Abstract
The Olfactory Receptor Database (ORDB; http://senselab.med.yale.edu/senselab/ordb) is a central repository of olfactory receptor (OR) and olfactory receptor-like gene and protein sequences. To deal with the very large OR gene family, we have constructed an algorithm that automatically downloads sequences from web sources such as GenBank and SWISS-PROT into the database. The algorithm uses hypertext markup language (HTML) parsing techniques that extract information relevant to ORDB. The information is then correlated with the metadata in the ORDB knowledge base to encode the unstructured text extracted into the structured format compliant with the database architecture, entity attribute value with classes and relationship (EAV/CR), which supports the SenseLab project as a whole. Three population methods: batch, automatic and semi-automatic population are discussed. The data is imported into the database using extensible markup language (XML).
INTRODUCTION
The publication of results of cloning rat olfactory receptors (ORs) (1) in 1991 spawned a strong research effort in the area of chemosensory reception (2,3). The Olfactory Receptor Database (ORDB) (4–6; http://senselab.med.yale.edu/senselab/ORDB) is a database in SenseLab (7; http://senselab.med.yale.edu/senselab) dedicated to archiving and disseminating chemosensory receptor protein and gene sequence information to researchers and the public. The database currently contains approximately 1750 OR sequences of more than 30 species and from more than 30 source tissues. ORDB serves 100 laboratories all over the world and is the primary repository for gene and protein sequences of ORs. Besides ORs and olfactory receptor-like sequences (ORLs), ORDB also houses Caenorhabditis elegans chemosensory receptors (CeCRs), insect olfactory receptors (IORs), fungal pheromone receptors (FPRs), taste papilla receptors (TPRs) and vomeronasal receptors (VNRs).
Closely related to ORs are odor ligands, molecules that elicit the sensation of smell and are experimentally determined as binding to an OR. OdorDB, a prototype database (http://senselab.med.yale.edu/senselab/odorDB) has been developed to operate in conjunction with ORDB as a repository for the odor ligands linked to the ORs to which they bind (8,9).
ORs constitute the largest family of genes in the human genome and belong to a superfamily of GTP-protein binding coupled receptors (GPCRs) (10). GPCRs are characterized by a seven trans-membrane helical structure.
It was initially estimated that there were approximately 1000 olfactory genes in the human genome (1). With the publication of the first draft of the human genome (11,12), two groups have separately identified and published OR sequences from the human genome. Of the greater than 900 sequences projected as ORs, these groups have identified 347 (13) and approximately 360 (14) complete, functional sequences as human OR genes. The remaining genes are either non-identifiable or have been identified as pseudogenes. Recent publications highlight the importance of recognizing pseudogenes as indicators of variation in chemosensory function with evolution (15,16).
The curators of ORDB are faced with a challenge due to the large number of OR and ORL sequences being identified. ORDB currently has approximately 296 human olfactory gene sequences and 472 mouse sequences. The reported mouse ORs are more numerous than those for humans since different strains of mice are being studied (http://www.jax.org/pub-cgi/imrpub.sh?objtype=stridx). When the mouse genome is published (17,18), several hundred additional olfactory gene sequences will become available.
ORDB is responsible for making the gene sequence information available to users and laboratories as quickly as possible. Efficient and rapid dissemination of information requires rapid population of ORDB. Previously used population methods, e.g. manually populating the database by filling in forms with information from GenBank and SWISS-PROT, were limited by time and the available manpower. With new data becoming available at unprecedented rates, it is incumbent upon ORDB to make sequences available as soon as they are cloned, expressed and deposited into GenBank and SWISS-PROT.
