Abstract
The Conserved Key Amino Acid Positions DataBase (CKAAPs DB) provides access to an analysis of structurally similar proteins with dissimilar sequences where key residues within a common fold are identified. CKAAPs may be important in protein folding and structural stability and function, and hence useful for protein engineering studies. This paper provides an update to the initial report of CKAAPs DB [Li et al. (2001) Nucleic Acids Res., 29, 329–331]. CKAAPs DB contains CKAAPs for the representative set of polypeptide chains derived from the CE and FSSP databases, as well as subdomains (conserved regions of the order of 100 residues within a domain) identified by CE. The new version now offers different perspectives on the CKAAPs. First, CKAAPs are mapped onto their respective Protein Data Bank (PDB) structures rendered by Molscript, providing a spatial context for the CKAAPs. Secondly, CKAAPs may be highlighted within a structure-based sequence alignment, as well as secondary structure alignment. Thirdly, the resulting sequence homologs from the structure alignment may be viewed in alignments colorized based on identities and property groups using Mview. New search capabilities have also been provided for searching by keyword combinations, PDB IDs, EC numbers, GI numbers, LocusLink ID, taxonomy, gene ontology and pathways. A new custom CKAAPs analysis interface has been implemented where a user may change the criteria for inclusion of chains, initiate CKAAPs analysis and retrieve results. CKAAPs DB is accessible through the web at http://ckaaps.sdsc.edu/. Plain text analysis results are available by FTP at ftp://ftp.sdsc.edu/pub/sdsc/biology/ckaap.
BACKGROUND
A Conserved Key Amino Acid Positions (CKAAPs) analysis attempts to better understand the relationship between protein sequence and protein structure. The CKAAP algorithm has been described previously (1). In summary, the algorithm analyzes the sequence relationship among representative polypeptides defined by structure alignment studies (2,3). Polypeptides (subsequences with <25% identity and at least 24 amino acids long) adopting a similar conformation are aligned to the master sequence based on the structural alignment in a pairwise fashion. The sequence space is expanded by obtaining the homologs of each subsequence from SWALL (combined SWISS-PROT, TrEMBL, TrEMBL New) using FASTA3 (4). Using a weighted scoring scheme, the algorithm attempts to provide an unbiased examination of the conservation of amino acid positions based on amino acid identities and property groups (1,5).
The importance of the CKAAPs identified for a number of common folds such as the immunoglobulin fold (IgFF) is well supported by existing experimental and theoretical studies found in the literature (1,6,7). CKAAPs are found not only within the expected hydrophobic core of proteins, but also in loops and turns. CKAAPs identify the majority of the nucleation/stabilization centers predicted by other methods (8,9). In cases where systematic mutation studies are available (10,11), CKAAPs are substantiated and may provide guidance for future studies. We have shown this in a recent study related to the Paracelsus Challenge (B.V.B.Reddy, W.W.Li and P.E.Bourne, submitted for publication).
The significance of CKAAPs is linked to the number of structures that can be aligned and the accuracy of that alignment. At present, no good statistical treatment for this exists. Our approach for the CKAAPs DataBase (CKAAPs DB) is to recalculate CKAAPs as more structures become available, as we improve our own alignment methodology (12) and to incorporate the alignment methodology of others, especially FSSP (3). CKAAPs DB is updated with each major release of the structural alignment databases, as well as sequence databases providing a means to easily retrieve and review a current list of those amino acid positions believed to be most crucial in a biological and structural role.
DATABASE
CKAAPs DB uses the Oracle 8i object relational database (http://ckaaps.sdsc.edu). It takes advantage of customized Oracle WebDB/Portal features to provide the query interface. New database server hardware has significantly improved the response time. Other custom interfaces are developed using PERL, DBI, javascript and XHTML.
At the time of writing, the database comprises the following features, including some beta features that are going into production shortly.
Content
1496 representative polypeptide chains determined by CE, which represent ∼40% of the 3800 CE representatives found in the June 2001 release. Each of the 1496 chains is similar to four or more other representative chains. The criteria for inclusion are a Z score >4.5 and an r.m.s.d. <3 Å (13,14). The minimal chain length is 30 residues for CE representatives.
