Abstract
Booster immunizations and breakthrough infections can elicit severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron subvariant neutralizing activity. However, the durability of the neutralization response is unknown. We characterize the sensitivity of BA.1, BA.2, BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB against neutralizing antibodies from vaccination, hybrid immunity, and breakthrough infections 4–6 months after vaccination and infection. We show that a two-dose CoronaVac or a third-dose ZF2001 booster elicits limited neutralization against Omicron subvariants 6 months after vaccination. Hybrid immunity as well as Delta, BA.1, and BA.2 breakthrough infections induce long-term persistence of the antibody response, and over 70% of sera neutralize BA.1, BA.2, BA.4/BA.5, and BF.7. However, BQ.1.1 and XBB, followed by BA.2.75, are more resistant to neutralization, with neutralizing titer reductions of ∼9- to 41-fold, ∼16- to 63-fold, and ∼4- to 25-fold, respectively. These data highlight additional vaccination in CoronaVac- or ZF2001-vaccinated individuals and provide insight into the durability of neutralization against Omicron subvariants.
Keywords: booster vaccination, hybrid immunity, breakthrough infection, Omicron subvariants, neutralization, immune evasion
Graphical abstract
Zhu et al. report that a two-dose CoronaVac or ZF2001 booster elicits limited neutralization against Omicron subvariants 6 months after vaccination. Hybrid immunity and Delta, BA.1, and BA.2 breakthrough infection induce neutralization against earlier Omicron variants, but not for BQ.1.1 and XBB, up to 5 months after vaccination or infection.
Introduction
As of October 2022, coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in a devastating global pandemic with over 624 million cases and 6.5 million deaths globally.1 SARS-CoV-2 continues to evolve and has generated five variants of concern (VOCs), including Alpha, Beta, Gamma, Delta, and Omicron.2 The Omicron variant (B.1.1.529 or BA.1) was first detected in South Africa in November 2021.3 Thereafter, Omicron subvariants, such as BA.2, BA.3, BA.2.12.1, BA.2.75, BA.4, BA.5 (hereafter BA.4/BA.5, share an identical spike sequence), BA.4.6, and BF.7, subsequently emerged,4 , 5 , 6 and BA.1, BA.2, and BA.5 became worldwide dominant. Currently, BA.5 subvariants BQ.1 and BQ.1.1 and XBB (a recombinant of BA.2 subvariants BJ.1 and BA.2.75) are appearing more frequently in sequenced SARS-CoV-2 infections in the USA, France, Singapore, India, and elsewhere.7 BQ.1.1 and XBB subvariants have the substitution R346T in the receptor-binding domain of their spike glycoprotein, which allows the virus to escape neutralization by a large group of neutralizing antibodies.8
The emerged Omicron subvariants share multiple mutations, but each also has unique mutations,4 which are expected to be associated with different antigenic properties. Previous studies have shown that the Omicron subvariants BA.1, BA.2, BA.3, BA.2.12.1, BA.2.75, BA.4/BA.5, BA.4.6, and BF.7 are substantially resistant to vaccine- and infection-induced serum-neutralizing activity.6 , 7 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 Booster immunizations and breakthrough infections can elicit Omicron-neutralizing activity immediately after vaccination or infection.18 , 20 , 21 , 22 , 23 , 24 , 25 , 26 However, these studies have largely focused on responses early in the course of vaccination or infection; the durability of the neutralization capability of antibodies following booster vaccination and breakthrough infection against Omicron subvariants remains poorly understood, which limits informed guidance on vaccination strategies. In addition, BQ.1.1 and XBB variants that are increasing in frequency raise the concern that the viruses have developed additional mechanisms to escape neutralization by antibodies elicited by vaccination or infection.
Inactivated vaccines, such as CoronaVac by Sinovac, China, and protein subunit vaccines, such as ZF2001 by Anhui Zhifei Longcom, China (a tandem repeat of the dimeric receptor-binding domain of the SARS-CoV-2 Wuhan-Hu-1 spike protein-based protein subunit vaccine), have been widely used in China and several other countries,27 and boosting is available in China. Therefore, we evaluated the susceptibility of earlier and currently circulating Omicron subvariants BA.1, BA.2, BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB to six panels of sera from SARS-CoV-2 infection-naive individuals with two doses of CoronaVac or heterologous ZF2001 booster vaccination, previously SARS-CoV-2 Alpha variant-infected individuals with booster vaccination, and fully vaccinated individuals with breakthrough infection of Delta, BA.1, or BA.2 and compared them with SARS-CoV-2 Wuhan-Hu-1 (WA1).
