Abstract
Background: Neurodegenerative diseases, such as Alzheimer's disease patients (AD), Huntington's disease (HD) and Parkinson’s disease (PD), are common causes of morbidity, mortality, and cognitive impairment in older adults.
Objective: We aimed to understand the transcriptome characteristics of the cortex of neurodegenerative diseases and to provide an insight into the target genes of differently expressed microRNAs in the occurrence and development of neurodegenerative diseases.
Methods: The Limma package of R software was used to analyze GSE33000, GSE157239, GSE64977 and GSE72962 datasets to identify the differentially expressed genes (DEGs) and microRNAs in the cortex of neurodegenerative diseases. Bioinformatics methods, such as GO enrichment analysis, KEGG enrichment analysis and gene interaction network analysis, were used to explore the biological functions of DEGs. Weighted gene co-expression network analysis (WGCNA) was used to cluster DEGs into modules. RNA22, miRDB, miRNet 2.0 and TargetScan7 databases were performed to predict the target genes of microRNAs.
Results: Among 310 Alzheimer's disease (AD) patients, 157 Huntington's disease (HD) patients and 157 non-demented control (Con) individuals, 214 co-DEGs were identified. Those co-DEGs were filtered into 2 different interaction network complexes, representing immune-related genes and synapse-related genes. The WGCNA results identified five modules: yellow, blue, green, turquoise, and brown. Most of the co-DEGs were clustered into the turquoise module and blue module, which respectively regulated synapse-related function and immune-related function. In addition, human microRNA-4433 (hsa-miR-4443), which targets 18 co-DEGs, was the only 1 co-up-regulated microRNA identified in the cortex of neurodegenerative diseases.
Conclusion: 214 DEGs and 5 modules regulate the immune-related and synapse-related function of the cortex in neurodegenerative diseases. Hsa-miR-4443 targets 18 co-DEGs and may be a potential molecular mechanism in neurodegenerative diseases' occurrence and development.
Keywords: Neurodegenerative diseases, differentially expressed genes, bioinformatics analysis, weighted gene co-expression network analysis, differentially expressed microRNA, risk factor
1. INTRODUCTION
Neurodegenerative diseases, such as Alzheimer's disease patients (AD), Huntington's disease (HD) and Parkinson’s disease (PD), are common causes of morbidity, mortality, and cognitive impairment in older adults [1]. The increasing prevalence of neurodegenerative diseases brings excellent social and economic burdens and has become a global problem [2]. Therefore, the pathology and therapy of neurodegenerative diseases urgently need to be studied.
Cortex atrophy and dysfunction are common pathological characteristics of neurodegenerative diseases and are associ-ated with behavioral and emotional symptoms [3, 4]. The development of high throughput sequencing and bioinformatics technology provides new ideas for the transcriptome of cortex in neurodegenerative diseases to study molecular mechanisms and therapeutic targets of neurodegenerative diseases [5-7]. Weighted gene co-expression network analysis (WGCNA) is an efficient statistical method to describe the correlation patterns of expressed genes to find highly correlated gene clusters, known as modules, and to identify the characteristics of gene modules to find the molecular mechanisms involved in the occurrence and development of diseases and to screen biomarkers or potential therapeutic targets [8].
MicroRNAs, small RNA molecules, are associated with numerous neurodegenerative diseases with their role in regulating gene expressions by binding to target mRNAs [9]. Therefore, microRNAs and their target genes are intensely studied as candidates for diagnostic and prognostic biomarkers, as predictors of drug response and as therapeutic agents of diseases, including neurodegenerative diseases [10]. For instance, miR-101 [11], miR-153 [12] and miR-339-5p [13] inhibit the expression of amyloid-β precursor protein. MicroRNA-346 upregulated amyloid-β precursor protein [14]. Prediction of target genes of differentially expressed microRNAs in neurodegenerative diseases could be performed by several databases, such as RNA22 (https://cm.jefferson.edu/rna22/Precomputed/) [15], miRDB (http://mirdb.org/) [16, 17], miRNet 2.0 (https://www.mirnet.ca/miRNet/home.xhtml) [18] and TargetScan7 (http://www.cuilab.cn/transmir) [19].
Here, in the present study, GSE33000, GSE157239, GSE64977 and GSE72962 datasets were analyzed to obtain more insight into the mRNA and microRNA transcriptome characteristics of the cortex of neurodegenerative diseases. GO and KEGG enrichment analyses, gene interaction network analysis and WGCNA were performed on the differentially expressed genes (DEGs) to profile functional and molecular mechanisms in the occurrence and development of neurodegenerative diseases. RNA22, miRDB, miRNet 2.0 and TargetScan7 databases were accustomed to recognize the target genes of differently expressed microRNAs to find biomarkers or potential therapeutic targets of neurodegenerative diseases.
2. MATERIALS AND METHODS
2.1. Data Set
The data discussed in this publication were deposited in NCBI’s Gene Expression Omnibus and accessible through GEO Series accession numbers GSE33000, GSE157239, GSE64977 and GSE72962 (https://www.ncbi.nlm.nih.gov/ geo). GSE33000 dataset, including 310 AD patients, 157 HD patients, and 157 non-demented control (Con) individuals, was a gene expression dataset of human prefrontal cortex brain tissues [5]. The GSE157239 dataset was a microRNA expression dataset from the post-mortem temporal cortex of 8 AD patients and 8 non-demented Con individuals [20]. GSE64977 dataset was a microRNA-seq expression from 28 HD patients and 36 normal human (Con) post-mortem prefrontal cortex brain samples [21]. The GSE72962 dataset was microRNA profiles in 29 PD patients and 33 non-demented Con individuals [22].
2.2. DEGs Analysis
The Limma package of R language was applied to identify the DEGs and differential expressed microRNAs [23]. The mRNA and microRNA meeting |FC| > 2 and P-value < 0.05 were the required data. The heat map and volcano plot were plotted using pheatmap and ggplot2 packages.
2.3. Functional and Pathway Enrichment Analyses
Both GO and KEGG pathways were the biological sequence analysis methods that could effectively cluster functional genes into different biological processes, mainly used to study DNA and protein-related issues [24]. Next, the DAVID database (https://david.ncifcrf.gov/tools.jsp) took advantage of by performing GO and KEGG analyses on differential expression genes. These analyses were mapped with Bioinformatics (http://www.bioinformatics.com.cn/). The P-value and false discovery rate (FDR) were controlled at the 0.05 threshold.
2.4. Gene Interaction Network Analysis
The gene interaction networks were analyzed by the STRING database version 11.0 (http://string-db.org) [25]. The interaction networks of differential expression genes were visualized by Cytoscape 3.6.0 software (http://www.cytoscape.org/).
2.5. Weighted Gene Co-Expression Network Analysis
The WGCNA package of R language was applied to perform weighted gene co-expression network analysis on the differential expression genes. In this study, 214 co-differentially expressed genes were included in the weighted gene co-expression network model. The optimal soft threshold (power) was selected as 10 and the min Module Size was [5]. The interaction networks of intramodular genes were visualized by Cytoscape 3.6.0 software.
2.6. Prediction of Target Genes of Differentially Expressed microRNAs
RNA22 (https://cm.jefferson.edu/rna22/Precomputed/) [15], miRDB (http://mirdb.org/) [16, 17], miRNet2.0 (https://www.mirnet.ca/miRNet/home.xhtml) [18] and TargetScan7 (http://www.cuilab.cn/transmir) [19] databases were accustomed to recognize the target genes of microRNAs.
