Key Points
-
1.
Platelet factor 4 (PF4) forms complexes with constituents of ChAdOx1‐S. Identification of the binding partners of PF4 is important for improving vaccine safety.
-
2.
No sulfated glycosaminoglycans are detectable in ChAdOx1‐S.
Alt-text: Unlabelled Box
1. INTRODUCTION
Vaccination against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is one of the most important measures to fight the coronavirus 2019 (COVID‐19) pandemic. One of the vaccines, ChAdOx1‐S (AstraZeneca), is a recombinant, replication‐deficient chimpanzee adenovirus encoding the SARS‐CoV‐2 spike glycoprotein. The vector is propagated in T‐REx‐293 human embryonic kidney (HEK) cells, a genetically modified derivative of the HEK 293 cell line.1 Hundreds of cases of unusual thrombotic events in combination with thrombocytopenia after vaccination with ChAdOx1‐S have been observed.2., 3., 4. This so‐called vaccine‐induced immune thrombotic thrombocytopenia (VITT; also known as thrombotic thrombocytopenia syndrome [TTS]) resembles a subtype of autoimmune heparin‐induced thrombocytopenia (aHIT) known as “spontaneous HIT syndrome.”5 Its pathogenesis remains unclear and many potential mechanisms have been discussed.6 It is unequivocally accepted that VITT is associated with high titers of IgG class antibodies directed against the cationic platelet chemokine, platelet factor 4 (PF4; CXCL4).7 These antibodies potently activate platelets via platelet FcγIIa receptors with platelet activation greatly enhanced by PF4.3., 8.
PF4 is a positively charged, compact homotetrameric globular protein, which binds to a wide array of polyanions, especially sulfated glycosaminoglycans (GAG) like heparins, heparan sulfates, and chondroitin sulfates, and other polyanions like polyphosphates and nucleic acids.9., 10., 11., 12. Recently, we have shown that constituents of the vaccine form complexes with PF4.7 These complexes contain adenoviral hexon proteins as shown by staining with a monoclonal anti‐hexon antibody. However, it is unresolved whether additional molecules are involved in PF4 complex formation. Potential binding partners are sulfated GAG13 from the T‐REx‐293 HEK cells. It is technically challenging to detect low concentrations of GAGs in a complex matrix such as a vaccine, which contains many proteins. We report the experimental approach by which we excluded relevant amounts of contaminating GAG in the vaccine.
2. METHODS
2.1. Anticoagulant activity
The anticoagulant activity of ChAdOx1‐S was determined by mixing 40 µl vaccine with 400 µl citrated pool human plasma and measuring the activated thromboplastin time (APTT; Actin FS, Siemens) and the anti‐factor Xa (aFXa) activity (Coamatic Heparin, Chromogenix) on a CS5100 (Siemens Healthcare).
2.2. ChAdOx1‐S vaccine sample preparation for 1H nuclear magnetic resonance spectroscopy
By collecting the residues of three batches of multiple‐dose vials of the ChAdOx1‐S vaccine, a sample of 30 ml was obtained. First, we incubated the vaccine with 0.4 mg/ml proteinase K (recombinant, PCR grade; Thermo Fisher Scientific No. EO0491) in the presence of 2 mM Ca2+ and 4 M urea at 37°C for 2 h to digest the proteins in the vaccine. The enzymatically digested vaccine was dialyzed (MW cut‐off 1 kDa) for 72 h to remove the digested small peptides and additives and then lyophilized. To remove the proteinase K, the dried sample was fractionated by strong‐anion exchange chromatography (SAX) at pH 8.9 corresponding to the isolectric point of the enzyme. The basic pH resulted in loss of charge of (the otherwise positively charged) proteinase K. The sample was dissolved in 1 ml of ammonium acetate buffer (0.05 M, pH 8.9; start buffer) and applied to a Mono Q™ 5/50 SAX column (Cytiva Europe GmbH) equilibrated with the start buffer and eluted with a linear NaCl gradient at a flow rate of 2.0 ml/min. After running the start buffer for 15 min (including 1 min for Vdead) to elute the unbound proteinase K, the NaCl concentration was continuously increased by 0.075 mol/L per minute until 3.0 mol/L NaCl was reached after 40 min. By preliminary experiments with proteinase K and chondroitin sulfate (Carl Roth, Cat. No. 5197.1) as exemplary low‐sulfated GAG, we determined the elution fractions containing protein or GAG, respectively. After fractionation of the vaccine, those fractions that according to the pilot experiments with spiked vaccine potentially contained GAG were pooled, dialyzed (MW cut‐off 1 kDa), concentrated by evaporation under reduced pressure, and lyophilized in the vial used for the nuclear magnetic resonsonance (NMR) analysis (to avoid any loss of GAG). After dissolving in 500 µl D2O, the sample was analyzed by NMR.
