Significance
Many embryonic and metastatic cell types undergo an epithelial-to-mesenchymal transition (EMT) to migrate throughout the body. This change in behavior requires reorganization of plasma membrane proteins; yet, how plasma membrane lipids control EMT during development is incompletely understood. Here, we find that membrane sphingolipid metabolism by the enzyme nSMase2 is spatiotemporally regulated and essential for avian neural crest EMT. Our results support a model in which nSMase2 acts to regulate the endocytosis and activation of developmental signaling pathways required for mesenchymalization. These findings illustrate the dynamic role of structural lipids in the plasma membrane in orchestrating cell signaling events and, in turn, influencing downstream transcriptional programs.
Keywords: neural crest, epithelial-to-mesenchymal transition, ceramide, cell signaling, endocytosis
Abstract
Epithelial-to-mesenchymal transition (EMT) is a dramatic change in cellular physiology during development and metastasis, which requires coordination between cell signaling, adhesion, and membrane protrusions. These processes all involve dynamic changes in the plasma membrane; yet, how membrane lipid content regulates membrane function during EMT remains incompletely understood. By screening for differential expression of lipid-modifying genes over the course of EMT in the avian neural crest, we have identified the ceramide-producing enzyme neutral sphingomyelinase 2 (nSMase2) as a critical regulator of a developmental EMT. nSMase2 expression begins at the onset of EMT, and in vivo knockdown experiments demonstrate that nSMase2 is necessary for neural crest migration. We find that nSMase2 promotes Wnt and BMP signaling and is required to activate the mesenchymal gene expression program. Mechanistically, we show that nSMase2-dependent ceramide production is necessary for and sufficient to up-regulate endocytosis and is required for Wnt co-receptor internalization. Finally, inhibition of endocytosis in the neural crest mimics the loss of migration and Wnt signaling observed following nSMase2 knockdown. Our results support a model in which nSMase2 is expressed at the onset of neural crest EMT to produce ceramide and facilitate receptor-mediated endocytosis of Wnt and BMP signaling complexes, thereby activating promigratory gene expression. These results highlight the critical role of plasma membrane lipid metabolism in regulating transcriptional changes during developmental EMT programs.
Epithelial-to-mesenchymal transition (EMT) is a critical process in both development and metastasis during which epithelial cells gain migratory capacity to disperse throughout the body. The transcriptional circuits that regulate these events have been at least partially elucidated and appear similar in both developmental and metastatic contexts (1–3).
Recent studies in cancer cells have revealed significant changes in the plasma membrane lipid content during metastasis (4, 5). These observations raise the intriguing possibility that lipid metabolism functions to promote the EMT process by exploiting the structural and bioactive features of membrane lipids. Accordingly, sphingolipid metabolism induces EMT in cancer cell lines by reducing ceramide content to suppress apoptosis and by producing the signaling lipids sphingosine-1-phosphate (S1P) and lysophosphatidic acid (LPA) which enhance proliferation and motility in metastatic cells (6–10). In addition to cancer, such lipids may also play a role in development—for example, signaling by S1P and LPA regulates migration of multiple embryonic cell populations (11–13), and ceramide induces neural tube closure defects when present at ectopic levels (14). Yet, the functions of plasma membrane lipids during developmental EMT in vivo remain less clear.
Avian neural crest cells undergo a stereotypical EMT shortly after neural tube closure (1, 15, 16), making them an excellent model to test how membrane lipid changes facilitate the transition from an epithelial to a mesenchymal state. Specified premigratory neural crest cells exhibit epithelial characteristics and embark upon a temporally regulated EMT via a well-characterized gene regulatory network to adopt a migratory, mesenchymal physiology (15, 16). Evidence from neuroblastoma, a neural crest-derived pediatric cancer, suggests that lipid metabolism may play a critical role in neural crest biology (17, 18). In neuroblastoma, receptor tyrosine kinase activation is achieved by organizing with Src kinases into ordered plasma membrane microdomains (17). Formation of such microdomains is largely driven by the thermodynamic features of sphingolipids and cholesterol (19), thus highlighting the importance of understanding lipid metabolism and plasma membrane organization during neural crest cell development.
