Abstract
Background: Heterogeneous nuclear ribonucleoproteins (hnRNPs), a large family of RNA-binding proteins, have been implicated in tumor progression in multiple cancer types. However, the expression pattern and prognostic value of hnRNPs in five gastrointestinal (GI) cancers, including gastric, colorectal, esophageal, liver, and pancreatic cancer, remain to be investigated.
Objective: The current research aimed to identify prognostic biomarkers of the hnRNP family in five major types of gastrointestinal cancer.
Methods: Oncomine, Gene Expression Profiling Interactive Analysis (GEPIA), and Kaplan-Meier Plotter were used to explore the hnRNPs expression levels concerning clinicopathological parameters and prognostic values. The protein level of hnRNPU was validated by immunohistochemistry (IHC) in human tissue specimens. Genetic alterations of hnRNPs were analyzed using cBioportal, and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to illustrate the biological functions of co-expressed genes of hnRNPs.
Results: The vast majority of hnRNPs were highly expressed in five types of GI cancer tissues compared to their adjacent normal tissues, and mRNA levels of hnRNPA2B1, D, Q, R, and U were significantly different in various GI cancer types at different stages. In addition, Kaplan-Meier analysis revealed that the increased hnRNPs expression levels were correlated with better prognosis in gastric and rectal cancer patients (log-rank p < 0.05). In contrast, patients with high levels of hnRNPs exhibited a worse prognosis in esophageal and liver cancer (log-rank p < 0.05). Using immunohistochemistry, we further confirmed that hnRNPU was overexpressed in gastric, rectal, and liver cancers. In addition, hnRNPs genes were altered in patients with GI cancers, and RNA-related processing was correlated with hnRNPs alterations.
Conclusion: We identified differentially expressed genes of hnRNPs in tumor tissues versus adjacent normal tissues, which might contribute to predicting tumor types, early diagnosis, and targeted therapies in five major types of GI cancer.
Keywords: RNA-binding proteins, early diagnosis, GI tract cancers, gene ontology, kyoto encyclopedia of genes and genomes
1. INTRODUCTION
The five major types of gastrointestinal cancer, including gastric, colorectal, esophageal, liver, and pancreatic cancer, account for 26.3% (4.8 million) of the global cancer incidence and 35.4% (3.4 million) of all cancer-related deaths [1]. Although most gastrointestinal cancers develop sporadically due to a combination of genetic predisposition and environmental influences, patients with hereditary gastrointestinal cancers also represent a substantial fraction of the overall affected population [2]. In recent years, biomarkers of gene mutations and epigenetic modifications have been discovered and applied in clinical practice [3-5]. However, patients with gastric, esophageal, liver, and pancreatic cancer are usually diagnosed at an advanced stage with a poor prognosis [6]. Diagnoses and treatment are still challenging due to the poor prognosis and late stage of most cancers.
Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a group of genes that encode a large family of RNA-binding proteins [7] and act as key proteins in nucleic acid metabolism, including alternative splicing, mRNA stabilization as well as transcriptional and translational regulation [8]. The hnRNPs include at least twenty-three members and are named alphabetically from hnRNP A1 to hnRNP U [9]. The expressions of hnRNPs are upregulated in many human malignancies, including glioblastoma [10], oral squamous cancer [11], myeloid leukemia [12], lung cancer [13], bladder cancer [14], breast cancer [15], ovarian cancer [16], prostate cancer [17], and gastrointestinal cancer [18-22]. To date, twenty-three hnRNPs named hnRNPA1, A3, A/B, A2B1, C, D, DL, E1, E2, F, G (RBMX), H1, H2, H3, I (PTBP1), K, L, LL, M, P (FUS), Q (SYNCRIP), R, and U have been identified [9]. HnRNPs have been reported to play important roles in human GI cancers [23]. However, to the best of our knowledge, the expression pattern, prognosis, and underlying mechanism of hnRNPs in gastrointestinal cancers have not been fully elucidated. Therefore, the role of hnRNPs in gastrointestinal cancers has yet to be explored by bioinformatics analysis.
We investigated the expressions and mutations of hnRNPs based on thousands of genes published online. Moreover, we confirmed the expression of hnRNPU by IHC in gastric, rectal, and liver cancers. The differentially expressed genes may serve as biomarkers for early clinical diagnosis of GI tract cancers. Among them, the prognosis-related genes, as well as their co-expression genes, were analyzed by GO functional annotation and KEGG pathway enrichment to find the underlying mechanisms and possible therapeutic targets.
2. MATERIALS AND METHODS
2.1. Patients and Samples
Twelve pairs of cancer and normal adjacent tissues from surgically resected stomach adenocarcinoma, rectal adenocarcinoma, and liver hepatocellular carcinoma samples were collected in the Affiliated Hospital of Qingdao University in 2021. Patients were included in this experiment upon giving informed consent. The study was carried out according to the principle of the declaration of Helsinki and approved by the Institutional Review Board of the Ethics Committee of the Affiliated Hospital of Qingdao University (QYFY WZLL 26689).
2.2. Oncomine Analysis
Oncomine (https://www.oncomine.org) is a publicly accessible online cancer microarray database providing genome-wide expression analysis, which is used to determine the mRNA levels of distinct hnRNPs in five major types of gastrointestinal cancers. The transcriptional levels of hnRNPs in gastrointestinal cancer and normal specimens were compared by Student’s t-test. The cut-offs of p-value and fold change were defined as 1E-4 (the minimum P-value that oncomine can select) and 2, respectively.
2.3. GEPIA Dataset
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn) is a newly developed interactive web server containing high-throughput RNA sequencing data from The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects [24]. Differentially expressed hnRNPs genes and pathological stages of cancers were analyzed using GEPIA.
2.4. The Kaplan-Meier Plotter
Kaplan Meier-plotter (KM-plotter, https://kmplot.com) is a meta-analysis web port for the discovery and validation of survival biomarkers [25]. Sources for the databases include Gene Expression Omnibus (GEO), the European Genome-phenome Archive (EGA), and TCGA. We used this online tool to evaluate the prognostic value of hnRNPs in GI tract cancers. Patient samples were divided into two groups by Auto-select the best cut-off values (high expression groups versus low expression groups). Only the JetSet best probe set of hnRNPs was chosen for our analysis. The hazard ratio (HR) with 95% confidence intervals (CIs) and log-rank p-value was calculated in each plot. P-value < 0.05 was considered statistically significant.
2.5. cBioPortal and TCGA
The cBio Cancer Genomics Portal (cBioPortal, http://www.cbioportal.org) is a publicly available resource for interactive exploration of multidimensional cancer genomics data sets, providing access to data from 20 cancer studies [26, 27]. The stomach adenocarcinoma (The Cancer Genome Atlas, Firehose Legacy) dataset, colorectal adenocarcinoma (The Cancer Genome Atlas, Firehose Legacy) dataset, esophageal adenocarcinoma (The Cancer Genome Atlas, PanCancer Atlas) dataset, and hepatocellular liver carcinoma (The Cancer Genome Atlas, Firehose Legacy) dataset were selected separately for further analyses. The genomic profiles included data for mutations, mRNA expression Z scores (RNAseq V2 RSEM), protein expression Z scores (reverse phase protein array [RPPA]), and putative copy number alterations (CNA) from genomic identification of significant targets in cancer (GISTIC). Co-expressed genes were calculated according to the cBioPortal’s online instructions, and the intersection was obtained based on the Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/).