2.7. Statistics
R 3.5.1 was used for statistical analysis. P values less than 0.05 were considered statistically significant.
3. RESULTS
3.1. DEGs in Prefrontal Cortex of Neurodegenerative Diseases
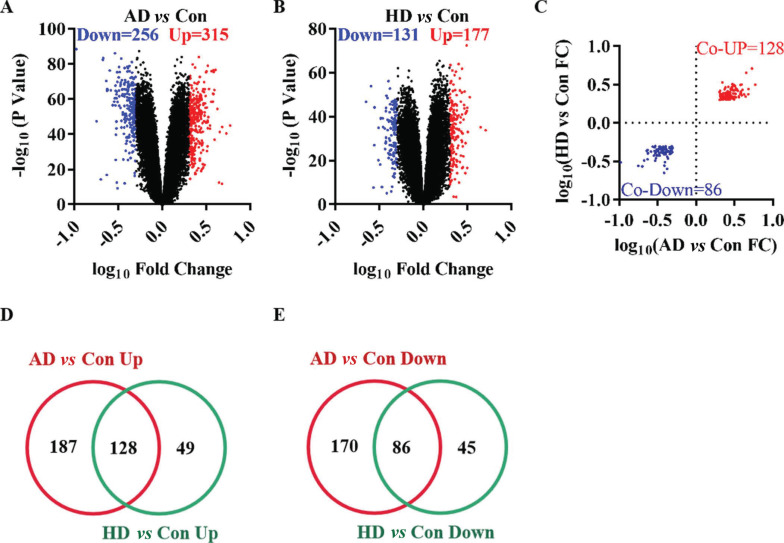
The GSE33000 dataset included three groups of samples: 310 AD patients, 157 HD patients and 157 non-demented Con individuals. Compared with the Con group, 571 DEGs were identified in the prefrontal cortex of the AD group, including 315 up-regulated genes and 256 down-regulated genes (Fig. 1A). In the HD group, 308 DEGs were identified compared with the Con group, including 177 up-regulated and 131 down-regulated genes (Fig. 1B). Among the total 214 co-DEGs in AD and HD (Fig. 1C), 128 co-up-regulated genes (Fig. 1D) and 86 co-down-regulated genes (Fig. 1E) were found to represent the effect of gene expressions on neurodegenerative diseases. These 214 co-DEGs, functional and pathway enrichment analyses, gene interaction network analyses and WGCNA were further performed.
Fig. (1).
The differentially expressed genes in Alzheimer's disease and Huntington's disease. (A) The volcano plot of AD group compared with control group. (B) The volcano plot of HD group compared with control group. (C) The fold change of co-GEGs. (D) The Veen diagram of up-regulated genes. (E) The Veen diagram of down-regulated genes.
Abbreviations: AD: Alzheimer's disease group. HD: Huntington's disease group. Con: control group. FC: fold change.
3.2. Functional, Pathway enrichment and Gene Interaction Network Analyses of DEGs in Neurodegenerative Diseases
GO and KEGG analyses were performed to summarize the functional and pathway enrichment of the co-DEGs. In GO analyses, co-DEGs were enriched in 80 biological processes (BP) terms, [25] cellular component (CC) terms and [17] molecular function (MF) terms (Fig. S1 (1.3MB, pdf) ). Among the BP terms, immune-related biological processes, such as innate immune response, immune response and complement activation, and synapse-related biological processes, such as synapse, synaptic vesicle, presynaptic membrane and synaptic vesicle membrane, were enriched. Enrichment analysis of the KEGG pathway showed that co-DEGs were mainly related to 19 pathways (Fig. S2 (1.3MB, pdf) ), such as the MAPK signaling pathway, staphylococcus aureus infection, and epithelial cell signaling in Helicobacter pylori infection.
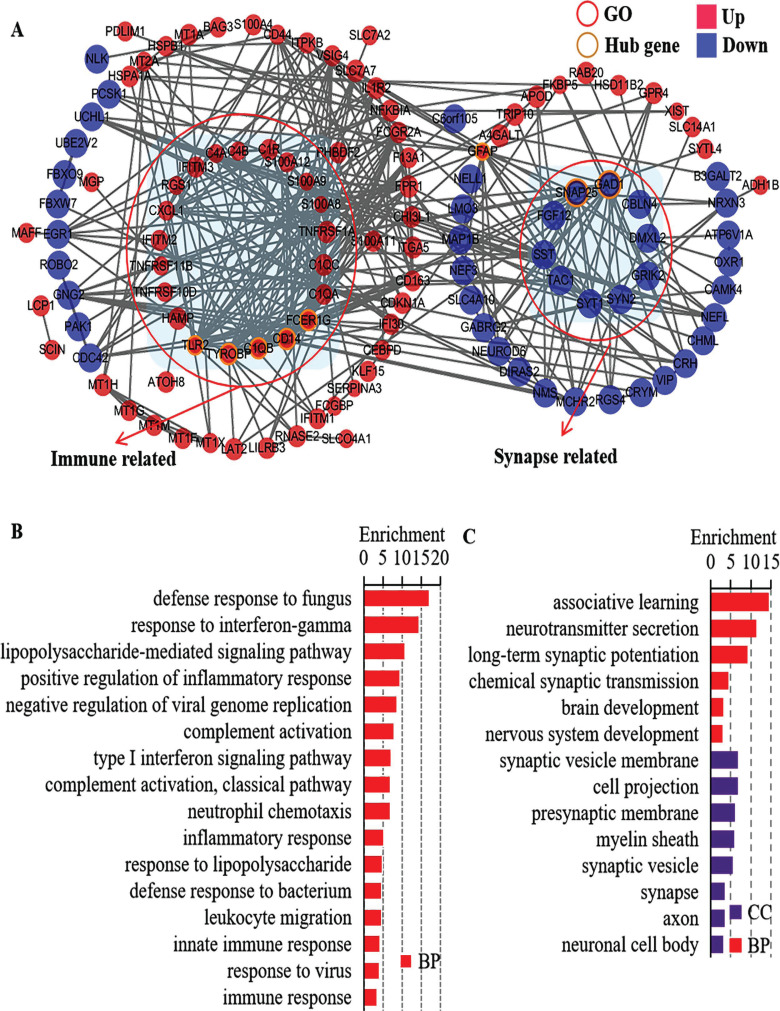
In the interactions network analyses, the 214 co-DEGs were filtered into 2 different interaction network complexes, containing 119 nodes and 349 edges (Fig. 2A and Table S1 (1.3MB, pdf) ). The two different interaction network complexes were immune-related genes and synapse-related genes. In addition, C1QB, CD14, FCER1G, GAD1, GFAP, SNAP25, TLR2 and TYROBP interacted with at least 16 co-DEGs, and were defined as the hub genes. The immune-related BP terms in GO analysis, such as defense response to fungus, response to interferon-gamma, and lipopolysaccharide-mediated signaling pathway, are presented in Fig. (2B). The synapse-related GO terms, such as long-term synaptic potentiation, chemical synaptic transmission, synaptic vesicle membrane, and presynaptic membrane, are presented in Fig. (2C).
Fig. (2).
Gene interaction network analyses and functional enrichment of the co-differentially expressed genes (co-DEGs) in neurodegenerative disease. (A) Gene interaction network analyses of co-DEGs. (B) Functional enrichment of the immune related co-DEGs. (C) Functional enrichment of the synapse related co-DEGs.