2.3. 1H NMR spectroscopy
The 1H NMR spectra were recorded at 300 K on a Fourier transform‐NMR (FT‐NMR) spectrometer Avance II by Bruker BioSpin AG. The instrument operates at a frequency of 600.13 MHz and is equipped with a cryogenically cooled triple resonance probehead. All samples were dissolved in 500 µl D2O before measurement (Sigma Aldrich). As internal standard, the sodium salt of trimethylsilyl propionic acid–d4 (TMSP) was used. One‐dimensional 1H NMR experiments were performed using excitation sculpting water suppression (zg30esgp) with 128 scans. A spectral width of 16 ppm, a repetition delay of 2 s, and an acquisition time of 2 s were used. The spectra were recorded, processed, and analyzed using the TopSpin 4.1.1 software (Bruker BioSpin AG). Sensitivity was controlled by spiking the sample with heparan sulfate.
3. RESULTS AND DISCUSSION
Addition of the vaccine to normal plasma neither prolonged the APTT nor resulted in any measurable aFXa activity (n = 3). Both results make it unlikely that ChAdOx1‐S contains larger amounts of sulfated GAG interacting with clotting factors.
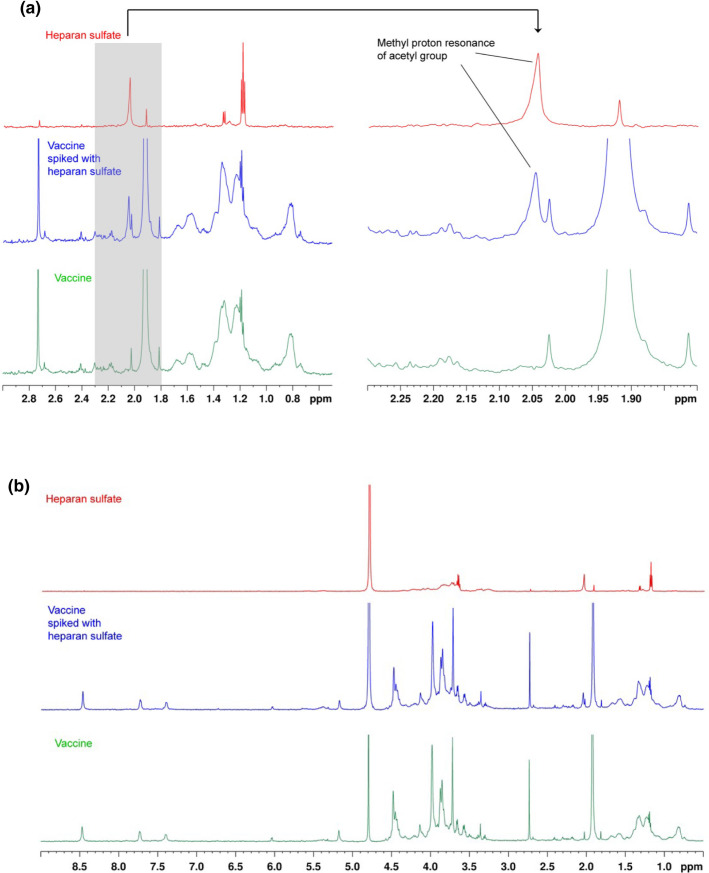
To directly detect/exclude GAG, we performed 1H‐NMR analysis. NMR detection of GAG is highly specific but rather insensitive.14 In addition, proteins show signals in the same range and thus impair GAG detection. Commercially available kits to detect GAG have a limit of detection of 5 µg/ml, which is too high for the purpose of our study. We aimed for a detection limit of <1.0 µg/ml to be sufficiently sensitive to detect GAG in concentrations relevant for forming immunogenic complexes with PF4, for which typically 2–3 µg/ml GAG are required.14 We overcame these problems by using 30 ml of the vaccine as start volume and by eliminating the proteins by proteinase K digestion. The digested vaccine was dialyzed with a 1 kDa exclusion membrane to remove peptides and small‐molecule excipients including the disaccharide sucrose and subsequently lyophilized. Removal of the saccharide sucrose and potentially other small oligosaccharides was aimed to avoid any NMR signals, which are irrelevant for complex formation with PF4, as previous studies have clearly shown that GAG consisting of less than five saccharide units barely interact with PF4 in a way that is causing conformational changes.15 We took advantage of the alkaline isoelectric point of proteinase K to separate it from potentially present GAG by performing SAX at pH 8.9. These experimental conditions at the isoelectic point of the protein additionally avoided any complex formation between the protein and negatively charged GAG and thereby the loss of GAG. Finally, we concentrated the original vaccine sample 60‐fold. By this procedure, we reached a sensitivity of detecting less than 0.83 µg/ml of the exemplary GAG heparan sulfate in the vaccine as judged by the signal of the positive control with spiked heparan sulfate (blue line in Figure 1 ). According to previous studies on detecting GAG contaminants,15 the NMR spectrum was analyzed at around 2 ppm, the shift range of GAG‐specific acetyl signals. The spectrum of the vaccine sample did not show any acetyl signal that would indicate the presence of GAG (Figure 1).