Here, we examined how changes in plasma membrane lipid composition affect EMT in developing neural crest cells. We find that the sphingolipid metabolizing enzyme nSMase2 is necessary for neural crest EMT. Our results support a mechanism wherein nSMase2 activity regulates the endocytosis of cell signaling receptors from the plasma membrane, thereby potentiating cell signaling by Wnts and bone morphogenetic proteins (BMPs) and activating the mesenchymal gene expression program critical to achieve a migratory phenotype.
Results
Neural Crest Cells Express Smpd3 during EMT and Migration.
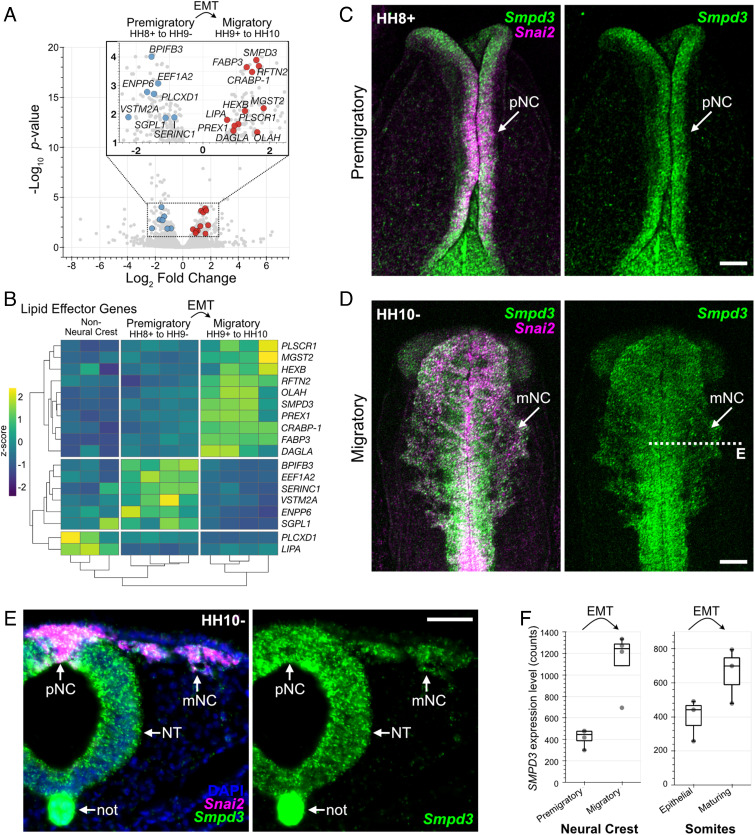
We hypothesized that temporally regulated changes in lipid content during EMT would result from the differential expression of lipid metabolic genes; thus, we explored transcriptome resources comparing chicken cranial neural crest cells at premigratory (epithelial) and migratory (mesenchymal) stages (20). We parsed these data for genes with Gene Ontology terms (21) containing “lipid” and found seven lipid-related genes significantly enriched in epithelial, premigratory neural crest, and 11 significantly enriched in mesenchymal, migratory neural crest (Fig. 1 A and B), consistent with a potential role for temporally regulated lipid modifications during neural crest EMT. The gene SMPD3, which encodes the enzyme neutral sphingomyelinase 2 (nSMase2), was of particular interest since it showed the most significant enrichment at migratory stages (Fig. 1 A and B). nSMase2 is a sphingolipid-metabolizing enzyme that functions at the plasma membrane to hydrolyze sphingomyelin and generate ceramide (22–24). Since sphingolipid metabolism is implicated in cancer EMT (4, 8, 25), we delved further into the role of nSMase2 during neural crest EMT and development.
Fig. 1.