Abbreviations: AD: Alzheimer's disease group. HD: Huntington's disease group. Con: Control group. CC: Cell component. BP: Biological process.
3.3. WGCNA of DEGs in Neurodegenerative Diseases
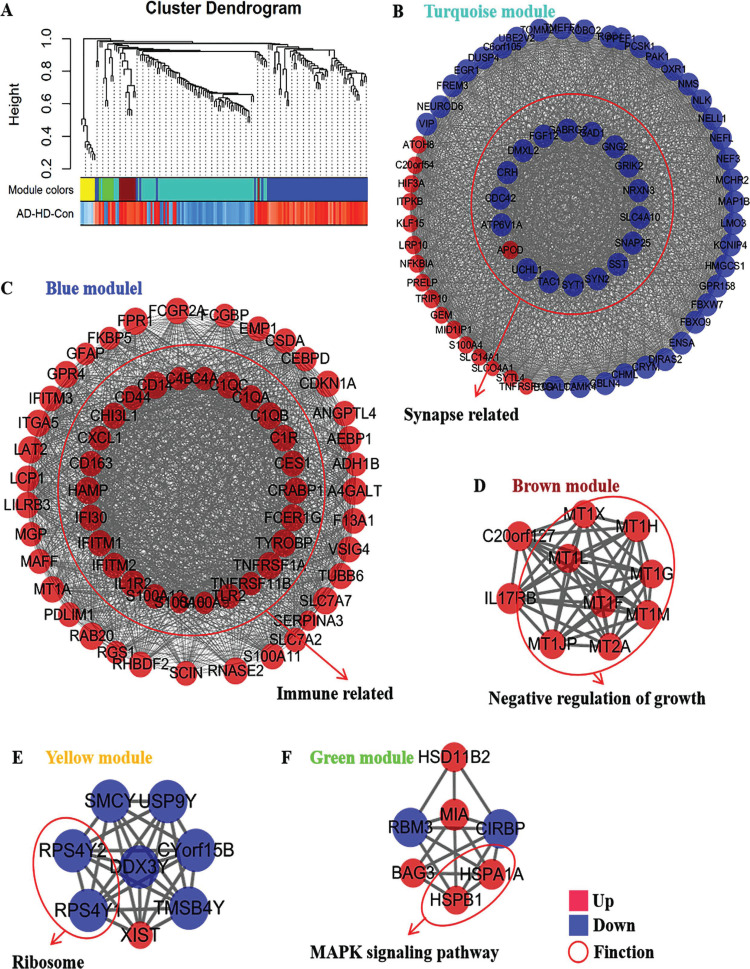
According to the expression trend of genes between the AD, HD and Con groups, 214 co-DEGs were included in the weighted gene co-expression network model. In this study, the optimal soft threshold (power) was selected as 10 and the min Module Size as 5. By average linkage hierarchical clustering, five modules were identified and represented in different colors, including yellow module, blue module, green module, turquoise module and brown module (Fig. 3A). Based on the WGCNA results, the interaction network of intramodular genes is presented in Figs. (3B-F). Most DEGs were clustered among the modules into turquoise and blue modules. The turquoise module was associated with the synapse-related function (Fig. 3B and Table S2 (1.3MB, pdf) ). The Blue module was associated with the immune-related function (Fig. 3C and Table S2 (1.3MB, pdf) ). Besides, the brown module negative regulated growth (Fig. 3D), the yellow module was associated with the ribosome (Fig. 3E), and the green module regulated MAPK signaling pathway (Fig. 3F).
Fig. (3).
Weighted gene co-expression network analysis of the co-differentially expressed genes in in neurodegenerative disease. (A) Cluster dendrogram of co-differentially expressed genes. (B) Interaction network of intramodular genes in turquoise module. (C) Interaction network of intramodular genes in blue module. (D) Interaction network of intramodular genes in brown module. (E) Interaction network of intramodular genes in yellow module. (F) Interaction network of intramodular genes in green module.
3.4. Hsa-miR-4443 Increased in Cortex of Neurodegenerative Diseases and Target 18 DEGs.
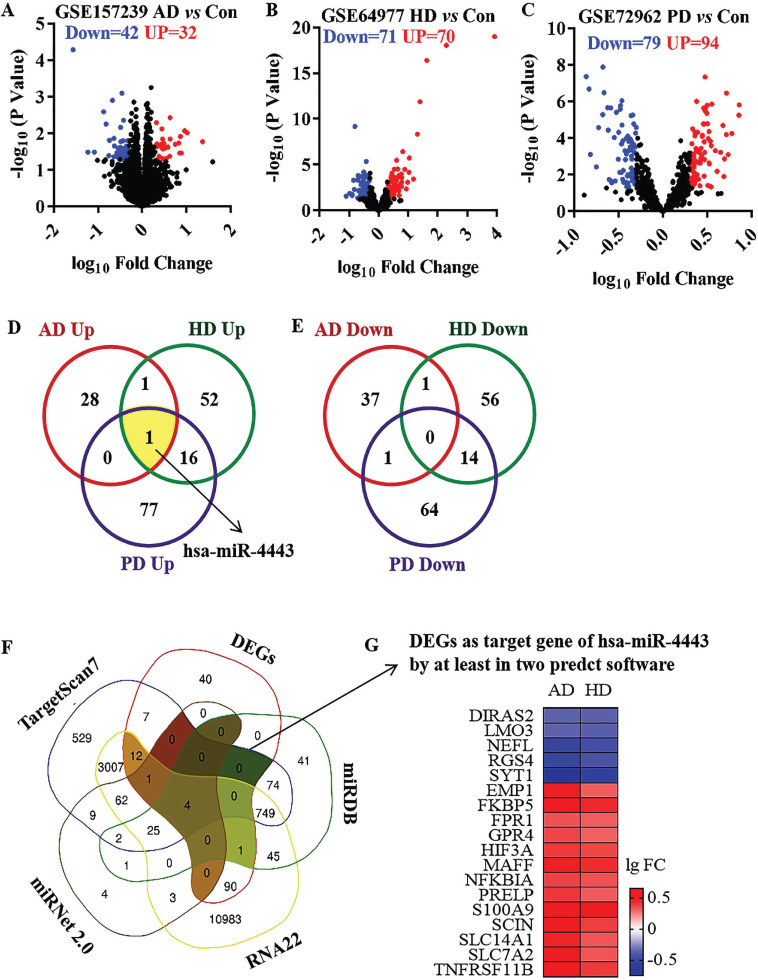
The GSE157239 dataset included 8 AD patients and 8 non-demented Con individuals. Compared with the Con group, 74 differentially expressed microRNAs were identified in the cortex of the AD group, including 32 up-regulated microRNAs and 32 down-regulated microRNAs (Fig. 4A). In GSE64977 dataset, 28 HD patients and 36 non-demented Con individuals were included. Compared with the Con group, 141 differentially expressed microRNAs were identified in the prefrontal cortex of the HD group, including 70 up-regulated microRNAs and 71 down-regulated microRNAs (Fig. 4B). The GSE72962 dataset included 29 PD patients and 33 non-demented Con individuals. 173 differentially expressed microRNAs were identified in the prefrontal cortex of the PD group, including 94 up-regulated microRNAs and 79 down-regulated microRNAs (Fig. 4C). Among the differentially expressed microRNAs, only 1 co-up-regulated microRNA, human microRNA-4433 (hsa-miR-4443), and no co-down-regulated microRNA were identified in neurodegenerative diseases (Figs. 4D and E). The present study was performed to predict the target genes of hsa-miR-4443, RNA22, miRDB, miRNet 2.0 and TargetScan7 databases. 18 co-DEGs were regulated by hsa-miR-4443 (Fig. 4F). Among the 18 target DEGs, 13 genes were up-regulated, and 5 genes were down-regulated in neurodegenerative diseases (Fig. 4G).