2.6. GO and KEGG Pathway Analysis
The Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.8, https://david.ncifcrf.gov) is a comprehensive set of functional annotation tools [28]. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of hnRNPs were analyzed using DAVID. FDR < 0.001 and P < 0.05 were considered as the cut-off criterion of GO and KEGG with significant differences, respectively. The horizontal bar chart and bubble chart of GO and KEGG from the DAVID database were performed by bioinformatics (http://www.bioinformatics.com.cn).
3. EXPERIMENTAL SECTION
Immunohistochemistry (IHC): Tissue sections were baked at 60ºC and deparaffinized with xylene, followed by rehydration and antigen retrieval using decreased gradient concentrations of alcohol and citrate (10 mM, pH 6) buffers, respectively. Sections blocked endogenous peroxidase by 3% hydrogen peroxidase/methanol solution. To decrease non-specific antibody binding, tissue sections were blocked with 10% serum for 30 minutes at 37ºC and incubated overnight at 4°C with hnRNPU antibody (1:500 dilution; Abcam #ab172608). Following washes in PBS, the tissues were incubated with a second antibody, detected with DAB and counterstained with hematoxylin. Then, the sections were dehydrated with increased gradient concentrations of alcohol, cleared in xylene and sealed with neutral resin.
4. RESULTS
4.1. Transcriptional Levels of hnRNPs in Patients with Five Major Types of Gastrointestinal Cancer
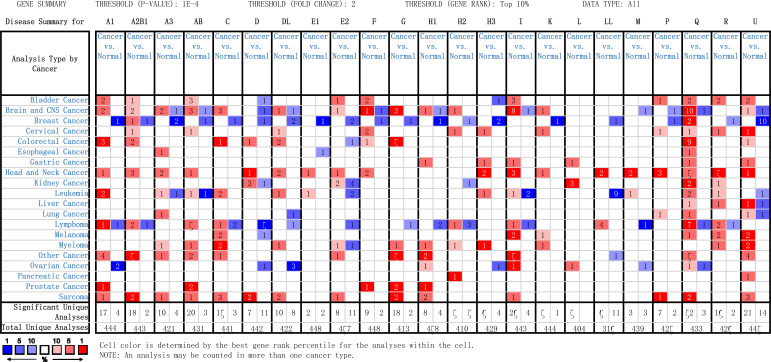
Transcriptional levels of hnRNPs were compared between five major GI cancer tissues and normal samples via Oncomine database, as shown in Fig. (1) (fold change > 2, p-value < 0.0001). We found that hnRNPs (except hnRNPQ and U) showed some degree of tumor-specificity in different types of GI cancer. In detail, the mRNA expression levels of hnRNPA1, A2B1, C, D, DL, F, and G increased in colorectal cancer, while hnRNPA3 was upregulated in esophageal cancer compared to corresponding normal tissues. HnRNPR was increased in liver cancer. In addition, hnRNPH1, H3, I, and L were found to be highly expressed in patients with gastric cancer, while hnRNPH2 levels increased in pancreatic cancer. Besides, hnRNPQ was highly expressed in colorectal, esophageal, gastric, and liver cancer tissues, and hnRNPU was increased in colorectal, gastric, liver and pancreatic cancer tissues (Fig. 1). We also found that the levels of hnRNPE1 and E2 decreased in cancer tissues compared to normal esophagus and colorectum, respectively (Fig. 1). Taken together, these results suggested that hnRNPs exhibit tumor specificity and vary in different gastrointestinal cancer types. As revealed by multiple datasets, the mRNA expression levels of most hnRNPs were significantly upregulated in patients with gastrointestinal cancers. Details, including specific tumor type, references, and p-value are listed in Table 1A-E.
Fig. (1).
Transcription levels of hnRNPs in five major types of gastrointestinal cancer (Oncomine). The color is determined by the best gene rank percentile for the analyses within the cell. The red color indicates gene expression upregulation, and the blue color indicates gene expression downregulation. The digits within the cell represent the number of studies.
Table 1A.
Changes and datasets of hnRNPs expression in transcription level between colorectal cancer tissues and normal colorectal tissues (Oncomine).
| Gene | Type of Colorectal Cancer versus Normal Colorectal Tissues | Fold Change | p-Value | t-Test | Source and/or References |
|---|---|---|---|---|---|
| A1 | Colon Adenocarcinoma vs. Normal | 2.424 | 3.47E-08 | 6.497 | Alon Colon [34] |
| Colon Adenocarcinoma vs. Normal | 2.534 | 1.32E-08 | 7.183 | Notterman Colon [35] | |
| Rectal Adenoma vs. Normal | 2.120 | 8.92E-06 | 8.530 | Sabates-Bellver Colon [36] | |
| A2B1 | Colon Carcinoma Epithelia vs. Normal | 2.064 | 2.59E-08 | 11.573 | Skrzypczak Colorectal 2 [37] |
| Colon Carcinoma vs. Normal | 4.242 | 5.98E-09 | 14.508 | Skrzypczak Colorectal 2 [37] | |
| C | Rectal Adenoma vs. Normal | 2.079 | 1.22E-11 | 13.006 | Sabates-Bellver Colon [36] |
| D | Colon Carcinoma vs. Normal | 2.033 | 3.65E-08 | 10.841 | Skrzypczak Colorectal 2 [37] |
| DL | Colon Carcinoma vs. Normal | 2.039 | 1.72E-09 | 15.329 | Skrzypczak Colorectal 2 [37] |
| Rectal Adenoma vs. Normal | 2.104 | 4.10E-06 | 7.425 | Sabates-Bellver Colon [36] | |
| F | Rectal Adenoma vs. Normal | 2.142 | 1.06E-05 | 8.500 | Sabates-Bellver Colon [36] |
| G | Rectosigmoid Adenocarcinoma vs. Normal | 3.269 | 1.81E-12 | 13.299 | TCGA Colorectal |
| Cecum Adenocarcinoma vs. Normal | 2.180 | 2.97E-13 | 10.328 | TCGA Colorectal | |
| Colon Mucinous Adenocarcinoma vs. Normal | 2.001 | 1.39E-11 | 8.988 | TCGA Colorectal | |
| Rectal Adenocarcinoma vs. Normal | 2.045 | 6.32E-13 | 10.948 | TCGA Colorectal | |
| Rectal Adenoma vs. Normal | 2.022 | 6.29E-05 | 6.957 | Sabates-Bellver Colon [36] | |
| Q | Rectosigmoid Adenocarcinoma vs. Normal | 2.663 | 2.91E-07 | 9.205 | Kaiser Colon [38] |
| Colon Mucinous Adenocarcinoma vs. Normal | 2.436 | 2.61E-07 | 8.746 | Kaiser Colon [38] | |
| Rectal Adenocarcinoma vs. Normal | 2.305 | 3.03E-05 | 6.427 | Kaiser Colon [38] | |
| Cecum Adenocarcinoma vs. Normal | 2.388 | 2.79E-07 | 9.517 | Kaiser Colon [38] | |
| Colon Adenocarcinoma vs. Normal | 2.425 | 7.13E-07 | 10.763 | Kaiser Colon [38] | |
| Colon Carcinoma vs. Normal | 2.174 | 6.99E-11 | 19.513 | Skrzypczak Colorectal 2 [37] | |
| Colon Adenoma vs. Normal | 2.000 | 7.67E-08 | 14.319 | Skrzypczak Colorectal 2 [37] | |
| Colorectal Carcinoma vs. Normal | 2.154 | 2.55E-11 | 10.589 | Hong Colorectal [39] | |
| Colon Adenoma vs. Normal | 2.204 | 6.66E-11 | 8.104 | Sabates-Bellver Colon [36] | |
| U | Rectal Adenoma vs. Normal | 2.151 | 3.41E-05 | 7.377 | Sabates-Bellver Colon [36] |
A1, hnRNPA1; A2B1, hnRNPA2B1; C, hnRNPC; D, hnRNPD; DL, hnRNPDL; F, hnRNPF; G, hnRNPG; Q, hnRNPQ; U, hnRNPU; TCGA, The Cancer Genome Atlas.
