 | 0.101 | 1.94e-02 | 0.076 | 1.01e-01 | ||
| CD38 | 0.213 | 6.74e-07 | 0.181 | 9.31e-05 | ||
| CD8+ T cell | CD8A | 0 | 9.96e-01 | −0.047 | 3.11e-01 | |
| CD8B | −0.047 | 2.84 e-01 | −0.099 | 3.32e-02 | ||
| T cell (general) | CD3D | 0.004 | 9.27e-01 | −0.053 | 2.52e-01 | |
| CD3E | 0.029 | 5.11e-01 | −0.025 | 5.85e-01 | ||
| CD2 | 0.054 | 2.16e-01 | −0.359 | 9.84e-01 | ||
| Monocyte | CD14 | 0.222 | 2.21e-07 | 0.184 | 7.34e-05 | |
| CD115 | 0.219 | 3.33e-07 | 0.177 | 1.34e-04 | ||
| Tfh | BCL6 | 0.176 | 4.23e-05 | 0.216 | 2.97e-06 | |
| ICOS | 0.135 | 1.73e-03 | 0.112 | 1.59e-02 | ||
| CXCR5 | 0.177 | 3.82e-05 | 0.154 | 9.31e-04 | ||
| Th1 | T-bet (TBX21) | 0.027 | 5.26e-01 | 0.011 | 8.18e-01 | |
| STAT1 | 0.227 | 1.22e-07 | 0.197 | 2.00e-05 | ||
| STAT4 | 0.194 | 6.25e-06 | 0.192 | 3.26e-05 | ||
| IL12RB2 | 0.118 | 6.43e-03 | 0.127 | 6.16e-03 | ||
| WSX1 (IL27RA) | 0.256 | 1.96e-09 | 0.262 | 1.07e-08 | ||
| IFN-γ (IFNG) | 0.024 | 5.79e-01 | −0.009 | 8.42e-01 | ||
| TNF-α (TNF) | 0.066 | 1.26e-01 | 0.047 | 3.10e-01 | ||
| Th2 | STAT6 | 0.047 | 2.75e-01 | 0.087 | 6.21e-02 | |
| GATA3 | 0.03 | 4.83e-01 | −0.006 | 8.92e-01 | ||
| STAT5A | 0.111 | 1.03e-02 | 0.071 | 1.27e-01 | ||
| CCR3 | 0.156 | 3.07e-04 | 0.129 | 5.43e-03 | ||
| Th9 | TGFBR2 | 0.21 | 1.02e-06 | 0.213 | 4.09e-06 | |
| PU.1 (SPI1) | 0.128 | 3.11e-03 | 0.075 | 1.09e-01 | ||
| IRF4 | 0.208 | 1.32e-06 | 0.192 | 3.34e-05 | ||
| Th17 | IL-17A (IL17A) | 0.083 | 5.42e-02 | 0.11 | 1.82e-02 | |
| IL-21R (IL21R) | 0.245 | 9.43e-09 | 0.186 | 5.71e-05 | ||
| IL-23R (IL23R) | 0.202 | 2.65e-06 | 0.202 | 1.27e-05 | ||
| STAT3 | 0.215 | 5.41e-07 | 0.197 | 2.09e-05 | ||
| Th22 | AHR | 0.242 | 1.49e-08 | 0.261 | 1.25e-08 | |
| CCR10 | −0.002 | 9.62e-01 | −0.031 | 5.11e-01 | ||
| Treg | CD25 (IL2RA) | 0.404 | 2.31e-22 | 0.383 | 1.37e-17 | |
| FOXP3 | 0.182 | 2.31e-05 | 0.164 | 4.18e-04 | ||
| CCR8 | 0.271 | 1.95e-10 | 0.271 | 3.45e-09 | ||
| T cell exhaustion | CTLA4 | 0.093 | 3.16e-02 | 0.074 | 1.12e-01 | |
| PD-1 (PDCD1) | −0.028 | 5.13e-01 | −0.063 | 1.76e-01 | ||
| TIM-3 (HAVCR2) | −0.023 | 5.96e-01 | −0.045 | 3.36e-01 | ||
| LAG3 | −0.007 | 8.75e-01 | −0.055 | 2.43e-01 | ||
| TAM | CD80 | 0.256 | 2.14e-09 | 0.238 | 2.21e-07 | |
| CD86 | 0.234 | 4.68e-08 | 0.202 | 1.27e-05 | ||
| CCL2 | 0.012 | 7.82e-01 | 0.029 | 5.35e-01 | ||
| CCR5 | 0.13 | 2.65e-03 | 0.089 | 5.71e-02 | ||