Fig. (4).
The differentially expressed microRNAs in neurodegenerative disease and the target DEGs of hsa-miR-4443. (A) The volcano plot of microRNAs in AD group compared with control group. (B) The volcano plot of microRNAs in HD group compared with control group. (C) The volcano plot of microRNAs in PD group compared with control group. (D) The Veen diagram of up-regulated microRNAs in neurodegenerative disease. (E) The Veen diagram of down-regulated microRNAs in neurodegenerative disease. (F) The Veen diagram of the target genes of hsa-miR-4443. (G) Heatmap of DEGs as target gene of hsa-miR-4443 by at least in two predct software.
Abbreviations: AD: Alzheimer's disease group. HD: Huntington's disease group. PD: Parkinson’s disease. Con: control group. FC: fold change.
4. DISCUSSION
In the present study, GSE33000 dataset was analyzed to get more insight into the transcriptome characteristics of the cortex of neurodegenerative diseases. Among 310 AD patients, 157 HD patients and 157 non-demented Con individuals, 214 co-DEGs, including 128 co-up-regulated genes and 86 co-down-regulated genes, were identified. Those co-DEGs were enriched into 80 biological processes (BP) terms, 25 cellular component (CC) terms, 17 molecular function (MF) terms and 19 KEGG pathways. In the interaction network analysis, the 214 co-DEGs were filtered into 2 different interaction network complexes, which represented immune-related genes and synapse-related genes. Based on the WGCNA results, a total of five modules, including yellow module, blue module, green module, turquoise module and brown module, were identified. Most co-DEGs were clustered into turquoise and blue modules, which respectively regulated synapse-related and immune-related functions. In addition, GSE157239, GSE64977 and GSE72962 datasets were analyzed to identify the differentially expressed microRNAs in neurodegenerative diseases. Only 1 co-up-regulated microRNA hsa-miR-4443 and no co-down-regulated microRNA were identified in neurodegenerative diseases. Predicted by RNA22, miRDB, miRNet 2.0 and TargetScan7 databases, 18 co-DEGs were regulated by hsa-miR-4443.
Neurodegenerative diseases are classified according to primary clinical features (e.g., dementia, parkinsonism, or motor neuron disease), anatomic distribution of neurodegeneration (e.g., frontotemporal degenerations, extrapyramidal disorders, or spinocerebellar degenerations), or principal molecular abnormality [26]. In the present study, three prevalent neurodegenerative diseases, AD, HD and Parkinson's syndrome, were included. 128 co-up-regulated genes and 86 co-down-regulated genes were identified in neurodegenerative diseases. Based on the functional enrichment and gene interaction network analyses, immune-related and synapse-related function was enriched by DEGs of neurodegenerative diseases. Interestingly, most immune-related DEGs were increased, and synapse-related DEGs were enriched in this study, indicating the serious neuroinflammation and synaptic impairments of the cortex in neurodegenerative diseases. The immune system is inextricably involved in shaping the brain during development by not only mediating damage but also regeneration and repair. The growing awareness has stimulated therapeutic approaches to modulate the immune system in neurodegenerative diseases [27]. In gene interaction network analyses, close connections were found between the immune-related DEGs and synapse-related DEGs. Then, implied neuroinflammation induces synaptic impairments to aggravate behavioral and emotional symptoms of neurodegenerative diseases.
Supplement to bioinformatics analysis which mainly focuses on strong effect genes and genes with known functions, WGCNA is to observe the function of genes with similar expression trend and weak effect genes [28]. WGCNA has been successfully applied in AD [29], Parkinson's syndrome [30], cancer [31], ischemic stroke [32] and other diseases [33] to find the molecular mechanisms involved in the occurrence and development of diseases and to screen biomarkers or potential therapeutic targets. In this study, five modules, including yellow, blue, green, turquoise, and brown, were identified based on the WGCNA results. Similarly, with gene interaction network analyses, most of the co-DEGs were clustered into synapse-related module (turquoise module) and immune-related module (blue module), which might be as a whole as the characteristic targets of neurodegenerative diseases.
MicroRNAs and their target genes are intensely studied as candidates for diagnostic and prognostic biomarkers. For instance, miR-146a and miR-155 are involved in brain development and neurodegeneration, and dysregulation of these two miRNAs in AD is a potential therapeutical implication [34]. In addition, down-regulation of MiR-107 worsens spatial memory through suppression of the NF-κB signaling pathway and SYK in AD model mice [35]. In the present study, hsa-miR-4443 (miRbase accession MI0016786, http://www.mirbase.org/cgibin/mirna_entry.pl?acc=MI0016786) was the only 1 co-differently expressed microRNA identified in this study. Hsa-miR-4443 was reported to be involved in several types of cancer and in the biology of the immune system, such as participating in the malignancy of breast cancer [36, 37], tumors of glial origin [38, 39] and head and neck squamous cell carcinoma [40], suppressing invasiveness of ovarian cancer [41] and colon cancer [42], promoting the resistance of non-small cell lung cancer cells to epirubicin [43, 44], downregulated in hepatocellular carcinoma [45] peripheral blood mononuclear cells of non-segmental vitiligo (NSV) patients [46] and acute Kawasaki disease [47, 48], and regulating T cell-mediated inflammatory processes [49] and monocyte activation [50]. Although no studies on the roles of hsa-miR-4443 in neurodegenerative diseases have been reported yet, the effects of hsa-miR-4443 on tumors of glial origin [38, 39] and immune cells [49, 50] indicated hsa-miR-4443 regulating glial cells and neuroimmunity to participate in occurrence of neurodegenerative diseases.