Table 1E.
Changes and datasets of hnRNPs expression in transcription level between pancreatic cancer tissues and normal pancreatic tissues (Oncomine).
| Gene | Type of Pancreatic Cancer versus Normal Pancreatic Tissues | Fold Change | p-Value | t-Test | Source and/or References |
|---|---|---|---|---|---|
| H2 | Pancreatic Ductal Adenocarcinoma vs. Normal | 2.066 | 5.52E-06 | 7.391 | Buchholz Pancreas [44] |
| U | Pancreatic Adenocarcinoma vs. Normal | 2.211 | 6.11E-05 | 5.271 | Iacobuzio-Donahue Pancreas 2 [45] |
H2, hnRNPH2; U, hnRNPU.
4.2. Correlation between the Expression of hnRNPs and Tumor Stage of Patients with Five Major Types of Gastrointestinal Cancer
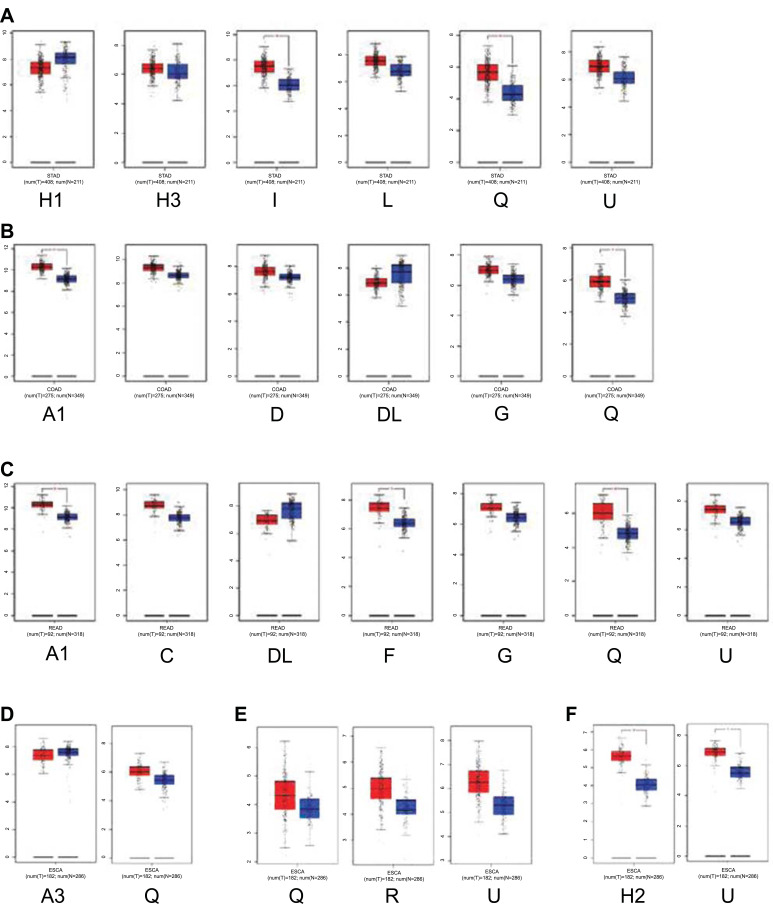
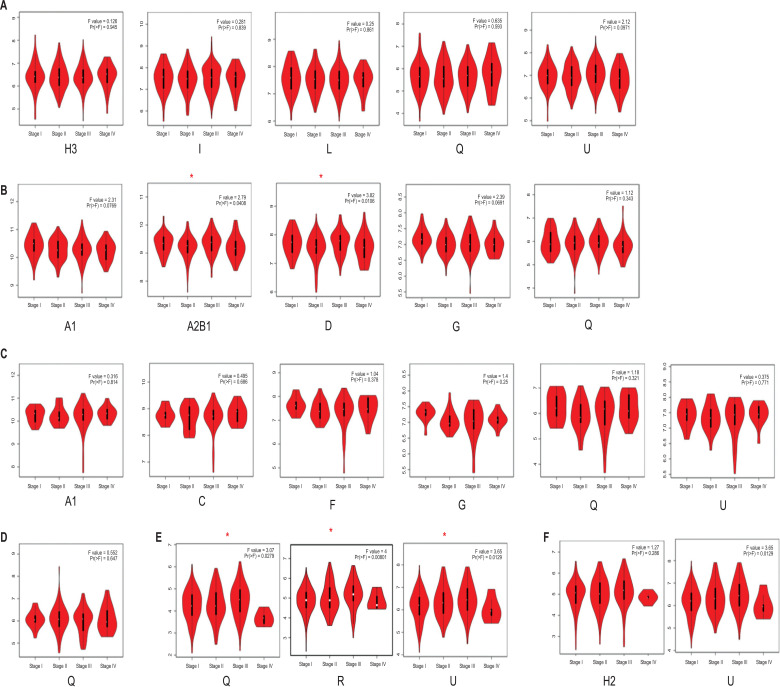
The mRNA levels of hnRNPs were then compared between GI tract cancer tissues and normal GI tract tissues by using the GEPIA database. We found that the mRNA expression levels of hnRNPA1, A2B1, C, D, F, G, H2, H3, I, L, Q, R, and U were higher in GI cancer tissues than in normal tissues; however, the expression level of hnRNPA3, DL, and H1 decreased in cancer tissues (Fig. 2A-E). Next, we analyzed the correlation between hnRNPs mRNA expression and tumor stage. We found that in colon cancer, hnRNPA2B1 and D increased in tumor stages I and III (Fig. 3B). In addition, hnRNPQ, R, and U increased in tumor stage III but decreased in tumor stage IV in liver cancer (Fig. 3E). However, in gastric, rectal, esophageal, and pancreatic cancer, no change was observed in the expression levels of hnRNPs at different tumor stages (Fig. 3A-F).
Fig. (2).
