Abstract
S/MARt DB, the S/MAR transaction database, is a relational database covering scaffold/matrix attached regions (S/MARs) and nuclear matrix proteins that are involved in the chromosomal attachment to the nuclear scaffold. The data are mainly extracted from original publications, but a World Wide Web interface for direct submissions is also available. S/MARt DB is closely linked to the TRANSFAC database on transcription factors and their binding sites. It is freely accessible through the World Wide Web (http://transfac.gbf.de/SMARtDB/) for non-profit research.
INTRODUCTION
Scaffold/matrix attached regions (S/MARs) are DNA elements of the eukaryotic genome that attach the chromatin fiber to the proteinaceous network of the nucleus, called either the nuclear matrix (1) or nuclear scaffold (2). S/MARs are found at the base of the chromatin loops into which the eukaryotic genome is organized. Since these loops appear to represent functional subunits, S/MARs are thought to be the tools which subdivide the eukaryotic genome into structural and functional domains. In order to obtain a systematic insight into S/MAR fine-structure and how it relates to function, we have developed a database on S/MARs and the proteins that have been implicated in nuclear scaffold attachment. This database will provide the basis for elucidating the nature of DNA/scaffold interaction. It will enhance the understanding of gene regulation with respect to functional domains. In this context it is important to note that there are hints that correct matrix attachment is a prerequisite for proper cell function (3–5). Besides that, a new generation of vectors tries to take advantage of S/MAR properties (6,7). First promising results show that these vectors are stably maintained without selection for several hundred generations (7).
STRUCTURE AND CONTENTS OF THE DATABASE
The S/MAR transaction database, S/MARt DB, is a compilation of S/MARs and nuclear matrix proteins that are implicated in chromosomal attachment to the nuclear scaffold. These data have been collected mainly from original publications and, wherever necessary, they were re-confirmed by contacting the authors.
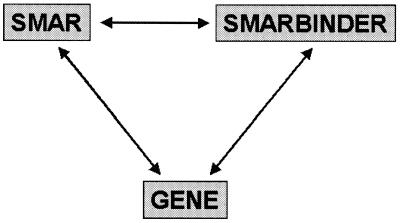
The database is organized in three hyperlinked flat files (Fig. 1) which are derived from the internal relational system consisting of more than 30 tables. Since the basic functions of S/MARs rely on DNA–protein interactions, the principal structure of a database handling these data comprises two linked flat files, SMAR and SMARBINDER. The SMAR flat file characterizes individual S/MAR elements by reference to the biological species from which they have been obtained, their chromosomal location, data on structural and functional properties and—where available—information about the binding potential in vitro. The methods by which a S/MAR has been identified are also provided since they indicate the reliability of the information given.
Figure 1.
Flat file structure of S/MARt DB.
At least some S/MARs are located next to known genes. Therefore, the SMAR flat file provides links to the GENE flat file (Fig. 1) which keeps information on genes. GENE also provides links to the SMARBINDER flat file.
The properties of individual nuclear matrix proteins that interact with S/MAR elements are described by the SMARBINDER flat file. These entries include the biological species from which these matrix proteins have been obtained and their known cell specificity. Data about their size and isoelectric point are collected in this table as well as information concerning structural and functional features. The SMARBINDER flat file provides links to the FACTOR table of the TRANSFAC database (8) since some proteins that are supposed to bind to S/MARs are known to act as transcription factors as well (5,9,10).
Often individual S/MARs as well as S/MAR binding proteins are given distinct names in different laboratories. These different names are collected as synonyms. Published interactions of S/MAR binding proteins with S/MARs are included both in the SMAR as well as in the SMARBINDER entries. Interactions are described by providing information on the experimental set-up, i.e. experimental methods and the cellular contexts. Finally, all bibliographic references of the evaluated publications are quoted.