| Macrophage | CD68 | 0.216 | 4.62e-07 | 0.217 | 2.57e-06 | |
| CD11b (ITGAM) | 0.235 | 4.06e-08 | 0.209 | 5.76e-06 | ||
| M1 | IRF5 | −0.037 | 3.98e-01 | −0.028 | 5.44e-01 | |
| COX2(PTGS2) | 0.36 | 8.93e-18 | 0.353 | 5.39e-15 | ||
| INOS (NOS2) | −0.037 | 3.94e-01 | −0.063 | 1.79e-01 | ||
| M2 | ARG1 | 0.014 | 7.43e-01 | 0.021 | 6.51e-01 | |
| CD16 (FCGR3A) | 0.192 | 7.83e-06 | 0.156 | 7.78e-04 | ||
| MRC1 | 0.328 | 7.29e-15 | 0.322 | 1.46e-12 | ||
| MS4A4A | 0.315 | 1.01e-13 | 0.294 | 1.25e-10 | ||
| Neutrophil | CD11b (ITGAM) | 0.235 | 4.06e-08 | 0.209 | 5.76e-06 | |
| CD15 (FUT4) | 0.239 | 2.37e-08 | 0.231 | 5.17e-07 | ||
| CD66b (CEACAM8) | 0.036 | 4.1e-01 | 0.049 | 2.89e-01 | ||
| Natural killer cell | CD7 | −0.026 | 5.43e-01 | −0.086 | 6.37e-02 | |
| XCL1 | 0.014 | 7.5e-01 | −0.033 | 4.76e-01 | ||
| KIR3DL1 | −0.007 | 8.69e-01 | −0.008 | 8.62e-01 | ||
| Dendritic cell | CD141 (THBD) | 0.216 | 4.81e-07 | 0.226 | 9.48e-07 | |
| CD11c (ITGAX) | 0.168 | 1.01e-04 | 0.16 | 5.72e-04 | ||
| CD1C | 0.107 | 1.37e-02 | 0.069 | 1.39e-01 | ||
| Monocyte | CD14 | 0.222 | 2.21e-07 | 0.184 | 7.34e-05 | |
| CD16 (FCGR3B) | 0.178 | 3.62e-05 | 0.197 | 2.01e-05 | ||
| CD115 | 0.219 | 3.33e-07 | 0.177 | 1.34e-04 | ||
KIRC, kidney renal clear cell carcinoma; TIMER, Tumor Immune Estimation Resource.
Table 3. Correlation analysis between SLC7A11 and markers of immune cells for KIRP in TIMER.
| Description | Gene markers | SLC7A11 | ||||
|---|---|---|---|---|---|---|
| None | Purity | |||||
| r | P | r | P | |||
| B cell | CD27 | 0.151 | 9.85e-03 | 0.204 | 9.89e-04 | |
| CD19 | 0.174 | 2.98e-03 | 0.207 | 8.37e-04 | ||
| CD38 | 0.319 | 2.69e-08 | 0.359 | 2.93e-09 | ||
| CD8+ T cell | CD8A | 0.163 | 5.43e-03 | 0.197 | 1.43e-03 | |
| CD8B | 0.166 | 4.69e-03 | 0.205 | 9.19e-04 | ||
| T cell (general) | CD3D | 0.115 | 5.11e-02 | 0.15 | 1.60e-02 | |
| CD3E | 0.100 | 8.77e-02 | 0.14 | 2.44e-02 | ||
| CD2 | 0.082 | 1.64e-01 | 0.122 | 5.07e-02 | ||
| Monocyte | CD14 | 0.149 | 1.13e-02 | 0.174 | 5.17e-03 | |
| CD115 | 0.139 | 1.79e-02 | 0.172 | 5.53e-03 | ||
| Tfh | BCL6 | −0.007 | 9.11e-01 | −0.014 | 8.21e-01 | |
| ICOS | 0.152 | 9.48e-03 | 0.193 | 1.87e-03 | ||
| CXCR5 | 0.206 | 4.24e-04 | 0.231 | 1.79e-04 | ||
| Th1 | T-bet (TBX21) | 0.003 | 9.6e-01 | 0.031 | 6.22e-01 | |
| STAT1 | 0.151 | 1.01e-02 | 0.144 | 2.07e-02 | ||