The mRNA expression of hnRNPs in five major types of gastrointestinal cancer (GEPIA). The red color represents tumor tissues, and the blue color represents normal tissues. (A) Gastric cancer. (B) Colon cancer. (C) Rectum cancer. (D) Esophageal cancer. (E) Liver cancer. (F) Pancreas cancer. (STAD, Stomach adenocarcinoma; COAD, Colon adenocarcinoma; READ, Rectum adenocarcinoma; ESCA, Esophageal carcinoma; LIHC, Liver hepatocellular carcinoma; PAAD, Pancreatic adenocarcinoma.)
Fig. (3).
Correlation between hnRNPs expression and tumor stage in patients with different types of gastrointestinal tract cancer (GEPIA). (A) Gastric cancer. (B) Colon cancer. (C) Rectal cancer. (D) Esophageal cancer. (E) Liver cancer. (F) Pancreatic cancer. (The red asterisk: P < 0.05)
4.3. Correlation between hnRNPs and Prognosis Alterations in Patients with Five Major Types of Gastrointestinal Cancer
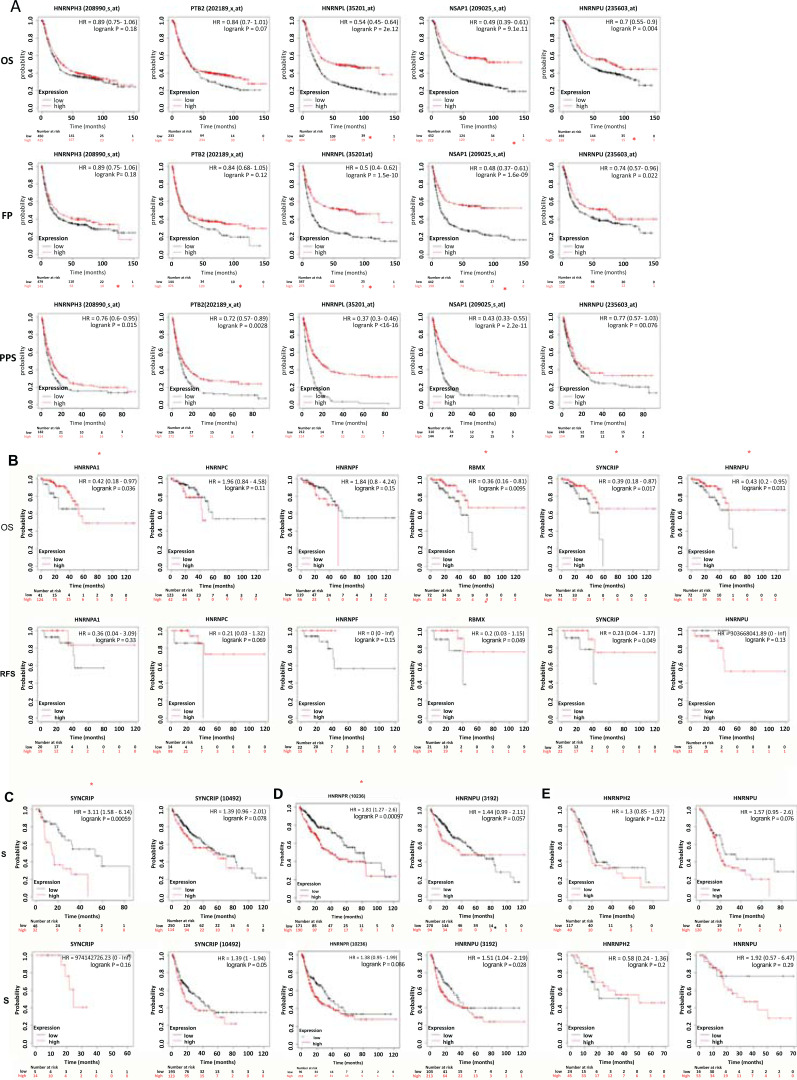
We further explored the impacts of hnRNPs on patient survival in different types of GI tract cancers. Kaplan-Meier Plotter database was used to analyze the relationship between the mRNA levels of hnRNPs and patient outcomes [29-31], except for colon cancer, due to lack of the survival data in the Kaplan-Meier Plotter database. The Kaplan-Meier curve and log-rank test analyses revealed that patients with increased hnRNPs mRNA levels had a better prognosis (log-rank p < 0.05) in gastric and rectal cancer (Fig. 4A and B). On the contrary, in esophageal and liver cancer, patients with upregulated hnRNPs were identified with a worse prognosis (log-rank p < 0.05) (Fig. 4C and D). Specifically, among patients with gastric cancer, hnRNPL and Q were significantly associated with the overall survival (OS), first progression (FP) and post-progression survival (PPS); hnRNPH3 and I were significantly associated with PPS; and hnRNPU was significantly associated with the OS and FP (log-rank p < 0.05) (Fig. 4A). Higher hnRNPU was related to the longer OS in gastric cancer patients [32]. In rectal cancer, hnRNPA1, G, Q, and U were related to OS and hnRNPG was associated with relapse-free survival (RFS) of patients (log-rank p < 0.05) (Fig. 4B). In addition, hnRNPQ was correlated with the OS in patients with esophageal cancer (log-rank p < 0.05) (Fig. 4C), while hnRNPR and U were associated with OS and RFS, respectively, in liver cancer patients (log-rank p < 0.05) (Fig. 4D). The upregulation of hnRNPU was related to poor prognosis in liver cancer patients [33]. In contrast, expression of hnRNPs did not correlate with survival prognosis in patients with pancreatic cancer (Fig. 4E). Taken together, in colorectal and gastric cancer, patients with increased hnRNPs had a better prognosis. On the contrary, in esophageal and liver cancer, patients with upregulated hnRNPs were identified with a worse prognosis (Table 1A-E).
Fig. (4).
The prognostic value of hnRNPs in a patient with different types of gastrointestinal tract cancer (Kaplan-Meier Plotter). The red line indicates gene expression upregulation, and the black line indicates gene expression downregulation. (A) Gastric cancer. (B) Rectal cancer. (C) Esophageal cancer. (D) Liver cancer. (E) Pancreatic cancer. (The red asterisk: Log-rank P < 0.05)
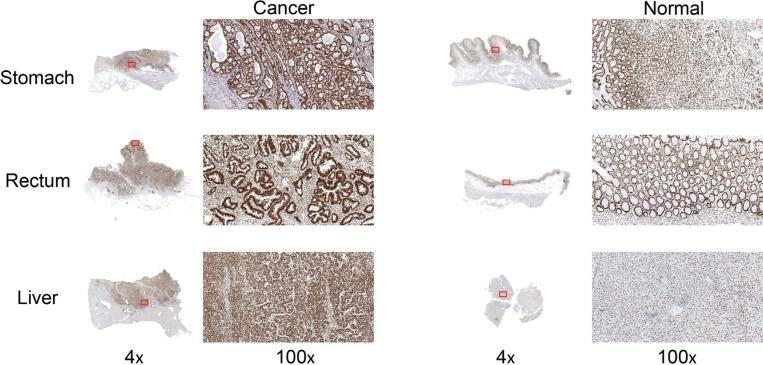
4.4. Validation of the Expression of hnRNPU Protein in Gastric, Rectal, and Liver Cancer by IHC
The mRNA expression levels of hnRNPQ and hnRNPU were increased in multiple GI tract cancers versus normal GI tract tissues (Oncomine). Besides, hnRNPQ was significantly associated with prognosis in gastric, rectal, and esophageal cancers and hnRNPU was significantly associated with prognosis in gastric, rectal, and liver cancers (Kaplan-Meier Plotter). Due to the higher morbidity and mortality of liver cancer than esophageal cancer [6], we validated the protein expression level of hnRNPU in gastric, rectal, and liver cancer tissues compared to normal adjacent tissues by IHC. We collected twelve pairs of cancer and normal adjacent tissues from surgically resected stomach adenocarcinoma, rectal adenocarcinoma, and liver hepatocellular carcinoma samples. IHC analysis showed that hnRNPU was expressed in both three types of GI cancers and normal tissues, however, the staining intensity was significantly higher in caner tissues than normal tissues (Fig. 5).
