Abstract
Sample pooling testing for SARS-COV-2 can be an effective tool in COVID-19 screening when resources are limited, yet it is important to assess the performance before implementation as pooling has its limitations. Our objective was to assess the efficacy of pooling samples for coronavirus 2019 (COVID-19) compared to an individual analysis by using commercial platforms for nucleic acid testing. A total of 2200 nasopharyngeal swabs for SARS-COV-2 were tested individually and in pools of 4, 8, and 10. The cycle threshold (Ct) values of the positive pooled samples were compared to their corresponding individual positive samples. In pool size 10 samples, an estimated increase of 3-Ct was obtained, which led to false negative results in low viral load positive samples. Pooling SARS COV-2 samples is an effective strategy of screening to increase laboratories’ capacity and reduce costs without affecting diagnostic performance. A pool size of 8 is recommended.
Keywords: Sample pooling, COVID-19, RT-PCR, SARS-COV-2, Screening, Limited resources
Introduction
The COVID-19 pandemic is caused by the SARS-CoV-2 virus and has spread worldwide (Mahmoud et al. 2021). The nature of the novel virus has made its prevention and control difficult. Rapid identification and isolation of individuals with SARS-CoV-2 infections, including asymptomatic carriers, is one of the main strategies for controlling the virus (Mutesa et al. 2021). The World Health Organization (WHO) has approved molecular reverse transcription PCR (RT-PCR) testing of respiratory samples as a gold standard method for the identification of SARS-CoV-2 infection. WHO suggests 10 negative tests to one positive test for adequate testing with a recommended positivity rate lower than 10% (Mahmoud et al. 2021; Mutesa et al. 2021; Sawicki et al. 2021; WHO 2020).
Given the importance of increasing the number of tested samples to detect SARS-CoV-2, sample pooling has proven to be effective in terms of maximizing resources and obtaining results without compromising testing performance (Costa et al. 2021; Barak et al. 2021). Sample pooling methods differ in the number and size of pools into which each sample is placed. Dorfman’s pooling is the most straightforward pooling technique: Here, each sample is assigned to a pool, and each pool contains an equal number of samples. Samples are retested individually only if the pool test result is positive. If the pool is negative, then all samples included in the pool are considered negative (Barak et al. 2021). In other pooling methods, samples are assigned to overlapping pools to reduce the number of repeated samples (Barak et al. 2021; Brault et al. 2021; Grobe et al. 2021).
The aim of the study for the testing of COVID-19 samples by pooling is to facilitate large-scale immense screening that serves the community and reduces the turnaround time. Pooling by groups of eight is recommended and can be efficient in situations with low prevalence after performing validation; the impact on cycle threshold was shown to be insignificant.
Materials and methods
Study design and setting
This study was conducted in Fayoum University Hospital Covid Molecular Laboratory over a period of 3 months from September 2021 to November 2021 and approved by the Fayoum University Research Ethics Committee—a member of the Egyptian Network Research Ethics Committee (ENREC). Informed consent was not required. Samples were collected from screening candidates and healthcare professionals in the hospital.
Specimen collection
The 2200 nasopharyngeal swabs were collected and transported in viral transport medium (VTM) using Dacron or polyester flock swabs. Samples were collected using the proper technique according to the manufacturer’s recommendations. They were then transported at 4 °C and processed within 24 h.
Equipment and reagents
We used the following equipment: AllSheng Autopure96 Nucleic Extraction System, ThermoScientific KingfisherFlex Nucleic Extraction System, CFX96 Touch™ Real-Time PCR Detection System, and 3DMED ANDiS FAST SARS-CoV-2 RT-qPCR Detection Kit. The latter was ready to use and included all reagents and enzymes for the detection of the virus including RT-PCR reaction mix (850 µl/tube), enzyme mix (150 µl/tube), and positive and negative controls (1000 µl/tube each).