| STAT4 | 0.164 | 5.1e-03 | 0.204 | 9.98e-04 | ||
| IL12RB2 | −0.281 | 1.16e-06 | −0.292 | 1.75e-06 | ||
| WSX1 (IL27RA) | 0.058 | 3.22e-01 | 0.082 | 1.89e-01 | ||
| IFN-γ (IFNG) | 0.144 | 1.44e-02 | 0.164 | 8.26e-03 | ||
| TNF-α (TNF) | −0.02 | 7.36e-01 | −0.018 | 7.79e-01 | ||
| Th2 | STAT6 | −0.225 | 1.1e-04 | −0.243 | 8.12e-05 | |
| GATA3 | 0.225 | 1.14e-04 | 0.23 | 2.00e-04 | ||
| STAT5A | −0.063 | 2.87e-01 | −0.046 | 4.60e-01 | ||
| CCR3 | 0.091 | 1.22e-01 | 0.087 | 1.65e-01 | ||
| Th9 | TGFBR2 | 0.02 | 7.36e-01 | 0.014 | 8.20e-01 | |
| PU.1 (SPI1) | 0.084 | 1.56e-01 | 0.096 | 1.24e-01 | ||
| IRF4 | 0.23 | 7.48e-05 | 0.261 | 2.19e-05 | ||
| Th17 | IL-17A (IL17A) | 0.026 | 6.65e-01 | 0 | 9.97e-01 | |
| IL-21R (IL21R) | 0.339 | 2.98e-09 | 0.372 | 6.94e-10 | ||
| IL-23R (IL23R) | 0.045 | 4.5e-01 | 0.011 | 8.61e-01 | ||
| STAT3 | 0.065 | 2.68e-01 | 0.053 | 3.97e-01 | ||
| Th22 | AHR | −0.25 | 1.71e-05 | −0.29 | 2.15e-06 | |
| CCR10 | −0.008 | 8.88e-01 | −0.055 | 3.83e-01 | ||
| Treg | CD25 (IL2RA) | 0.336 | 4.17e-09 | 0.359 | 2.87e-09 | |
| FOXP3 | 0.28 | 1.32e-06 | 0.281 | 4.44e-06 | ||
| CCR8 | 0.334 | 5.51e-09 | 0.349 | 8.56e-09 | ||
| T cell exhaustion | CTLA4 | 0.129 | 2.76e-02 | 0.148 | 1.77e-02 | |
| PD-1 (PDCD1) | 0.032 | 5.84e-01 | 0.046 | 4.61e-01 | ||
| TIM-3 (HAVCR2) | 0.055 | 3.48e-01 | 0.052 | 4.07e-01 | ||
| LAG3 | 0.181 | 1.94e-03 | 0.202 | 1.13e-03 | ||
| TAM | CD80 | 0.329 | 9.74e-09 | 0.353 | 5.32e-09 | |
| CD86 | 0.144 | 1.38e-02 | 0.17 | 6.12e-03 | ||
| CCL2 | 0.155 | 8.04e-03 | 0.159 | 1.07e-02 | ||
| CCR5 | 0.179 | 2.19e-03 | 0.224 | 2.94e-04 | ||
| Macrophage | CD68 | 0.031 | 5.97e-01 | 0.036 | 5.63e-01 | |
| CD11b (ITGAM) | 0.161 | 5.85e-03 | 0.157 | 1.18e-02 | ||
| M1 | IRF5 | −0.138 | 1.88e-02 | −0.144 | 2.05e-02 | |
| COX2 (PTGS2) | 0.164 | 5.12e-03 | 0.167 | 7.32e-03 | ||
| INOS (NOS2) | −0.023 | 7.02e-01 | −0.001 | 9.85e-01 | ||
| M2 | ARG1 | −0.048 | 4.16e-01 | −0.05 | 4.26e-01 | |
| CD16 (FCGR3B) | 0.258 | 8.91e-06 | 0.295 | 1.37e-06 | ||
| MRC1 | 0.321 | 2.36e-08 | 0.325 | 9.34e-08 | ||
| MS4A4A | 0.182 | 1.9e-03 | 0.21 | 6.90e-04 | ||
| Neutrophil | CD11b (ITGAM) | 0.161 | 5.85e-03 | 0.157 | 1.18e-02 | |
| CD15 (FUT4) | −0.056 | 3.43e-01 | −0.076 | 2.23e-01 | ||
| CD66b (CEACAM8) | −0.01 | 8.71e-01 | 0.034 | 5.87e-01 | ||
| Natural killer cell | CD7 | 0.134 | 2.24e-02 | 0.167 | 7.01e-03 | |
