Abstract
MicroRNAs (miRNAs) are about 22-nucleotide (nt) noncoding RNAs forming the effector complexes with Argonaute (AGO) proteins to repress gene expression. Although tiny RNAs (tyRNAs) shorter than 19 nt have been found to bind to plant and vertebrate AGOs, their biogenesis remains a long-standing question. Here, our in vivo and in vitro studies show several 3′→5′ exonucleases, such as interferon-stimulated gene 20 kDa (ISG20), three prime repair exonuclease 1 (TREX1), and ERI1 (enhanced RNAi, also known as 3′hExo), capable of trimming AGO-associated full-length miRNAs to 14-nt or shorter tyRNAs. Their guide trimming occurs in a manganese-dependent manner but independently of the guide sequence and the loaded four human AGO paralogs. We also show that ISG20-mediated guide trimming makes Argonaute3 (AGO3) a slicer. Given the high Mn2+ concentrations in stressed cells, virus-infected cells, and neurodegeneration, our study sheds light on the roles of the Mn2+-dependent exonucleases in remodeling gene silencing.
Keywords: RNAi, non-coding RNAs, exonucleases, immunology, neurodegeneration
In the canonical microRNA (miRNA) biogenesis, Dicer processes the hairpin-structured precursor miRNAs into about 22-nucleotide (nt) miRNA duplexes (1). Argonautes (AGOs) load the duplexes and eject the passenger strand, forming the effector complexes of the RNA interference (RNAi) (2, 3). Previous RNA sequencing studies identified AGO-associated 10- to 18-nt tiny RNAs (tyRNAs) in plants and vertebrates that mapped onto tRNAs and miRNAs (4). However, how those tyRNAs are synthesized remains unknown. The present study focused on the biogenesis of miRNA-derived tyRNAs.
Results
We raised several hypothetical pathways toward AGO-associated tyRNAs (Fig. 1A). If short single-stranded (ss) RNAs are directly loaded into AGOs, they must exist stably in the cell. To test the idea, we transfected 14- or 23-nt ss miR-20a or their small interfering RNA (siRNA)-like duplex into HEK293T cells. Only the guide RNA was radiolabeled at its 5′ end. Both 14- and 23-nt ssRNAs and the 14-nt siRNA-like duplex were degraded (Fig. 1B), suggesting that neither has a chance of being loaded into AGOs efficiently. In contrast, the 23-nt siRNA-like duplex remained 19 to 23 nt, and no tyRNA was detected, indicating that a detectable level of tyRNAs is not generated in normal conditions. These results prompted us to think that after the canonical RNA-induced silencing complex (RISC) assembly, the AGO-associated miRNAs are trimmed to tyRNAs by yet unidentified 3′→5′ exonucleases (Fig. 1A, Bottom pathway). Previous studies reported the expression of interferon-stimulated gene 20 kDa (ISG20), a 3′→5′ exonuclease, induced by interferon upon viral infection and stress and by estrogen hormone (5, 6). When ISG20 was exogenously expressed, a small amount of 13 to 14 nt was detected (Fig. 1C). To test whether the generated tyRNA is associated with AGO, FLAG-AGO2 was coexpressed with ISG20 in HEK293T cells (Fig. 1D, Bottom) that were also transfected with an siRNA duplex whose 23-nt miR-20a guide is radiolabeled at its 5′ end. After 48 h, FLAG-AGO2 was immunopurified, and the associated RNAs were resolved on a denaturing gel. Coexpression of ISG20 generated AGO-associated tyRNAs (Fig. 1D, Top). tyRNA generation was also observed when three prime repair exonuclease 1 (TREX1) or enhanced RNAi (ERI1) was coexpressed instead of ISG20. ERI1 is known to negatively regulate global miRNA abundance in mouse lymphocytes (7), but the mechanism remains unclear. Poly(A)-specific ribonuclease (PARN) shortened the 23-nt guide down to 19 nt, whereas exonuclease 5 (EXO5) did not trim the guide at all. The experiment was repeated with their catalytically dead mutants to confirm that the observed trimming was due to their exonuclease activity. As a result, none of the mutants generated tyRNA (Fig. 1D), proving that the catalytic center of the exonucleases is essential for tyRNA generation. ISG20, TREX1, and ERI1 also generated tyRNAs from AGO1-, AGO3-, and AGO4-associated miR-20a (Fig. 1E).
Fig. 1.
Specific 3′→5′ exonucleases convert miRNAs to tyRNAs. (A) Possible pathways toward AGO-associated tyRNAs. (B) In vivo stability of 14- and 23-nt ss miR-20a and their siRNA-like duplexes (ds). (C) tyRNA accumulation upon ISG20 expression. (D) In vivo trimming of FLAG-AGO2-associated miR-20a by 3′→5′ exonucleases and their catalytic mutants. Top: a representative gel image. Bottom: Western blots with antibodies for each protein. (E) tyRNA synthesis on four human AGOs by ISG20, TREX1, ERI1, and PARN. Mean ± SD.