Among the 18 target DEGs, EMP1 [51], FKBP5 [52, 53], FPR1 [54, 55], GPR4 [56], MAFF [57], NFKBIA [58], S100A9 [59, 60], SLC14A1 [61], SLC7A2 [62], TNFRSF11B [63], LMO3 [64], NEFL [65], RGS4 [66], and SYT1 [67] were reported associated with neurodegenerative diseases. In a previous study, EMP1, the A2-astrocytes marker, was significantly decreased in Aβ-activated microglia conditioned medium-treated astrocytes, indicating Aβ could indirectly activate A1 astrocytes by Aβ-activated microglia [68]. FKBP5, the FKB506 binding protein 51 gene, is a critical regulator of glucocorticoid receptor activity and HPA function. FKBP5 has been found regulative for tau oligomerisation and Aβ toxicity [69]. While, lowering the levels of FKBP5 reduced HTT in HD models both in vitro and in vivo [70]. FKBP5 knockout AD models exhibit few phenotypic changes or behavioral alterations [71, 72] and, tau levels were reduced throughout the brains of Fkbp5-/- mice [73, 74]. These results supported the modulation of FKBP5 as a therapeutic target for neurological diseases. FPR1/2 was strongly increased in the cortex and hippocampus of APP/PS1 transgenic mice [75]. While, the FPR1/2 antagonist Boc2-treatment significantly improved spatial memory performance, reduced neuronal pathology, induced the expression of homeostatic growth factors, ameliorated microglia reactivity and reduced the elevated levels of amyloid plaques in the hippocampus [54]. The expression of MAFF was increased in the hippocampus of AD patients and significantly increased in patients with Braak stage V-VI compared to those with Braak stage III-IV. While, MAFF knockdown suppressed the increase of glutathione induced by Aβ, suggesting MAFF as a potential therapeutic target in AD [76]. NFKBIA, nuclear factor-κB inhibitor alpha, is the primary negative regulator of NF-κB, an important transcription factor that has been suggested to have a role in synaptic plasticity by affecting LTP and neuronal health [77-79]. NFKBIA was reported upregulated in AD patients and could be used as a potential biomarker for AD [80, 81]. Previous studies found the pro-inflammatory protein S100A9 was correlated with Aβ(1-42) levels in cerebrospinal fluid samples of stable mild cognitive impairment individuals [82]. While, knockdown of S100A9 increased spatial reference memory in the Morris water maze task and Y-maze task, decreased Aβ neuropathology and phosphorylated tau, increased expression of anti-inflammatory IL-10, and also decreased expression of inflammatory IL-6 and TNF-α in AD model mice [82, 83]. In a study of RNA-sequencing on striatal tissue from a cohort of 5-y-old OVT73-line sheep expressing a human CAG-expansion HTT cDNA transgene, the levels of the urea transporter SLC14A1 significantly increased in the OVT73 striatum, indicating that aberrant urea metabolism could be the primary biochemical disruption initiating neuropathogenesis in HD [84]. RGS4 was reported as a biomarker of schizophrenia [85]. Overexpression of RGS4 caused a comparable reduction in tau [86]. Neurofilament light (NEFL) and synaptotagmin 1 (SYT1) were all significantly reduced in the hippocampus of 12-month-old AD mice, which expressed mutant (V717I) AβPP [87]. NEFL is also an established biomarker of early neuronal injury and axonal degeneration in preclinical AD [88] and correlated with tau inclusion formation at transcriptomic and proteomic levels [89]. While, NEFL deficiency significantly increased neocortical DN pathology, Aβ deposition, synapse vulnerability, and microgliosis in APP/PS1 mice. Thus, EEFL may have a role in protecting neurites from dystrophy and regulating cellular pathways related to the generation of Aβ plaques [90]. SYT1 have a regulatory role in membrane interactions during synaptic vesicle trafficking in the active synapse zone [91]. SYT1 positive correlated with the hippocampal expression of BDNF [92], and could regulate synaptic Aβ, Aβ level, and the Aβ42/40 ratio [93].
Besides, HIF3A, PRELP, SCIN and DIRAS2 were novel discovered genes that also played an important role in the occurrence and development of neurodegenerative diseases. Future research should focus on the potential mechanisms of those genes in neuron cells, gail cells, C.elegans or mice models to identify therapeutic targets for reducing the risk of neurodegenerative diseases.
In the present study, the DEGs were analyzed from the prefrontal cortex brain tissues of 310 AD patients, 157 HD patients, and 157 Con individuals. The differentially expressed microRNAs were identified from the temporal cortex of 8 AD patients compared to 8 non-demented Con individuals, the prefrontal cortex of 28 HD patients compared to 36 Con individuals, and the prefrontal cortex of 29 PD patients compared to 33 non-demented Con individuals. Although all those diseases presented cortex atrophy, and the sub-regional atrophy differed [94]. Therefore, spatial transcriptomics should be used in future studies [95]. Besides, we cannot confirm the cell type of the DEGs in the present study. Single-cell sequencing could be an effective method to find disease-related genes and RNA signatures of single cell type in the brain [96].
CONCLUSION
In conclusion, 214 differentially expressed genes and 5 modules regulate the immune-related and synapse-related function of the cortex in neurodegenerative diseases. Hsa-miR-4443 targeting 18 co-DEGs may be a potential molecular mechanism in the occurrence and development of neurodegenerative diseases. Our results contribute to a better understanding of the molecular events of neurodegenerative diseases. Future research should focus on the potential mechanisms of Hsa-miR-4443 and the target genes to identify therapeutic targets for neurodegenerative diseases.
ACKNOWLEDGEMENTS
Declared none.
LIST OF ABBREVIATIONS
- Hsa-miR-4443
Human microRNA-4433
- AD
Alzheimer's Disease
- HD
Huntington's Disease
- PD
Parkinson’s Disease
- DEGs
Differentially Expressed Genes
- WGCNA
Weighted Gene Co-expression Network Analysis
- FDR
False Discovery Rate
- CC
Cellular Component
- MF
Molecular Function
- BP
Biological Processes
- NSV
Non-segmental Vitiligo
- NEFL
Neurofilament Light
- SYT1
Synaptotagmin 1
AUTHORS’ CONTRIBUTION
LCX designed the study and reviewed the manuscript; XG performed the statistical analysis and wrote the manuscript; TTY performed the statistical analysis; CRZ and XXW coordinated the work. We thank QQW for helpful comments on the use of the WGCNA package.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data discussed in this publication were deposited in NCBI’s Gene Expression Omnibus and accessible through GEO series accession numbers GSE33000, GSE157239, GSE64977 and GSE72962 (https://www.ncbi.nlm.nih.gov/ geo).
FUNDING
This study was supported by the National Natural Science Foundation of China (No. 81872647), Natural Science Research of Jiangsu Higher Education Institutions of China (No. 19KJB330007), Scientific Research Projects for Outstanding Teachers of Xuzhou Medical University (No. D2019011) and the Post-graduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX20_2445).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
REFERENCES
- 1.Nazam F., Shaikh S., Nazam N., Alshahrani A.S., Hasan G.M., Hassan M.I. Mechanistic insights into the pathogenesis of neuro-degenerative diseases: Towards the development of effective therapy. Mol. Cell. Biochem. 2021;476(7):2739–2752. doi: 10.1007/s11010-021-04120-6. [DOI] [PubMed] [Google Scholar]
- 2.Marešová P., Mohelská H., Dolejš J. Kuča K. Socio-economic aspects of Alzheimer’s disease. Curr. Alzheimer Res. 2015;12(9):903–911. doi: 10.2174/156720501209151019111448. [DOI] [PubMed] [Google Scholar]
