Significance
Recent studies revealed that mismatch repair (MMR)-deficient cancer cells require Werner (WRN) helicase for survival. However, how WRN loss triggers death of MMR-deficient cancer cells remains unclear. We show that WRN depletion selectively induces p53/PUMA-mediated cell death program in MMR-deficient colorectal cancer cells. The in vitro and in vivo effects of WRN depletion or inhibition are dependent on p53/PUMA-mediated apoptosis. Our findings provide a mechanistic insight on the vulnerability of MMR-deficient cancer cells to WRN loss and support WRN as a promising therapeutic target in p53-wildtype and MMR-deficient colorectal cancer.
Keywords: Werner, mismatch repair, synthetic lethal, p53, PUMA
Abstract
Synthetic lethality is a powerful approach for targeting oncogenic drivers in cancer. Recent studies revealed that cancer cells with microsatellite instability (MSI) require Werner (WRN) helicase for survival; however, the underlying mechanism remains unclear. In this study, we found that WRN depletion strongly induced p53 and its downstream apoptotic target PUMA in MSI colorectal cancer (CRC) cells. p53 or PUMA deletion abolished apoptosis induced by WRN depletion in MSI CRC cells. Importantly, correction of MSI abrogated the activation of p53/PUMA and cell killing, while induction of MSI led to sensitivity in isogenic CRC cells. Rare p53-mutant MSI CRC cells are resistant to WRN depletion due to lack of PUMA induction, which could be restored by wildtype (WT) p53 knock in or reconstitution. WRN depletion or treatment with the RecQ helicase inhibitor ML216 suppressed in vitro and in vivo growth of MSI CRCs in a p53/PUMA-dependent manner. ML216 treatment was efficacious in MSI CRC patient-derived xenografts. Interestingly, p53 gene remains WT in the majority of MSI CRCs. These results indicate a critical role of p53/PUMA-mediated apoptosis in the vulnerability of MSI CRCs to WRN loss, and support WRN as a promising therapeutic target in p53-WT MSI CRCs.
DNA mismatch repair (MMR) is an evolutionarily conserved system for recognizing and repairing erroneous insertion, deletion, and misincorporation of bases generated during DNA replication and recombination (1, 2). In mammalian cells, there are five key MMR proteins including MSH2, MSH3, MSH6, MLH1, and PMS2. These proteins form heterodimers to repair base–base mismatches and single nucleotide or larger insertion–deletions (indels) (1, 3). MMR genes play an important role in the maintenance of genomic stability. Deficiency in MMR genes contributes to pathogenesis of colorectal cancer (CRC) and other types of cancer (4).
CRC is the second leading cause of cancer-related deaths in the United States (5). A key factor driving colorectal tumor initiation and progression is genomic instability, including chromosomal instability and microsatellite instability (MSI) (4). MSI is characterized by increased rates of small indels and point mutations in short-tandem microsatellite repeat sequences (6), which exist in ~15% of CRCs (3). MSI in CRCs is caused by genetic and epigenetic silencing of MMR genes, such as mutations of MSH2, MLH1, and MSH6 (7–9), as well as promoter hypermethylation of MLH1 (1). Compared with microsatellite stable (MSS) CRCs, MSI CRCs typically have high levels of tumor-infiltrating lymphocytes (10) and also have sustained responses to immune checkpoint inhibitors (ICIs), such as the anti-PD-1 antibodies (11, 12). However, a substantial fraction (~60%) of MSI CRCs do not respond or ultimately develop resistance to ICIs. Therefore, there is an unmet need for novel therapies against MSI CRCs.
Synthetic lethality, referring to loss of cell viability due to a combination of two separate nonlethal mutations, has emerged as a powerful approach for developing new anticancer therapies (13). Recent studies identified Werner (WRN), a RecQ helicase functioning in replication fork remodeling and protection (14), as a synthetic lethal target in MMR-deficient cancer cells (15–17). WRN was initially identified as the gene mutated in Werner syndrome, a genetic disease characterized by premature aging (18). It promotes unwinding of non–B-DNA structures including those that can form in microsatellite repeats (19). WRN depletion induces massive DNA damage in MSI cancer cells (20), resulting in cell death with features of apoptosis (15–17). However, how WRN loss triggers death of MSI cancer cells remains unclear.
The tumor suppressor p53 is a critical regulator of stress-induced apoptosis in mammalian cells. Upon DNA damage, p53 is stabilized to inhibit cell growth by activating p21 and other cell cycle regulators, or induce apoptosis through proapoptotic targets, such as PUMA, Noxa, and Bax (21). PUMA is a BH3-only Bcl-2 family member that functions as a critical regulator of apoptosis in CRC cells (22). Upon its induction by p53 or other stress, PUMA potently induces apoptosis in CRC cells by inhibiting antiapoptotic Bcl-2 proteins to activate Bax and/or Bak, resulting in mitochondrial dysfunction, cytosolic release of cytochrome c, and activation of caspases to execute cell death (22). p53 mutations are highly prevalent (>50%) in MSS CRCs but are infrequent (<20%) in MSI CRCs (4, 23).
In this study, we found that WRN loss selectively induced p53/PUMA-mediated apoptosis in MSI CRC cells. The in vitro and in vivo effects of WRN depletion or inhibition are dependent on p53/PUMA-mediated apoptosis in MSI CRCs, providing mechanistic insight on the vulnerability of MSI cancer cells to WRN loss.
Results
WRN Depletion Induces Mitochondrion-Mediated Apoptosis in MSI but Not MSS CRC Cells.
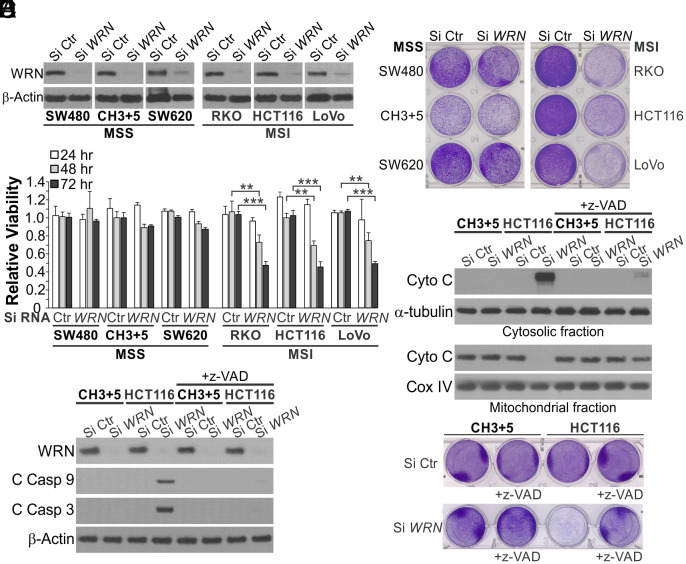
To determine how WRN loss affects CRC cells with different MMR status, we used small-interfering RNA (siRNA) to knock down (KD) WRN in 3 MSI CRC cell lines, including HCT116, RKO, and LoVo, and 2 MSS CRC cell lines, including SW480 and SW620 (Fig. 1A). Isogenic HCT116 cells corrected for MMR deficiency by transferring chromosomes 3 and 5 carrying wildtype (WT) MLH1 and MSH3, respectively (HCT116 CH3+5) (24) were used to further control the effects from genetic background. WRN depletion markedly reduced the viability of MSI cells as shown by MTS analysis of cell viability (Fig. 1B), crystal violet staining of viable cells (Fig. 1C), and long-term cell survival assayed by colony formation (SI Appendix, Fig. S1A). These effects were significantly blocked in MSS CRC cells (Fig. 1 B and C and SI Appendix, Fig. S1A). Markers of mitochondrion-mediated apoptosis were detected only in MSI CRC cells with WRN KD, including Annexin V-positive staining (SI Appendix, Fig. S1 B and C), cytosolic release of mitochondrial cytochrome c (Fig. 1D), and caspases 3 and 9 cleavage (Fig. 1E). Pretreating cells with the pancaspase inhibitor z-VAD-fmk (z-VAD) suppressed cell viability loss and apoptosis induced by Si WRN in HCT 116 cells (Fig. 1 D–F and SI Appendix, Fig. S1D). These results indicate that WRN loss selectively kills MSI CRC cells by inducing mitochondrion-mediated apoptosis and caspase activation.