Results and discussion
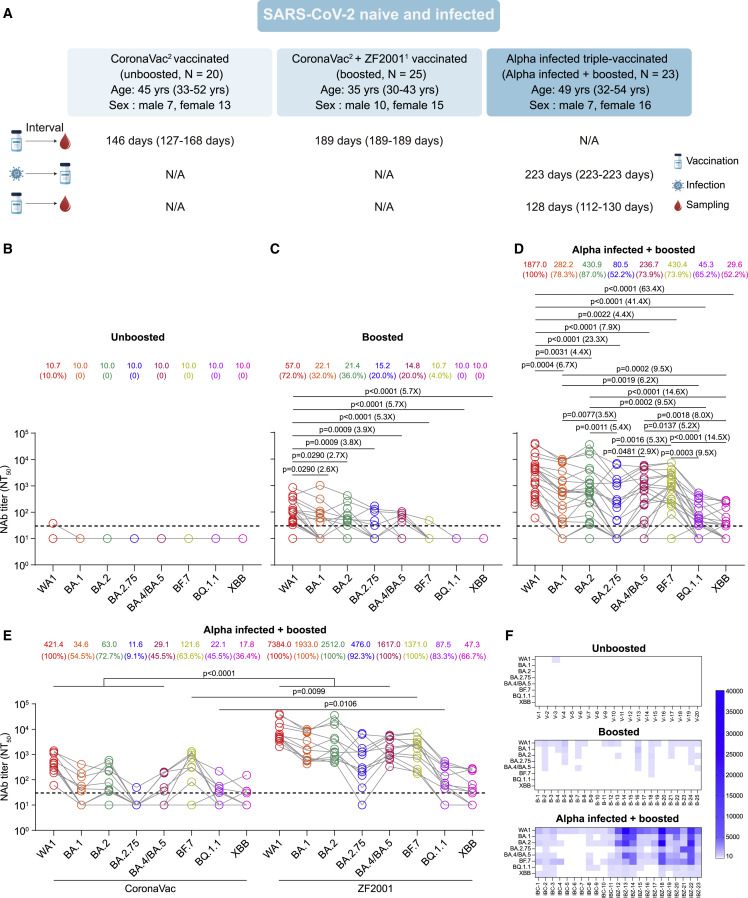
We first examined the neutralizing activity of sera collected from SARS-CoV-2 infection-naive individuals who were fully vaccinated with two doses of CoronaVac (unboosted) or a third dose of heterologous ZF2001 booster (boosted) against WA1, BA.1, BA.2, BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB using a pseudovirus neutralization assay (Figure 1A). Unboosted sera were collected from 20 individuals approximately 5 months (median 146 days, interquartile range [IQR] 127–168 days) after the second dose (Figure 1A; Table 1 ). We observed that only one of 20 unboosted sera had neutralization titers of 38 against WA1, and none neutralized Omicron subvariants (Figures 1B and 1F). Boosted sera were collected from 25 individuals approximately 6 months (median 189 days, IQR 189–189 days) after the third ZF2001 dose (Figure 1A; Table 1). We observed that 18 (72%) of 25 boosted sera had neutralizing antibody titers >30 against WA1 with a geometric mean titer (GMT) of 57 (95% confidence interval [CI] 32.3–100.6), whereas only 20%–36% of sera neutralized BA.1, BA.2, BA.2.75, and BA.4/BA.5 variants with a GMT decrease of 2.6- to 3.9-fold compared with WA1 (Figures 1C and 1F). However, only one of the 25 sera neutralized BF.7, and none neutralized BQ.1.1 and XBB. These data suggest that two doses of CoronaVac or heterologous ZF2001 booster vaccination elicited minimal or absent neutralizing antibodies against Omicron subvariants, especially for BQ.1.1 and XBB, 6 months after vaccination.
Figure 1.
Serum neutralization of SARS-CoV-2 Wuhan-Hu-1 (WA1) and Omicron subvariants in SARS-CoV-2 infection-naive and Alpha variant-infected individuals after vaccination
(A) Characteristics of vaccinated SARS-CoV-2 infection-naive and Alpha variant-infected individuals and sampling. Serum samples were collected from SARS-CoV-2 infection-naive individuals with two doses of CoronaVac (CoronaVac2, unboosted), SARS-CoV-2 infection-naive individuals with two doses of CoronaVac following heterologous ZF2001 booster (CoronaVac2 + ZF20011, boosted), and previously SARS-CoV-2 Alpha variant-infected individuals with three doses of CoronaVac or two doses of CoronaVac and one dose of ZF2001 (Alpha infected, triple vaccinated, and Alpha infected + boosted). All values are presented as the median and interquartile range. The number in the top right corner of the vaccines shows the number of doses. N/A, not available.
(B) Serum neutralization of WA1 and Omicron subvariants in 20 sera collected from unboosted individuals with two CoronaVac doses approximately 5 months after vaccination.
(C) Serum neutralization of WA1 and Omicron subvariants in 25 sera from boosted individuals with two doses of CoronaVac following heterologous ZF2001 booster vaccination approximately 6 months after vaccination.
(D) Serum neutralization of WA1 and Omicron subvariants in 23 sera from previous Alpha infections with boosted vaccination 4 months after the third vaccination dose.
(E) Serum NT50 values for homologous CoronaVac booster-vaccinated (n = 11) and heterologous booster-vaccinated (n = 12) individuals with SARS-CoV-2 Alpha variant infection are plotted according to vaccine type.
(F) Heatmaps showing vaccine NT50 values against each variant with subject numbers identified as “V” for two doses of CoronaVac vaccination, “B” for CoronaVac vaccination and ZF2001 booster vaccination, and “IBC” and “IBZ” for previous Alpha variant infection with homologous CoronaVac and heterologous ZF2001 booster vaccination, respectively.
All serum samples were tested in duplicates. The horizontal dotted line represents the limit of detection (LOD) of 30. The geometric mean titers (GMTs) are shown at the top of the plots along with the percentage of individuals with NT50 values above the LOD. A two-tailed Friedman test with a false discovery rate for multiple comparisons was performed in (C) and (D) and a two-tailed Wilcoxon rank-sum test was used in (E). A p value less than 0.05 is shown, and the fold change of GMT is also denoted.
Table 1.