- 3.Beagle A.J., Zahir A., Borzello M., et al. Amount and delay insensitivity during intertemporal choice in three neurodegenerative diseases reflects dorsomedial prefrontal atrophy. Cortex. 2020;124:54–65. doi: 10.1016/j.cortex.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanton B.R., Leigh P.N., Howard R.J., Barker G.J., Brown R.G. Behavioural and emotional symptoms of apathy are associated with distinct patterns of brain atrophy in neurodegenerative disorders. J. Neurol. 2013;260(10):2481–2490. doi: 10.1007/s00415-013-6989-9. [DOI] [PubMed] [Google Scholar]
- 5.Narayanan M., Huynh J.L., Wang K., et al. Common dysregulation network in the human prefrontal cortex underlies two neurodegenerative diseases. Mol. Syst. Biol. 2014;10(7):743. doi: 10.15252/msb.20145304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathys H., Davila-Velderrain J., Peng Z., et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570(7761):332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alieva A.K., Filatova E.V., Kolacheva A.A., et al. Transcriptome profile changes in mice with MPTP-induced early stages of Parkinson’s disease. Mol. Neurobiol. 2017;54(9):6775–6784. doi: 10.1007/s12035-016-0190-y. [DOI] [PubMed] [Google Scholar]
- 8.Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(1):559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juźwik CA, S Drake S, Zhang Y, et al. microRNA dysregulation in neurodegenerative diseases: A systematic review. Prog Neurobiol. 2019. p. 182: 101664. [DOI] [PubMed]
- 10.Ridolfi B., Abdel-Haq H. Neurodegenerative disorders treatment: The MicroRNA role. Curr. Gene Ther. 2017;17(5):327–363. doi: 10.2174/1566523218666180119120726. [DOI] [PubMed] [Google Scholar]
- 11.Vilardo E., Barbato C., Ciotti M., Cogoni C., Ruberti F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 2010;285(24):18344–18351. doi: 10.1074/jbc.M110.112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long J.M., Ray B., Lahiri D.K. MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer’s disease patients. J. Biol. Chem. 2012;287(37):31298–31310. doi: 10.1074/jbc.M112.366336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long J.M., Ray B., Lahiri D.K. MicroRNA-339-5p down-regulates protein expression of β-site amyloid precursor protein-cleaving enzyme 1 (BACE1) in human primary brain cultures and is reduced in brain tissue specimens of Alzheimer’s disease subjects. J. Biol. Chem. 2014;289(8):5184–5198. doi: 10.1074/jbc.M113.518241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long J.M., Maloney B., Rogers J.T., Lahiri D.K. Novel upregulation of amyloid-β precursor protein (APP) by microRNA-346 via targeting of APP mRNA 5′-untranslated region: Implications in Alzheimer’s disease. Mol. Psychiatry. 2019;24(3):345–363. doi: 10.1038/s41380-018-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda K.C., Huynh T., Tay Y., et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y., Wang X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20(1):18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang L., Zhou G., Soufan O., Xia J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020;48(W1):W244-51. doi: 10.1093/nar/gkaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabaie H., Talebi M., Gharesouarn J., et al. Identification and analysis of BCAS4/hsa-miR-185-5p/SHISA7 competing endogenous RNA axis in late-onset Alzheimer’s disease using bioinformatic and experimental approaches. Front. Aging Neurosci. 2022;14:812169. doi: 10.3389/fnagi.2022.812169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoss A.G., Labadorf A., Latourelle J.C., et al. miR-10b-5p expression in Huntington’s disease brain relates to age of onset and the extent of striatal involvement. BMC Med. Genomics. 2015;8(1):10. doi: 10.1186/s12920-015-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wake C., Labadorf A., Dumitriu A., et al. Novel microRNA discovery using small RNA sequencing in post-mortem human brain. BMC Genomics. 2016;17(1):776. doi: 10.1186/s12864-016-3114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L., Zhang Y-H., Wang S., Zhang Y., Huang T., Cai Y-D. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017;12(9):e0184129. doi: 10.1371/journal.pone.0184129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D., Gable A.L., Lyon D., et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dugger B.N., Dickson D.W. Pathology of neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 2017;9(7):a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephenson J., Nutma E., van der Valk P., Amor S. Inflammation in CNS neurodegenerative diseases. Immunology. 2018;154(2):204–219. doi: 10.1111/imm.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farhadian M., Rafat S.A., Panahi B., Mayack C. Weighted gene co-expression network analysis identifies modules and functionally enriched pathways in the lactation process. Sci. Rep. 2021;11(1):2367. doi: 10.1038/s41598-021-81888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rangaraju S., Dammer E.B., Raza S.A., et al. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Mol. Neurodegener. 2018;13(1):24. doi: 10.1186/s13024-018-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chuang Y.H., Lu A.T., Paul K.C., et al. Longitudinal epigenome-wide methylation study of cognitive decline and motor progression in Parkinson’s disease. J. Parkinsons Dis. 2019;9(2):389–400. doi: 10.3233/JPD-181549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Z., He W., Tang J., et al. Identification of important modules and biomarkers in breast cancer based on WGCNA. OncoTargets Ther. 2020;13:6805–6817. doi: 10.2147/OTT.S258439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M., Wang L., Pu L., et al. LncRNAs related key pathways and genes in ischemic stroke by weighted gene co-expression network analysis (WGCNA). Genomics. 2020;112(3):2302–2308. doi: 10.1016/j.ygeno.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Dai Y., Sun X., Wang C., et al. Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization. BMC Genomics. 2021;22(1):236. doi: 10.1186/s12864-021-07510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arena A., Iyer A.M., Milenkovic I., et al. Developmental expression and dysregulation of miR-146a and miR-155 in down’s syndrome and mouse models of down’s syndrome and Alzheimer’s disease. Curr. Alzheimer Res. 2017;14(12):1305–1317. doi: 10.2174/1567205014666170706112701. [DOI] [PubMed] [Google Scholar]
- 35.Hu W., Wen L., Cao F., Wang Y. Down-Regulation of Mir-107 worsen spatial memory by suppressing SYK expression and inactivating NF-KB signaling pathway. Curr. Alzheimer Res. 2019;16(2):135–145. doi: 10.2174/1567205016666181212154347. [DOI] [PubMed] [Google Scholar]
- 36.Chen X., Zhong S.L., Lu P., et al. miR-4443 participates in the malignancy of breast cancer. PLoS One. 2016;11(8):e0160780. doi: 10.1371/journal.pone.0160780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Zhang Q., Wang D., et al. Microenvironment-induced TIMP2 loss by cancer-secreted exosomal miR-4443 promotes liver metastasis of breast cancer. J. Cell. Physiol. 2020;235(7-8):5722–5735. doi: 10.1002/jcp.29507. [DOI] [PubMed] [Google Scholar]