Fig. (5).
The expression of hnRNPU protein in gastric, rectal, and liver cancer by IHC.
4.5. HnRNPs-related Genes and Pathways in Patients with Five Major Types of Gastrointestinal Cancer
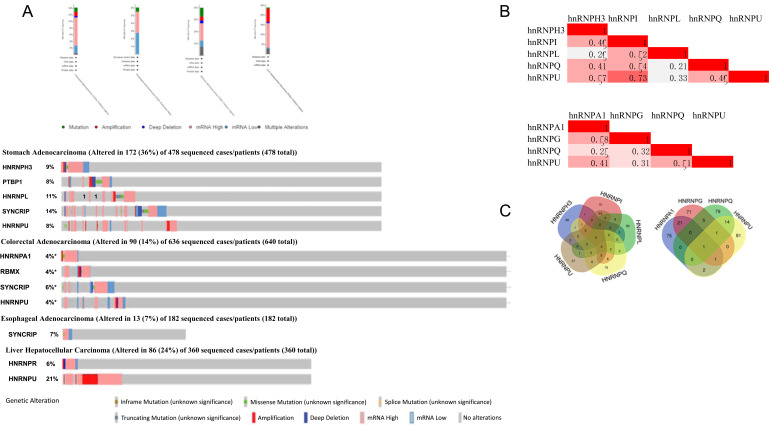
We analyzed the hnRNPs levels whose alterations were closely correlated with prognosis in patients with gastric, colorectal, esophageal, and liver cancer using the cBioPortal online tool. HnRNPH3, I (PTBP1), L, Q, and U were altered in 172 samples of 478 patients with gastric adenocarcinoma (36%); hnRNPA1, G (RBMX), Q (SYNCRIP), and U were altered in 90 samples of 636 patients with colorectal adenocarcinoma (14%); hnRNPQ were altered in 13 samples of 182 patients with esophageal adenocarcinoma (7%); hnRNPR and U were altered in 86 samples of 360 patients with liver hepatocellular carcinoma (24%) (Fig. 6A). We then evaluated the correlation of mRNA expressions (RNA seq V2 RSEM) of hnRNPH3, I, L, Q, and U in gastric cancer and hnRNPA1, G, Q, and U in colorectal cancer via the cBioPortal online tool, followed by Pearson’s correlation analysis. The results indicated significant correlations (pearson ≥ 0.4) between hnRNPs and different types of cancers in the following: hnRNPH3 with I, Q, and U, hnRNPI with L, Q, and U, hnRNPQ with U in gastric cancer; hnRNPA1 with G and U, hnRNPQ with U in colorectal cancer (Fig. 6B). We then performed gene co-expression analysis for functional classification by identifying 100 highly ranked co-expressed genes of hnRNPH3, I, L, Q, and U (spearman > 0.5, p < 5.5e-4) in stomach adenocarcinoma and 100 most highly co-expressed genes with hnRNPA1, G, Q and U (spearman > 0.4, p < 1.7e-16) in colorectal cancer using cBioPortal, followed by the intersection of all above-mentioned genes, as shown in Venn diagram (Fig. 6C).
Fig. (6).
(A) HnRNPs gene expression and mutation analysis in gastrointestinal tract cancers (cBioPortal). (B) Correction between different hnRNPs in gastric cancer (left) and colorectal (right) (cBioPortal). (C) Venn diagram of co-expression genes (cBioPortal) with each hnRNPs in gastric cancer (Left) and colorectal (right).
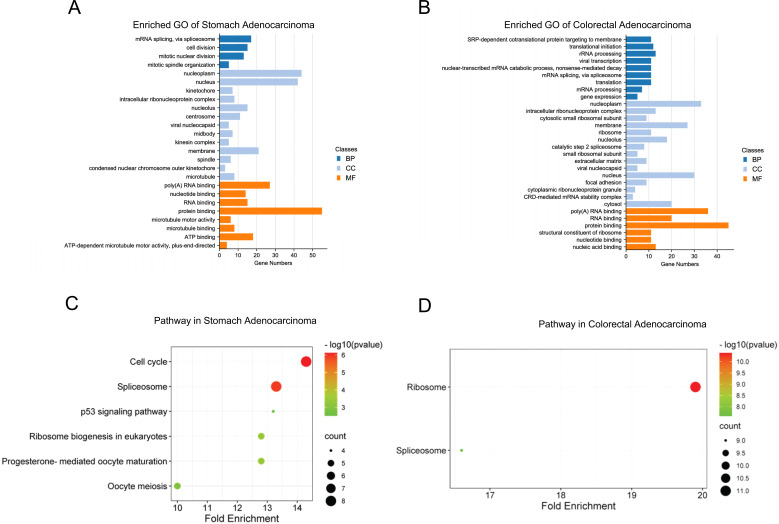
DAVID database was then used to predict the functions of hnRNPs and their highly ranked co-expressed genes. GO annotations include three aspects, such as biological processes (BP), cellular components (CC), and molecular functions (MF). We found that mRNA splicing, cell division, mitotic division, RNA, protein and ATP binding, etc., were associated with hnRNPH3, I, L, Q, U and their co-expressed genes in gastric cancer (Fig. 7A). Translation, RNA-related processing and RNA and protein binding processes were correlated with hnRNPA1, G, Q, U, and their co-expressed genes in colorectal cancer (Fig. 7B). Furthermore, KEGG pathway analysis revealed related functions and pathways due to hnRNPs expression alterations. In gastric cancer, six KEGG pathways were significantly involved, including cell cycle, spliceosome, P53 signaling pathway, ribosome biogenesis in eukaryotes, progesterone-mediated oocyte maturation, and oocyte meiosis (Fig. 7C), while in colorectal cancer, hnRNPs were significantly correlated with functions of ribosome and spliceosome (Fig. 7D).
Fig. (7).
(A) GO enrichment of hnRNPs and genes associated with hnRNPs alterations in gastric adenocarcinoma (p < 0.001). (B) GO enrichment of hnRNPs and genes associated with hnRNPs alterations in colorectal adenocarcinoma (p < 0.001) (C) KEGG pathway of hnRNPs and genes associated with hnRNPs alterations in gastric adenocarcinoma. (D) KEGG pathway of hnRNPs and genes associated with hnRNPs alterations in colorectal adenocarcinoma.