Performance characteristics
Fayoum University Hospital Molecular Laboratory evaluated and validated the performance of the 3DMED ANDiS FAST SARS-CoV-2 RT-qPCR Detection Kit assay using the Dorfman pooling strategy according to FDA’s guidance in its Molecular Diagnostic Template for Laboratories for Emergency Use Authorization (EUA) (U. S. Food and Drug Administration 2021).
Analytical sensitivity/limit of detection
The data demonstrate that the 3DMED ANDiS FAST SARS-CoV-2 RT-qPCR Detection Kit assay detects ≤ 0.20 copies/µl of SARS-CoV-2 whole viral genome RNA ≥ 95% of the time. This concentration is thus considered the limit of detection of the assay.
Accuracy
We performed a clinical evaluation of the pooled sample testing for 3DMED ANDiS FAST SARS-CoV-2 RT-qPCR Detection Kit with 15 nasopharyngeal swabs (10 positive pools and 5 negative pools) before starting to use the kit. Positive pooled samples were generated with the following strategy (Table 1):
Table 1.
Positive pooled samples strategy
| Known positive | Ct value | |
|---|---|---|
| Pool 1 | 1 | Ct ≥ 31 |
| Pool 2 | 1 | Increase Ct by 2 versus pool 1 |
| Pool 3 | 1 | Increase Ct by 2 versus pool 2 |
| Pool 4 | 1 | Increase Ct by 2 versus pool 3 |
| Pool 5 | 1 | Increase Ct by 2 versus pool 4 |
| Pool 6 | 1 | Increase Ct by 2 versus pool 5 |
| Pool 7 | 1 | Increase Ct by 2 versus pool 6 |
| Pool 8 | 2 | 1 × high Ct + 1 × low Ct |
| Pool 9 | 3 | 1 × high Ct + 2 × low Ct |
| Pool 10 | 3 | 2 × high Ct + 1 × low Ct |
Specificity and sensitivity
The 3DMED ANDiS FAST SARS-CoV-2 RT-qPCR Detection Kit Assay demonstrated 100% specificity and 100% sensitivity (Table 2).
Table 2.
Specificity and sensitivity of the amplification kit
| Pools by 5 s | Individual results | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | # True positive (TP) (15) | # False negative (FN) (0) | TP + FN (15) |
| Negative |
# False positive (FP) (0) |
# True negative (TN) (35) |
FP + TN (35) |
| Total | TP + FP (15) | FN + TN (35) | N |
| Concordance | 100% | ||
| True positive | # True positive (TP) (100%) | ||
| True negative | # True negative (TN) (100%) | ||
| False positive | # False positive (FP) (0%) | ||
| False negative | # False negative (FN) (0%) | ||
Study procedure
Selection criteria and considerations for pooled sample testing
Screening for COVID-19 for individual infections in a group even if there is no reason to suspect that those individuals are infected.
Screening for asymptomatic individuals who do not have known exposures with the intent of making individual decisions based on the test results.
Screening with the goal of identifying infected people before they develop symptoms or identifying infected people who may not develop symptoms, so that measures can be taken to prevent further spread.
Samples of suspected positive patients, patients having symptoms, and known or diagnosed positive cases are run individually without the use of pooling.
The laboratory does not report positive or indeterminate results of a pooled test collectively to the individuals in the pool.
Each positive pooled sample is repeated and run individually for re-extraction to identify which sample/samples were positive.
Laboratories using pooling must follow the manufacturer’s authorized instructions for use (IFU).
When introducing new detection methods, the laboratory validates pooling prior to use.
Sample pooling preparation procedure
One hundred positive samples with different Ct values and 1900 negative samples were included in the study.
One positive sample was included in each of the 4, 8, and 10 pools for a total of 100 pools. Each pool contained one known positive and 3 known negative samples in the fourfold pools. There were one known positive and 7 known negative samples in the eightfold pools; each well contained one known positive and 9 known negative samples in the tenfold pools.