Towards this goal, we have designed an automatic population algorithm AUTO/POP that imports data from GenBank (http://www.ncbi.nlm.nih.gov/) and SWISS-PROT (http://www.expasy.ch/sprot/sprot-top.html). The algorithm makes use of hypertext markup language (HTML) parsing techniques to extract information relevant to ORDB from GenBank and SWISS-PROT from automatically downloaded sequence files. This information is then filtered through the ORDB knowledge base to correlate relevant data from the HTML files to the ORDB metadata. The automatic population program encodes the unstructured text into ORDB structure by building an extensible markup language (XML)-encoded document that is embedded in an HTML form. The program, developed to aid registered users and curators, can extract a single or multiple appropriate sequences from each GenBank entry. The program is available on the Internet at http://chutney.med.yale.edu/autoput/readme.htm. The only requirement is a GenBank accession number for a chemosensory receptor nucleotide sequence.
MATERIALS AND METHODS
ORDB architecture
ORDB’s architecture is based on the entity attribute value with classes and relationship (EAV/CR) database architecture, the details of which have been published previously (19). This structure stores information describing all the data as ‘metadata.’ This metadata in turn can serve as a knowledge base to assist in the automated population of ORDB. The EAV/CR database representation is an extension of the EAV schema (20,21). The EAV/CR storage design of SenseLab allows a flexible and extensible data storage approach.
Design of the automatic population algorithm
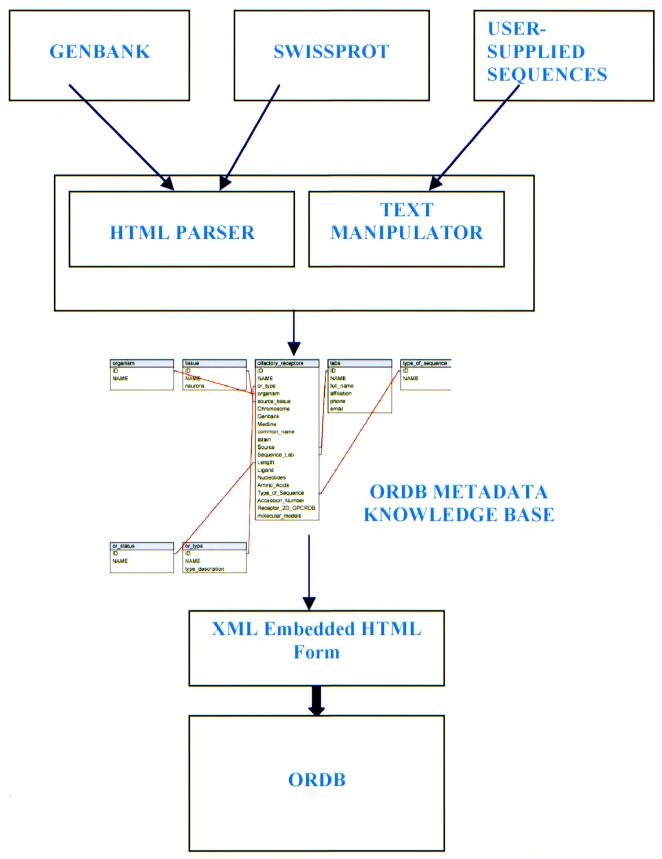
The steps in the automatic population of ORDB are illustrated in Figure 1. They include: (i) downloading a GenBank or SWISS-PROT entry as an HTML file automatically; (ii) parsing this HTML file to extract information relevant to ORDB; (iii) filtering each datum extracted against ORDB knowledge base to match the unstructured data with terms (objects and attributes) that ORDB uses to store this information; (iv) creating an XML-encoded file that contains the data extracted in a format compliant with the SenseLab database architecture; and (v) importing the information into ORDB using the XML ‘presentation-transfer’ module.
Figure 1.
Schematic outline of the automatic population algorithm, AUTO/POP. Relevant HTML files from GenBank and SWISS-PROT and user-supplied text files are parsed to extract relevant data. The extracted data is filtered through ORDB knowledge base. ORDB structure-equivalent information is then used to build an XML-encoded file that is entered into the database via an HTML submission form.