Fig. 1.
Knockdown of WRN induces apoptosis in MSI but not MSS CRC cells. (A) Western blotting of WRN in indicated MSI and MSS CRC cell lines transfected with control scrambled (Si Ctr) or WRN (Si WRN) siRNA for 72 h. (B) MTS analysis of viability of indicated MSI and MSS cell lines transfected as in (A) for 24, 48 and 72 h. Results were expressed as mean ± SD of three independent experiments. **P < 0.01; ***P < 0.001. (C) Crystal violet staining of cell lines transfected as in (A) for 72 h. (D–F) Parental and CH3+5 HCT116 cells with or without pretreatment with the pancaspase inhibitor z-VAD-fmk (z-VAD; 10 μM) for 4 h were transfected with Si Ctr or Si WRN for 72 h. (D) Western blotting of cytochrome c (Cyto C) in cytosolic and mitochondrial fractions isolated from transfected cells. Cytosolic α-tubulin and mitochondrial cyclooxygenase IV (Cox IV) were used as controls for loading and fractionation. (E) Western blotting of WRN and cleaved caspases (C Casp) 3 and 9. (F) Analysis of cell viability by crystal violet staining.
WRN Depletion Activates the p53 Apoptosis Pathway in MSI CRC Cells.
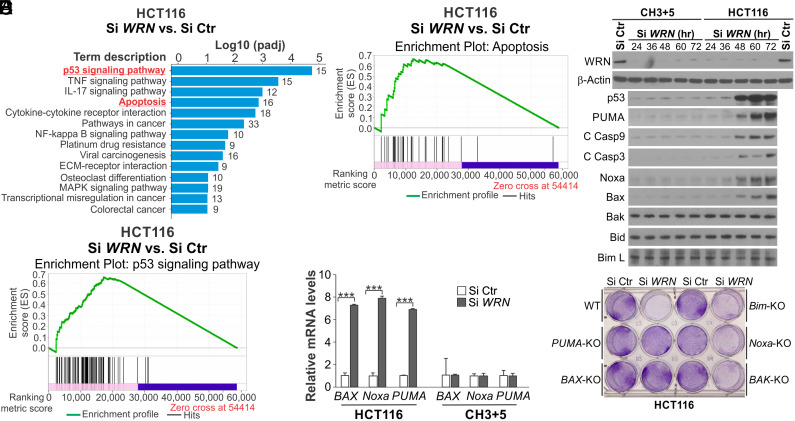
To determine the mechanism by which WRN depletion kills MSI CRC cells, we performed mRNA sequencing (RNA-Seq) on parental and control CH3+5 HCT116 cells transfected with Si WRN. We identified over 3,000 differentially expressed genes, including 1,085 up-regulated and 1,997 down-regulated genes, in HCT116 cells, compared with only 990 altered genes in CH3+5 cells (SI Appendix, Fig. S2A). Gene Set Enrichment Analysis (GSEA) revealed that p53 and apoptosis pathways are among the most significantly activated pathways by WRN KD (Fig. 2 A–C). The identified changes included well-known p53 downstream targets, such as p21 (CDKN1A), PUMA (BBC3), Noxa (PMAIP1), Gadd45 A and B, 14-3-3σ, and Thrombospondin 1 (SI Appendix, Fig. S2B), as well as other apoptosis regulators, such as BAK1, DR4, FAS, cIAP2, and FLIP (SI Appendix, Fig. S2C). Interestingly, serpine-1 (PAI-1), a p53 downstream target involved in induction of replicative senescence (25), was identified as the most up-regulated (>14 fold) p53 target gene (SI Appendix, Fig. S2B).
Fig. 2.
Knockdown of WRN induces p53 and its apoptosis targets in HCT116 cells. (A–C) HCT116 cells transfected with Si Ctr or Si WRN for 36 h were analyzed by RNA-Seq. Gene expression was determined by calculating Fragments Per Kilobase of transcript per Million mapped reads, and differentially expressed genes were identified. (A) GSEA of most significantly up-regulated pathways with indicated Log P values. (B) GSEA of p53 pathway genes. (C) GSEA of apoptosis pathway genes. (D) Western blotting of indicated proteins in parental and CH3+5 HCT116 cells at indicated time points after Si WRN transfection. (E) Real-time RT-PCR analysis of BAX, Noxa, and PUMA mRNA expression in parental and CH3+5 HCT116 cells transfected as in (A) for 36 h. Results were expressed as mean ± SD of three independent experiments. ***P < 0.001. (F) Crystal violet staining of viable cells in WT, PUMA-KO, BAX-KO, Noxa-KO, BAK-KO, and Bim-KO HCT116 cells transfected with Si Ctr or Si WRN for 72 h.
These data prompted us to further investigate p53-mediated apoptotic signaling. We found p53 was strongly induced upon WRN KD for 48 h in HCT116 cells (Fig. 2D). Consistent with the RNA-Seq data, WRN KD markedly induced the mRNA and protein expression of p53 downstream apoptosis targets, including PUMA, Noxa, and Bax, in HCT116 cells, coinciding with p53 induction and caspase activation (Fig. 2 D and E). These effects were largely absent in CH3+5 cells (Fig. 2 D and E). Robust induction of p53 and its downstream apoptosis regulators was also detected in other MSI CRC cells with WRN depletion but not in MSS CRC cells (SI Appendix, Fig. S3 A and B). These findings suggest that WRN loss triggers DNA damage-induced and p53-mediated apoptosis selectively in MSI CRC cells.
p53-Mediated PUMA Induction is Critical for Killing of MSI CRC Cells by WRN Depletion.