Out of the one hundred positive samples, we used 25 with a high viral load (Ct 20), 25 with a medium viral load (Ct 21–30), and 50 with a low viral load (Ct 31).
The pooled sample was prepared by pipetting the samples into an RNAse-free microcentrifuge tube to make up one 1000-µl pooled sample.
Each tube was labeled with the source plate number and plate position number. Each pooled sample was considered one sample and processed for extraction.
We included the positive and negative controls each time an extraction was performed.
Extraction elusion was collected for the amplification process according to the manufacturer’s instructions.
Interpretation of results
We first verified the success of the run before interpreting sample results. In the positive control, internal control (IC)-specific amplifications were detected with a Ct value of ≤ 40 for the ORF1ab gene as well as the N and E genes. In the negative control, any specific amplification signal should not be detected.
Each patient sample from the pooled samples that yielded negative results based on the manufacturer’s interpretation was reported as SARS-COV-2-negative. Each patient sample from the pooled samples with a positive or indeterminate result was re-extracted to identify the sample(s) that is/are positive and reported as SARS-COV-2- positive. Individual samples with negative results are reported as SARS-COV-2-negative.
Data analysis
Differences in Ct values, means, and standard deviations were calculated. The delta Ct value (ΔCt) was defined as the absolute change in Ct value when the pooled sample was tested versus a positive sample that composed the pool when tested individually. Therefore, a positive ΔCt value (i.e., an increase in the Ct value of the pooled sample) represents the loss of rRT-PCR sensitivity as a consequence of individual sample dilution within the pool.
Results
The average positivity rate during the 3 months of the study period was calculated as shown in Table 3.
Table 3.
The average of positivity rate in screening population over 3 months
| Number of positive | Number of negative | Positive rate | |
|---|---|---|---|
| September | 97 | 3463 | 2.72% |
| October | 122 | 4146 | 2.86% |
| November | 136 | 3790 | 3.46% |
| Total | 355 | 11399 | 3.01% |