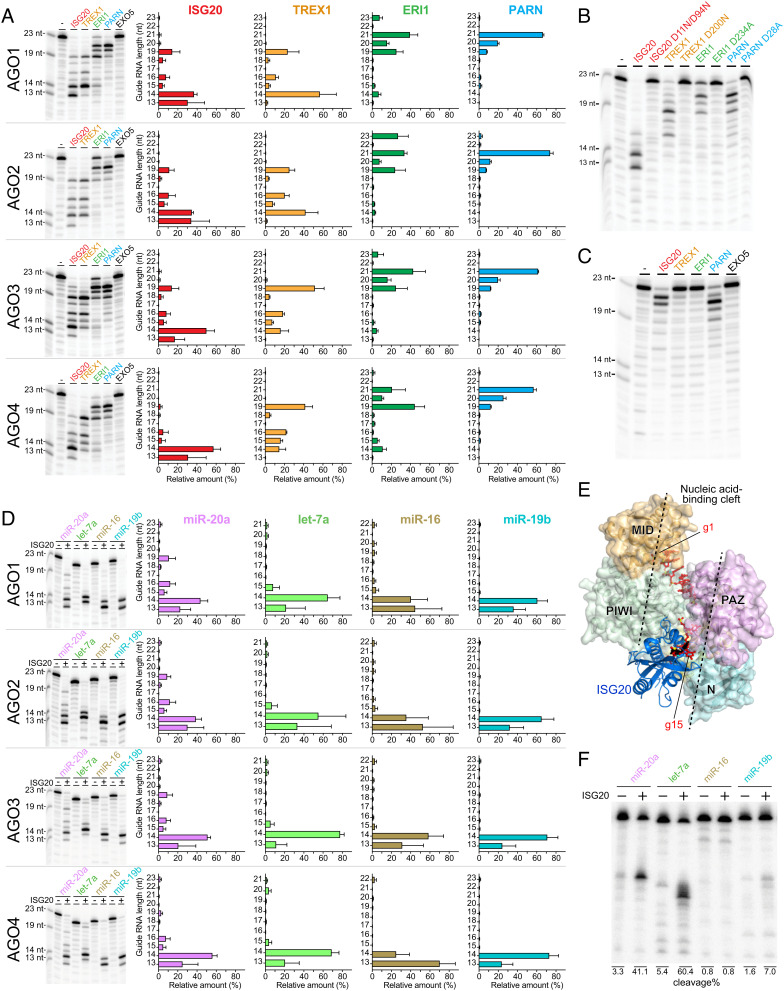
To characterize their guide trimming, we reconstituted an in vitro trimming system (SI Appendix). FLAG-tagged human AGO1, AGO2, AGO3, and AGO4 were programmed with a 5′-end radiolabeled 23-nt miR-20a, immobilized on anti-FLAG beads to wash out free guide RNAs, and incubated with one of the abovementioned exonucleases in the presence of manganese. All exonucleases, except for EXO5, trimmed miR-20a that had been loaded onto slicer-deficient AGO1 and AGO4 as well as slicing-competent AGO2 and AGO3 (8) (Fig. 2A, Left). In contrast, the catalytic mutants of the exonucleases showed no trimming activity (Fig. 2B), further supporting our conclusion that the observed guide trimmings are solely due to the catalytic activity of the exonucleases, not that of the AGOs. Each exonuclease similarly trimmed the loaded miR-20a across the four AGOs, but the trimming patterns differed between the exonucleases (Fig. 2A, Right). PARN generated an abundance of 21 nt. ERI1 ceased trimming when the guide length became 19 nt while developing a small population of 14 to 15-nt tyRNAs. In contrast, ISG20 and TREX1 shortened miR-20a to 13 and 14 nt, respectively. Replacing manganese with magnesium drastically reduced the trimming activities of ISG20, TREX1, and ERI1, but not PARN (Fig. 2C). These results suggest that these exonucleases can synthesize tyRNAs from miRNAs in a Mn2+-dependent manner with different processing rates.
Fig. 2.
ISG20, TREX1, and ERI1 generate tyRNAs. (A) In vitro guide trimming by 3′→5′ exonucleases at 2 mM MnCl2. Left: a representative gel image for each AGO. Right: the relative amount of each guide length after incubation with either 3′→5′ exonuclease. Mean ± SD. (B) In vitro trimming of AGO2-associated miR-20a by the catalytically dead exonuclease mutants at 2 mM MnCl2. (C) In vitro trimming of AGO2-associated miR-20a by 3′→5′ exonucleases at 2 mM MgCl2. (D) In vitro trimming of different miRNAs by ISG20. Left: a representative gel image. Right: the relative amount of each guide length after ISG20 incubation. Mean ± SD. (E) Docking model of ISG20 (blue) on a guide (red)-bound AGO3 (surface model). (F) In vitro trimming of AGO3-associated different miRNAs, followed by target cleavage.