An overview of the amount of data stored in S/MARt DB is given in Table 1. Since its first introduction to the public in October 1999, the data content has increased by ∼38% (S/MARs) or ∼20% (S/MAR binders). In particular, the number of entries that include a sequence has been enhanced (from 64 to 78% of all S/MARs), and the mean sequence length per entry has nearly been doubled. The data derive from a comparable number of animal and plant species (Table 1). To the best of our knowledge, we have close to 100% coverage as far as publicly available data about S/MARs are concerned.
Table 1. Content of S/MARt DB releases 1.0 (when it was published first) and 1.1 (actual status).
| Table | Entries(release 1.0, Oct 1999) | Entries(release 1.1, Jan 2001) |
|---|---|---|
| S/MAR (total) | 192 | 313 |
| From animals (mammalia/insecta) | 135 (107/20) | 240 (144/80) |
| From plant species (dicotylidonae/monocotylidonae) | 49 | 63 (50/13) |
| From other species (i.e. yeast, viridae, artificial sequences) | 8 | 9 |
| Sequences | 123 | 245 |
| Mean length of sequence | 1142 bp | 2128 bp |
| Genes assigned to a S/MAR | 110 | 124 |
| Species | 28 | 31 |
| Animals (mammalia/insecta) | 12 (10/1) | 13 (10/1) |
| Plants (dicotylidonae/monocotylidonae) | 11 | 12 (9/3) |
| Other | 5 | 6 |
| SMARBINDER (total) | 51 | 61 |
| Known transcription factors | 8 | 8 |
| References | 150 | 180 |
LINKS TO OTHER DATABASES
As mentioned above, the SMARBINDER flat file provides links to the TRANSFAC database. This also holds true for the GENE flat file, which connects S/MARt DB to Transcription Regulatory Region Database (TRRD) as well (11). Besides that, S/MARt DB contains hyperlinked cross-references to MEDLINE/PubMed, to the EMBL data library (12) and, where appropriate, entries are also linked to SWISS-PROT and PIR (13,14).
SUBMISSION OF DATA AND ACCESS TO DATA
In order to keep S/MARt DB as up-to-date as possible we strongly encourage all the experts in the field to directly submit relevant data via the Internet. For this purpose, we have developed HTML-based forms that enable electronic submission of S/MAR sequences and S/MAR binding proteins. Data that are submitted to the database are marked accordingly and become publicly available after passing through an expert reviewing procedure comparable to that which is common for paper publications. Submission forms can be found on the S/MARt DB homepage (http://transfac.gbf.de/SMARtDB/).
The ASCII flat file version of S/MARt DB is freely accessible through the Internet for non-profit research. The flat files are organized in a manner similar to those displayed by the EMBL or SWISS-PROT databases. S/MARt DB is connected to a search engine that enables the user to browse the database through all fields of the flat files. Documentation explaining the data content, the data format of the flat files and introduced changes may be called from the S/MARt DB homepage.
FUTURE PROSPECTS
In addition to maintaining and updating the S/MAR database described above, its structure and properties will be continuously extended. In addition to pure textual descriptions, a next step will be the inclusion into the SMAR flat files of graphic representations for the regions surrounding the S/MAR elements as well as standardized biomathematical computations of S/MAR activities along the published sequence. Extending the possibilities for direct updating by the community, it will be possible in the future to report S/MAR–protein interactions to the S/MARt DB via a separate form. This becomes increasingly important as more and more S/MARs and S/MAR binding proteins are included in the database, and new reports demonstrating the interaction between these already known components become available.
In order to establish a useful network of information resources for researchers we shall enhance cross-referencing between databases. For instance, pointers to FlyBase and OMIM will be inserted into S/MARt DB (15,16).
Classifications of S/MARs and S/MAR binding proteins will be developed once the data sets appear sufficiently comprehensive.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Holger Karas for fruitful discussions. This work was supported by grants of the Federal Ministry of Education and Research (BMBF, project no. 0311640 and 01SF9988/4).