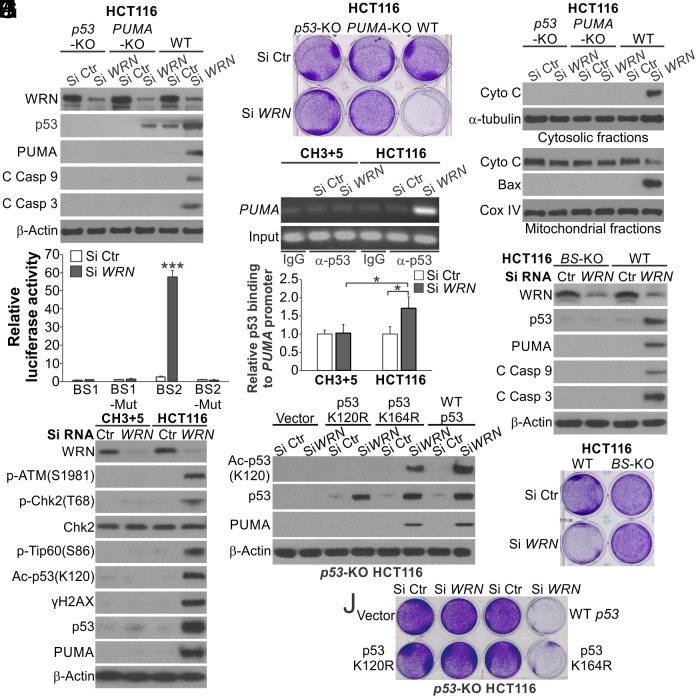
We then investigated the functional role of the p53 downstream apoptosis regulators. Knockout (KO) of PUMA or BAX, but not Bim, Noxa or BAK, rescued the viability of HCT116 cells transfected with Si WRN (Fig. 2F and SI Appendix, Fig. S3C), suggesting a critical role of the p53/PUMA/Bax axis in apoptosis induction. Indeed, p53 or PUMA KO completely suppressed cell viability loss, cytochrome c release, Bax mitochondrial translocation, and caspases 3 and 9 cleavage in HCT116 cells with WRN KD (Fig. 3 A–C and SI Appendix, Fig. S4A). Similar effects of p53 and PUMA KO were also observed in MSI LoVo cells with WRN KD (SI Appendix, Fig. S4 B–E). These findings are consistent with a critical role of the p53/PUMA/Bax axis in apoptosis induction of colon cancer cells and intestinal epithelial cells (26, 27).
Fig. 3.
p53 and PUMA are required for apoptosis induced by WRN KD in MSI CRC cells. (A) Western blotting of indicated proteins in WT, p53-KO and PUMA-KO HCT116 cells transfected with Si Ctr or Si WRN for 72 h. (B) Crystal violet staining of viable cells in cells transfected as in (A) for 72 h. (C) Western blotting of cytochrome c in cytosolic and mitochondrial fractions isolated from cells transfected with siRNA as in (A) for 72 h. (D) Luciferase reporters containing four copies of either the p53 binding site BS1 or BS2 or mutant versions of these sites were transfected into HCT116 cells for 36 h. Reporter activities were normalized to the luciferase/β-galactosidase ratio of the mutant reporter. (E) Chromatin immunoprecipitation (ChIP) analysis of the binding of p53 to the PUMA promoter in parental and CH3+5 HCT116 cells transfected with indicated siRNA for 36 h. ChIP was done using a p53 antibody for pull-down and anti-IgG as a negative control, followed by PCR analysis of the PUMA promoter region covering the p53 binding site. Upper, representative gel pictures of PCR products; Lower, quantification of p53 binding to the PUMA promoter by NIH ImageJ program. (F) Western blotting of indicated proteins in WT and PUMA p53 binding site knockout (BS-KO) HCT116 cells transfected with indicated siRNA for 72 h. (G) Crystal violet staining of WT and BS-KO HCT116 cells transfected as in (F) for 72 h. (H) Western blotting of indicated proteins in parental and CH3+5 HCT116 cells transfected with Si Ctr or Si WRN for 72 h. (I and J) p53-KO HCT116 cells were reconstituted with WT p53, K164R, or K120R mutant p53 by transfection. Cells transfected with Si Ctr or Si WRN for 72 h were analyzed by (I) Western blotting of indicated proteins and (J) crystal violet staining of viable cells. Bar graphs in (D and E) were expressed as mean ± SD of three independent experiments. *P < 0.05; ***P < 0.001.
p53 directly binds to two p53 binding sites, BS1 and BS2, in the PUMA promoter to activate its transcription (SI Appendix, Fig. S5A) (28). WRN KD strongly activated PUMA promoter reporters (SI Appendix, Fig. S5B) and the BS2 site in a sequence-specific manner (Fig. 3D), consistent with the finding that BS2 is the major and evolutionarily conserved p53 binding site (28). Chromatin immunoprecipitation (ChIP) revealed markedly increased binding of p53 to the PUMA promoter in the parental, but not CH3+5, HCT116 cells upon WRN depletion (Fig. 3E). Furthermore, KO of the p53 binding sites in the PUMA promoter in HCT116 cells (BS-KO) (29) blocked cell viability loss and caspase activation (Fig. 3 F and G). These results indicate that upon WRN depletion, p53 directly activates PUMA transcription to induce apoptosis in MSI CRC cells.
WRN KD activated DNA damage signaling upstream of p53 in the parental, but not CH3+5 HCT116 cells, such as phosphorylation of ATM (Ser1981) and Chk2 (Thr68) (Fig. 3H and SI Appendix, Fig. S5C). The ATM kinase inhibitor Ku55933 treatment suppressed the induction of p53 and its upstream and downstream signaling induced by WRN KD (SI Appendix, Fig. S5 D and E), indicating ATM/Chk2/p53-mediated DNA damage response signaling. Upon DNA damage, p53 induces either cell cycle arrest or apoptosis depending on cellular context, and cell fate determination is mediated by specific p53 posttranslational modifications, such as lysine acetylation (30). We detected increased p53 K120 acetylation, which was shown to be critical for p53 to induce PUMA and apoptosis (31), as well as activating phosphorylation of Tip60 (S86), the acetyltransferase that mediates p53 K120 acetylation (32), in WRN-KD HCT116 cells (Fig. 3H). To determine if p53 K120 acetylation is critical for apoptosis induced by WRN KD, p53-KO HCT116 cells were reconstituted with WT or acetylation-deficient K120R or control K164R mutant at a similar level (SI Appendix, Fig. S5F). WT p53 and K164R mutant, but not K120R mutant, could restore PUMA induction and cell viability loss in p53-KO HCT116 cells with WRN KD (Fig. 3 I and J and SI Appendix, Fig. S5G). Together, these results indicate that WRN loss triggers DNA damage-induced and p53/PUMA-mediated apoptosis to kill MSI CRC cells, which requires p53 K120 acetylation.
p53-Mutant MSI CRC Cells Are Resistant to WRN Depletion Due to Lack of PUMA Induction.