Characteristics of study subjects
| Characteristics | CoronaVac2 vaccinated (unboosted) | CoronaVac2 and ZF2001 vaccinated (boosted) | Alpha variant infected, triple vaccinated (Alpha infected + boosted) | Fully vaccinated, Delta infected (Delta breakthrough) | CoronaVac3 vaccinated, BA.1 infected (BA.1 breakthrough) | CoronaVac3 vaccinated, BA.2 infected (BA.2 breakthrough) |
|---|---|---|---|---|---|---|
| No. of participants | 20 | 25 | 23 | 23 | 24 | 24 |
| Age (median, IQR) | 45 (32.5–51.3) | 35 (29.5–42.5) | 49 (32–54) | 47 (40–51) | 50 (41.5–57.5) | 38.5 (32–46.8) |
| Sex (%) | ||||||
| Male | 7 (35) | 10 (40) | 7 (30.4) | 8 (34.8) | 11 (45.8) | 11 (45.8) |
| Female | 13 (65) | 15 (60) | 16 (69.6) | 15 (65.2) | 13 (54.2) | 13 (54.2) |
| Vaccine type (n, %) | ||||||
| Two doses CoronaVac | 20 (100) | 25 (100) | 12 (52.2) | 10 (43.5) | NA | NA |
| Three doses CoronaVac | NA | NA | 11 (47.8) | NA | 24 (100) | 24 (100) |
| One dose ZF2001 | NA | 25 (100) | 12 (52.2) | NA | NA | NA |
| Three doses ZF2001 | NA | NA | NA | 13 (56.5) | NA | NA |
| Sample collection timing | ||||||
| Days from second vaccine dose to sampling (median, IQR) | 146 (127–167.8) | NA | NA | NA | NA | NA |
| Days from third vaccine dose to sampling (median, IQR) | NA | 189 (189–189) | 128 (112–130) | NA | NA | NA |
| Days from positive PCR test to sampling (median, IQR) | NA | NA | NA | 158 (156–159) | 114.5 (111–116) | 147 (145–148) |
IQR, interquartile range; NA, not available; PCR, polymerase chain reaction. The number in the top right corner of the vaccines shows the number of doses.
We further examined the neutralizing activity of sera collected from previously SARS-CoV-2 Alpha variant-infected individuals with booster vaccination (Alpha infected + boosted) against Omicron subvariants along with WA1 (Figure 1A; Table 1). Sera were collected from 23 previously Alpha variant-infected individuals approximately 4 months (median 128 days, IQR 112–130 days) after the third homologous CoronaVac (n = 11) or heterologous ZF2001 (n = 12) dose (Table 1). We found that previous Alpha variant infection following vaccination not only increased the neutralizing antibody titers against all tested variants but also restored neutralization of BA.1, BA.2, BA.4/BA.5, and BF.7 4 months after vaccination, with only a 4.4- to 7.9-fold reduction in GMT relative to WA1 (Figure 1D). In contrast, only approximately 50% of sera neutralized BA.2.75, BQ.1.1, and XBB, with a 23.3-, 41.4-, and 63.4-fold reduction in GMT compared with WA1, respectively. Moreover, GMTs against BA.2.75, BQ.1.1, and XBB were significantly lower than GMTs against BA.1, BA.2, BA.4/BA.5, and BF.7 (Figures 1D and 1F). Homologous CoronaVac-vaccinated individuals exhibited significantly lower neutralizing antibody titers than heterologous ZF2001-vaccinated individuals after the booster vaccination but a comparable neutralizing titer against XBB between the two groups (Figure 1E). Interestingly, several ZF2001-vaccinated individuals exhibited broad neutralizing activity that was effective against Omicron subvariants (Figure 1F). These data indicate that hybrid immunity by ZF2001 booster dose administration in previously Alpha variant-infected individuals enhances neutralizing antibody titers and increases the breadth of the neutralizing antibody response, especially against Omicron subvariants.
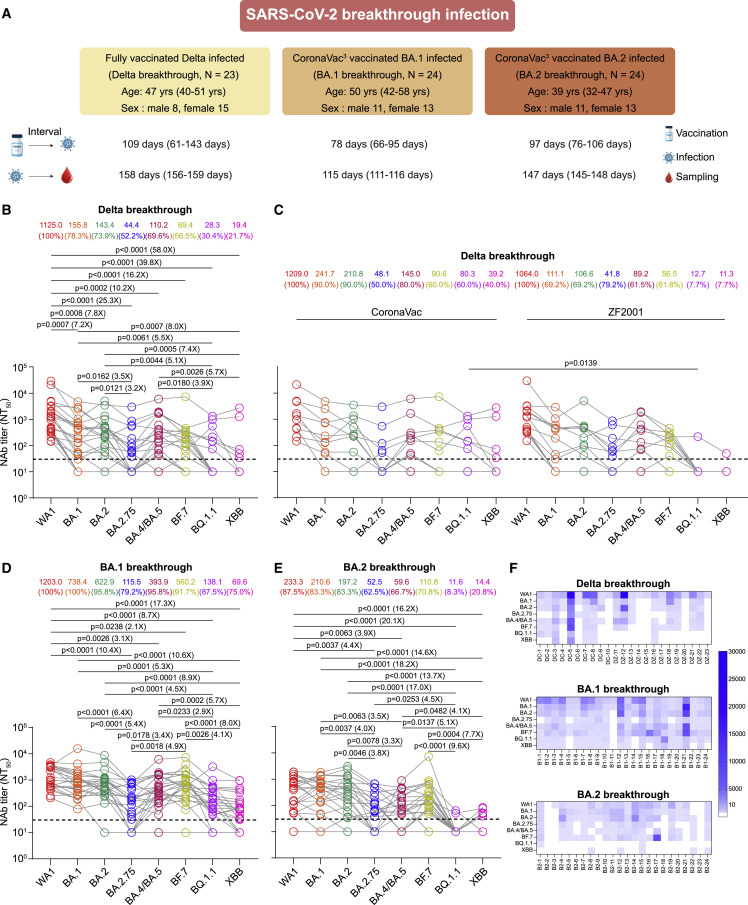
We next investigated neutralizing antibody responses and the extent of cross-neutralizing immunity in a cohort of 71 individuals who were vaccinated and then experienced Delta (n = 23), BA.1 (n = 24), or BA.2 (n = 24) breakthrough infections (Delta breakthrough, BA.1 breakthrough, or BA.2 breakthrough) (Figure 2A; Table 1). Serum samples were collected approximately 5 (median 158 days, IQR 156.0–159.0), 4 (median 114.5 days, IQR 111–116), and 5 months (median 147 days, IQR 145–148) after symptom onset or PCR test positivity for Delta, BA.1, and BA.2, respectively. We observed that all sera from Delta breakthrough infection neutralized WA1, and 78.3% (18/23), 73.9% (17/23), 52.2% (12/23), and 69.6% (16/23) of sera neutralized BA.1, BA.2, BA.2.75, and BA.4/BA.5, respectively (Figures 2B and 2F). Cross-neutralizing activity against Omicron subvariants was comparable but was limited by the 7.2-, 7.8-, 25.3-, 10.2-, and 16.2-fold GMT reductions in BA.1 (155.8), BA.2 (143.4), BA.2.75 (44.4), BA.4/BA.5 (110.2), and BF.7 (69.4) neutralization relative to WA1 (1,125), respectively (Figure 2B). In addition, GMT against BA.2.75 was significantly reduced compared with GMT against BA.1 and BA.2. However, ∼30% and ∼20% of sera neutralized BQ.1.1 and XBB, and neutralization titers were far lower against BQ.1.1 and XBB, with reductions of 39.8- and 58-fold compared with WA1, respectively. Moreover, neutralization titers against BQ.1.1 and XBB were significantly lower than neutralizing antibody titers against BA.1, BA.2, and BA.4/BA.5. Among these Delta breakthrough individuals, two doses of CoronaVac-vaccinated individuals (n = 10) induced a neutralizing antibody titer comparable to that of three doses of ZF2001-vaccinated individuals (n = 13) before infection except for a higher neutralizing titer against BQ.1.1 in CoronaVac-vaccinated individuals (Figure 2C).
Figure 2.
Neutralization of Omicron subvariants by sera from breakthrough infections after vaccination
(A) Characteristics of Delta, BA.1, and BA.2 breakthrough-infected individuals and sampling. Serum samples were collected from individuals vaccinated with two doses of CoronaVac or three doses of the ZF2001 vaccine (fully vaccinated) who subsequently had a breakthrough infection with Delta (Delta breakthrough) and individuals vaccinated with three doses of CoronaVac (CoronaVac3) who subsequently had a breakthrough infection with BA.1 (BA.1 breakthrough) or BA.2 (BA.2 breakthrough). All values are presented as the median and interquartile range. The number in the top right corner of the vaccines shows the number of doses. N/A, not available.
(B) Neutralization of SARS-CoV-2 Wuhan-Hu-1 (WA1) and Omicron subvariants by 23 sera collected from individuals with Delta breakthrough infection 5 months after infection.
(C) Serum NT50 values for two doses of CoronaVac-vaccinated (n = 10) and three doses of ZF2001-vaccinated (n = 13) individuals before Delta infection are plotted according to vaccine type.
(D) Neutralization of WA1 and Omicron subvariants by 24 sera collected from individuals with BA.1 breakthrough infection 4 months after infection.
(E) Neutralization of WA1 and Omicron subvariants by 24 sera collected from individuals with BA.2 breakthrough infection 5 months after infection.
(F) Heatmaps showing patient NT50 values against each variant. Patients were identified as “DC” or “DZ” for the Delta breakthrough infection following CoronaVac or ZF2001 vaccination, “B1” for BA.1 breakthrough infection, and “B2” for BA.2 breakthrough infections.
All serum samples were tested in duplicates. The horizontal dotted line represents the LOD of 30. The GMTs are shown at the top of the plots along with the percentage of individuals with NT50 values above the LOD. A two-tailed Friedman test with a false discovery rate for multiple comparisons was performed in (B), (D), and (E) and a two-tailed Wilcoxon rank-sum test was used in (C). A p value less than 0.05 is shown, and the fold change of GMT is also denoted.
Interestingly, sera from BA.1 breakthrough infection approximately 4 months after infection not only efficiently neutralized WA1 (100%), BA.1 (100%), BA.2 (95.8%), BA.4/BA.5 (95.8%), and BF.7 (91.7%) but also relatively efficiently neutralized BA.2.75 (79.2%), BQ.1.1 (87.5%), and XBB (75%) (Figures 2D and 2F). However, neutralizing antibody titers against BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB were substantially lower than those against WA1, with a 10.4-, 3.1-, 2.1-, 8.7-, and 17.3-fold reduction in GMT compared with WA1, and BQ.1.1 and XBB still exhibited strong resistance. Moreover, the GMTs of BQ.1.1 and XBB were significantly lower than those of BA.1, BA.2, BA.4/BA.5, and BF.7 (Figure 2D). Unexpectedly, sera from 5 months after BA.2 breakthrough infection, in contrast to BA.1 breakthrough infection, exhibited relatively small increases in neutralizing titers against WA1 and tested variants, and approximately only 80% of sera neutralized WA1, BA.1, and BA.2 and approximately 60% of sera neutralized BA.4/BA.5 and BA.2.75 (Figure 2E). Neutralizing antibody titers against BA.1, BA.2, and BF7 were comparable to those against WA1, but neutralizing titers against BA.2.75 (4.4-fold) and BA.4/BA.5 (3.9-fold) were significantly decreased (Figure 2E). In line with other cohorts, neutralization titers were far lower against BQ.1.1 and XBB, with reductions of 20.1- and 16.2-fold compared with WA1. Moreover, only two (8.3%) and five (20.8%) sera neutralized BQ.1.1 and XBB, respectively, with the lowest titers observed (Figures 2E and 2F).