| XCL1 | 0.195 | 8.58e-04 | 0.235 | 1.35e-04 | ||
| KIR3DL1 | 0.139 | 1.78e-02 | 0.164 | 8.35e-03 | ||
| Dendritic cell | CD141 (THBD) | −0.016 | 7.86e-01 | −0.062 | 3.18e-01 | |
| CD11c (ITGAX) | 0.159 | 6.68e-03 | 0.165 | 8.01e-03 | ||
| CD1C | 0.089 | 1.3e-01 | 0.085 | 1.76e-01 | ||
| Monocyte | CD14 | 0.149 | 1.13e-02 | 0.174 | 5.17e-03 | |
| CD16 (FCGR3B) | 0.048 | 4.19e-01 | 0.06 | 3.37e-01 | ||
| CD115 | 0.139 | 1.79e-02 | 0.172 | 5.53e-03 | ||
KIRP, kidney renal papillary cell carcinoma; TIMER, Tumor Immune Estimation Resource.
Discussion
It has been reported that KIRC is highly susceptible to reduced glutathione, which leads to the accumulation of reactive oxygen species and ferroptosis activation (18-20). Ferroptosis is a type of regulated cell death that is triggered by iron-dependent excessive lipid peroxidation (5). The overexpression of SLC7A11 is common in a variety of human malignancies and plays a role in importing cysteine for glutathione production and antioxidant defense (6). A recent study reported that SLC7A11 overexpression enhances cancer progression and induces resistance to chemoradiotherapy partly through suppressing ferroptosis (21). Previous research has also reported that the differential expression of SLC7A11 in KIRC is involved in the onset and progression of RCC (7). In this study, we further investigated the prognostic value of SLC7A11 and the potential ceRNA regulatory network in 3 types of RCC. By differential gene expression analysis, we found upregulation of SLC7A11 in most human cancers, including RCC. High expression of SLC7A11 was linked to a higher risk of KIRC, KICH, and KIRP. Moreover, the Kaplan-Meier survival plot strongly illustrated a significantly shorter OS and DFS in patients with KIRC and elevated expression level of SLC7A11 compared with those with lower expression, with similar conclusions for the 3 types of RCCs. We further compared SLC7A11 expression in KIRC, KICH, and KIRP samples with the clinical characteristics (different grades and stages). The results showed that upregulation of SLC7A11 is associated with higher malignancy or later stages of KIRC. Similar results were also shown in KIRP. However, no correlation was found between SLC7A11 expression and the clinical characteristics of patients with KICH, which may be a result of the smaller number of samples.