Real-time PCR for SARS-CoV-2 targeting the N-gene, E-GENE, and Orf1ab gene (3DMed ANDiS FAST SARS-CoV-2 RT-qPCR Detection) used the extracted nucleic acid from pooled and individual samples; Ct values up to 40 with amplification curves were considered positive.
Our study found that the sensitivity of four-pool samples was 100%, eight-pool samples was 99%, and ten-pool samples was 91%. In weak positive samples with Ct values ≥ 31, sensitivity was decreased in 8 pools to 98% and 88% in the 10-pool approach (Table 4).
Table 4.
Sensitivity of pooling according to pooling size
| Ct value for individual positive samples | Number of positive samples | Clinical agreement between individual testing and pool testing | ||
|---|---|---|---|---|
| 4 samples | 8 samples | 10 samples | ||
| ≤ 20 cycles | 25 | 25/25 (100%) | 25/25 (100%) | 24/25 (96%) |
| 21–30 cycles | 25 | 25/25 (100%) | 25/25 (100%) | 23/25 (94%) |
| ≥ 31 cycles | 50 | 50/50 (100%) | 49/50 (98%) | 44/50 (88%) |
| Total | 100 | 100/100 (100%) | 99/100 (99%) | 91/100 (91%) |
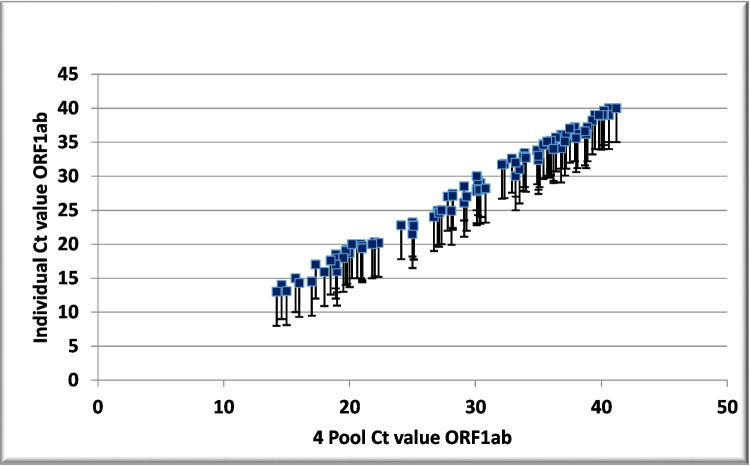
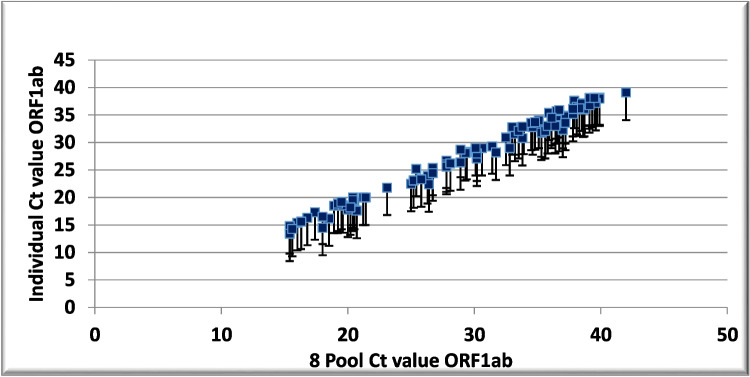
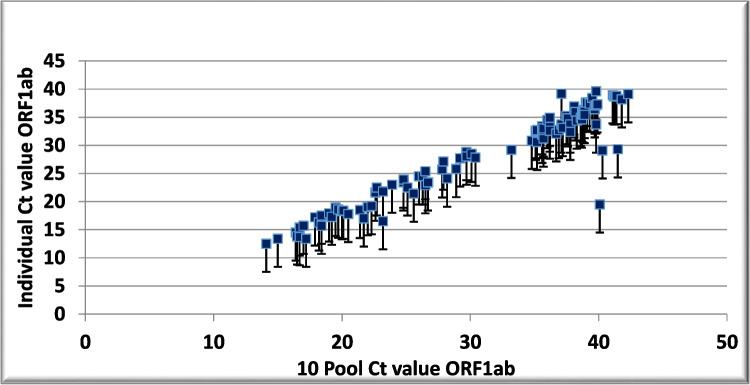
The average Ct values were compared for individual and pooled samples with different viral loads (Table 5). In the 4-, 8-, and 10-sample pools, the Ct values exceeded the individual Ct value of the corresponding samples by 1.45, 2.5, and 4.16, respectively, with a higher increase in low viral load samples (Figs. 1, 2, and 3).
Table 5.
Comparison between the mean Ct values of Orf1ab of individual test and pool testing of different sizes
| Ct value for individual positive samples |
Ct mean (± SD) Orf1ab |
|||
|---|---|---|---|---|
| Individual | 4 samples | 8 samples | 10 samples | |
| ≤ 20 cycles | 18.2 (± 1.8) | 19.4 (± 1.8) | 20.15 (± 0.9) | 22.51 (± 1.4) |
| 21–30 cycles | 27.31 (± 2.5) | 28.5 (± 2.8) | 30.3 (± 1.5) | 31.4 (± 2.3) |
| ≥ 31 cycles | 35.13 (± 0.8) | 37.1 (± 0.9) | 37.7 (± 2.1) | 39.2 (± 2.2) |
| Total | 26.88 (± 5.1) | 28.33 (± 5.5) | 29.38 (± 4.5) | 31.04 (± 5.9) |
Fig. 1.
Scatter plots showing cycle threshold (Ct) values and Ct shifts detected by rRT-PCR in four sample pools, composed of three negative and one positive sample, with respect to the individual positive samples for the ORF1ab gene
Fig. 2.
