Abstract
The current release of ProTherm, Thermodynamic Database for Proteins and Mutants, contains more than 10 000 numerical data (300% of the first version) of several thermodynamic parameters, experimental methods and conditions, reversibility of folding, details about the surrounding residues in space for all mutants, structural, functional and literature information. In the current version, we have added information about the source of each protein, identification codes for SWISS-PROT and Protein Information Resource and unique Protein Data Bank (PDB) code for proteins with relevant source. We have also provided additional options to search for data based on PDB code, number of states and reversibility. ProTherm is cross-linked with other sequence, structural, functional and literature databases, and the mutant sites and surrounding residues are automatically mapped on the structure. The ProTherm database is freely available at http://www.rtc.riken.go.jp/jouhou/protherm/protherm.html.
INTRODUCTION
Thermodynamic data for proteins are essential for understanding the mechanism of protein folding and stability, and for designing stable mutants. As complete genome sequences of many organisms become available, proteins are the next target for intensive study, as evidenced by emerging fields such as structural genomics and proteomics. The compilation of thermodynamic data along with the sequence, structural, functional and literature information would be a valuable resource for such studies. Pfeil (1) collected a set of several thermodynamic parameters from experimental studies. We have designed an electronically accessible database, ProTherm (2,3), including several thermodynamic parameters (unfolding Gibbs free energy change, enthalpy change, heat capacity change, transition temperature, etc.), along with sequence and structural information, experimental methods and conditions and literature information. Furthermore, we have provided a World Wide Web interface to facilitate searching the database, sorting and visualizing the results.
ORGANIZATION OF ProTherm
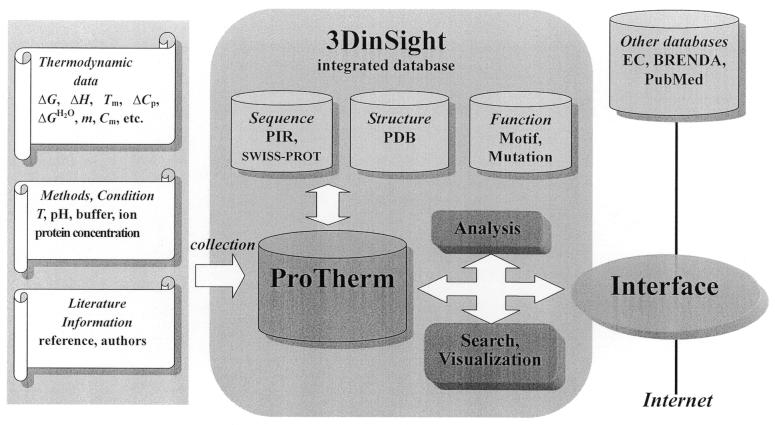
ProTherm is implemented into 3DinSight, a relational database system for structure, function and property of biomolecules (4). This facilitates more efficient search and retrieval of data by flexible queries, and enables users to gain insight into the relationship among structure, thermodynamics and function of proteins. Figure 1 illustrates the organization of ProTherm. The information about the name and source of the protein, details about thermodynamic data and other supplementary information are put together in ProTherm. The data in ProTherm can be retrieved through a form-based interface with several search, display and sorting options. The details about search options, tutorials and database statistics are described in the homepage.
Figure 1.
Organization of ProTherm. Content of data and links to other databases are shown.
RECENT DEVELOPMENTS
1. Release 3.0 contains more than 10 000 entries, 300% of the release 1.0 (3317 entries).
2. Protein Data Bank (PDB) code can be used for searching thermodynamic data associated with the structure.
3. The source of the protein is included.
4. Enthalpy change, ΔH, from calorimetric measurements is classified into van’t Hoff enthalpy change, ΔHvH, and calorimetric enthalpy change, ΔHcal.
5. Concentration of protein for each experiment is added.
6. Protein Information Resource [PIR (5)] and SWISS-PROT (6) codes are provided.
7. Unique PDB codes (7) are assigned for proteins with relevant source.
8. NMR category is added to the ‘Measure’ item and can be searched.
9. Quick search is provided, covering all the text fields.
DATA CHECKING AND REFINEMENT
We make every effort to minimize errors in registered data. However, errors can still occur. In order to improve the checking process, we send emails to the corresponding authors of each paper cited in ProTherm every time we add new data to ProTherm, through which the authors can check their data by themselves immediately. We have received feedback from authors, which helps us to maintain the data quality. We also receive suggestions and comments on ProTherm, and refine the functionality of ProTherm according to them.
CITATION OF ProTherm
The users of ProTherm are asked to cite this article in their publication including the URL http://www.rtc.riken.go.jp/jouhou/protherm/protherm.html. Suggestions and other materials for inclusion in the database are welcome and should be sent to protherm@rtc.riken.go.jp.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs K. Yutani, S. Kidokoro, K. Takahashi, M. Oobatake and H. Kono for their advice, and K. Kitajima for his help. The database is partially supported by the Grant-in-Aid for Publication Scientific Research Results from the Japan Society for the Promotion of Sciences (JSPS). M.M.G. is a recipient of RIKEN researcher fellowship.
REFERENCES
- 1.Pfeil W. (1998) Protein Stability and Folding: A Collection of Thermodynamic Data. Springer, NY.
- 2.Gromiha M.M., An,J., Kono,H., Oobatake,M, Uedaira,H. and Sarai,A. (1999) ProTherm: Thermodynamic Database for Proteins and Mutants. Nucleic Acids Res., 27, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gromiha M.M., An,J., Kono,H., Oobatake,M., Uedaira,H., Prabakaran,P. and Sarai,A (2000) ProTherm, version 2.0: thermodynamic database for proteins and mutants. Nucleic Acids Res., 28, 283–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An J., Nakama,T., Kubota,Y. and Sarai,A. (1998) 3DinSight: an integrated relational database and search tool for structure, function and property of biomolecules. Bioinformatics, 14, 188–195. [DOI] [PubMed] [Google Scholar]
- 5.Barker W.C., Garavelli,J.S., Huang,H., McGarvey,P.B., Orcutt,B.C., Srinivasarao,G.Y., Xiao,C., Yeh,L.S., Ledley,R.S., Janda,J.F. et al. (2000) The Protein Information Resource (PIR). Nucleic Acids Res., 28, 41–44. Updated article in this issue: Nucleic Acids Res. (2002), 30, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook,J., Feng,Z, Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2002), 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]