miRNAs belong to the small ncRNA family constituting 20–25 nucleotides. They attach to the target 3' untranslated region (3'-UTR) of mRNAs and initiate gene silencing by inhibiting translation and increasing mRNA degradation (22). Amassed evidence reveals that SLC7A11 mRNA is derepressed by reduced miR-26b in human breast malignancy, miR-27a in bladder malignancy, and miR-375 in oral squamous cell carcinoma (7). Bioinformatics has shown that SLC7A11 is targeted by various miRNAs, such as has-mir-373 and has-mir-372 related to B cell infiltration in lung adenocarcinoma (23), miR-374b-5p and miR-26b-5p in collecting duct carcinoma (24), and miRNA-126-3p/5p in lung adenocarcinoma (25). By using various target gene prediction programs, we inferred the upstream miRNAs of SLC7A11 to be miR-26a-5p for KICH and miR-27b-3p for KIRC and KIRP. Upon conducting expression analysis and survival analysis for these miRNAs, miR-26a-5p and miR-27b-3p were identified in RCC as potential upstream miRNAs. Numerous studies have shown that decreased expression of miR-26a-5p and miR-27b-3p is linked to the occurrence and progression of RCC (26-28). The ceRNA hypothesis has suggested a novel mechanism for interactions of ncRNAs and mRNAs, where lncRNAs, miRNAs, and mRNAs are involved (29). The miRNA can bind to the 3'-UTRs of RNAs to suppress the translation of target genes in regulatory ceRNA networks (30). By sharing miRNA response elements with reverse complementary binding seed regions, lncRNAs can compete for miRNA binding and indirectly impact translational control and mRNA stabilization (31). Accumulated research has demonstrated that the ceRNA gene interaction network plays a crucial function in tumorigenesis, progression, and prognosis (32). Consequently, we further investigated the binding lncRNAs of miR-26a-5p and miR-27b-3p using starBase and miRNet. By a cascade of in silico analyses, including survival analysis, expression analysis, and correlation analysis, we identified 1 negatively correlated lncRNA–miRNA pair (SNHG6/miR-26a-5p), 3 lncRNA–miRNA pairs (CASC19, CYTOR, LINC00997/miR-27b-3p) whose expression was negatively correlated to KIRC, and 3 lncRNA–miRNA pairs (CASC19, CYTOR, PVT1/miR-27b-3p) whose expression was negatively correlated to KIRP (Figure 9D). In addition, a few of the lncRNAs recorded in these lncRNA–miRNA pairs have previously been linked to the tumorigenesis of RCC. For instance, An et al. confirmed that elevated SNHG6 expression was associated with poor OS in RCC (33). Moreover, CASC19 was found to be overexpressed in KIRC tissues in vivo and in vitro, while the high expression of CASC19 was closely linked to unfavorable clinicopathological parameters and poor clinical outcomes among individuals with KIRC (34). CYTOR has been shown to play a vital part in the carcinogenesis of several forms of malignancy in previous research, but its role in RCC has not been established (35,36). Furthermore, it was discovered that overexpression of LINC00997 in KIRC is linked to poor prognosis, which plays an essential function in enhancing KIRC metastasis by combining with STAT3, thus elevating the S100A11 expression level (37). Finally, it has also been reported that PVT1 was overexpressed in KIRC and KIRP, and the upregulated PVT1 was negatively correlated with OS (38).
It has been established that RCC is among the major immune-infiltrating cancers, and the clinical use of PD-1/PD-L1 antibody has been accepted as front-line management of metastatic RCC (39). Recently, the link between immune response and tumor prognosis has received increasing research attention (4,40). However, the objective response rate is still less than 30%. The response to immunotherapy can be predicted using lymphocyte infiltration as a marker (41). Furthermore, the presence of intratumoral tertiary lymphoid structures has been linked to positive clinical outcomes and immunotherapy responses in cancer. In our study, TIMER was used to evaluate the link between SLC7A11 and 6 immune-infiltrating cells in RCC. Our results indicated that in KICH, SLC7A11 expression was linked to the infiltration of B cells and myeloid DCs. In KIRC, SLC7A11 expression was significantly linked to the infiltration of CD4+ T cells, macrophages, neutrophils, and myeloid DCs. Finally, in KIRP, our results demonstrated that SLC7A11 expression was linked to the infiltration of B cells, CD8+ T cells, and myeloid DCs. These findings revealed that SLC7A11 might play a vital role in the progression of RCCs by controlling the ferroptosis of a variety of immune cells. These findings are in agreement with those of another study (40). Further investigation of infiltrated lymphocyte markers demonstrated that the levels of the majority of the immune marker sets, including T cells, B cells, Th17, Th22, Treg, macrophages, and TAMs, were linked to SLC7A11 expression in KICH. SLC7A11 expression also showed a positive correlation with the infiltrating levels of CD4+ T cells, macrophages, neutrophils, M1/M2 macrophages, and DCs in KIRC. Lastly, the levels of most immune marker sets, including B cells, different T cells, TAMs, M2 macrophages, and natural killer cells were linked to SLC7A11 expression in KIRP.