Next, we investigated the susceptivity of different miRNAs to the 3′→5′ exonucleases. To this end, FLAG-tagged AGOs were programmed with a 5′-end radiolabeled 23-nt miR-20a, 21-nt let-7a, 22-nt miR-16, or 23-nt miR-19b, followed by incubation with ISG20. Although miR-20a was trimmed slower than the others, all the tested miRNAs were shortened to 13 to 14 nt, regardless of which AGO was loaded (Fig. 2D). This suggests that ISG20 can generate tyRNAs from a variety of miRNAs that are incorporated into any AGO. Our docking model indicates that the nucleic acid-binding cleft of AGO sequesters the guide nt 1 to 14 (g1 to g14) positions while the remaining 3′ half is accessible to ISG20 (Fig. 2E). This could explain why trimming stops when the guide length becomes 13 to 14 nt. Our previous study revealed that 14-nt variants of miR-20a and let-7a, but not of miR-16 or miR-19b, whose 3′ 7 to 9 nt were trimmed from their mature miRNAs, conferred a decent slicing activity on AGO3 (8). These results prompted us to test whether trimming of AGO3-associated specific miRNAs makes the RISC a slicer. To this end, FLAG-AGO3 was programmed with the same full-length miRNAs used in Fig. 2D, incubated with ISG20, and mixed with a cap-labeled target RNA containing a complementary sequence to each guide. Cleavage was observed only when FLAG-AGO3 was programmed with miR-20a and let-7a (Fig. 2F), demonstrating that ISG20 catalytically activates AGO3.
Discussion
Given the significance of the 3′ supplementary region, 14-nt or shorter tyRNAs would change the target specificity from their full length. ISG20, TREX1, and ERI1 convert AGO-associated miRNAs to tyRNAs, which requires Mn2+, an essential transition metal for human health. Dysregulation of the cellular Mn2+ concentration has been implicated in neurodegenerative diseases, such as Parkinson’s disease, Alzheimer’s disease, Huntington’s disease, and manganism (9). Notably, the ISG20 level is elevated in neurodegenerative disease models (10) and brain injury (5), while the malfunction of TREX1 causes autoimmune diseases such as Aicardi–Goutieres syndrome (11). Natural killer cells and T cells deficient in ERI1 enhance the RNAi (7). These results suggest a possible correlation between the Mn2+-dependent guide-trimming and neurodegenerative diseases.
Materials and Methods
Details are provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by Pelotonia Fellowships (to G.Y.S. and M.S.P.), a Center for RNA Fellowship (to G.Y.S.), and the NIH (R01GM138997 to K.N. and S10OD023582).
Author contributions
K.N. designed research; G.Y.S., A.C.K., M.S.P., J.S., C.D., H.Z., N.M., D.B., and E.A.E.-W. performed research; G.Y.S. and K.N. analyzed data; and G.Y.S. and K.N. wrote the paper.
Competing interest
Yes, the authors have patent filings to disclose. K.N. has filed a patent application related to this work.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Ambros V., et al. , A uniform system for microRNA annotation. RNA 9, 277–279 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakanishi K., Anatomy of RISC: How do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip. Rev. RNA 7, 637–660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakanishi K., Anatomy of four human Argonaute proteins. Nucleic Acids Res. 50, 6618–6638 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakanishi K., Are argonaute-associated tiny RNAs junk, inferior miRNAs, or a new type of functional RNAs? Front. Mol. Biosci. 8, 795356 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stankiewicz A. M., Goscik J., Majewska A., Swiergiel A. H., Juszczak G. R., The effect of acute and chronic social stress on the hippocampal transcriptome in mice. PLoS One 10, e0142195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deymier S., Louvat C., Fiorini F., Cimarelli A., ISG20: An enigmatic antiviral RNase targeting multiple viruses. FEBS Open Bio. 12, 1096–1111 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas M. F., et al. , Eri1 regulates microRNA homeostasis and mouse lymphocyte development and antiviral function. Blood 120, 130–142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park M. S., Sim G., Kehling A. C., Nakanishi K., Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proc. Natl. Acad. Sci. U.S.A. 117, 28576–28578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budinger D., Barral S., Soo A. K. S., Kurian M. A., The role of manganese dysregulation in neurological disease: Emerging evidence. Lancet. Neurol. 20, 956–968 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Friedman B. A., et al. , Diverse brain myeloid expression profiles reveal distinct microglial activation states and aspects of Alzheimer’s disease not evident in mouse models. Cell Rep. 22, 832–847 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Crow Y. J., et al. , Mutations in the gene encoding the 3’-5’ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 38, 917–920 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.