Scatter plots showing cycle threshold (Ct) values and Ct shifts detected by rRT-PCR in eight sample pools, composed of seven negative and one positive sample, with respect to the individual positive samples for the ORF1ab gene
Fig. 3.
Ct value shift due to sample pooling. Scatter plots showing cycle threshold (Ct) values and Ct shifts detected by rRT-PCR in ten sample pools, composed of nine negative and one positive sample, with respect to the individual positive samples for the ORF1ab gene
Discussion
Pooled testing is a known procedure that is usually used for the screening of a large number of samples to reduce the cost and facilitate the detection of infectious diseases especially in the sera of blood donors (Bilder and Tebbs 2012; Wein and Zenios 1996). In its most basic and simple form, pooled testing works by mixing individual samples together into one pool. If the pool test result is negative, then all samples within the pool are considered negative. If the pool test result is positive, then retesting of all samples individually is needed to identify the positive sample (Bilder and Tebbs 2012).
Monitoring the spread of COVID-19 is very important to contain the disease in its early stages and to ensure that the healthcare system is ready and prepared for proper control of the pandemic (de Salazar et al. 2020; Narayanan et al. 2020). Reverse transcription PCR is recommended for SARS-COV-2 diagnosis for its high sensitivity and specificity (Ouma et al. 2021; Qaqish et al. 2022). The establishment of molecular laboratories to cover the testing capacity required for massive screening to help the community is a real challenge, especially in low-resource settings, due to its high cost, limited number of available facilities, complex infrastructure requirements, and lack of trained personnel (Qaqish et al. 2022; Maniruzzaman et al. 2022).
The pooling method is critical in COVID-19 laboratories because it can increase testing capacity to meet the high demand for RT-PCR during population screening (de Salazar et al. 2020; Costa et al. 2021; Barathidasan et al. 2022; Praharaj et al. 2020). However, pooling SARS-COV-2 samples carries the risk of missing low viral loads due to sample dilution (Sawicki et al. 2021; Bateman et al. 2020; Hueda-Zavaleta et al. 2022).
In our study with a 3.01% prevalence out of 2200 samples tested, we aimed to evaluate the efficiency of the pooling strategy for SARS-COV-2 in nasopharyngeal samples in pool sizes of 4, 8, and 10 samples. Our study showed the false negative rate was 0% for pools with 4 samples and 1% for pools with 8 samples—even with low viral load samples of Ct > 31.
Pooled testing affected the sensitivity of the test in the low viral load samples for pools of 10 with a false negative rate of 9%; however, pooling remains an efficient and cost-effective method to contain the epidemic in developing countries at low prevalence.
Conclusion
Based on our study, we can summarize the strengths and weaknesses of the pooling technique as follows:
The numbers of results obtained per day are high, that allow the screening of large populations; false positive results are low because the sample was run more than once, individually and in a pool. Increased throughput by increasing the number of samples processed in a given time, in addition to cost reduction by conserving materials.
The study showed limitations to the pooling technique due to the decreased sensitivity of low viral load samples due to dilution. Pooling increased procedure complexity by adding more steps for pooling and individual samples to be repeated and by taking strict measures to prevent the possible cross-contamination of mixing samples together.
In conclusion, and after weighing advantages against limitations considering the low resource setting of the study, applying the pooling technique is better than not performing the test at all by expanding the testing capacity and overcoming the bottleneck and struggles of mass testing, in addition to reducing the cost, time, and manpower needs. Before implementing the pooling strategy, all laboratories must conduct validation studies with their kits and platforms for extraction and amplification at a known COVID-19 prevalence rate.
Acknowledgements
We thank all technologists of the COVID Laboratory, Fayoum University Hospital, for technical assistance and data collection during the current SARS-CoV-2 pandemic.
Author contribution
AMA: conceptualization; data curation; investigation; methodology; project administration; resources; writing—original draft; writing—review and editing. MNA: project administration; resources; supervision; writing—original draft; writing—review and editing. HMA: conceptualization; methodology; project administration; resources and investigation.