The automatic population program is written in a practical extraction and report language (PERL) script. In the case of user-supplied data, the HTML parsing steps are not required. The data supplied can be manipulated to create the XML-encoded file that will be recognized by the XML transfer script.
The presentation-transfer module, coded in visual basic (VB) script, in the active server pages (ASP) web design software, is used either (i) to present data from an ORDB entry as an XML-encoded document or (ii) to import information as a structured XML file into ORDB. Each datum of information is stored as a string or an object related to an attribute that describes an ORDB entry encoded in XML. These attributes have been tabulated previously (5).
Filtering information through the ORDB knowledge base converts unstructured textual data from the HTML files into objects, attributes and strings within their intrinsic relationships that constitute the structure of ORDB.
For example, if the HTML parsing program identifies that the OR sequence was cloned for the organism Mus musculus, it matches this string (mus musculus) against the ORDB knowledge base. AUTO/POP accesses the ORDB knowledge base and recognizes the following line: ‘30:144:mus musculus│mouse:mus musculus’. The somewhat cryptic metadata is stored in the knowledge base in the same structural hierarchy as the ORDB. In the above example, the program would also recognize the metadata (object and attribute) as identical to mus musculus if the text string denoting organism was ‘mouse’. This representation of the knowledge base also allows for introduction of synonyms for mouse. The program determines that mus musculus corresponds to the attribute ‘a30’, the attribute for ‘organism’, and that mus musculus is stored in ORDB as object ‘o144’. The program automatically creates the following XML line: <a30 object_name=‘mus musculus’>o144</a30>. If the information is to be entered as a string data type instead of an object, the string is matched with the knowledge base as ‘a106:strain:Strain’ to determine the attribute to which the string corresponds. The line <a106 value=‘Strain’> 129/SvJ </a106> indicates that the value of the mouse strain ‘129/SVJ’ extracted from the GenBank or SWISS-PROT HTML file will be entered as a string and the knowledge base ‘informs’ the program that ORDB attribute assignation for Strain is ‘a106.’
In this way, the XML-encoded document with the relevant attributes and objects and strings is created to comply with the structured SenseLab architecture. In the final step, the XML document is embedded in an HTML submission form for entry into ORDB.
Semi-automatic population of ORDB from GenBank
To use AUTO/POP to populate the ORDB from GenBank, the user/curator first searches GenBank for relevant entries (ORs, VNRs, TPRs, IORs, FPRs and CeCRs). Each GenBank entry returned may contain single or multiple gene sequences. The user enters the relevant GenBank accession number in AUTO/POP. A query is automatically created which automatically downloads this GenBank entry as an HTML file. An HTML parsing sub-routine extracts the relevant information from this file, uses ORDB knowledge base to convert into the structured format of ORDB architecture and builds the XML-encoded submission file.
Automatic population from SWISS-PROT
The automatic population algorithm was tested for ORs in SWISS-PROT. 199 OR sequences were automatically downloaded from http://bioinf.man.ac.uk/dbbrowser/gpcrPRINTS/PR00245.html. Each protein sequence was compared to a list of protein sequences and their MEDLINE entries in ORDB. The SWISS-PROT files with ‘new’ sequences were then downloaded as HTML files and parsed to extract information relevant to ORDB in a similar fashion to GenBank. Automatic population from SWISS-PROT is more facile than GenBank because of its higher level of consistency in annotations for data objects.
Batch population from user-supplied sequences
Any sequences submitted by users can be entered into ORDB en masse by building an XML file embedded into an HTML submission form without the use of HTML parsers, since the information is available in text format. ORDB was batch-populated by 213 CeCR sequences in this fashion. Professor Hugh Robertson from the University of Illinois supplied the protein and nucleotide sequences along with common names for each.