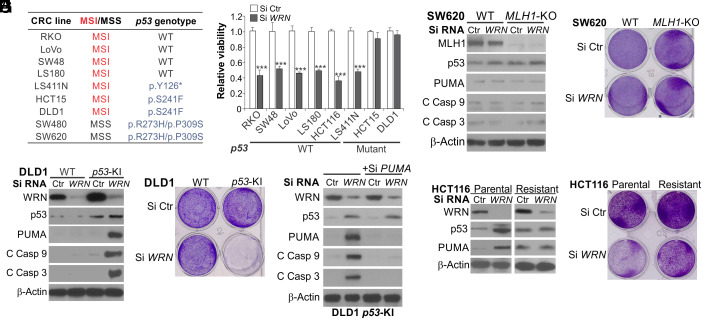
p53 mutations are relatively infrequent in MSI tumors compared with MSS tumors (23). Widely used MSI CRC cell lines mostly express WT p53; but some express mutant p53, including DLD1 and HCT15, which share the same genetic origin and p53 mutation (p.S241F) (Fig. 4A) but have different karyotypes (33), and KM12 and LS411N (15, 16). A previous study showed that among 18 MSI cell lines analyzed, only DLD1 and HCT15 were not affected by WRN KD (16). Indeed, WRN KD failed to induce viability loss and p53/PUMA expression in DLD1 cells (Fig. 4 B and C). In contrast, isogenic DLD1 cells with knock in (KI) of WT p53 by homologous recombination (34) showed completely restored p53 and PUMA induction, cell viability loss, and caspase 9 and 3 activation upon WRN KD (Fig. 4 C–E and SI Appendix, Fig. S6A). PUMA KD by siRNA suppressed caspase activation induced by WRN KD in p53-KI DLD1 cells (Fig. 4E). Similarly, transfecting WT p53 into HCT15 cells restored the induction of PUMA and apoptosis by WRN KD (SI Appendix, Fig. S6 B–D). Lack of PUMA and apoptosis induction by WRN KD was also observed in p53-mutant SW620 cells with CRISPR KO of MLH1 (Fig. 4 F and G and SI Appendix, Figs. S1B and S6E). Furthermore, we generated MSI cell lines with acquired resistance to WRN KD by continuously exposing HCT116 and RKO cells to Si WRN for 2 wk and then expanding the remaining viable cells. The resistant HCT116 and RKO cells were found to express a high level of endogenous p53 but lose the ability to induce p53 and PUMA by WRN KD (Fig. 4 H and I and SI Appendix, Fig. S6 F–I). These results support that p53 mutations and oncogenic alterations inhibiting p53 signaling in some MSI cancer cells can cause resistance to WRN depletion by blocking PUMA and apoptosis induction. Consistent with previous studies (15, 16), p53-mutant MSI LS411N cells were sensitive to WRN KD (Fig. 4 A and B), which involved PUMA induction and PUMA-mediated caspase activation (SI Appendix, Fig. S6J), suggesting p53-independent but PUMA-dependent cell death (22).
Fig. 4.
WT p53 is required for WRN-KD-induced apoptosis in some MSI CRC cells. (A) MSI/MSS status and p53 genotypes of the listed CRC cell lines. (B) MTS analysis of viability of indicated MSI CRC cell lines transfected with Si Ctr or Si WRN for 72 h. Results were expressed as mean ± SD of three independent experiments. ***P < 0.001. (C and D) Parental and WT-p53-knockin (p53-KI) DLD1 cells were transfected with Si Ctr or Si WRN for 72 h and analyzed by (C) Western blotting of indicated proteins and (D) crystal violet staining of viable cells. (E) Western blotting of indicated proteins in p53-KI DLD1 cells transfected with Si Ctr or Si WRN for 72 h, with or without cotransfection with PUMA siRNA. (F and G) WT and MLH1-KO SW620 cells were transfected with Si Ctr or Si WRN for 72 h and analyzed by (F) Western blotting of indicated proteins and (G) crystal violet staining of viable cells. (H and I) HCT116 cells resistant to WRN knockdown were generated by continuously exposing parental HCT116 cells to Si WRN for 2 wk and expanding the remaining viable cells. Parental and resistant HCT116 cells were transfected with Si Ctr or Si WRN for 72 h and analyzed by (H) Western blotting of indicated proteins and (I) crystal violet staining of viable cells.
PUMA is Required for the In Vivo Therapeutic Effects of WRN KD on MSI CRC Tumors.
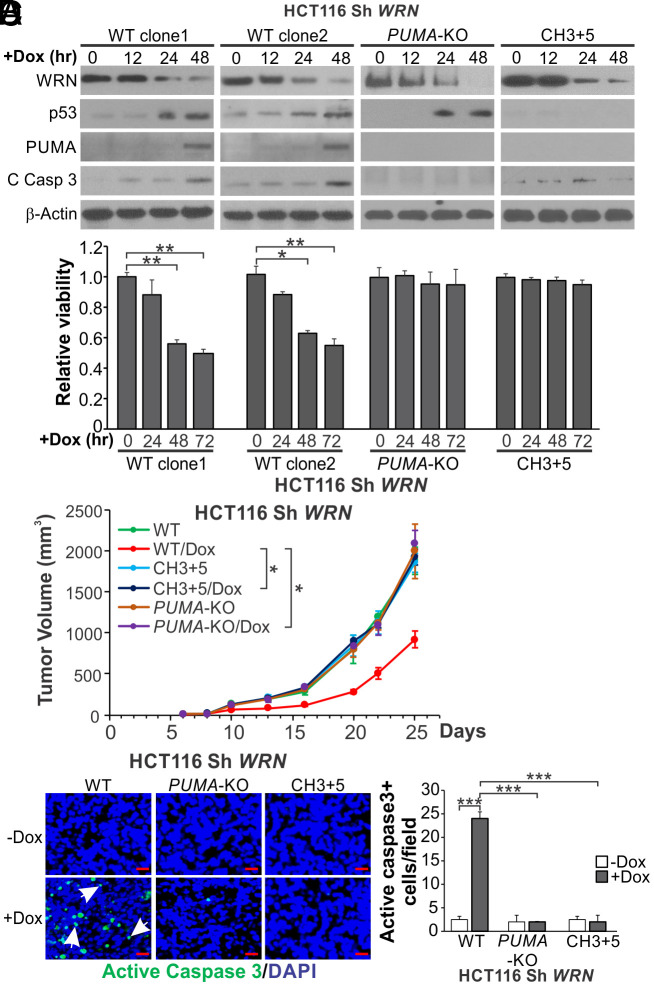
To study the effects of WRN depletion in vivo, we generated stable WT, PUMA-KO, and CH3+5 HCT116 cell lines with doxycycline (Dox)-inducible expression of WRN small-hairpin RNA (Sh WRN). As expected, depleting WRN by Dox treatment for over 48 h markedly induced p53 and PUMA expression, caspase activation, and viability loss in two independent WT HCT116 clones, but not in PUMA-KO and CH3+5 HCT116 cells (Fig. 5 A and B and SI Appendix, Fig. S7A). We then implanted these cell lines into nude mice to establish xenograft tumors and induce WRN KD using a Dox-containing diet. Induction of WRN KD significantly impaired the growth of WT but not PUMA-KO and CH3+5, HCT116 xenograft tumors (Fig. 5C). Tumor growth increased at later time points likely due to recovery of WRN expression. WRN KD did not affect the body weight in different groups (SI Appendix, Fig. S7B). Immunostaining of tumor tissues showed robust induction of γH2AX and p53 in WT and PUMA-KO but not in CH3+5, HCT116 tumors (SI Appendix, Fig. S7 C and D). Strong induction of apoptosis was further confirmed by TUNEL and active caspase 3 staining in WT but not in PUMA-KO and CH3+5, HCT116 tumors (Fig. 5D and SI Appendix, Fig. S7E). These results demonstrate that WRN KD suppresses the growth of MSI HCT116 xenograft tumors through p53/PUMA-mediated apoptosis.