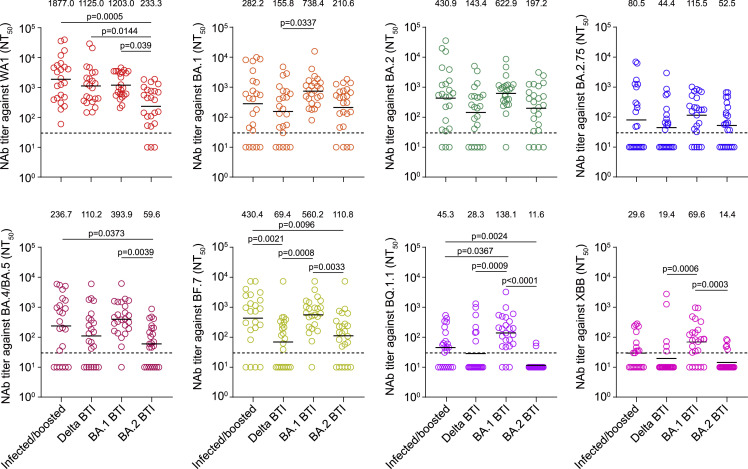
We further compared neutralization antibody titers against WA1 and Omicron subvariants among sera from Alpha infected + boosted as well as Delta, BA.1, and BA.2 breakthrough infections. We did not include sera from individuals vaccinated with two doses of the CoronaVac or ZF2001 booster, as these sera lacked neutralization capacity against Omicron subvariants 6 months after vaccination. We found that Delta breakthrough infections elicited reduced neutralizing antibody titers against BA.1, BF.7, BQ.1.1, and XBB compared with BA.1 breakthrough infection or Alpha infected + boosted (Figure 3 ). BA.2 breakthrough infections led to a significantly lower neutralizing antibody titer against WA1 compared with Alpha infected + boosted, Delta breakthrough infection, and BA.1 breakthrough infection; against BA.4/BA.5 compared with BA.1 breakthrough infection and Alpha infected + boosted; against BF.7 or BQ.1.1 compared with BA.1 breakthrough infection and Alpha infected + boosted; and against XBB compared with BA.1 breakthrough infection (Figure 3). It should be noted that the Alpha infected + boosted and BA.1 breakthrough infection samples were collected approximately 4 months after the third vaccination or infection. In contrast, Delta and BA.2 breakthrough infection samples were collected approximately 5 months after infection following infection, which might partially explain the lower levels of neutralizing antibodies for the Delta and BA.2 breakthrough infection samples. Comparable neutralization levels for other Omicron subvariants were observed among sera from Alpha infected + boosted and breakthrough infections.
Figure 3.
Neutralizing antibody titers against Omicron subvariants among prior SARS-CoV-2 infection and breakthrough infections
Comparisons of neutralizing antibody titers against SARS-CoV-2 Wuhan-Hu-1 (WA1), BA.1, BA.2, BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB among prior SARS-CoV-2 Alpha variant infections with booster vaccination (infected/boosted) and Delta, BA.1, and BA.2 breakthrough infections (BTIs). All serum samples were tested in duplicates. The GMTs are shown above each column. The horizontal dotted line represents the LOD of 30. A two-tailed Kruskal-Wallis test with a false discovery rate was performed for multiple comparisons. A p value less than 0.05 is shown.
Our data demonstrate that two doses of CoronaVac and ZF2001 booster vaccination elicit limited or absent neutralizing activity against Omicron subvariants 6 months after vaccination, suggesting that an additional dose is likely required to maintain effectiveness against Omicron subvariants. In contrast, while previous SARS-CoV-2 infection with booster vaccination, especially for ZF2001 booster vaccination, and BA.1 breakthrough infection elicited substantial neutralization activity against all tested Omicron subvariants, including BQ.1.1 and XBB, 4 months after vaccination or infection, neutralization of BQ.1.1 and XBB 5 months after Delta and BA.2 breakthrough infection was significantly impaired. However, the duration of protection, the potential need for an additional dose, and whether an Omicron-modified vaccine is needed should be further evaluated.