- 38.Drusco A., Fadda P., Nigita G., et al. Circulating micrornas predict survival of patients with tumors of glial origin. EBioMedicine. 2018;30:105–112. doi: 10.1016/j.ebiom.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y., Xu Y., Wang J., Yang X., Wen L., Feng J. lncRNA MNX1-AS1 promotes glioblastoma progression through inhibition of miR-4443. Oncol. Res. 2019;27(3):341–347. doi: 10.3727/096504018X15228909735079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M., Zhang X., Ding X., et al. Long Noncoding RNA LINC00460 promotes cell progression by sponging miR-4443 in head and neck squamous cell carcinoma. Cell Transplant. 2020;29:963689720927405. doi: 10.1177/0963689720927405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ebrahimi S.O., Reiisi S. Downregulation of miR-4443 and miR-5195-3p in ovarian cancer tissue contributes to metastasis and tumorigenesis. Arch. Gynecol. Obstet. 2019;299(5):1453–1458. doi: 10.1007/s00404-019-05107-x. [DOI] [PubMed] [Google Scholar]
- 42.Meerson A. Leptin-responsive MiR-4443 is a small regulatory RNA independent of the canonic MicroRNA biogenesis pathway. Biomolecules. 2020;10(2):E293. doi: 10.3390/biom10020293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W., Qiao B., Fan J. Overexpression of miR-4443 promotes the resistance of non-small cell lung cancer cells to epirubicin by targeting INPP4A and regulating the activation of JAK2/STAT3 pathway. Pharmazie. 2018;73(7):386–392. doi: 10.1691/ph.2018.8313. [DOI] [PubMed] [Google Scholar]
- 44.Song Z., Jia G., Ma P., Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021;276:119399. doi: 10.1016/j.lfs.2021.119399. [DOI] [PubMed] [Google Scholar]
- 45.Gong J., Wang J., Liu T., Hu J., Zheng J. lncRNA FEZF1 AS1 contributes to cell proliferation, migration and invasion by sponging miR 4443 in hepatocellular carcinoma. Mol. Med. Rep. 2018;18(6):5614–5620. doi: 10.3892/mmr.2018.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z., Yang X., Liu O., et al. Differentially expressed microRNAs in peripheral blood mononuclear cells of non-segmental vitiligo and their clinical significance. J. Clin. Lab. Anal. 2021;35(2):e23648. doi: 10.1002/jcla.23648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y., Ding Y.Y., Ren Y., et al. Identification of differentially expressed microRNAs in acute Kawasaki disease. Mol. Med. Rep. 2018;17(1):932–938. doi: 10.3892/mmr.2017.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma J., Gui H., Tang Y., et al. In silico Identification of 10 hub genes and an miRNA-mRNA regulatory network in acute kawasaki disease. Front. Genet. 2021;12:585058. doi: 10.3389/fgene.2021.585058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shefler I., Salamon P., Levi-Schaffer F., Mor A., Hershko A.Y., Mekori Y.A. MicroRNA-4443 regulates mast cell activation by T cell-derived microvesicles. J. Allergy Clin. Immunol. 2018;141(6):2132–2141.e4. doi: 10.1016/j.jaci.2017.06.045. [DOI] [PubMed] [Google Scholar]
- 50.Li S., Lu G., Wang D., et al. MicroRNA-4443 regulates monocyte activation by targeting tumor necrosis factor receptor associated factor 4 in stroke-induced immunosuppression. Eur. J. Neurol. 2020;27(8):1625–1637. doi: 10.1111/ene.14282. [DOI] [PubMed] [Google Scholar]
- 51.Ghani M., Sato C., Lee J.H., et al. Evidence of recessive Alzheimer’s disease loci in a Caribbean Hispanic data set: Genome-wide survey of runs of homozygosity. JAMA Neurol. 2013;70(10):1261–1267. doi: 10.1001/jamaneurol.2013.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aminyavari S., Zahmatkesh M., Khodagholi F., Sanati M. Anxiolytic impact of Apelin-13 in a rat model of Alzheimer’s disease: Involvement of glucocorticoid receptor and FKBP5. Peptides. 2019;118:170102. doi: 10.1016/j.peptides.2019.170102. [DOI] [PubMed] [Google Scholar]
- 53.Arlt S., Demiralay C., Tharun B., et al. Genetic risk factors for depression in Alzheimer’s disease patients. Curr. Alzheimer Res. 2013;10(1):72–81. doi: 10.2174/156720513804871435. [DOI] [PubMed] [Google Scholar]
- 54.Schröder N., Schaffrath A., Welter J.A., et al. Inhibition of formyl peptide receptors improves the outcome in a mouse model of Alzheimer disease. J. Neuroinflammation. 2020;17(1):131. doi: 10.1186/s12974-020-01816-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Y., Ye R.D., Gong W., Li J., Iribarren P., Wang J.M. Identification of functional domains in the formyl peptide receptor-like 1 for agonist-induced cell chemotaxis. FEBS J. 2005;272(3):769–778. doi: 10.1111/j.1742-4658.2004.04514.x. [DOI] [PubMed] [Google Scholar]
- 56.Haque M.E., Azam S., Akther M., Cho D.Y., Kim I.S., Choi D.K. The neuroprotective effects of gpr4 inhibition through the attenuation of caspase mediated apoptotic cell death in an MPTP induced mouse model of Parkinson’s disease. Int. J. Mol. Sci. 2021;22(9):4674. doi: 10.3390/ijms22094674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Q., Li W.X., Dai S.X., et al. Meta-analysis of parkinson’s disease and Alzheimer’s disease revealed commonly impaired pathways and dysregulation of NRF2-dependent genes. J. Alzheimers Dis. 2017;56(4):1525–1539. doi: 10.3233/JAD-161032. [DOI] [PubMed] [Google Scholar]
- 58.Baltus T.H.L., Morelli N.R., de Farias C.C., et al. Association of -94 ATTG insertion/deletion NFkB1 and c.*126G>A NFkBIA genetic polymorphisms with oxidative and nitrosative stress biomarkers in Brazilian subjects with Parkinson’s disease. Neurosci. Lett. 2021;740:135487. doi: 10.1016/j.neulet.2020.135487. [DOI] [PubMed] [Google Scholar]
- 59.Horvath I., Iashchishyn I.A., Moskalenko R.A., et al. Co-aggregation of pro-inflammatory S100A9 with α-synuclein in Parkinson’s disease: Ex vivo and in vitro studies. J. Neuroinflammation. 2018;15(1):172. doi: 10.1186/s12974-018-1210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leri M., Chaudhary H., Iashchishyn I.A., et al. Natural compound from olive oil inhibits S100A9 amyloid formation and cytotoxicity: Implications for preventing Alzheimer’s disease. ACS Chem. Neurosci. 2021;12(11):1905–1918. doi: 10.1021/acschemneuro.0c00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Recabarren D., Alarcón M. Gene networks in neurodegenerative disorders. Life Sci. 2017;183:83–97. doi: 10.1016/j.lfs.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 62.Leandro G.S., Evangelista A.F., Lobo R.R., Xavier D.J., Moriguti J.C., Sakamoto-Hojo E.T. Changes in expression profiles revealed by transcriptomic analysis in peripheral blood mononuclear cells of Alzheimer’s disease patients. J. Alzheimers Dis. 2018;66(4):1483–1495. doi: 10.3233/JAD-170205. [DOI] [PubMed] [Google Scholar]
- 63.Willmann G., Schäferhoff K., Fischer M.D., et al. Gene expression profiling of the retina after transcorneal electrical stimulation in wild-type Brown Norway rats. Invest. Ophthalmol. Vis. Sci. 2011;52(10):7529–7537. doi: 10.1167/iovs.11-7838. [DOI] [PubMed] [Google Scholar]
- 64.Zenchak J.R., Palmateer B., Dorka N., et al. Bioluminescence-driven optogenetic activation of transplanted neural precursor cells improves motor deficits in a Parkinson’s disease mouse model. J. Neurosci. Res. 2020;98(3):458–468. doi: 10.1002/jnr.24237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang S., Ji D., Yang Q., et al. NEFLb impairs early nervous system development via regulation of neuron apoptosis in zebrafish. J. Cell. Physiol. 2019;234(7):11208–11218. doi: 10.1002/jcp.27771. [DOI] [PubMed] [Google Scholar]
- 66.Emilsson L., Saetre P., Jazin E. Low mRNA levels of RGS4 splice variants in Alzheimer’s disease: Association between a rare haplotype and decreased mRNA expression. Synapse. 2006;59(3):173–176. doi: 10.1002/syn.20226. [DOI] [PubMed] [Google Scholar]
- 67.Shi Z., Zhang K., Zhou H., et al. Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer’s disease. Aging Cell. 2020;19(3):e13125. doi: 10.1111/acel.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Yang X, Zhuang J, Zhang H, Gao C. β-Amyloid activates reactive astrocytes by enhancing glycolysis of astrocytes. Mol. Biol. Rep. 2022;49:4699–4707. doi: 10.1007/s11033-022-07319-y. [DOI] [PubMed] [Google Scholar]