5. DISCUSSION
In this study, we comprehensively analyzed the expression level, tumor stage, prognostic values, gene mutations, and biological function of hnRNPs in five GI cancers. Studies from Oncomine (Fig. 1) showed that, excluding hnRNPQ and U, each isoform of hnRNPs that upregulated in five major gastrointestinal cancers exhibited cancer type-specific expression pattern. Whereas no tumor-specific hnRNPs expression was found in other studies, for example, hnRNPA1 promoted the aggressiveness of colorectal, gastric, and liver cancer [46-48]. We suggested that the reason behind this phenomenon may lie in the significant difference among each hnRNPs in different types of gastrointestinal cancers, as we defined the cut-offs of p-value and fold change as 1E-4 and 2 in Oncomine, respectively. Therefore, by setting up a certain threshold, hnRNPs may predict the exact GI cancer types.
Interestingly, although most hnRNPs were increased in five GI cancer tissues, the expression levels of hnRNPE1 and E2 were higher in the normal esophagus and colorectum tissues, respectively, which have been reported to show tumor-suppressing rather than tumor-promoting effects [49-52]. We explored the possible reason for this phenomenon and found that hnRNPE1 and E2 were structurally composed of three homologous K-homology (KH) domains, lacking RNA recognition motif (RRM) that ubiquitously expressed in hnRNPA1, A2B1, A3, C, D, DL, F, G, H1, H2, H3, I, L, and Q [9]. HnRNPU was another structurally unique molecule in the hnRNPs family, which lacked both RRM and KH domain but was also highly expressed in gastric, rectal, and liver cancer, possibly due to the that hnRNPU had an arginine and glycine-rich RGG box at the C-terminus. Taken together, RRM and arginine and glycine-rich RGG box may play a key role in the progress of GI cancer.
We found that most hnRNPs, including hnRNPA1, A2B1, C, D, F, G, H2, H3, I, L, Q, R, and U were highly expressed in GI cancer tissues than in normal tissues by GEPIA, which was consistent with our results from Oncomine and existing literature reports [18, 33, 46, 53-58]. The vast majority of increased hnRNPs were correlated with patient survival. In our study, the increased hnRNPs were correlated with better prognosis in patients with gastric or rectal cancer. However, authors have reported the contrary results [54, 59, 60]. In liver cancer, hnRNPQ, R, and U were significantly different at the tumor stage, and hnRNPR and U were related to survival, hnRNPR and U may play an important role in tumor progression. Pancreatic cancer is a highly lethal type of cancer [61, 62], despite advances in surgery and immunotherapy [63, 64]. We attempted to find key hnRNPs, which might be involved in pancreatic cancer, yet no prognostic targets of hnRNPs were identified in this study. Whereas, in other reports, increased hnRNPH2 and U were correlated with worse prognosis in patients with pancreatic cancer [57, 58].
In the present study, IHC staining was performed to confirm the expression of hnRNPU based on our bioinformatics analysis in gastric, rectal, and liver cancer tissues compared to normal adjacent tissues. These results indicated that hnRNPU preotein was highly expressed in caner tissues compared to normal tissues, which was consistent with data mining results in Oncomine and GEPIA, indicating the accuracy and reliability of our bioinformatic study. In addition, we analyzed the gene alterations in GI tract cancer, the results indicated that hnRNPs may be associated with heredity and represent a substantial fraction of patients. Besides, there may exist interactions between hnRNPs molecules. The co-expresssed genes of hnRNPs with functional prediction showed that the biological process mainly enriched in RNA-related processing. These results showed that the alterations in hnRNPs and hnRNPs-mediated RNA metabolism might play important roles in GI tract cancer.
CONCLUSION
We conducted a systemic analysis of the expression and prognostic value of hnRNPs in five major types of GI cancer and gained comprehensive insights into the molecular features of hnRNPs. Novel molecular biomarkers were also identified with the potential to specifically diagnose five GI cancers in their early stages. Simultaneously, we predicted the possible underlying molecular mechanism. Taken together, we hope that our study offers new perspectives in identifying new biomarkers of hnRNPs family for early diagnosis and targeted therapy.
Table 1B.
Changes and datasets of hnRNPs expression in transcription level between gastric cancer tissues and normal gastric tissues (Oncomine).
| Gene | Type of Gastric Cancer versus Normal Gastric Tissues | Fold Change | p-Value | t-Test | Source and/or References |
|---|---|---|---|---|---|
| H1 | Gastric Mixed Adenocarcinoma vs. Normal | 2.933 | 6.05E-06 | 5.186 | DErrico Gastric [40] |
| H3 | Gastric Mixed Adenocarcinoma vs. Normal | 2.289 | 4.00E-06 | 8.904 | DErrico Gastric [40] |
| I | Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.011 | 4.06E-10 | 7.408 | DErrico Gastric [40] |
| L | Gastric Intestinal Type Adenocarcinoma vs. Normal | 5.148 | 4.74E-11 | 8.045 | DErrico Gastric [40] |
| Q | Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.116 | 1.74E-10 | 7.808 | DErrico Gastric [40] |
| U | Gastric Intestinal Type Adenocarcinoma vs. Normal | 2.173 | 1.96E-10 | 7.600 | DErrico Gastric [40] |
H1, hnRNPH1; H3, hnRNPH3; I, hnRNPI; L, hnRNPL; Q, hnRNPQ; U, hnRNPU.
Table 1C.
Changes and datasets of hnRNPs expression in transcription level between esophageal cancer tissues and normal esophageal tissues (Oncomine).
| Gene | Type of Esophageal Cancer versus Normal Esophageal Tissues | Fold Change | p-Value | t-Test | Source and/or References |
|---|---|---|---|---|---|
| A3 | Esophageal Adenocarcinoma vs. Normal | 3.266 | 9.07E-06 | 5.708 | Hao Esophagus [41] |
| Q | Barrett's Esophagus vs. Normal | 2.789 | 1.29E-06 | 5.648 | Hao Esophagus [41] |
A3, hnRNPA3; Q, hnRNPQ.
Table 1D.
Changes and datasets of hnRNPs expression in transcription level between liver cancer tissues and normal liver tissues (Oncomine).
| Gene | Type of Liver Cancer versus Normal Liver Tissues | Fold Change | p-Value | t-Test | Source and/or References |
|---|---|---|---|---|---|
| Q | Hepatocellular Carcinoma vs. Normal | 2.031 | 6.48E-46 | 16.136 | Roessler Liver 2 [42] |
| R | Hepatocellular Carcinoma vs. Normal | 2.019 | 6.05E-53 | 17.700 | Roessler Liver 2 [42] |
| U | Hepatocellular Carcinoma vs. Normal | 3.064 | 5.62E-11 | 8.445 | Wurmbach Liver [43] |
Q, hnRNPQ; R, hnRNPR; U, hnRNPU
ACKNOWLEDGEMENTS
The authors would like to thank members of the Center of Tumor Immunology and Cytotherapy for their support and help.