Data Availability
The data and materials that support the findings of the study are available from the corresponding author on reasonable request.
Compliance with ethical standard
Funding
This study was not supported by any funding.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of Fayoum University Research ethics committee, which is a member of the Egyptian Network Research Ethics Committee (ENREC) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
For this type of study, informed consent is not required.
Consent for publication
For this type of study, consent for publication is not required.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Abeer Mohamed Abdelrazik, Email: abdelrazik50@hotmail.com.
Manal Niazi El Said, Email: mniazy@gmail.com.
Hossam M. Abdelaziz, Email: hma05@fayoum.edu.eg
References
- Barak N, Ben-Ami R, Sido T, Perri A, Shtoyer A, Rivkin M, Licht T, Peretz A, Magenheim J, Fogel I, Livneh A, Daitch Y, Oiknine-Djian E, Benedek G, Dor Y, Wolf DG, Yassour M (2021) Hebrew University-Hadassah COVID-19 Diagnosis Team. Lessons from applied large-scale pooling of 133,816 SARS-CoV-2 RT-PCR tests. Sci Transl Med 13(589):eabf2823. 10.1126/scitranslmed.abf2823. Epub 2021 Feb 22. PMID: 33619081; PMCID: PMC8099176 [DOI] [PMC free article] [PubMed]
- Barathidasan R, Sharmila FM, Raj RV, Dhanalakshmi G, Anitha G, Dhodapkar R (2022) Pooled sample testing for COVID-19 diagnosis: evaluation of bi-directional matrix pooling strategies. J Virol Methods 304:114524. 10.1016/j.jviromet.2022.114524. Epub 2022 Mar 15. PMID: 35301022; PMCID: PMC8920575 [DOI] [PMC free article] [PubMed]
- Bateman AC, Mueller S, Guenther K, Shult P. Assessing the dilution effect of specimen pooling on the sensitivity of SARS-CoV-2 PCR tests. J Med Virol. 2020 doi: 10.1002/jmv.26519. [DOI] [PubMed] [Google Scholar]
- Bilder CR, Tebbs JM (2012) Pooled-testing procedures for screening high volume clinical specimens in heterogeneous populations. Stat Med 31(27):3261–8. 10.1002/sim.5334. Epub 2012 Mar 13. PMID: 22415972; PMCID: PMC3500568 [DOI] [PMC free article] [PubMed]
- Brault V, Mallein B, Rupprecht J-F (2021) Group testing as a strategy for COVID-19 epidemiological monitoring and community surveillance. PLoS Comput Biol 17(3):e1008726. 10.1371/journal.pcbi.1008726 [DOI] [PMC free article] [PubMed]
- Costa MS, Guimarães NS, Andrade AB, Vaz-Tostes LP, Oliveira RB, Simões MDS, Gelape GO, Alves CRL, Machado EL, Fonseca FGD, Teixeira SMR, Sato HI, Takahashi RHC, Tupinambás (2021) Detection of SARS-CoV-2 through pool testing for COVID-19: an integrative review. Rev Soc Bras Med Trop 12(54):e0276. 10.1590/0037-8682-0276-2021. PMID: 34787261; PMCID: PMC8582953 [DOI] [PMC free article] [PubMed]
- de Salazar A et al (2020) Sample pooling for SARS-CoV-2 RT-PCR screening.” Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases 26(12):1687.e1–1687.e5. 10.1016/j.cmi.2020.09.008 [DOI] [PMC free article] [PubMed]