FIGURE 1.
A, Analysis of the 1H nuclear magnetic resonsonance (NMR) spectrum of ChAdOx1‐S regarding presence of sulfated glycosaminoglycans (GAG). The gray shaded area of the spectrum in the left panel, shown in magnification in the right panel, is the informative segment of the NMR spectrum for detection of GAG. To detect any GAG, an overlay of 1H‐NMR spectra was analyzed in the aliphatic region (i.e., low‐shift ppm range, left panel), specifically at around 2 ppm (right panel). The spectrum of heparan sulfate (25 µg in 500 µl D2O, red spectrum) as positive control showed the methyl proton resonance of the typical GAG acetyl group at 2.05 ppm. The processed and 60‐fold concentrated vaccine sample (green spectrum) did not show any signal indicative of GAG‐specific acetyl groups. After spiking the vaccine with heparan sulfate corresponding to a concentration in the vaccine of 0.83 µg/ml, this signal was clearly visible (blue spectrum). The overall spectra of heparan sulfate, the vaccine and the spiked vaccine are shown in (B). B, 1H NMR spectra of ChAdOx1‐S after protein digestion, strong‐anion exchange chromatography (SAX)‐Ch purification, and 60‐fold concentration (green); vaccine supplemented with 25 µg heparan sulfate (blue), and 25 µg heparan sulfate (red)
In principle, treatment of vaccines with enzymes degrading GAG would reduce any risk of GAG‐PF4 complex formation. However, one issue is that treatment of such a complex protein‐rich vaccine does not ensure that all the enzymes (several heparanases and chondroitinases) are active. In addition, it has to be considered that such enzymes might cause problems of adverse immune reactions if they cannot be completely removed from the vaccine during the final production steps.
In conclusion, we found no evidence for the presence of >0.5 µg/ml sulfated glycosaminoglycans with a molecular weight >1 kDa in ChAdOx1‐S. These data support that the search for PF4 interaction partners in the vaccine should focus on other vaccine constituents beside the hexon protein, which we7 and Baker et al.16 have already demonstrated to be part of PF4 complexes.
CONFLICTS OF INTEREST
AG reports grants and non‐financial support from Aspen, Boehringer Ingelheim, MSD, Bristol Myers Squibb (BMS), Paringenix, Bayer Healthcare, Gore Inc., Rovi, Sagent, Biomarin/Prosensa; personal fees from Aspen, Boehringer Ingelheim, MSD, Macopharma, BMS, Chromatec, Instrumentation Laboratory; non‐financial support from Boehringer Ingelheim, Portola, Ergomed, GTH e.V. outside the submitted work. SA reports a grant and personal fees from Bayer Healthcare, and personal fees from Aspen, Boehringer Ingelheim, and Leo Pharma outside the submitted work. None of the other authors has any conflicts to declare.
AUTHOR CONTRIBUTIONS
SA planned the experiments, analyzed and interpreted the data, and wrote the manuscript. SN performed the experiments, the data analysis, and prepared the figures. FDS and UG performed the NMR experiments and analysis. AG initiated the study, interpreted results, and wrote the manuscript.
ACKNOWLEDGMENTS
We thank Dr. Juliane Schöpfel for performing the APTT and aFXa measurements. The study has been supported by Deutsche Forschungsgemeinschaft, Grant/Award Number: 374031971‐TRR 240. Open access funding enabled and organized by ProjektDEAL.