The tumor microenvironment (TME) of RCCs and that of other tumors types are different. Furthermore, the impact of tumor-infiltrating CD8+ T cells is remarkably different between tumor types. In a great number of tumors, the elevation in CD8+ density is linked to a better prognosis (42), whereas previous data suggested that in RCC, elevated CD8+ T cell density is linked to a worse outcome (43,44). KICH had lower CD8+ T cell frequencies relating to immune cell infiltration than did KIRC and KIRP. A high tumor grade and shorter survival in patients have been linked to an abundance of intratumoral CD8+ and CD4+ T cells in RCC (41). The tumor-suppressive roles of B-lymphocytes have been reported by reputable studies, which have noted a correlation between B cells and prolonged survival in various cancers (45). On the contrary, another study found B cells to be linked to reduced OS in RCC, ovarian cancer, and primary cutaneous melanoma (46). DCs are important in triggering antitumor immune responses and in maintaining immune homeostasis (47). Normal kidney tissue is infiltrated by a large population of highly diverse DCs, demonstrating active immune monitoring within the renal parenchyma (48). In RCC, a buildup of tolerogenic DCs within the TME has been linked to poor outcomes (49) and probably promotes the overall immunosuppressed state of the RCC TME. The RCC parenchyma is also colonized by TAMs, which were first described as having an M2 phenotype, that plays a role in the secretion of pro-inflammatory cytokines related to immune modulation and wound healing (50,51), whereas an M1 phenotype produces cytokines that enhance TH1 responses. RCC TAMs, on the other hand, have been shown to have a mixed M1 and M2 phenotype and are one of the primary sources of vascular endothelial growth factor (VEGF) and proinflammatory cytokines, including interleukin 6 (IL-6), tumor necrosis factor (TNF), and IL-1β inside the TME (50). Myeloid-derived suppressor cells play a significant role in tumor growth as well. Patients with cancer, particularly KIRC, experience high levels of circulating myeloid-derived suppressor cells which rise with disease stage and decrease with tumor regression (41). According to the majority of research, high amounts of neutrophils are linked to a poorer prognosis in a variety of malignancies, including RCC. The link between neutrophils and patient prognosis is less obvious in other malignancies; however, studies have found that neutrophils are linked to a better outcome (52). Taken together, our findings suggested that SLC7A11 is a crucial molecule bridging the gap between ferroptosis and immunotherapy. A recent study has shown the vital role of ferroptosis in immunotherapy based on the interaction of ferroptosis with tumor immunotherapy, chemotherapy and radiotherapy (53). Therefore, our results suggest that inhibitors targeting SLC7A11 may enhance the effect of cancer treatment in RCC.
Conclusions
We conducted a systemic evaluation of SLC7A11 expression and its prognostic value in RCC. Our findings revealed that SLC7A11 overexpression is linked to favorable survival and has prognostic value in patients with KICH, KIRC, or KIRP. As a result, our research not only elucidated a crucial function of SLC7A11 in RCC but also partially described the molecular mechanism of SLC7A11 in RCC. Ferroptosis may be a potential modality for developing potent combinational therapy approaches in the advanced stage of anti-cancer therapy. Meanwhile, SLC7A11 expression could be a potential new factor for the stratification of patients with RCC in guiding ferroptosis and immunotherapy. These findings, however, need to be experimentally validated in future studies.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank Bullet Edits (www.bulletedits.cn) for editing and proofreading this manuscript.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tau.amegroups.com/article/view/10.21037/tau-22-663/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tau.amegroups.com/article/view/10.21037/tau-22-663/coif). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol 2015;67:85-97. 10.1016/j.eururo.2014.04.029 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 4.Chen S, Su X, Mi H, et al. Comprehensive analysis of glutathione peroxidase-1 (GPX1) expression and prognostic value in three different types of renal cell carcinoma. Transl Androl Urol 2020;9:2737-50. 10.21037/tau-20-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 2012;149:1060-72. 10.1016/j.cell.2012.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021;12:599-620. 10.1007/s13238-020-00789-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin W, Wang C, Liu G, et al. SLC7A11/xCT in cancer: biological functions and therapeutic implications. Am J Cancer Res 2020;10:3106-26. [PMC free article] [PubMed] [Google Scholar]