Fig. 5.
Inducible knockdown of WRN suppresses xenograft growth in a PUMA-dependent fashion. Parental (WT), CH3+5, and PUMA-KO HCT116 cells with Dox inducible expression of WRN short hairpin RNA (Sh WRN) were established as described in Materials and Methods. (A) Western blotting of indicated proteins in two independent clones of WT, CH3+5, and PUMA-KO Sh WRN HCT116 cells with induction of Sh WRN by Dox (0.2 μg/mL) treatment for indicated time. (B) MTS analysis of viability of indicated cells with induction of Sh WRN as in (A) at indicated time points. Results were expressed as mean ± SD of three independent experiments. (C and D) Nude mice were implanted with 2 × 106 of WT (clone 1), CH3+5, or PUMA-KO Sh WRN HCT116 cells to establish xenograft tumors. Tumor bearing mice were randomized and treated with 18% Protein Rodent Diet +/− Dox hyclate (625 mg/kg). (C) Tumor volume at indicated time points after treatment was measured and plotted with P values for indicated comparisons (N = 6 in each group). (D) Tumor sections from mice treated as in (C) for 13 d were analyzed by immunostaining for active caspase 3 expression. Left, representative staining pictures with arrows indicating examples of positive signals (scale bars: 25 μm); Right, quantification of positive signals showing means ± SEM of each field in 3 fields per mouse (N = 3 in each group). *P < 0.05; **P < 0.01; ***P < 0.001.
WRN Inhibitors Suppress MSI CRC Growth In Vitro and In Vivo via p53/PUMA-Mediated Apoptosis.
To explore WRN targeting as a therapeutic approach for MSI CRCs, we first tested a small-molecule WRN helicase inhibitor NSC617145 described previously (35). NSC617145 at 6 μM, but not higher concentrations, could selectively inhibit parental HCT116 relative to CH3+5 cells (SI Appendix, Fig. S8A), which was accompanied by p53/PUMA induction and p53/PUMA-dependent cell viability loss and apoptosis induction (SI Appendix, Fig. S8 B–E). However, NSC617145 did not show in vivo activity.
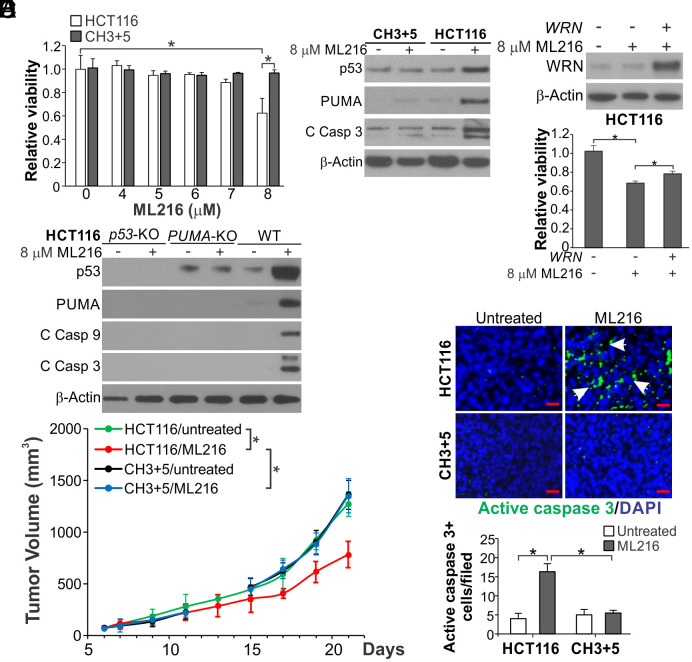
ML216 was initially identified as an inhibitor of Bloom (BLM) helicase, and could inhibit full-length WRN and a truncated derivative that lacks the N-terminal exonuclease domain at IC50s of 5 and 12.6 μM, respectively (36). Although human fibroblasts proficient or deficient in WRN were equally sensitive to ML216 (36), this inhibitor had not been tested in MSI cancer cell lines. Treatment of ML216 at 8 μM for 72 h, but not higher concentrations, resulted in a higher loss of cell viability, PUMA induction, and apoptosis in parental compared with CH3+5 HCT116 cells (Fig. 6 A and B and SI Appendix, Fig. S9A). Re-expressing WRN modestly rescued the viability loss of parental cells (Fig. 6C), while p53 KO or PUMA KO completely blocked cell death and caspase activation (Fig. 6D and SI Appendix, Fig. S9B), supporting WRN targeting and p53 pathway activation by ML216 in MSI CRC cells.
Fig. 6.
ML216 inhibits MSI CRC xenograft growth via apoptosis induction. (A) MTS analysis of parental and CH3+5 HCT116 cells treated with ML216 at indicated concentrations for 72 h. (B) Western blotting of indicated proteins in parental and CH3+5 HCT116 cells treated with 8 μM ML216 for 72 h. (C) HCT116 cells in 12-well plate transfected with 0.2 μg/well control empty or WRN expression vector (pLX209-neo-active WRN) were treated with 8 μM ML216. Upper, Western blotting of WRN after transfection for 24 h; Lower, MTS analysis of viability of cells after ML216 treatment for 72 h. (D) Western blotting of indicated proteins in WT, p53-KO, and PUMA-KO HCT116 cells treated with 8 μM ML216 for 72 h. (E and F) Nude mice with established parental and CH3+5 HCT116 xenograft tumors were randomized and treated with control vehicle or ML216 (i.p; 1.5 mg/kg daily in first 7 d and every other day after day 7). (E) Tumor volume at indicated time points was calculated and plotted with P values for indicated comparisons (N = 6 in each group). (F) Tumor sections from mice treated for 13 d were analyzed for apoptosis by active caspase 3 staining. Upper, representative staining pictures (scale bars: 25 μm); Lower, quantification of positive signals showing means ± SEM of each field in 3 fields per mouse (N = 3 in each group). Bar in (A and C) were expressed as mean ± SD of three independent experiments. *P < 0.05.
We then determined the antitumor effects of ML216 in vivo by treating nude mice bearing xenograft tumors established from the parental and CH3+5 HCT116 cells with ML216 (i.p; 1.5 mg/kg daily in first 7 d and every other day after day 7). ML216 treatment more effectively suppressed the growth of HCT116 tumors compared with CH3+5 tumors or untreated tumors (Fig. 6E), and markedly increased TUNEL and active caspase 3 staining only in HCT116 tumors (Fig. 6F and SI Appendix, Fig. S9C). These results suggest that WRN inhibition by ML216 has in vitro and/or in vivo antitumor effects against MSI tumor cells by inducing p53/PUMA-mediated apoptosis.
ML216 Inhibits MSI Patient-Derived Xenografts (PDX) Tumor Growth and Induces p53/PUMA, as Well as Apoptosis.