LIST OF ABBREVIATIONS
- CBioPortal
cBio Cancer Genomics Portal
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- FP
First progression
- GEPIA
Gene Expression Profiling Interactive Analysis
- GI cancer
Gastrointestinal cancer
- GO
Gene Ontology
- HnRNPs
Heterogeneous nuclear ribonucleoproteins
- IHC
Immunohistochemistry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KH
K-homology
- OS
Overall survival
- PPS
Post-progression survival
- RFS
Relapse-free survival
- RRM
RNA recognition motif
- TCGA
The Cancer Genome Atlas
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the Institutional Review Board of the Ethics Committee of the Affiliated Hospital of Qingdao University (QYFY WZLL 26689).
HUMAN AND ANIMAL RIGHTS
No animals were used in this study. All human experiments were performed according to the Declaration of Helsinki.
CONSENT FOR PUBLICATION
Patients were included in this experiment upon giving informed consent.
AVAILABILITY OF DATA AND MATERIALS
All data of this study are derived from publicly available databases. Detailed websites and methods have been explained in this article.
FUNDING
This work was financially supported by the Tianjin Medical University Cancer Institute and Hospital (18JCJQJC 47800).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Vangala D.B., Cauchin E., Balmaña J., et al. Screening and surveillance in hereditary gastrointestinal cancers: Recommendations from the European Society of Digestive Oncology (ESDO) expert discussion at the 20th European Society for Medical Oncology (ESMO)/World Congress on Gastrointestinal Cancer, Barcelona, June 2018. Eur. J. Cancer. 2018;104:91–103. doi: 10.1016/j.ejca.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Badreddine R., Wang K.K. Biomarkers in gastrointestinal cancers. Am. J. Gastroenterol. 2008;103(8):2106–2110. doi: 10.1111/j.1572-0241.2008.01994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong C.C., Li W., Chan B., Yu J. Epigenomic biomarkers for prognostication and diagnosis of gastrointestinal cancers. Semin. Cancer Biol. 2019;55:90–105. doi: 10.1016/j.semcancer.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 5.Duffy M.J., Lamerz R., Haglund C., et al. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int. J. Cancer. 2014;134(11):2513–2522. doi: 10.1002/ijc.28384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold M., Abnet C.C., Neale R.E., et al. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology. 2020;159(1):335–349.e15. doi: 10.1053/j.gastro.2020.02.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geuens T., Bouhy D., Timmerman V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016;135(8):851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dreyfuss G., Matunis M.J., Piñol-Roma S., Burd C.G. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993;62(1):289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 9.Bampton A., Gittings L.M., Fratta P., Lashley T., Gatt A. The role of hnRNPs in frontotemporal dementia and amyotrophic lateral sclerosis. Acta Neuropathol. 2020;140(5):599–623. doi: 10.1007/s00401-020-02203-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park Y.M., Hwang S.J., Masuda K., et al. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol. Cell. Biol. 2012;32(20):4237–4244. doi: 10.1128/MCB.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu C., Guo J., Liu Y., Jia J., Jia R., Fan M. Oral squamous cancer cell exploits hnRNP A1 to regulate cell cycle and proliferation. J. Cell. Physiol. 2015;230(9):2252–2261. doi: 10.1002/jcp.24956. [DOI] [PubMed] [Google Scholar]
- 12.Vu L.P., Prieto C., Amin E.M., et al. Functional screen of MSI2 interactors identifies an essential role for SYNCRIP in myeloid leukemia stem cells. Nat. Genet. 2017;49(6):866–875. doi: 10.1038/ng.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tockman M.S., Mulshine J.L., Piantadosi S., et al. Prospective detection of preclinical lung cancer: Results from two studies of heterogeneous nuclear ribonucleoprotein A2/B1 overexpression. Clin. Cancer Res. 1997;3(12 Pt 1):2237–2246. [PubMed] [Google Scholar]
- 14.Chen X., Gu P., Xie R., et al. Heterogeneous nuclear ribonucleoprotein K is associated with poor prognosis and regulates proliferation and apoptosis in bladder cancer. J. Cell. Mol. Med. 2017;21(7):1266–1279. doi: 10.1111/jcmm.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Li Y., Fan Y., Yu X., Mao X., Jin F. PTBP1 promotes the growth of breast cancer cells through the PTEN/Akt pathway and autophagy. J. Cell. Physiol. 2018;233(11):8930–8939. doi: 10.1002/jcp.26823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y., Wei Q., Tang Y., et al. Loss of hnRNPA2B1 inhibits malignant capability and promotes apoptosis via down-regulating Lin28B expression in ovarian cancer. Cancer Lett. 2020;475:43–52. doi: 10.1016/j.canlet.2020.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Fei T., Chen Y., Xiao T., et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl. Acad. Sci. USA. 2017;114(26):E5207–E5215. doi: 10.1073/pnas.1617467114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J.M., Jiang H., Yuan T., Zhou G.X., Li X.B., Wen K.M. High hnRNP AB expression is associated with poor prognosis in patients with colorectal cancer. Oncol. Lett. 2019;18(6):6459–6468. doi: 10.3892/ol.2019.11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuyama S., Goto Y., Sueoka N., et al. Heterogeneous nuclear ribonucleoprotein B1 expressed in esophageal squamous cell carcinomas as a new biomarker for diagnosis. Jpn. J. Cancer Res. 2000;91(6):658–663. doi: 10.1111/j.1349-7006.2000.tb00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang R., Zeng Y., Xu H., et al. Heterogeneous nuclear ribonucleoprotein K is overexpressed and associated with poor prognosis in gastric cancer. Oncol. Rep. 2016;36(2):929–935. doi: 10.3892/or.2016.4845. [DOI] [PubMed] [Google Scholar]
- 21.Guo Y., Zhao J., Bi J., Wu Q., Wang X., Lai Q. Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a tissue biomarker for detection of early hepatocellular carcinoma in patients with cirrhosis. J. Hematol. Oncol. 2012;5(1):37. doi: 10.1186/1756-8722-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Zhang J., Chen Q., et al. Long noncoding RNA SOX2OT promotes the proliferation of pancreatic cancer by binding to FUS. Int. J. Cancer. 2020;147(1):175–188. doi: 10.1002/ijc.32827. [DOI] [PubMed] [Google Scholar]
- 23.Masuda K., Kuwano Y. Diverse roles of RNA-binding proteins in cancer traits and their implications in gastrointestinal cancers. Wiley Interdiscip. Rev. RNA. 2019;10(3):e1520. doi: 10.1002/wrna.1520. [DOI] [PubMed] [Google Scholar]
- 24.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerami E., Gao J., Dogrusoz U., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao X., Sherman B.T., Huang W., et al. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szász A.M., Lánczky A., Nagy Á., et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menyhárt O., Nagy Á., Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018;5(12):181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy Á., Munkácsy G., Győrffy B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021;11(1):6047. doi: 10.1038/s41598-021-84787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing C., Cai Z., Gong J., Zhou J., Xu J., Guo F. Identification of potential biomarkers involved in gastric cancer through integrated analysis of Non-Coding RNA associated competing endogenous RNAs network. Clin. Lab. 2018;64(10):1661–1669. doi: 10.7754/Clin.Lab.2018.180419. [DOI] [PubMed] [Google Scholar]