- 8.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21:22-36. 10.1038/s41568-020-00306-0 [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569:270-4. 10.1038/s41586-019-1170-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang X, Green MD, Wang W, et al. Radiotherapy and Immunotherapy Promote Tumoral Lipid Oxidation and Ferroptosis via Synergistic Repression of SLC7A11. Cancer Discov 2019;9:1673-85. 10.1158/2159-8290.CD-19-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. 10.1593/neo.07112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. 10.1093/nar/gkx1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu TP, Lee CY, Tsai MH, et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One 2012;7:e42390. 10.1371/journal.pone.0042390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang L, Zhou G, Soufan O, et al. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res 2020;48:W244-51. 10.1093/nar/gkaa467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miess H, Dankworth B, Gouw AM, et al. The glutathione redox system is essential to prevent ferroptosis caused by impaired lipid metabolism in clear cell renal cell carcinoma. Oncogene 2018;37:5435-50. 10.1038/s41388-018-0315-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou Y, Palte MJ, Deik AA, et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun 2019;10:1617. 10.1038/s41467-019-09277-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou Y, Henry WS, Ricq EL, et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 2020;585:603-8. 10.1038/s41586-020-2732-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong Y, Xiao C, Li Z, et al. Engineering nanomedicine for glutathione depletion-augmented cancer therapy. Chem Soc Rev 2021;50:6013-41. 10.1039/D0CS00718H [DOI] [PubMed] [Google Scholar]
- 22.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol 2019;20:21-37. 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei B, Kong W, Mou X, et al. Comprehensive analysis of tumor immune infiltration associated with endogenous competitive RNA networks in lung adenocarcinoma. Pathol Res Pract 2019;215:159-70. 10.1016/j.prp.2018.10.032 [DOI] [PubMed] [Google Scholar]
- 24.Wach S, Taubert H, Weigelt K, et al. RNA Sequencing of Collecting Duct Renal Cell Carcinoma Suggests an Interaction between miRNA and Target Genes and a Predominance of Deregulated Solute Carrier Genes. Cancers (Basel) 2019;12:64. 10.3390/cancers12010064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P, Gu YY, Ma FC, et al. Expression levels and cotargets of miRNA1263p and miRNA1265p in lung adenocarcinoma tissues: Alphan exploration with RTqPCR, microarray and bioinformatic analyses. Oncol Rep 2019;41:939-53. 10.1007/s12094-018-02012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng C, Guo L, Ma Y, et al. Up-Regulation of miR-26a-5p Inhibits E2F7 to Regulate the Progression of Renal Carcinoma Cells. Cancer Manag Res 2020;12:11723-33. 10.2147/CMAR.S271710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedrich M, Heimer N, Stoehr C, et al. CREB1 is affected by the microRNAs miR-22-3p, miR-26a-5p, miR-27a-3p, and miR-221-3p and correlates with adverse clinicopathological features in renal cell carcinoma. Sci Rep 2020;10:6499. 10.1038/s41598-020-63403-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu L, Li H, Chen L, et al. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget 2015;6:32545-60. 10.18632/oncotarget.5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 2007;27:91-105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Z, Pan X, Yang Y, et al. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 2018;34:2185-94. 10.1093/bioinformatics/bty085 [DOI] [PubMed] [Google Scholar]
- 32.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 2016;17:272-83. 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 33.An HX, Xu B, Wang Q, et al. Up-regulation of long non-coding RNA SNHG6 predicts poor prognosis in renal cell carcinoma. Eur Rev Med Pharmacol Sci 2018;22:8624-9. [DOI] [PubMed] [Google Scholar]