RESULTS AND DISCUSSION
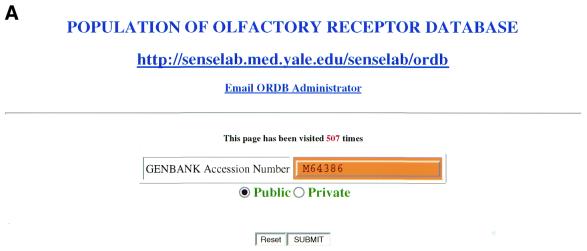
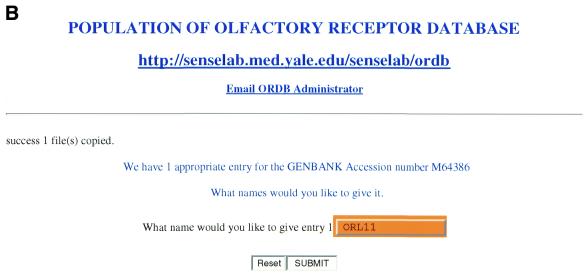
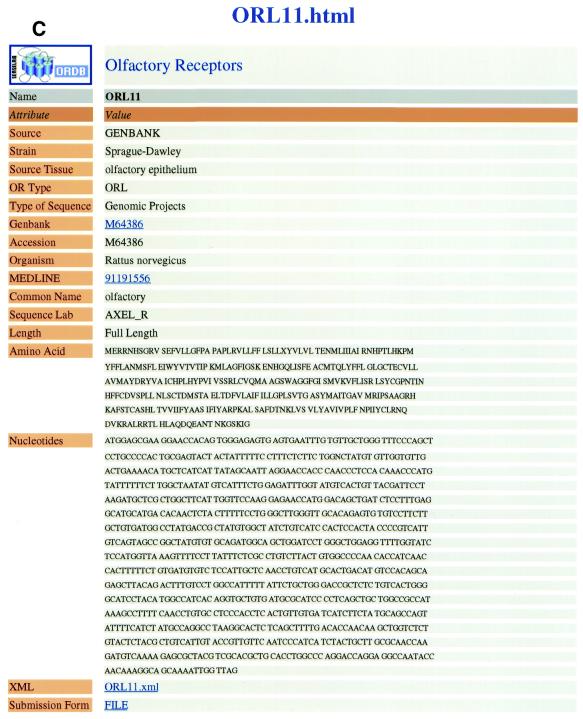
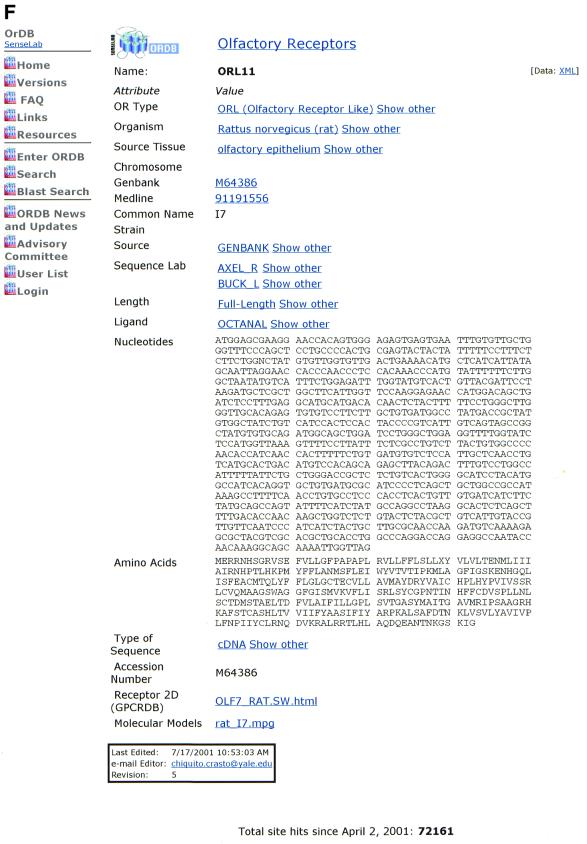
This section illustrates AUTO/POP’s operation, discusses specific examples of its use and discusses certain issues that are used in building the AUTO/POP program. Figure 2A–F illustrates steps in the operation of the automatic population program AUTO/POP for the entry M64386 from GenBank. This entry corresponds to the first OR cloned (1) and returns that single protein and nucleotide sequence.