Deutsche Forschungsgemeinschaft374031971‐TRR 240
Footnotes
Manuscript handled by: Ton Lisman
Final decision: Ton Lisman, 30 December 2021
REFERENCES
- 1.European Medicines Agency (EMA). Vaxzevria (previously COVID‐19 Vaccine AstraZeneca): EPAR ‐ Public assessment report (EMA/94907/2021, first published 18.02.2021, last updated 13.12.2021). https://www.ema.europa.eu/en/medicines/human/EPAR/vaxzevria‐previously‐covid‐19‐vaccine‐astrazeneca. Accessed December 29, 2021.
- 2.Pavord S., Scully M., Hunt B.J., et al. Clinical features of vaccine‐induced immune thrombocytopenia and thrombosis. N Engl J Med. 2021;385:1680–1689. doi: 10.1056/NEJMoa2109908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic thrombocytopenia after ChAdOx1 nCov‐19 vaccination. N Engl J Med. 2021;384:2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Medicines Agency (EMA). COVID‐19 vaccine safety update for Vaxzevria (previously COVID‐19 Vaccine AstraZeneca): 21.05.2021. https://www.ema.europa.eu/en/medicines/human/EPAR/vaxzevria‐previously‐covid‐19‐vaccine‐astrazeneca. Accessed May 31, 2021.
- 5.Greinacher A., Selleng K., Warkentin T.E. Autoimmune heparin‐induced thrombocytopenia. J Thromb Haemost. 2017;15:2099–2114. doi: 10.1111/jth.13813. [DOI] [PubMed] [Google Scholar]
- 6.Gresele P., Momi S., Marcucci R., Ramundo F., de Stefano V., Tripodi A. Interactions of adenoviruses with platelets and coagulation and the vaccine‐associated autoimmune thrombocytopenia thrombosis syndrome. Haematologica. 2021;106:3034–3045. doi: 10.3324/haematol.2021.279289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greinacher A., Selleng K., Wesche J., et al. Towards understanding ChAdOx1 nCov‐19 vaccine‐induced immune thrombotic thrombocytopenia (VITT) Blood. 2021;138:2256–2268. doi: 10.1182/blood.2021013231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz N.H., Sørvoll I.H., Michelsen A.E., et al. Thrombosis and thrombocytopenia after ChAdOx1 nCoV‐19 vaccination. N Engl J Med. 2021;384:2124–2130. doi: 10.1056/NEJMoa2104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cines D.B., Yarovoi S.V., Zaitsev S.V., et al. Polyphosphate/platelet factor 4 complexes can mediate heparin‐independent platelet activation in heparin‐induced thrombocytopenia. Blood Adv. 2016;1:62–74. doi: 10.1182/bloodadvances.2016000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandt S., Krauel K., Jaax M., et al. Polyphosphates form antigenic complexes with platelet factor 4 (PF4) and enhance PF4‐binding to bacteria. Thromb Haemost. 2015;114:1189–1198. doi: 10.1160/TH15-01-0062. [DOI] [PubMed] [Google Scholar]
- 11.Alban S., Krauel K., Greinacher A. In: Heparin‐Induced Thrombocytopenia. 5th. Warkentin TE, Greinacher A, editors. Taylor & Francis; 2012. Role of sulfated polysaccharides in the pathogenesis of heparin induced thrombocytopenia; pp. 181–208. [Google Scholar]
- 12.Jaax M.E., Krauel K., Marschall T., et al. Complex formation with nucleic acids and aptamers alters the antigenic properties of platelet factor 4. Blood. 2013;122:272–281. doi: 10.1182/blood-2013-01-478966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holen H.L., Zernichow L., Fjelland K.E., et al. Ephrin‐B3 binds to a sulfated cell‐surface receptor. Biochem J. 2011;433:215–223. doi: 10.1042/BJ20100865. [DOI] [PubMed] [Google Scholar]
- 14.Alban S., Lühn S., Schiemann S., et al. Comparison of established and novel purity tests for the quality control of heparin by means of a set of 177 heparin samples. Anal Bioanal Chem. 2011;399:605–620. doi: 10.1007/s00216-010-4169-7. [DOI] [PubMed] [Google Scholar]
- 15.Kreimann M., Brandt S., Krauel K., et al. Binding of anti‐platelet factor 4/heparin antibodies depends on the thermodynamics of conformational changes in platelet factor 4. Blood. 2014;124:2442–2449. doi: 10.1182/blood-2014-03-559518. [DOI] [PubMed] [Google Scholar]
- 16.Baker A.T., Boyd R.J., Sarkar D., et al. ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome. Sci Adv. 2021;7:eabl8213. doi: 10.1126/sciadv.abl8213. [DOI] [PMC free article] [PubMed] [Google Scholar]