- 69.Blair L.J., Baker J.D., Sabbagh J.J., Dickey C.A. The emerging role of peptidyl-prolyl isomerase chaperones in tau oligomerization, amyloid processing, and Alzheimer’s disease. J. Neurochem. 2015;133(1):1–13. doi: 10.1111/jnc.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bailus B.J., Scheeler S.M., Simons J., et al. Modulating FKBP5/FKBP51 and autophagy lowers HTT (huntingtin) levels. Autophagy. 2021;17(12):4119–4140. doi: 10.1080/15548627.2021.1904489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Leary J.C., III, Dharia S., Blair L.J., et al. A new anti-depressive strategy for the elderly: Ablation of FKBP5/FKBP51. PLoS One. 2011;6(9):e24840. doi: 10.1371/journal.pone.0024840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheung-Flynn J., Prapapanich V., Cox M.B., Riggs D.L., Suarez-Quian C., Smith D.F. Physiological role for the cochaperone FKBP52 in androgen receptor signaling. Mol. Endocrinol. 2005;19(6):1654–1666. doi: 10.1210/me.2005-0071. [DOI] [PubMed] [Google Scholar]
- 73.Blair L.J., Nordhues B.A., Hill S.E., et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Invest. 2013;123(10):4158–4169. doi: 10.1172/JCI69003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Qiu B., Zhong Z., Righter S., et al. FKBP51 modulates hippocampal size and function in post-translational regulation of Parkin. Cell. Mol. Life Sci. 2022;79(3):175. doi: 10.1007/s00018-022-04167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slowik A., Merres J., Elfgen A., et al. Involvement of formyl peptide receptors in receptor for advanced glycation end products (RAGE)--and amyloid beta 1-42-induced signal transduction in glial cells. Mol. Neurodegener. 2012;7(1):55. doi: 10.1186/1750-1326-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X., Zhang Y., Wan X., et al. Responsive expression of MafF to β-amyloid-induced oxidative stress. Dis. Markers. 2020;2020:8861358. doi: 10.1155/2020/8861358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Neill L.A., Kaltschmidt C. NF-kappa B: A crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20(6):252–258. doi: 10.1016/S0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- 78.Oikawa K., Odero G.L., Platt E., et al. NF-κB p50 subunit knockout impairs late LTP and alters long term memory in the mouse hippocampus. BMC Neurosci. 2012;13(1):45. doi: 10.1186/1471-2202-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boersma M.C., Dresselhaus E.C., De Biase L.M., Mihalas A.B., Bergles D.E., Meffert M.K. A requirement for nuclear factor-kappaB in developmental and plasticity-associated synaptogenesis. J. Neurosci. 2011;31(14):5414–5425. doi: 10.1523/JNEUROSCI.2456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu L., Wu Q., Zhong W., et al. Microarray analysis of differential gene expression in Alzheimer’s disease identifies potential biomarkers with diagnostic value. Med. Sci. Monit. 2020;26:e919249. doi: 10.12659/MSM.919249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Palomino-Alonso M., Lachén-Montes M., González-Morales A., et al. Network-driven proteogenomics unveils an aging-related imbalance in the olfactory IκBα-NFκB p65 complex functionality in Tg2576 Alzheimer’s disease mouse model. Int. J. Mol. Sci. 2017;18(11):E2260. doi: 10.3390/ijms18112260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Horvath I., Jia X., Johansson P., et al. Pro-inflammatory S100A9 protein as a robust biomarker differentiating early stages of cognitive impairment in Alzheimer’s disease. ACS Chem. Neurosci. 2016;7(1):34–39. doi: 10.1021/acschemneuro.5b00265. [DOI] [PubMed] [Google Scholar]
- 83.Kim H.J., Chang K.A., Ha T.Y., et al. S100A9 knockout decreases the memory impairment and neuropathology in crossbreed mice of Tg2576 and S100A9 knockout mice model. PLoS One. 2014;9(2):e88924. doi: 10.1371/journal.pone.0088924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Handley R.R., Reid S.J., Brauning R., et al. Brain urea increase is an early Huntington’s disease pathogenic event observed in a prodromal transgenic sheep model and HD cases. Proc. Natl. Acad. Sci. USA. 2017;114(52):E11293–E11302. doi: 10.1073/pnas.1711243115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohammadi A., Rashidi E., Amooeian V.G. Brain, blood, cerebrospinal fluid, and serum biomarkers in schizophrenia. Psychiatry Res. 2018;265:25–38. doi: 10.1016/j.psychres.2018.04.036. [DOI] [PubMed] [Google Scholar]
- 86.Bender K., Nasrollahzadeh P., Timpert M., Liu B., Pott L., Kienitz M.C. A role for RGS10 in beta-adrenergic modulation of G-protein-activated K+ (GIRK) channel current in rat atrial myocytes. J. Physiol. 2008;586(8):2049–2060. doi: 10.1113/jphysiol.2007.148346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jęśko H, Wieczorek I, Wencel PL, Gąssowska-Dobrowolska M, Lukiw WJ, Strosznajder RP. Age-related transcriptional deregulation of genes coding synaptic proteins in Alzheimer’s disease murine model: Potential neuroprotective effect of fingolimod. Front. Mol. Neurosci. 2021;14:660104. doi: 10.3389/fnmol.2021.660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khalil M., Teunissen C.E., Otto M., et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 2018;14(10):577–589. doi: 10.1038/s41582-018-0058-z. [DOI] [PubMed] [Google Scholar]
- 89.Ficulle E., Kananathan S., Airey D., et al. A human tau seeded neuronal cell model recapitulates molecular responses associated with Alzheimer’s disease. Sci. Rep. 2022;12(1):2673. doi: 10.1038/s41598-022-06411-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fernandez-Martos C.M., King A.E., Atkinson R.A., Woodhouse A., Vickers J.C. Neurofilament light gene deletion exacerbates amyloid, dystrophic neurite, and synaptic pathology in the APP/PS1 transgenic model of Alzheimer’s disease. Neurobiol. Aging. 2015;36(10):2757–2767. doi: 10.1016/j.neurobiolaging.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 91.Sap K.A., Guler A.T., Bury A., Dekkers D., Demmers J.A.A., Reits E.A. Identification of full-length wild-type and mutant huntingtin interacting proteins by crosslinking immunoprecipitation in mice brain cortex. J. Huntingtons Dis. 2021;10(3):335–347. doi: 10.3233/JHD-210476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu S., Fan M., Xu J.X., et al. Exosomes derived from bone-marrow mesenchymal stem cells alleviate cognitive decline in AD-like mice by improving BDNF-related neuropathology. J. Neuroinflammation. 2022;19(1):35. doi: 10.1186/s12974-022-02393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuzuya A., Zoltowska K.M., Post K.L., et al. Identification of the novel activity-driven interaction between synaptotagmin 1 and presenilin 1 links calcium, synapse, and amyloid beta. BMC Biol. 2016;14(1):25. doi: 10.1186/s12915-016-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fathy Y.Y., Hoogers S.E., Berendse H.W., et al. Differential insular cortex sub-regional atrophy in neurodegenerative diseases: A systematic review and meta-analysis. Brain Imaging Behav. 2020;14(6):2799–2816. doi: 10.1007/s11682-019-00099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen W.T., Lu A., Craessaerts K., et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell. 2020;182(4):976–991.e19. doi: 10.1016/j.cell.2020.06.038. [DOI] [PubMed] [Google Scholar]
- 96.Olah M., Menon V., Habib N., et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 2020;11(1):6129. doi: 10.1038/s41467-020-19737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.
Data Availability Statement
The data discussed in this publication were deposited in NCBI’s Gene Expression Omnibus and accessible through GEO series accession numbers GSE33000, GSE157239, GSE64977 and GSE72962 (https://www.ncbi.nlm.nih.gov/ geo).