Figure 2.
(A) The web page used to enter the appropriate GenBank accession number. In this case the number being entered (M64386) is for the first OR cloned, for illustration purposes. (B) The web page seen when the program automatically downloads the HTML file and prompts the user to enter an ORDB entry name. In ORDB the sequence corresponding to GenBank entry M64386 is named and stored as ORL11. (C) The visual representation of extracted data enables the curator to assess the data for completeness and accuracy. (D) The XML file, showing the attributes and objects for each datum extracted in compliance with ORDB architecture. (E) The HTML submission form with the XML file (D) embedded in it. On clicking the ‘Submit’ button, the data is entered into ORDB. (F) The web page in ORDB where the sequence ORL11 has been entered.
A recent submission of mouse and human open reading frames contained multiple olfactory sequences for humans and mouse (22) all of which were identified by the AUTO/POP program. The program can also sort through several sequences in an entry identifying only the ORs while ignoring the rest. For example, in the C.elegans GenBank entry (AF067942) only one of several sequences is an OR, and AUTO/POP identifies each appropriately.
From SWISS-PROT, 79 sequences of the 199 downloaded, which had not been manually entered previously, were input into the database in a single step. A single XML file, created for all the entries, was embedded into an HTML submission form and ORDB names for each entry were automatically generated.
An intermediate step in the population from GenBank prompts the user to input an ORDB entry name, the naming based on the chronology of availability of the sequences from the sources. For curators and users, this step is important for verification of the accuracy of the extracted data. The annotations for relevant data in SWISS-PROT are more specific than in GenBank, given the nature of information supplied by the two databases. For the SWISS-PROT protein sequences, ORDB names were automatically generated as each relevant HTML sequence file was being parsed and built into the XML-encoded file. The same automatic naming was employed for the batch population of ORDB with 213 C.elegans sequences, since the source and the information were very specific.
In our experience, manual population takes ∼5–10 min per ORDB entry for an experienced curator. With automatic population, the process can typically be completed (depending on the number of sequences per GenBank entry) in one-tenth of the time or less. The total time for downloading 199 sequences, creating the XML file with 79 sequences and entering into ORDB was <4 min. The batch population of 213 CeCR sequences (CeCR157–CeCR369 in ORDB) was also achieved in a few minutes.
The establishment of the batch population technique bodes well for entering the recently available human and mouse OR sequences into ORDB. The key features of the automatic population program are the correlation of each datum extracted to the ORDB knowledge base and the XML presentation-transfer module. The knowledge base metadata is stored in the same top-down architecture as the ORDB: classes–objects–attributes. Each ORDB entry belongs to the class of OR stored as an object, described by attributes. Each attribute itself is stored as a different data type: objects, strings, memos and integers. The ease of transfer of information into the ORDB is possible because the presentation-transfer module accepts the data for transfer into the database or presents the data as ORDB metadata.
Any information that is a potential ORDB entry from text, HTML files or any other source can be manipulated by comparing with the knowledge base, updating the knowledge base and creating the XML encoded file. Simultaneously, this XML file is also automatically embedded into an HTML submission form that serves as a transfer file of single and multiple entries into ORDB.