997 representative polypeptide chains determined by FSSP, which represent ∼40% of the 2600 FSSP representatives in the June 2001 release. Each representative is similar to four or more other representatives. The criteria for inclusion in FSSP is based on a Z score >5.0 and an r.m.s.d. <3.0 Å (15). The minimal chain length is 31 residues for FSSP representatives.
From the combined set of CE and FSSP representatives, only 340 have identical Protein Data Bank (PDB) and chain identifiers.
Query and report features
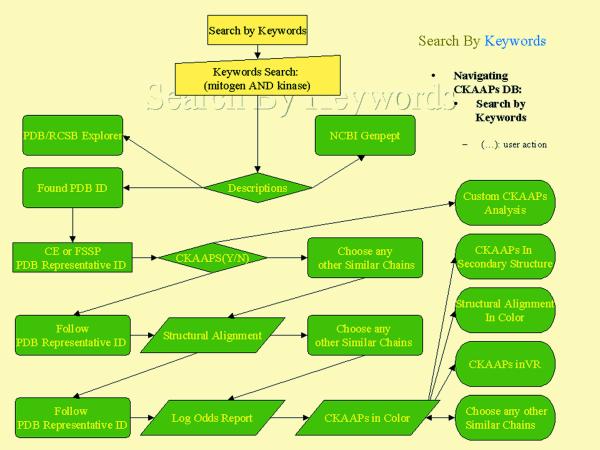
General queries are based on the following options: representative PDB IDs from CE or FSSP, any PDB ID, protein names, protein function, protein classification, enzyme classification numbers, GI number, sequence motif and keywords. Figure 1 illustrates a query flow chart using the keywords search feature. Searching by PDB ID remains the most direct approach.
A custom analysis interface is now available for users to specify only desired PDB IDs for custom CKAAPs analysis. The default is all available structures. Additionally, users may specify source (CE or FSSP), r.m.s.d., Z score, sequence length, sequence identity and alignment length as additional filters. The custom search is submitted, and results will be available at the web site or emailed to the user. The turn-around time depends on the number of chains chosen, the sequence length and the database size, and currently averages 1–2 min per chain.
The report interface has been significantly improved. The user may take the results from any of the search steps above and analyze a representative PDB ID from CE and FSSP. This includes a review of the CKAAPs with respect to location in the sequence, physicochemical properties and location within secondary structures as determined by Kabsch and Sander (16). If a representative PDB ID is not found, typically because there are less than four similar chains, there are two options. Either the user may perform a custom analysis with a different set of criteria or choose an alternative structure from a list of structural homologs (if they exist).
Queries by LocusLink ID, taxonomy, gene ontology, SCOP classifications and KEGG pathways are under development and will be available at the time of publication.
Figure 1.
Flow chart of searching CKAAPs DB by keyword. Alternative searches by PDB ID, sequence motif, etc. (data not shown) all converge on finding a CE or FSSP representative PDB and polypeptide chain ID. With this ID the user may review structure alignments, log odds report, properties of CKAAPs and locations within secondary and tertiary structure.
Display features
The number of CKAAPs displayed by default is set to be 20% of the average sequence length of the set of aligned sequences. The combined score from the amino acid identity and property group conservation is used to rank the amino acids within this cutoff. The rank of ‘a’ is the highest scoring position, ‘b–z’ ranks lower in that order; ‘A–Z’ is used for ranks lower than 26, and so on. A lookup table of a rank and its respective character designation is provided.
A confidence level is calculated based on the random withdrawal of 20% of the represented sequences. Currently 50–500 iterations are performed based on the number of aligned chains multiplied by 10, not to exceed 500. CKAAPs that are present 100% of the time are given a confidence level of 9, with a range down to 0 (present <20% of the time). A cumulative rank score is calculated such that a rank of ‘a’ is representative of the whole iterative process, and not just a single run.
A profile or log odds matrix is also provided with each structure-sequence alignment. The log odds matrix provides a complete picture of the potential residues that may occur at each position. This allows the user to combine the information from the property grouping to determine which group of amino acids are most likely to occur at each position.
Rendering of the CKAAPs using the molecular graphics program Molscript (17) is available and provides a spatial representation for the assessment of CKAAPs. The CKAAPs may be colored by rank or confidence. All atoms of the CKAAP residues may also be drawn using van der Waals radii.