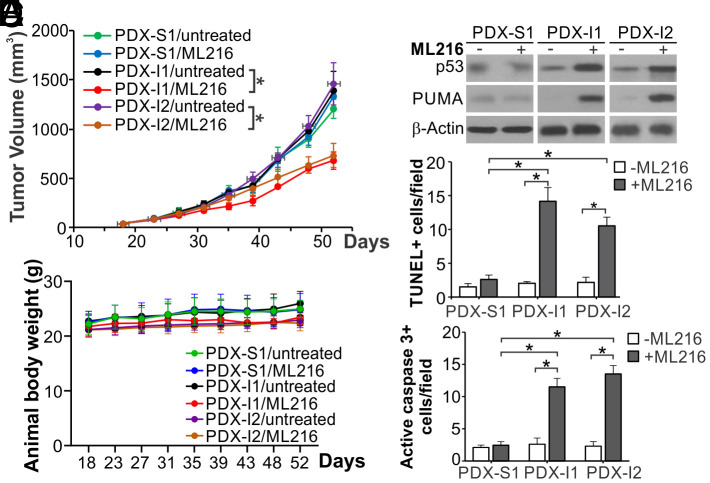
To explore the translational potential of WRN targeting for MSI CRCs, we analyzed the antitumor effects of ML216 on PDX models, which better recapitulate histology, heterogeneity, and molecular alterations of original patient tumors than cell line xenografts (37). Three PDX models from National Cancer Institute (NCI) were expanded and analyzed, including the MSS PDX-S1 model and two MSI PDX-I1 and PDX-I2, which both harbor MSH2 and MSH6 mutations and/or deletion (SI Appendix, Table S1). The MSS/MSI status of these PDX models was verified by analyzing the Bethesda panel of five microsatellite markers (SI Appendix, Fig. S10A). ML216 treatment significantly suppressed the growth of PDX-I1 and PDX-I2 tumors, but not PDX-S1 tumors, without affecting body weight (Fig. 7 A and B). Tissue analysis revealed that ML216 treatment induced p53 and PUMA (Fig. 7C), along with increased TUNEL and active caspase 3 staining (Fig. 7 D and E and SI Appendix, Fig. S10 B and C) in PDX-I1 and PDX-I2, but not in PDX-S1 tumors. These results suggest that WRN inhibitors have potential efficacy and could be further developed as therapeutic agents against p53-WT MSI tumors.
Fig. 7.
ML216 suppresses MSI PDX tumor growth with apoptosis induction. NSG mice were subcutaneously implanted with three different PDX tumor models, including an MSS model (PDX-S1) and two MSI models (PDX-I1 and PDX-I2), and were treated with ML216 (i.p; 1.5 mg/kg daily in first 7 d and every other day after day 7). (A) Tumor volume at indicated time points after treatment was calculated and plotted with P values for indicated comparisons (N = 6 in each group). (B) Animal body weight at indicated time points. (C) Western blotting of p53 and PUMA in representative PDX tumors from mice treated as in (A). (D and E) Apoptosis was analyzed by (D) TUNEL and (E) active caspase 3 staining of tumor sections from mice treated as in (A) for 35 d. Quantification of positive signals is shown. Results were expressed as mean ± SEM of each field in 3 fields per mouse (N = 3 in each group). *P < 0.05.
Discussion
Several recent studies identified WRN as a synthetic lethal target of MSI cancers (15–17), while little is known about how the cell death is triggered. Our results show that WRN depletion triggers a robust DNA damage response, progressing from ATM activation, p53 accumulation, and induction of p53 targets, leading to the onset of mitochondrion-mediated apoptosis in p53-WT MSI CRC cells. Apoptosis induction is selective and p53-dependent in p53-WT and some p53-mutant MSI CRC cells, compared with MSS CRC cells, and blocked by p53 KO or inactivating mutations. Among the p53 downstream targets, PUMA has the most prominent role in apoptosis induction. Upon WRN depletion, p53 directly binds to the PUMA promoter to strongly activate its transcription. Deletion of PUMA or the p53 binding sites in the PUMA promoter phenocopies p53 KO in blocking apoptosis induction. These results demonstrate that the p53/PUMA axis is largely responsible for WRN-KD-induced apoptosis of MSI CRC cells. However, some p53-mutant MSI CRC cells, such as KM12 and LS411N cells, remain sensitive to WRN depletion via the induction of p53-independent cell death (15, 16).
The robust induction of p53-mediated cell death could be explained by the level of DNA damage caused by a concomitant deficiency in both MMR and WRN. MMR not only corrects DNA replication errors, but also modulates DNA damage response (38). For example, MMR has been implicated in the repair and cytotoxicity of some DNA lesions generated by DNA damaging agents (39), and defective MMR could promote tolerance to cytotoxic DNA lesions (38). The helicase activity of WRN promotes unwinding of DNA that is frequently involved in DNA repair including MMR. Inhibition of WRN helicase could lead to replication stress, stalled replication forks, and genome fragility (40). A close functional interaction of WRN and MMR is revealed by a recent study showing that expanded TA-dinucleotide repeats in MSI cells form DNA secondary structures that stall replication forks and need to be unwound by WRN helicase (20). In the absence of WRN, these repeats can be cleaved by the MUS81 nuclease, resulting in extensive chromosome shattering. Therefore, WRN loss in MSI cells results in a high level of DNA damage that triggers cells to undergo apoptosis.
Induction of p53 by DNA damage leads to either cell cycle arrest or apoptosis. The predominant induction of apoptosis in MSI cells with WRN depletion cannot solely be explained by p53 induction or cellular context. It was shown that cell cycle arrest is the predominant response of HCT116 cells with p53 overexpression (41), or treatment with DNA damaging drugs (26). Our results suggest that p53 K120 acetylation functions as a critical switch for MSI cells to choose apoptosis as the dominant and ultimate fate following WRN depletion (Fig. 3 I and J). The connection between the excessive DNA damage induced by WRN depletion and p53 K120 acetylation has remained unclear. A previous study showed that glycogen synthase kinase-3 (GSK-3) promotes the p53 acetyltransferase Tip60 phosphorylation at S86, which mediates PUMA and apoptosis induction (32). We also observed Tip60 S86 phosphorylation in HCT116 with WRN KD (Fig. 3H). The role of GSK-3 and Tip60 in mediating apoptosis induced by WRN depletion will be further addressed in our future studies.
The key role of p53 in killing WRN-deficient MSI cells is further indicated by the inability of p53-mutant DLD1 and HCT15 cells to undergo apoptosis, and by the lack of p53 and PUMA induction in HCT116 and RKO cells with acquired resistance to WRN depletion (Fig. 4 and SI Appendix, Fig. S6). A functional interaction of WRN and p53 is indicated by a study showing that compounded deletion of WRN and p53 in mice led to an increased mortality rate (42). p53 mutations are most prevalent (>50%) in MSS CRCs, but still exist in a substantial fraction (<20%) of MSI-high CRCs (23). Our results indicate that the status of p53 mutations can be used as a biomarker for targeting MSI CRCs with WRN inhibitors. The selective pressure imposed by WRN deficiency in MSI cancer cells could enrich for resistant cells with p53 mutations, or compromised p53 signaling due to mechanisms such as MDM2 overexpression, p14ARF inactivation, or enhanced p53 nuclear export (43). A p53-based therapeutic strategy could be explored to overcome acquired resistance to WRN inhibition in MSI cells.