Limitations of the study
The current study has several limitations. First, our cohort was cross-sectional, which limits our ability to determine the dynamics of neutralizing antibody titers to variants across single individuals and assess the potential confounding effect on neutralizing antibody titers, and the durability of neutralization against the Omicron variant remains to be determined beyond 6 months after infection or vaccination. Second, the relatively small sample sizes and cohorts were not fully matched in terms of the interval between vaccination or infection and sampling and demographic characteristics such as the age of individuals. Third, we did not assess long-lived plasma cell, memory B cell, and T cell immunity, which could provide further insights into the mechanisms underlying the broad neutralizing activity associated with hybrid immunity and breakthrough infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| SARS-CoV-2 pseudovirus for WA1 | This study | N/A |
| SARS-CoV-2 pseudovirus for BA.1 | This study | N/A |
| SARS-CoV-2 pseudovirus for BA.2 | This study | N/A |
| SARS-CoV-2 pseudovirus for BA.2.75 | This study | N/A |
| SARS-CoV-2 pseudovirus for BA.4/BA.5 | This study | N/A |
| SARS-CoV-2 pseudovirus for BF.7 | This study | N/A |
| SARS-CoV-2 pseudovirus for BQ.1.1 | This study | N/A |
| SARS-CoV-2 pseudovirus for XBB | This study | N/A |
| E.coli DH5α Competent Cells | TaKaRa | Cat# 9057 |
| Biological samples | ||
| Serum samples from two doses CoronaVac vaccinated individuals | This study | N/A |
| Serum samples from ZF2001 booster vaccinated individuals | This study | N/A |
| Serum samples from previous Alpha variant infections with booster vaccination | This study | N/A |
| Serum samples from Delta breakthrough infections | This study | N/A |
| Serum samples from BA.1 breakthrough infections | This study | N/A |
| Serum samples from BA.2 breakthrough infections | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Phosphate buffered saline (PBS) | Gibco | Cat# C10010500BT |
| Dulbecco’s modified eagle medium (DMEM) | Gibco | Cat# C111995500BT |
| Trypsin-EDTA (0.25%) | Solarbio | Cat# T1300 |
| HEPES | Gibco | Cat# 15630-080 |
| Fetal bovine serum (FBS) | Gibco | Cat# 10099-141C |
| Penicillin/streptomycin | Gibco | Cat# 15140-122 |
| PEI MAX (MW 40000) | Polysciences | Cat# 24765-1 |
| Luciferase Assay Reagent | Vazyme | Cat# DD1201-01 |
| Experimental models: Cell lines | ||
| HeLa-hACE2 cells | Tsinghua University | N/A |
| HEK-293T cells | ATCC | Cat# CRL-3216 |
| Recombinant DNA | ||
| WA1 spike plasmid | This study | N/A |
| Omicron BA.1 spike plasmid | This study | N/A |
| Omicron BA.2 spike plasmid | This study | N/A |
| Omicron BA.2.75 spike plasmid | This study | N/A |
| Omicron BA.4/BA.5 spike plasmid | This study | N/A |
| Omicron BF.7 spike plasmid | This study | N/A |
| Omicron BQ.1.1 spike plasmid | This study | N/A |
| Omicron XBB spike plasmid | This study | N/A |
| Firefly luciferase encoding lentivirus backbone plasmid | Tsinghua University | N/A |
| Software and algorithms | ||
| GraphPad Prism | Version 9.0.0 | GraphPad |
| Adobe Illustrator | Version 23.0.1 | Adobe |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mai-Juan Ma (mjma@163.com).
Materials availability
All unique reagents generated during this study are available from the lead contact with a completed materials transfer agreement.
Experimental model and subject details
Study design, participants, and sample collection
We performed a cross-sectional study to investigate the effect of vaccination and breakthrough infection on the cross-variant neutralization capacity of human sera 4–6 months after vaccination or infection. We enrolled SARS-CoV-2 infection-naïve individuals with two doses of CoronaVac or two doses of CoronaVac + one dose of ZF2001, previously SARS-CoV-2 Alpha variant-infected individuals with three doses of CoronaVac or two doses of CoronaVac + one dose of ZF2001, two doses of CoronaVac or three doses of ZF2001-vaccinated individuals with a confirmed subsequent SARS-CoV-2 Delta breakthrough infection, and three doses of CoronaVac-vaccinated individuals with a confirmed subsequent SARS-CoV-2 BA.1 or BA.2 breakthrough infection.
Vaccine sera of 20 individuals (median 45.0 years, interquartile range [IQR] 32.5–51.3 years; 35.0% male) who had two doses of CoronaVac were collected at a median of 146 days after the second dose. Vaccine sera of 25 individuals who were boosted with the third dose of CoronaVac or ZF2001 (median 35.0 years, IQR 29.5–42.5 years; 40.0% male) were collected at a median of 189 days after the third dose of vaccination. Vaccine sera of 23 individuals who were previously infected with the SARS-CoV-2 Alpha variant and boosted with homologous CoronaVac or heterologous ZF2001 were collected at a median of 223 days after the third dose vaccination. Convalescent sera were collected from 23, 24, and 24 fully vaccinated individuals with Delta (10 two-dose CoronaVac vaccinated and 13 three-dose ZF2001 vaccinated; median 47.0 years, IQR 40.0–51.0 years, 34.8% male), BA.1 (three-dose CoronaVac vaccinated; median 50.0 years, IQR 41.5–57.5 years, 45.8% male) or BA.2 (three-dose CoronaVac vaccinated; median 38.5 years, IQR 32.0–46.8 years, 45.8% male) breakthrough infection at a median of 158, 115, and 147 days after symptom onset or PCR positivity, respectively. Sera were isolated by centrifugation at 2000 rpm for 10 min and cryopreserved at −80°C until use. The serum-neutralizing activity was measured using pseudovirus neutralization assays. The details of the demographic information and sample collection time points of different cohorts are shown in Figures 1A and 2A and Table 1.
Full vaccination was defined as when the second or third shot of the CoronaVac vaccination or the third shot of the ZF2001 vaccination was administered at least 14 days before symptom onset or a positive PCR test for SARS-CoV-2.28 , 29 Breakthrough infection was defined as fully vaccinated individuals being diagnosed with SARS-CoV-2 infection.28 , 29 All individuals with breakthrough infection had a sequence-confirmed infection or PCR-confirmed symptomatic disease occurring while in isolation and direct contact with sequence-confirmed cases.
This study was conducted following the Declaration of Helsinki and approved by the Institutional Review Board of the Beijing Institute of Microbiology and Epidemiology (IRB number: AF/SC-08/02.60 and AF/SC-08/02.124). All participants provided written consent.
Cell lines
Human embryonic kidney HEK-293T cells were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS, Gibco) and supplemented with 1% penicillin–streptomycin (Gibco). Cells were disrupted at confluence with 0.25% trypsin in 1 mM EDTA (Solarbio) every 48–72 h. HeLa-hACE2 cells were provided by Prof. Lin-Qi Zhang from Tsinghua University and were cultured under the same conditions.