- 34.Luo Y, Liu F, Yan C, et al. Long Non-Coding RNA CASC19 Sponges microRNA-532 and Promotes Oncogenicity of Clear Cell Renal Cell Carcinoma by Increasing ETS1 Expression. Cancer Manag Res 2020;12:2195-207. 10.2147/CMAR.S242472 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Tian Q, Yan X, Yang L, et al. lncRNA CYTOR promotes cell proliferation and tumor growth via miR-125b/SEMA4C axis in hepatocellular carcinoma. Oncol Lett 2021;22:796. 10.3892/ol.2021.13057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang G, Yu H, Li Z, et al. lncRNA cytoskeleton regulator reduces nonsmall cell lung cancer radiosensitivity by downregulating miRNA206 and activating prothymosin alpha. Int J Oncol 2021;59:88. 10.3892/ijo.2021.5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang Y, Li N, Yuan W, et al. LINC00997, a novel long noncoding RNA, contributes to metastasis via regulation of S100A11 in kidney renal clear cell carcinoma. Int J Biochem Cell Biol 2019;116:105590. 10.1016/j.biocel.2019.105590 [DOI] [PubMed] [Google Scholar]
- 38.Yan YJ, Zhang L, Zhou JJ, et al. Comprehensive Characterization of Common and Cancer-Specific Differently Expressed lncRNAs in Urologic Cancers. Comput Math Methods Med 2021;2021:5515218. 10.1155/2021/5515218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motzer RJ, Escudier B, McDermott DF, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 2015;373:1803-13. 10.1056/NEJMoa1510665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi ZZ, Tao H, Fan ZW, et al. Prognostic and Immunological Role of Key Genes of Ferroptosis in Pan-Cancer. Front Cell Dev Biol 2021;9:748925. 10.3389/fcell.2021.748925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Díaz-Montero CM, Rini BI, Finke JH. The immunology of renal cell carcinoma. Nat Rev Nephrol 2020;16:721-35. 10.1038/s41581-020-0316-3 [DOI] [PubMed] [Google Scholar]
- 42.Fridman WH, Pages F, Sautes-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298-306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 43.Nakano O, Sato M, Naito Y, et al. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res 2001;61:5132-6. [PubMed] [Google Scholar]
- 44.Giraldo NA, Becht E, Pages F, et al. Orchestration and Prognostic Significance of Immune Checkpoints in the Microenvironment of Primary and Metastatic Renal Cell Cancer. Clin Cancer Res 2015;21:3031-40. 10.1158/1078-0432.CCR-14-2926 [DOI] [PubMed] [Google Scholar]
- 45.Kim SS, Sumner WA, Miyauchi S, et al. Role of B Cells in Responses to Checkpoint Blockade Immunotherapy and Overall Survival of Cancer Patients. Clin Cancer Res 2021;27:6075-82. 10.1158/1078-0432.CCR-21-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sjöberg E, Frodin M, Lovrot J, et al. A minority-group of renal cell cancer patients with high infiltration of CD20+B-cells is associated with poor prognosis. Br J Cancer 2018;119:840-6. 10.1038/s41416-018-0266-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Nat Rev Cancer 2016;16:447-62. 10.1038/nrc.2016.54 [DOI] [PubMed] [Google Scholar]
- 48.Soos TJ, Sims TN, Barisoni L, et al. CX3CR1+ interstitial dendritic cells form a contiguous network throughout the entire kidney. Kidney Int 2006;70:591-6. 10.1038/sj.ki.5001567 [DOI] [PubMed] [Google Scholar]
- 49.Toma M, Wehner R, Kloss A, et al. Accumulation of tolerogenic human 6-sulfo LacNAc dendritic cells in renal cell carcinoma is associated with poor prognosis. Oncoimmunology 2015;4:e1008342. 10.1080/2162402X.2015.1008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamada I, Kato M, Yamasaki T, et al. Clinical effects of tumor-associated macrophages and dendritic cells on renal cell carcinoma. Anticancer Res 2002;22:4281-4. [PubMed] [Google Scholar]
- 51.Toge H, Inagaki T, Kojimoto Y, et al. Angiogenesis in renal cell carcinoma: the role of tumor-associated macrophages. Int J Urol 2009;16:801-7. 10.1111/j.1442-2042.2009.02377.x [DOI] [PubMed] [Google Scholar]
- 52.Shaul ME, Fridlender ZG. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol 2019;16:601-20. 10.1038/s41571-019-0222-4 [DOI] [PubMed] [Google Scholar]
- 53.Zhao L, Zhou X, Xie F, et al. Ferroptosis in cancer and cancer immunotherapy. Cancer Commun (Lond) 2022;42:88-116. 10.1002/cac2.12250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as