MSI CRCs often have durable responses to ICIs, such as the anti-PD-1 and anti-CTLA-4 antibodies (11, 12). However, a majority (~60%) of MSI CRCs do not respond or ultimately develop resistance to ICIs. Use of ICIs is also limited by immune-related side effects. WRN might be another promising target in MSI cancers, including those resistant to chemotherapy, targeted therapies, and immunotherapy, which have been shown to retain WRN dependency for survival (44). From analysis of Sh WRN inducible cell lines (Fig. 5) and small-molecule WRN inhibitors on xenografts and PDX models (Figs. 6 and 7), our results further support WRN as a promising therapeutic target on MSI CRCs in vivo. WRN inhibition by NSC617145 induces p53/PUMA-dependent apoptosis (SI Appendix, Fig. S8). The BLM/WRN inhibitor ML216 has both in vitro and in vivo therapeutic activities against MSI CRC cells and PDX models (Figs. 6 and 7). The activity of ML216 on MSI CRC cells is at least in part mediated by WRN (Fig. 6C) but may involve inhibition of other RecQ helicases such as BLM. WRN inhibitors with improved in vivo efficacy and selectivity are needed to further advance WRN inhibitors to clinical development. WRN inhibitors may also have potentiating effects on ICIs, including the anti-PD-1 antibody, due to the immunogenic effects of apoptosis induced by DNA damage (45).
In conclusion, our results demonstrate a critical role of p53/PUMA-mediated apoptosis in mediating the synthetic lethality of WRN and MMR deficiency. Inhibiting WRN elicits in vitro and in vivo antitumor effects on MSI CRCs via p53/PUMA-mediated apoptosis, supporting WRN as a promising therapeutic target in p53-WT MSI cancers.
Materials and Methods
Cell Culture and Treatment.
Parental human CRC cell lines, including HCT116, RKO, DLD1, LoVo, SW48, LS411N, LS180, HCT15, SW480, and SW620, were purchased from the American Type Culture Collection. HCT116 cells with transfer of chromosomes 3 and 5 (CH3+5) for correcting MMR deficiency caused by endogenous MLH1 and MSH3 mutations were previously described (24). Isogenic HCT116 derivatives, including p53-KO (46), BAX-KO (47), and PUMA-KO (26), were obtained from Dr. Bert Vogelstein (Johns Hopkins). HCT116 with KO of the p53 binding sites in the PUMA promoter (BS-KO), Noxa-KO and Bim-KO were previously described (29, 48).
Cell lines, chemicals, cell culture media, and supplements are listed in SI Appendix, Table S2. All cell lines were maintained at 37 °C and 5% CO2 atmosphere, and cultured in McCoy’s 5A modified media supplemented with 10% defined FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin. Cells plated at 20 to 30% density were treated with inhibitors including NSC617145, ML216 (Apexbio), Ku55933 (Sigma), and z-VAD-fmk (z-VAD; Bachem), which were solubilized in DMSO and diluted to appropriate concentrations with the cell culture medium before use.
Transfection and Small Interfering RNA (siRNA) Knockdown.
Expression constructs and siRNAs are described in SI Appendix, Table S2. Transfection was performed using Lipofectamine 2,000 (Invitrogen) for expression constructs, or Lipofectamine RNAiMAX (Invitrogen) for siRNAs, according to the manufacturer’s instructions.
Western Blotting.
Western blotting was performed as previously described (48) using antibodies listed in SI Appendix, Table S2.
Analysis of Cell Viability and Apoptosis.
Cells seeded in 96-well plates at a density of 4,000 cells/well were transfected with control scrambled or WRN siRNA for 24, 48, and 72 h. MTS assay was performed using the MTS assay kit (Promega) according to the manufacturer’s instructions. Chemiluminescence was measured using a Wallac Victor 1420 Multilabel Counter (Perkin Elmer). Each assay was conducted in triplicate and repeated 3 times. Cells plated in 12-well plates at 20 to 30% density and transfected with control scrambled or WRN siRNA were stained for viable cells by crystal violet. Long-term cell survival was analyzed by colony formation assays by plating treated cells in 6-well plates at appropriate dilutions, followed by crystal violet staining 14 d after plating as described (49).
Apoptosis was analyzed by annexin V (Alexa Fluor 488)/propidium iodide (PI) (Invitrogen) staining followed by flow cytometry as described (50). Cytochrome c release was analyzed by western blotting of cytochrome c in cytoplasmic and mitochondrial fractions prepared from treated cells using the Mitochondrial Fractionation Kit (Active Motif) according to the manufacturer’s instructions.
Analysis of Cell Lines Resistant to WRN Knockdown.
HCT116 and RKO cells were continuously exposed to 0.1 μM WRN siRNA (Si WRN) for 2 wk. Remaining viable cells were expanded and analyzed.
Animal Experiments.
All animal experiments were approved by the University of Pittsburgh Institutional Animal Care and Use Committee. Mice were housed in micro isolator cages in a sterile environment and allowed access to water and chow ad libitum.
Cell line xenograft tumors.
Xenograft tumors with Dox-inducible WRN knockdown were established by subcutaneously injecting 2 × 106 WT, PUMA-KO, CH3+5 Sh WRN HCT116 cells into right flank of female 5 to 6-wk-old Nu/Nu mice (Charles River). When tumor size reached ~50 mm3, mice were randomized and treated with Teklad Global 18% Protein Rodent Diet +/− Dox hyclate (625 mg/kg) throughout the study.
To analyze the in vivo activity of ML216, xenograft tumors were established by subcutaneously injecting parental and CH3+5 HCT116 cells into right flank of Nu/Nu mice. When tumor size reached ~50 mm3, mice were randomized and treated with ML216 (1.5 mg/kg by i.p.) as described in Fig. 6E.
PDX tumors.
PDX tumors were propagated in 5 to 6-wk-old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (Jackson Laboratory). For PDX passage, tumor tissues were cut into 25-mg pieces and implanted subcutaneously into both flanks of NSG mice as described (51, 52). Tumor fragments from NCI Patient-Derived Models Repository passaged and expanded for one generation (P4 for PDX-S1 and P2 for PDX-I1 and PDX-I2) in NSG mice were used for the described experiments. When PDX tumor size reached ~50 mm3, mice were randomized into different groups and treated with ML216 (1.5 mg/kg by i.p.) as described in Fig. 7A.
Analysis of tumor growth and immunostaining of tumor tissues.
Animal body weight was recorded twice weekly during the course of the study for body condition scoring. Tumor volume was measured by calipers and calculated according to the formula 1/2 × length × width2. Ethical end point represents a timepoint when tumors reached 2 cm or more in any dimension. After tumors reached 1.0 cm3 in size, mice were euthanized, and tumors were dissected and fixed in 10% formalin and embedded in paraffin. Immunostaining was performed using a TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling) staining kit (EMD Millipore), and primary antibodies for active caspase 3 (Cell Signaling) on 5-μm paraffin-embedded tumor sections as described (50). Signals were detected using AlexaFluor 488-conjugated secondary antibody (Invitrogen) with nuclear counter staining by 4′ 6-diamidino-2-phenylindole (DAPI).
Statistical Analysis.