REFERENCES
- 1.Berezney R. and Coffey,D.S. (1974) Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun., 60, 1410–1417. [DOI] [PubMed] [Google Scholar]
- 2.Mirkovitch J., Mirault,M.-E. and Laemmli,U.K. (1984) Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell, 39, 223–232. [DOI] [PubMed] [Google Scholar]
- 3.Yanagisawa J., Ando,J., Nakayama,J., Kohwi,Y. and Kohwi-Shigematsu,T. (1996) A matrix attachment region (MAR)-binding activity due to a p114 kilodalton protein is found in human breast carcinomas and not in normal and benign breast disease tissues. Cancer Res., 56, 457–462. [PubMed] [Google Scholar]
- 4.Kramer J.A., Zhang,S., Yaron,Y., Zhao,Y. and Krawetz,S.A. (1997) Genetic testing for male infertility: a postulated role for mutations in sperm nuclear matrix attachment regions. Genetic Testing, 1, 125–129. [DOI] [PubMed] [Google Scholar]
- 5.Deppert W. (2000) The nuclear matrix as a target for viral and cellular oncogenes. Crit. Rev. Eukaryot. Gene Expr., 10, 45–61. [PubMed] [Google Scholar]
- 6.Piechaczek C., Fetzer,C., Baiker,A., Bode,J. and Lipps,H.J. (1999) A vector based on the SV40 origin of replication and chromosomal S/MARs replicates episomally in CHO cells. Nucleic Acids Res., 27, 426–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baiker A., Maercker,C., Piechaczek,C., Schmidt,S.B., Bode,J., Benham,C. and Lipps,H.J. (2000) Mitotic stability of an episomal vector containing a human scaffold/matrix-attached region is provided by association with nuclear matrix. Nat. Cell Biol., 2, 182–184. [DOI] [PubMed] [Google Scholar]
- 8.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhäuser,R., Prüß, M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banan M., Rojas,I.C., Lee,W.H., King,H.L., Harriss,J.V., Kobayashi,R., Web,C.F. and Gottlieb,P.D. (1997) Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J. Biol. Chem., 272, 18440–18452. [DOI] [PubMed] [Google Scholar]
- 10.Dworetzky S.I., Wright,K.L., Fey,E.G., Penman,S., Lian,J.B., Stein,J.L. and Stein,G.S. (1992) Sequence-specific DNA-binding proteins are components of a nuclear matrix-attachment site. Proc. Natl Acad. Sci. USA, 89, 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolchanov N.A., Podkolodnaya,O.A., Ananko,E.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busygina,T.V. et al. (2000) Transcription regulatory regions database (TRRD): its status in 2000. Nucleic Acids Res., 28, 298–301. Updated article in this issue: Nucleic Acids Res. (2002), 30, 312–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoesser G., Baker,W., van den Broek,A., Camon,E., Garcia-Pastor,M., Kanz,C., Kulikova,T., Lombard,V., Lopez,R., Parkinson,H., Redaschi,N., Sterk,P., Stoehr,P. and Tuli,M.A. (2001) The EMBL nucleotide sequence database. Nucleic Acids Res., 29, 17–21. Updated article in this issue: Nucleic Acids Res. (2002), 30, 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barker W.C., Garavelli,J.S., Hou,Z., Huang,H., Ledley,R.S., McGarvey,P.B., Mewes,H.-W., Orcutt,B.C., Pfeiffer,F., Tsugita,A., Vinayaka,C.R., Xiao,C., Yeh,L.S. and Wu,C. (2001). Protein Information Resource: a community resource for expert annotation of protein data. Nucleic Acids Res., 29, 29–32. Updated article in this issue: Nucleic Acids Res. (2002), 30, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The FlyBase Consortium (1999) The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res., 27, 85–88. Updated article in this issue: Nucleic Acids Res. (2002), 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. Updated article in this issue: Nucleic Acids Res. (2002), 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]