- 33.Liang Y., Fan Y., Liu Y., Fan H. HNRNPU promotes the progression of hepatocellular carcinoma by enhancing CDK2 transcription. Exp. Cell Res. 2021;409(1):112898. doi: 10.1016/j.yexcr.2021.112898. [DOI] [PubMed] [Google Scholar]
- 34.Alon U., Barkai N., Notterman D.A., et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. USA. 1999;96(12):6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Notterman D.A., Alon U., Sierk A.J., Levine A.J. Transcriptional gene expression profiles of colorectal adenoma, adenocarcinoma, and normal tissue examined by oligonucleotide arrays. Cancer Res. 2001;61(7):3124–3130. [PubMed] [Google Scholar]
- 36.Sabates-Bellver J., Van der Flier L.G., de Palo M., et al. Transcriptome profile of human colorectal adenomas. Mol. Cancer Res. 2007;5(12):1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 37.Skrzypczak M., Goryca K., Rubel T., et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5(10):e13091. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser S., Park Y.K., Franklin J.L., et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8(7):R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong Y., Downey T., Eu K.W., Koh P.K., Cheah P.Y.A. ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin. Exp. Metastasis. 2010;27(2):83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 40.D’Errico M., de Rinaldis E., Blasi M.F., et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur. J. Cancer. 2009;45(3):461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 41.Hao Y., Triadafilopoulos G., Sahbaie P., Young H.S., Omary M.B., Lowe A.W. Gene expression profiling reveals stromal genes expressed in common between Barrett’s esophagus and adenocarcinoma. Gastroenterology. 2006;131(3):925–933. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roessler S., Jia H.L., Budhu A., et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wurmbach E., Chen Y.B., Khitrov G., et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 44.Buchholz M., Braun M., Heidenblut A., et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene. 2005;24(44):6626–6636. doi: 10.1038/sj.onc.1208804. [DOI] [PubMed] [Google Scholar]
- 45.Iacobuzio-Donahue C.A., Maitra A., Olsen M., et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am. J. Pathol. 2003;162(4):1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patry C., Bouchard L., Labrecque P., et al. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 2003;63(22):7679–7688. [PubMed] [Google Scholar]
- 47.Chen Y., Liu J., Wang W., et al. High expression of hnRNPA1 promotes cell invasion by inducing EMT in gastric cancer. Oncol. Rep. 2018;39(4):1693–1701. doi: 10.3892/or.2018.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S., Wang W., Ding H., et al. Aptamer BC15 against heterogeneous nuclear ribonucleoprotein A1 has potential value in diagnosis and therapy of hepatocarcinoma. Nucleic Acid Ther. 2012;22(6):391–398. doi: 10.1089/nat.2012.0363. [DOI] [PubMed] [Google Scholar]
- 49.Wang H., Vardy L.A., Tan C.P., et al. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell. 2010;18(1):52–62. doi: 10.1016/j.ccr.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 50.Ji F.J., Wu Y.Y., An Z., et al. Expression of both poly r(C) binding protein 1 (PCBP1) and miRNA-3978 is suppressed in peritoneal gastric cancer metastasis. Sci. Rep. 2017;7(1):15488. doi: 10.1038/s41598-017-15448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W., Shi H., Zhang M., et al. Poly C binding protein 1 represses autophagy through downregulation of LC3B to promote tumor cell apoptosis in starvation. Int. J. Biochem. Cell Biol. 2016;73:127–136. doi: 10.1016/j.biocel.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Roychoudhury P., Paul R.R., Chowdhury R., Chaudhuri K. HnRNP E2 is downregulated in human oral cancer cells and the overexpression of hnRNP E2 induces apoptosis. Mol. Carcinog. 2007;46(3):198–207. doi: 10.1002/mc.20265. [DOI] [PubMed] [Google Scholar]
- 53.Honoré B., Baandrup U., Vorum H. Heterogeneous nuclear ribonucleoproteins F and H/H′ show differential expression in normal and selected cancer tissues. Exp. Cell Res. 2004;294(1):199–209. doi: 10.1016/j.yexcr.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 54.Yu X., Cao F., Yu Y., et al. HNRNPL is identified and validated as a prognostic biomarker associated with microsatellite instability in human gastric cancer. DNA Cell Biol. 2021;40(10):1251–1260. doi: 10.1089/dna.2021.0165. [DOI] [PubMed] [Google Scholar]
- 55.Tian X.Y., Li J., Liu T.H., et al. The overexpression of AUF1 in colorectal cancer predicts a poor prognosis and promotes cancer progression by activating ERK and AKT pathways. Cancer Med. 2020;9(22):8612–8623. doi: 10.1002/cam4.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y.C., Chang K.C., Lin B.W., et al. The EGF/hnRNP Q1 axis is involved in tumorigenesis via the regulation of cell cycle-related genes. Exp. Mol. Med. 2018;50(6):1–14. doi: 10.1038/s12276-018-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X., Qian X., Peng L.X., et al. A splicing switch from ketohexokinase-C to ketohexokinase-A drives hepatocellular carcinoma formation. Nat. Cell Biol. 2016;18(5):561–571. doi: 10.1038/ncb3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutaria D.S., Jiang J., Azevedo-Pouly A.C.P., et al. Expression profiling identifies the noncoding processed transcript of HNRNPU with proliferative properties in pancreatic ductal adenocarcinoma. Noncoding RNA. 2017;3(3):E24. doi: 10.3390/ncrna3030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y., Liu S.X., Zhou Y.N., Wang J., Ji R. Research on the relationship between RAGE and its ligand HMGB1, and prognosis and pathogenesis of gastric cancer with diabetes mellitus. Eur. Rev. Med. Pharmacol. Sci. 2021;25(3):1339–1350. doi: 10.26355/eurrev_202102_24841. [DOI] [PubMed] [Google Scholar]
- 60.Ma Y.L., Peng J.Y., Zhang P., et al. Heterogeneous nuclear ribonucleoprotein A1 is identified as a potential biomarker for colorectal cancer based on differential proteomics technology. J. Proteome Res. 2009;8(10):4525–4535. doi: 10.1021/pr900365e. [DOI] [PubMed] [Google Scholar]
- 61.Sung H., Ferlay J., Siegel R.L., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 62.Wu W., Liu Q., Zhang J., Zhao Y. Survey on the current status of the diagnosis and treatment of pancreatic cancer in public tertiary hospitals in China: A cross-sectional questionnaire-based, observational study. J. Pancreatol. 2021;4(4):164–169. doi: 10.1097/JP9.0000000000000079. [DOI] [Google Scholar]
- 63.Pu N., Lou W., Yu J. PD-1 immunotherapy in pancreatic cancer: Current status. J. Pancreatol. 2019;2(1):6–10. doi: 10.1097/JP9.0000000000000010. [DOI] [Google Scholar]
- 64.Wu W., Jin G., Wang C., et al. The current surgical treatment of pancreatic cancer in China: A national wide cross-sectional study. J. Pancreatol. 2019;2(1):16–21. doi: 10.1097/JP9.0000000000000012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data of this study are derived from publicly available databases. Detailed websites and methods have been explained in this article.