Method details
Spike plasmid pseudovirus production
Pseudovirus particles were generated as previously described30 , 31 by cotransfecting HEK-293T cells (ATCC, CRL-3216) with human immunodeficiency virus backbones expressing firefly luciferase (pNL4-3-R-E-luciferase) and the pcDNA3.1 vector encoding either SARS-CoV-2 Wuhan-Hu-1 (WA1) or mutated S protein (BA.1, BA.2, BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB) plasmids. Codon-optimized, full-length open reading frames of the spike genes of WA1, BA.1, BA.2, BA.2.75, BA.4/BA.5, BF.7, BQ.1.1, and XBB were synthesized by GenScript (Nanjing, China). The mutations in the S protein of variants are shown in Table S1. All plasmid spike sequences were verified by Sanger sequencing.
Pseudovirus particles were generated by cotransfecting HEK-293T cells (ATCC) with human immunodeficiency virus backbones expressing firefly luciferase (pNL4-3-R-E-luciferase) and the pcDNA3.1 vector encoding either WA1 or mutated S protein (Delta and Omicron subvariants) plasmids. The medium was replaced with fresh medium at 24 h, and the supernatants were harvested at 48 h post-transfection and clarified by centrifugation at 300 × g for 10 min before being aliquoted and stored at −80°C until use.
Pseudovirus neutralization assay
A SARS-CoV-2 pseudovirus neutralization assay (pVNT) was performed as described,32 with the target cell line HeLa overexpressing hACE2 orthologs. All viruses were first titrated to normalize the viral input between assays. Duplicate 3-fold 8-point serial dilutions of heat-inactivated sera (starting at 1:30) were incubated with 500-1000 TCID50 of SARS-CoV-2 pseudotyped virus for 1 h at 37°C and 5% CO2. Subsequently, 1x104 HeLa-ACE2 cells per well were added and incubated at 37°C and 5% CO2 for 48 h. Afterward, the supernatant was removed, and the cells were lysed using passive lysis buffer (Vazyme) for 3 min at room temperature. The lysates were transferred to an opaque white 96-well plate, and reconstituted luciferase assay buffer (Vazyme) was added and mixed with each lysate. Luminescence was measured immediately after mixing using a GloMax 96 Microplate Luminometer (Promega). The neutralization titer (NT50) was determined by luciferase activity with a four-parameter nonlinear regression inhibitor curve in GraphPad Prism 8.4.2 (GraphPad Software). NT50 was reported as the reciprocal serum dilution causing a 50% reduction in relative light units. A sample with NT50 values no more than 30 (the detectable limit) was considered negative for neutralizing antibodies and was assigned a nominal value of 10 in geometric mean titer (GMT) calculations, which is the lowest serum dilution factor used in the pseudovirus neutralization assay.
Quantification and statistical analysis
The Friedman and Kruskal–Wallis tests with the false discovery rate method were used for multiple comparisons where it appreciates. The Wilcoxon rank-sum test was used for comparisons between the two groups. All statistical analyses were performed using GraphPad Prism (version 8.4.2, La Jolla, California, USA), and all statistical tests were 2-sided with a significance level of 0.05. Details are additionally provided in the Figure legends.
Acknowledgments
We thank all participants for providing blood samples. This work was supported by grants from the National Natural Science Foundation of China (82273692 and 92169207), the Beijing Natural Science Foundation (L202038), the Quzhou Science and Technology Bureau (2021K12), and the Natural Science Foundation of Shandong Province, China (ZR2022MH198).
Author contributions
M.-J.M. conceived and supervised the study. K.-L.Z., X.-L.J., X.X., B.-D.Z., G.-P.C., W.-K.S., P.-X.H., J.-Z.Z., H.-X.G., and E.-H.D. collected blood samples. X.-J.W. and K.-L.Z. constructed the spike expression plasmids, produced pseudoviruses, and conducted the pseudovirus neutralization experiments. M.-J.M. drafted the manuscript. All authors reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112075.