Statistical analysis was performed using Prism 9 software (GraphPad). For cell culture and immunostaining experiments, P values were calculated using Student’s t test. Mean ± SD are indicated in the figures. For animal treatment experiments, P values were calculated by an ANOVA with Fisher’s LSD post hoc test for tumor volume analysis. Mean ± SEM are indicated in the figures. Differences were considered significant if P < 0.05.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Kaylee Ermine and other lab members for discussion and critical reading. This work was supported by the U.S. National Institutes of Health grants (R01CA236271, R01CA247231, and R01CA248112 to L.Z.; R01CA215481 and R01CA260900 to J.Y.). This project used the Hillman Cancer Center Animal Facility, Cytometry Facility, and Tissue and Research Pathology Services, which are supported in part by P30CA047904.
Author contributions
S.H., J.T., and L.Z. designed research; S.H., J.T., A.J., D.R., D.L., and X.L. performed research; S.H., J.T., A.G., P.L.O., J.Y., and L.Z. contributed new reagents/analytic tools; S.H., P.L.O., J.Y., and L.Z. analyzed data; and S.H., J.Y., and L.Z. wrote the paper.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Jiricny J., The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 7, 335–346 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Goellner E. M., Putnam C. D., Kolodner R. D., Exonuclease 1-dependent and independent mismatch repair. DNA Repair (Amst) 32, 24–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boland C. R., Goel A., Microsatellite instability in colorectal cancer. Gastroenterology 138, 2073–2087 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogelstein B., et al. , Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., Cancer statistics, 2021. CA Cancer J. Clin. 71, 7–33 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Richard G. F., Kerrest A., Dujon B., Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 72, 686–727 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishel R., et al. , The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75, 1027–1038 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Nicolaides N. C., et al. , Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature 371, 75–80 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Miyaki M., et al. , Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat. Genet. 17, 271–272 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Lizardo D. Y., et al. , Immunotherapy efficacy on mismatch repair-deficient colorectal cancer: From bench to bedside. Biochim. Biophys. Acta Rev. Cancer 1874, 188447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le D. T., et al. , Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overman M. J., et al. , Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J. Clin. Oncol. 36, 773–779 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Huang A., Garraway L. A., Ashworth A., Weber B., Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug. Discov. 19, 23–38 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Dhar S., Datta A., Brosh R. M. Jr., DNA helicases and their roles in cancer. DNA Repair (Amst) 96, 102994 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Chan E. M., et al. , WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature 568, 551–556 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieb S., et al. , Werner syndrome helicase is a selective vulnerability of microsatellite instability-high tumor cells. Elife 8, e43333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kategaya L., Perumal S. K., Hager J. H., Belmont L. D., Werner syndrome helicase is required for the survival of cancer cells with microsatellite instability. iScience 13, 488–497 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu C. E., et al. , Positional cloning of the Werner’s syndrome gene. Science 272, 258–262 (1996). [DOI] [PubMed] [Google Scholar]
- 19.van Wietmarschen N., Nathan W. J., Nussenzweig A., The WRN helicase: Resolving a new target in microsatellite unstable cancers. Curr. Opin. Genet. Dev. 71, 34–38 (2021). [DOI] [PubMed] [Google Scholar]
- 20.van Wietmarschen N., et al. , Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature 586, 292–298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J., Zhang L., The transcriptional targets of p53 in apoptosis control. Biochem. Biophys. Res. Commun. 331, 851–858 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Yu J., Zhang L., PUMA, a potent killer with or without p53. Oncogene 27, S71–S83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samowitz W. S., et al. , Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am. J. Pathol. 158, 1517–1524 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haugen A. C., et al. , Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res. 68, 8465–8472 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kortlever R. M., Higgins P. J., Bernards R., Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 8, 877–884 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J., Wang Z., Kinzler K. W., Vogelstein B., Zhang L., PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc. Natl. Acad. Sci. U.S.A. 100, 1931–1936 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu W., et al. , PUMA regulates intestinal progenitor cell radiosensitivity and gastrointestinal syndrome. Cell Stem Cell 2, 576–583 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B., PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Wang P., Yu J., Zhang L., The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc. Natl. Acad. Sci. U.S.A. 104, 4054–4059 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang Y., Luo J., Zhang W., Gu W., Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Sykes S. M., et al. , Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24, 841–851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charvet C., et al. , Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol. Cell 42, 584–596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T. R., Dorotinsky C. S., McGuire L. J., Macy M. L., Hay R. J., DLD-1 and HCT-15 cell lines derived separately from colorectal carcinomas have totally different chromosome changes but the same genetic origin. Cancer Genet. Cytogenet. 81, 103–108 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Sur S., et al. , A panel of isogenic human cancer cells suggests a therapeutic approach for cancers with inactivated p53. Proc. Natl. Acad. Sci. U.S.A. 106, 3964–3969 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal M., et al. , Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Res. 73, 5497–5507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen G. H., et al. , A small molecule inhibitor of the BLM helicase modulates chromosome stability in human cells. Chem. Biol. 20, 55–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tentler J. J., et al. , Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 9, 338–350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z., Pearlman A. H., Hsieh P., DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 38, 94–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu D., Calvo J. A., Samson L. D., Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 12, 104–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basile G., Leuzzi G., Pichierri P., Franchitto A., Checkpoint-dependent and independent roles of the Werner syndrome protein in preserving genome integrity in response to mild replication stress. Nucleic Acids Res. 42, 12628–12639 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polyak K., Waldman T., He T.-C., Kinzler K. W., Vogelstein B., Genetic determinants of p53 induced apoptosis and growth arrest. Genes Dev. 10, 1945–1952 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Lombard D. B., et al. , Mutations in the WRN gene in mice accelerate mortality in a p53-null background. Mol. Cell Biol. 20, 3286–3291 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogelstein B., Lane D., Levine A. J., Surfing the p53 network. Nature 408, 307–310 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Picco G., et al. , Werner helicase is a synthetic-lethal vulnerability in mismatch repair-deficient colorectal cancer refractory to targeted therapies, chemotherapy, and immunotherapy. Cancer Discov. 11, 1923–1937 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Wang Y. J., Fletcher R., Yu J., Zhang L., Immunogenic effects of chemotherapy-induced tumor cell death. Genes Dis. 5, 194–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bunz F., et al. , Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Zhang L., Yu J., Park B. H., Kinzler K. W., Vogelstein B., Role of BAX in the apoptotic response to anticancer agents. Science 290, 989–992 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Tong J., et al. , Mcl-1 phosphorylation without degradation mediates sensitivity to HDAC inhibitors by liberating BH3-only proteins. Cancer Res. 78, 4704–4715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dudgeon C., et al. , PUMA induction by FoxO3a mediates the anticancer activities of the broad-range kinase inhibitor UCN-01. Mol. Cancer Ther. 9, 2893–2902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong J., et al. , Mcl-1 degradation is required for targeted therapeutics to eradicate colon cancer cells. Cancer Res. 77, 2512–2521 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan X., et al. , BET inhibitors potentiate chemotherapy and killing of SPOP-mutant colon cancer cells via induction of DR5. Cancer Res. 79, 1191–1203 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song X., et al. , Mcl-1 inhibition overcomes intrinsic and acquired regorafenib resistance in colorectal cancer. Theranostics 10, 8098–8110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.