Other updates and improvements to ORDB
A description of ORDB as a repository of the largest eukaryotic gene family (5) and a chemosensory receptor (6) has been published previously. These publications highlighted the features of ORDB, including (i) the EAV/CR architecture and (ii) extension of ORDB to include other receptors besides ORs. ORDB has now grown substantially and contains 1750 public and private sequences (990 ORLs, 478 CeCRs, 13 FPRs, 24 IORs, 97 TPRs and 124 VNRs, besides private sequences that do not follow ORDB nomenclature). It represents the work of 98 laboratories and from 34 organisms and expressed in 33 tissues. An NIH-sponsored advisory committee oversees ORDB. A user can search for any sequence (or representative sequences) in the database for all the attributes or for specific objects. The object search reveals more focused results. A recently added feature to ORDB is an attribute called molecular models. Visual graphical displays (movie and picture files) of computationally derived models of the seven helical trans-membrane domains with or without the interacting ligands can be stored in the database. A sample molecular model of the I7 ORs in rat interacting with an octanal molecule (23) can be seen at http://senselab.med.yale.edu/senselab/ORDB/molmodels/rat_I7.mpg. Users can also download each ORDB entry encoded in XML. The XML file presents the data for viewing by using the presentation side of the presentation-transfer module in the same ORDB metadata structure as would be necessary to transfer data into the ORDB.
The EAV/CR metadata has been recently enhanced to store semantic relationships that qualify Class (OR)–Attribute (Descriptors) dependencies and common external vocabulary identifiers for any possible element in the data store. A unified medical language system (UMLS; http://www.nlm.nih.gov/pubs/factsheets/umlssemn.html) concept identifier (whenever available) describes every element (Class, Object and Attribute) in ORDB, facilitating terminological interpretation of each element across databases and among research groups. ORDB monthly updates are now available to users and advisory committee members.
CONCLUSIONS AND FUTURE WORK
The automatic population of ORDB illustrates one step towards making large amounts of data accessible, and is especially topical due to the availability of drafts of the human and mouse genome.
The central role that ORDB plays in the field of chemosensory receptors provides further impetus to develop bioinformatics tools to serve the chemoreceptor community. The ORDB project has been given the task of creating a gold standard for gene sequences of human and mouse ORs. This implies linking the work of various groups that have identified human and mouse ORs and creating a medium where different databases of chemosensory receptors like GenBank, Human Olfactory Receptor Database Exploratorium (HORDE; http://bioinformatics.weizmann.ac.il/HORDE/) (21) and G-Protein Coupled Receptor Database (GPCRDB; http://www.gpcr.org/7tm/) (22) can interoperate.
Acknowledgments
ACKNOWLEDGEMENTS
The authors acknowledge the efforts of Mr Gautam Bahl and Mr Rishikesh Dalal for their efforts in testing the AUTO/POP program. We thank Professor Hugh Robertson at the University of Illinois, Urbana-Champagne, for making the C.elegans sequences available to test the batch population algorithm. NIH Grants P01 DC 04732 and G08 LM05583 from the National Library of Medicine support this work.
REFERENCES
- 1.Buck L. and Axel,R. (1991) A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell, 65, 175–181. [DOI] [PubMed] [Google Scholar]
- 2.Mombaerts P. (1999) Seven-transmembrane proteins as odorant and chemosensory receptors. Science, 286, 707–711. [DOI] [PubMed] [Google Scholar]
- 3.Dryer L. and Berghard,A. (1999) Odorant receptors: a plethora of G-protein coupled receptors. Trends Pharmacol. Sci., 20, 413–417. [DOI] [PubMed] [Google Scholar]
- 4.Healey M.D., Smith,J.E., Singer,M.S., Nadkarni,P.M., Skoufos,E., Miller,P.L. and Shepherd,G.M. (1997) Olfactory receptor database (ORDB): a resource for sharing and analyzing published and unpublished data. Chem. Senses, 22, 321–326. [DOI] [PubMed] [Google Scholar]