- Grobe N, Cherif A, Wang X, Dong Z, Kotanko P. Sample pooling: burden or solution? Clin Microbiol Infect. 2021;27(9):1212–1220. doi: 10.1016/j.cmi.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueda-Zavaleta M, Copaja-Corzo C, Benites-Zapata VA, et al. Diagnostic performance of RT-PCR-based sample pooling strategy for the detection of SARS-CoV-2. Ann Clin Microbiol Antimicrob. 2022;21:11. doi: 10.1186/s12941-022-00501-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud SA, Ibrahim E, Thakre B, Teddy JG, Raheja P, Ganesan S, Zaher WA. Evaluation of pooling of samples for testing SARS-CoV- 2 for mass screening of COVID-19. BMC Infect Dis. 2021;21(1):360. doi: 10.1186/s12879-021-06061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniruzzaman M, Islam M, Ali M, Mukerjee N, Maitra S, Kamal MA, Rahman M (2022) COVID-19 diagnostic methods in developing countries. Environ Sci Pollut Res 1–14 [DOI] [PMC free article] [PubMed]
- Mutesa L, Ndishimye P, Butera Y, et al. A pooled testing strategy for identifying SARS-CoV-2 at low prevalence. Nature. 2021;589:276–280. doi: 10.1038/s41586-020-2885-5. [DOI] [PubMed] [Google Scholar]
- Narayanan KR, Frost I, Heidarzadeh A, Tseng K, Banerjee S, John J, Laxminarayan R (2020) Pooling RT-PCR or NGS samples has the potential to cost-effectively generate estimates of COVID-19 prevalence in resource limited environments. 10.1101/2020.04.03.20051995
- Pan Y, Zhang D, Yang P, Poon LLM, Wang Q (2020) Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis 20:411e2 [DOI] [PMC free article] [PubMed]
- Praharaj I, Jain A, Singh M, Balakrishnan A, Dhodapkar R, Borkakoty B. Pooled testing for COVID-19 diagnosis by real-time RT-PCR: a multi-site comparative evaluation of 5- & 10-sample pooling. Indian J Med Res. 2020;152:88–94. doi: 10.4103/ijmr.IJMR_2304_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaqish B, Sallam M, Al-Khateeb M, Reisdorf E, Mahafzah A. Assessment of COVID-19 molecular testing capacity in Jordan: a cross-sectional study at the country level. Diagnostics (basel) 2022;12(4):909. doi: 10.3390/diagnostics12040909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez MT, Rosario CD, Contreras E, Cabrera J, Degaudenzi AV, Ramírez RP (2021) Evaluation of sample pooling for the detection of SARS-CoV-2 in a resource-limited setting, Dominican Republic. Enferm Infecc Microbiol Clin (Engl Ed). 10.1016/j.eimc.2021.07.004. Epub ahead of print. PMID: 34334859; PMCID: PMC8310719 [DOI] [PMC free article] [PubMed]
- Sawicki R, Korona-Glowniak I, Boguszewska A, et al. Sample pooling as a strategy for community monitoring for SARS-CoV-2. Sci Rep. 2021;11:3122. doi: 10.1038/s41598-021-82765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouma OK, Ephraim K, Loyce N et al (2021) Role and utility of COVID-19 laboratory testing in low-income and middle-income countries: protocol for rapid evidence synthesis. MJ Open 11:e050296. 10.1136/bmjopen-2021-050296 [DOI] [PMC free article] [PubMed]
- U. S. Food and Drug Administration (FDA) (2021) Molecular diagnostic template for laboratories for Emergency Use Authorization (EUA)
- Wein LM, Zenios SA. Pooled Testing for HIV Screening: Capturing the Dilution Effect. Oper Res. 1996;44(4):543–569. doi: 10.1287/opre.44.4.543. [DOI] [Google Scholar]
- WHO (2020) COVID-19 - virtual press conference - 30 March 2020. Available at: https://www.who.int/docs/default-source/coronaviruse/transcripts/who-audio-emergencies-coronavirus-press-conference-full-30mar2020.pdf?sfvrsn=6b68bc4a_2
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z et al (2020) SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 382:1177e9 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and materials that support the findings of the study are available from the corresponding author on reasonable request.