Rendering of structure and sequences alignments colored by sequence identity and property groups is done using Mview (18).
The PDB (http://www.rcsb.org) maintains secondary structure assignments according to the method of Kabsch and Sander (16) for all polypeptide chains within the PDB. CKAAPs may be highlighted in this context.
All reports contain links to the NCBI (http://www.ncbi.nlm.nih.gov) GenPept database and the RCSB PDB explorer (http://www.rcsb.org/pdb/cgi/explore.cgi).
Maintenance
1. SWALL is updated monthly, CE is updated every 3 months, FSSP is updated every month and CKAAPs DB is updated approximately every 2 months.
2. Currently, database searches are performed using FASTA3 parallel programmed for high-performance computing (4). For the current release, it takes ~1000 h of computer time at the current cutoff specified above. PSI-BLAST searches are being made available as a user requested option. We are currently using Linux clusters to parallelize PSI-BLAST searches of the NCBI nr database.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the National Biomedical Computation Resource (NIH P41 RR 08605-06), the National Science Foundation grant DBI 9808706 and the Microstructure Image-Based Collaboratory grant (NCRR 5 P41 RR04050-10S1).
REFERENCES
- 1.Reddy B.V., Li,W.W., Shindyalov,I.N. and Bourne,P.E. (2001) Conserved key amino acid positions (CKAAPs) derived from the analysis of common substructures in proteins. Proteins, 42, 148–163. [DOI] [PubMed] [Google Scholar]
- 2.Shindyalov I.N. and Bourne,P.E. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 3.Holm L. and Sander,C. (1996) The FSSP database: fold classification based on structure–structure alignment of proteins. Nucleic Acids Res., 24, 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearson W.R. (1994) Using the FASTA program to search protein and DNA sequence databases. Methods Mol. Biol., 24, 307–331. [DOI] [PubMed] [Google Scholar]
- 5.Taylor W.R. (1986) The classification of amino acid conservation. J. Theor. Biol., 119, 205–218. [DOI] [PubMed] [Google Scholar]
- 6.Mirny L.A. and Shakhnovich,E.I. (1999) Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J. Mol. Biol., 291, 177–196. [DOI] [PubMed] [Google Scholar]
- 7.Clarke J., Cota,E., Fowler,S.B. and Hamill,S.J. (1999) Folding studies of immunoglobulin-like β-sandwich proteins suggest that they share a common folding pathway. Struct. Fold Des., 7, 1145–1153. [DOI] [PubMed] [Google Scholar]
- 8.Demirel M.C., Atilgan,A.R., Jernigan,R.L., Erman,B. and Bahar,I. (1998) Identification of kinetically hot residues in proteins. Protein Sci., 7, 2522–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michnick S.W. and Shakhnovich,E. (1998) A strategy for detecting the conservation of folding-nucleus residues in protein superfamilies. Fold Des., 3, 239–251. [DOI] [PubMed] [Google Scholar]
- 10.Brown B.M. and Sauer,R.T. (1999) Tolerance of Arc repressor to multiple-alanine substitutions. Proc. Natl Acad. Sci. USA, 96, 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalal S., Balasubramanian,S. and Regan,L. (1997) Transmuting α helices and β sheets. Fold Des., 2, R71–R79. [DOI] [PubMed] [Google Scholar]
- 12.Guda C., Scheeff,E., Bourne,P. and Shindyalov,I. (2001) A new algorithm for alignment of multiple protein structures using Monte Carlo optimization. Pacific Symp. Biocomp., 275–286. [DOI] [PubMed] [Google Scholar]
- 13.Li W.W., Reddy,B.V., Shindyalov,I.N. and Bourne,P.E. (2001) CKAAPs DB: a conserved key amino acid positions database. Nucleic Acids Res., 29, 329–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shindyalov I.N. and Bourne,P.E. (2000) An alternative view of protein fold space. Proteins, 38, 247–260. [PubMed] [Google Scholar]
- 15.Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. and Sander,C. (1993) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 17.Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr., 55, 938–940. [DOI] [PubMed] [Google Scholar]
- 18.Brown N.P., Leroy,C. and Sander,C. (1998) MView: a web-compatible database search or multiple alignment viewer. Bioinformatics, 14, 380–381. [DOI] [PubMed] [Google Scholar]