- 5.Skoufos E., Healey,M.D., Singer,M.S., Nadkarni,P.M., Miller,P.L. and Shepherd,G.M. (1999) Olfactory Receptor Database: a database of the largest eukaryotic gene family. Nucleic Acids Res., 27, 343–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skoufos E., Marenco,L., Nadkarni,P.M., Miller,P. and Shepherd,G.M. (2000) Olfactory Receptor Database: a sensory chemoreceptor resource. Nucleic Acids Res., 28, 341–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepherd G.M., Mirsky,J.S., Healy,M.D., Singer,M.S., Skoufos,E., Hines M.S., Nadkarni,P.M. and Miller,P.L. (1998) The Human Brain Project: neuroinformatics tools for integrating, searching and modeling multidisciplinary neuroscience data. Trends Neurosci., 21, 460–468. [DOI] [PubMed] [Google Scholar]
- 8.Touhara K. (2001) Functional cloning and reconstitution of vertebrate odorant receptors. Life Sci., 68, 2199–2206. [DOI] [PubMed] [Google Scholar]
- 9.Malnic B., Hirono,J., Sato,T. and Buck,L.B. (1999) Combinatorial receptor codes for odors. Cell, 96, 713–723. [DOI] [PubMed] [Google Scholar]
- 10.Bockaert J. and Pin,J.P. (1999) Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J., 18, 1723–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 12.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 13.Zozulya S., Echeverri,F. and Nguyen,T. (2001) The human olfactory receptor repertoire. Genome Biol., 2, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glusman G., Yanai,I., Rubin,I. and Lancet,D. (2001) The complete human olfactory subgenome. Genome Res., 108, 685–702. [DOI] [PubMed] [Google Scholar]
- 15.Buettner J.A., Glusman,G., Ben-Arie,N., Ramos,P., Lancet,D. and Evans,G.A. (1999) Organization and evolution of olfactory receptor genes on human chromosome 11. Genomics, 53, 56–68. [DOI] [PubMed] [Google Scholar]
- 16.Roquier S., Blancher,A. and Giorgi,D. (2000) The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl Acad. Sci. USA, 97, 2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denny P. and Justice,M.J. (2000) Mouse as the measure of man? Trends Genet., 16, 283–287. [DOI] [PubMed] [Google Scholar]
- 18.Copeland N.G., Gilbert,D.J., Jenkins,N.A., Nadeau,J.H., Eppig,J.T., Maltais,L.J., Miller,J.C., Dietrich,W.F., Steen,R.G., Lincoln,S.E. et al. (1993) Genome Maps IV. Science, 262, 67. [DOI] [PubMed] [Google Scholar]
- 19.Nadkarni P.M., Marenco,L., Chen R., Skoufos,E., Shepherd,G. and Miller,P. (1999) Organization of heterogenous scientific data using EAV/CR representation. J. Am. Med. Inform. Assoc., 6, 478–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadkarni P.M., Brandt,C., Frawley S., Sayward,F.G., Einbinder,R., Zelterman,D., Schacter,L. and Miller P.L. (1998) Managing attribute–value clinical trials data using ACT/DB client-server database system. J. Am. Med. Inform. Assoc., 5, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadkarni P.M., Brandt,C. and Marenco,L. (2000) WebEAV: automatic metadata-driven generation of web interfaces to entity-attribute-value databases. J. Am. Med. Inform. Assoc., 7, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane R.P., Cutforth,T., Young,J., Athanasiou,M., Friedman,C., Rowen,L., Evans,G., Axel,R., Hood,L. and Trask,B.J. (2001) Genomic analysis of orthologous mouse and human olfactory receptor loci. Proc. Natl Acad. Sci. USA, 98, 7390–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer M.S. (2000) Analysis of the molecular basis for octanal interactions in the expressed rat I7 olfactory receptor. Chem. Senses, 25, 155–165. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs T., Glusman,G., Horn-Saban,S., Lancet,D. and Pilpel,Y. (2001) The human olfactory subgenome: from sequence to structure to evolution. Hum. Genet., 108, 1–13. [DOI] [PubMed] [Google Scholar]
- 25.Horn F., Weare,J., Beukers,M.W., Horsch,S., Bairoch,A., Chen,W., Edvardsen,O. and Vriend,G. (1998) GPCRDB: an information system for G protein-coupled receptors. Nucleic Acids Res., 26, 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]