Supplemental information
Data and code availability
-
•
All data reported in this paper are available within the main manuscript and the supplemental information.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.WHO . 2022. COVID-19 weekly epidemiological update.file:///C:/Users/mjma_/AppData/Local/Temp/MicrosoftEdgeDownloads/b76aaa79-187c-468f-8360-e8b5e1d0c1cd/20221005_Weekly_Epi_Update_112.pdf [Google Scholar]
- 2.WHO . 2022. Tracking SARS-CoV-2 Variants. [Google Scholar]
- 3.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/d41586-021-03832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., Subramoney K., Makatini Z., Moyo S., Amoako D.G., et al. Emergence of SARS-CoV-2 omicron lineages BA.4 and BA.5 in South Africa. Nat. Med. 2022;28:1785–1790. doi: 10.1038/s41591-022-01911-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jian F., Yu Y., Song W., Yisimayi A., Yu L., Gao Y., Zhang N., Wang Y., Shao F., Hao X., et al. Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect. Dis. 2022;22:1535–1537. doi: 10.1016/s1473-3099(22)00642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora P., Nehlmeier I., Kempf A., Cossmann A., Schulz S.R., Dopfer-Jablonka A., Baier E., Tampe B., Moerer O., Dickel S., et al. Lung cell entry, cell-cell fusion capacity, and neutralisation sensitivity of omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1537–1538. doi: 10.1016/s1473-3099(22)00591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Sander L.E., Kurth F., Klein F. Neutralisation sensitivity of the SARS-CoV-2 omicron BA.2.75 sublineage. Lancet Infect. Dis. 2022;22:1422–1423. doi: 10.1016/s1473-3099(22)00580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen X., Chalkias S., Feng J., Chen X., Zhou H., Marshall J.C., Girard B., Tomassini J.E., Aunins A., Das R., Montefiori D.C. Neutralization of SARS-CoV-2 omicron BA.2.75 after mRNA-1273 vaccination. N. Engl. J. Med. 2022;387:1234–1236. doi: 10.1056/NEJMc2210648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheward D.J., Kim C., Fischbach J., Muschiol S., Ehling R.A., Björkström N.K., Karlsson Hedestam G.B., Reddy S.T., Albert J., Peacock T.P., Murrell B. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1421–1422. doi: 10.1016/s1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan C.W., Lim B.L., Young B.E., Yeoh A.Y.Y., Yung C.F., Yap W.C., Althaus T., Chia W.N., Zhu F., Lye D.C., Wang L.F. Comparative neutralisation profile of SARS-CoV-2 omicron subvariants BA.2.75 and BA.5. Lancet. Microbe. 2022;3:e898. doi: 10.1016/s2666-5247(22)00220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022;30:1512–1517.e4. doi: 10.1016/j.chom.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan L.J., Jiang W.G., Wang Z.Y., Yao L., Zhu K.L., Meng Q.C., Wang B.S., Li L.B., Wang G.L., Ma M.J. Neutralizing immunity against SARS-CoV-2 Omicron BA.1 by infection and vaccination. iScience. 2022;25:104886. doi: 10.1016/j.isci.2022.104886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao L., Zhu K.L., Jiang X.L., Wang X.J., Zhan B.D., Gao H.X., Geng X.Y., Duan L.J., Dai E.H., Ma M.J. Omicron subvariants escape antibodies elicited by vaccination and BA.2.2 infection. Lancet Infect. Dis. 2022;22:1116–1117. doi: 10.1016/s1473-3099(22)00410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu K.L., Gao H.X., Yao L., Rong J., Yang L., Zhang Z., Jiang P., Duan L.J., Wang G.L., Dai E.H., Ma M.J. Delta infection following vaccination elicits potent neutralizing immunity against the SARS-CoV-2 Omicron. J. Infect. Dis. 2022;226:1551–1555. doi: 10.1093/infdis/jiac149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muik A., Lui B.G., Bacher M., Wallisch A.K., Toker A., Finlayson A., Kruger K., Ozhelvaci O., Grikscheit K., Hoehl S., et al. Omicron BA.2 breakthrough infection enhances cross-neutralization of BA.2.12.1 and BA.4/BA.5. Sci Immunol. 2022;7:eade2283. doi: 10.1126/sciimmunol.ade2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uraki R., Ito M., Furusawa Y., Yamayoshi S., Iwatsuki-Horimoto K., Adachi E., Saito M., Koga M., Tsutsumi T., Yamamoto S., et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. Lancet Infect. Dis. 2023;23:30–32. doi: 10.1016/s1473-3099(22)00816-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angyal A., Longet S., Moore S.C., Payne R.P., Harding A., Tipton T., Rongkard P., Ali M., Hering L.M., Meardon N., et al. T-cell and antibody responses to first BNT162b2 vaccine dose in previously infected and SARS-CoV-2-naive UK health-care workers: a multicentre prospective cohort study. Lancet. Microbe. 2022;3:e21–e31. doi: 10.1016/S2666-5247(21)00275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lozano-Ojalvo D., Camara C., Lopez-Granados E., Nozal P., Del Pino-Molina L., Bravo-Gallego L.Y., Paz-Artal E., Pion M., Correa-Rocha R., Ortiz A., et al. Differential effects of the second SARS-CoV-2 mRNA vaccine dose on T cell immunity in naive and COVID-19 recovered individuals. Cell Rep. 2021;36:109570. doi: 10.1016/j.celrep.2021.109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatatos L., Czartoski J., Wan Y.H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussières G., Brassard N., Laumaea A., Vézina D., Prévost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urbanowicz R.A., Tsoleridis T., Jackson H.J., Cusin L., Duncan J.D., Chappell J.G., Tarr A.W., Nightingale J., Norrish A.R., Ikram A., et al. Two doses of the SARS-CoV-2 BNT162b2 vaccine enhance antibody responses to variants in individuals with prior SARS-CoV-2 infection. Sci. Transl. Med. 2021;13:eabj0847. doi: 10.1126/scitranslmed.abj0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans J.P., Zeng C., Carlin C., Lozanski G., Saif L.J., Oltz E.M., Gumina R.J., Liu S.-L. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 2022;14:eabn8057. doi: 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X.-L., Zhu K.-L., Wang X.-J., Wang G.-L., Li Y.-K., He X.-J., Sun W.-K., Huang P.-X., Zhang J.-Z., Gao H.-X., et al. Omicron BQ.1 and BQ.1.1 escape neutralisation by omicron subvariant breakthrough infection. Lancet Infect. Dis. 2023;23:28–30. doi: 10.1016/S1473-3099(22)00805-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu K., Dai L., Gao G.F. Humoral and cellular immunity and the safety of COVID-19 vaccines: a summary of data published by 21 May 2021. Int. Immunol. 2021;33:529–540. doi: 10.1093/intimm/dxab061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juthani P.V., Gupta A., Borges K.A., Price C.C., Lee A.I., Won C.H., Chun H.J. Hospitalisation among vaccine breakthrough COVID-19 infections. Lancet Infect. Dis. 2021;21:1485–1486. doi: 10.1016/S1473-3099(21)00558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15:3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- 31.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper are available within the main manuscript and the supplemental information.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.