Identification of Smpd3 expression during neural crest epithelial-to-mesenchymal transition. A, Volcano plot showing genes significantly enriched within premigratory (negative fold change values) and migratory (positive fold change values) neural crest cells. Lipid-related genes with significant differential gene expression are labeled (blue, premigratory enrichment; red, migratory enrichment). Data analysis was performed using previously published RNA seq reads (20). EMT, epithelial-to-mesenchymal transition; HH, Hamburger–Hamilton stage. B, Heatmap displaying complete Euclidean clustering shows changes in gene expression levels for the lipid-related genes labeled in A between nonneural crest, premigratory, and migratory neural crest samples. Each column represents a single biological replicate. C and D, Hybridization chain reaction shows Smpd3 and neural crest EMT marker Snai2 expression at premigratory (C) and migratory (D) stages. pNC, premigratory neural crest; mNC, migratory neural crest. E, Transverse section through D (dashed line) shows strong Smpd3 expression in the neural crest, as well as in the ventral neural tube (NT) and notochord (not). F, Box plots display Smpd3 expression levels as counts determined from transcriptome profiling in the chicken cranial neural crest (Left, (20)) and developing chicken somites (Right, (26)). (Scale bars represent 100 µm (C and D) and 50 µm (E).) See also SI Appendix, Fig. S1.
We first examined the spatiotemporal expression pattern of Smpd3, together with the neural crest marker Snai2, using hybridization chain reaction (HCR), and found that Smpd3 expression initiated prior to EMT in the specified neural crest cells at premigratory stages (HH8+) and was then highly expressed following EMT in neural crest cells at migratory stages (HH10-, Fig. 1 C–E). We also observed strong expression of Smpd3 in the more ventral neural tube and in the notochord (Fig. 1E). Chromogenic in situ hybridization confirmed these observations and revealed that Smpd3 expression was not detectable at stages earlier than stage HH8 (SI Appendix, Fig. S1 A–E), indicating that Smpd3 expression is initiated during neural crest specification and just prior to EMT. To ask whether elevated Smpd3 expression may be common across developmental EMTs and thus reflect a generalizable molecular mechanism, we examined transcriptomes describing somite maturation during which the cells that comprise the sclerotome undergo EMT (26). The results show that Smpd3 expression levels increase as somites mature and mesenchymalize (Fig. 1F), similar to the increase observed during neural crest EMT.
The SMPD3 gene product, nSMase2, catalyzes the hydrolysis of sphingomyelin within the plasma membrane to produce ceramide (22, 24); thus, we asked whether ceramide is produced in Smpd3-expressing neural crest cells. Immunolabeling of stage HH11 chicken embryos with antibodies against ceramide and the neural crest marker Pax7 revealed strong enrichment of ceramide on the apical surface of the neural tube (SI Appendix, Fig. S1F), correlating with the enriched Smpd3 gene expression in this tissue (Fig. 1E). Importantly, ceramide labeling also appeared enriched on the surface of migrating neural crest cells (SI Appendix, Fig. S1F), indicating that ceramide is indeed produced in the cranial neural crest. Together, these results raise the intriguing possibility that temporally regulated sphingomyelin metabolism by nSMase2 may play a critical role during developmental EMTs.
Ceramide Production Is Required for Cranial Neural Crest EMT.
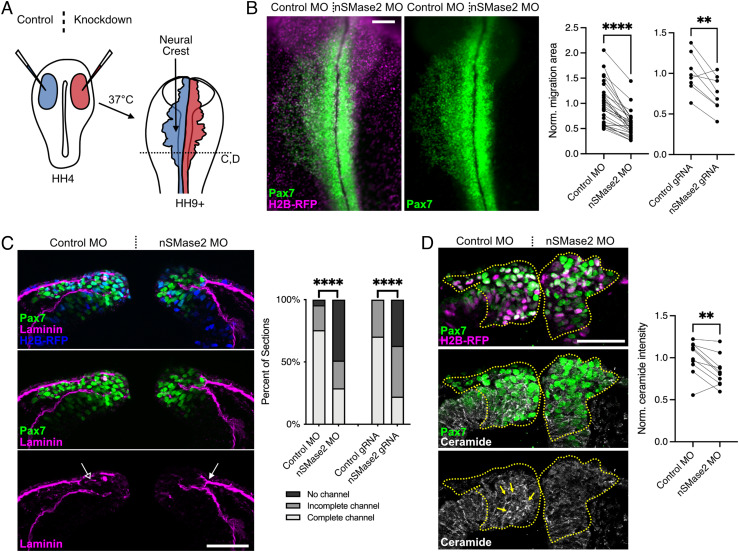
The coexpression of Smpd3 and ceramide during EMT led us to hypothesize that ceramide production by nSMase2 is required for neural crest EMT. To test this, we performed nSMase2 knockdown through electroporation of a nonbinding control antisense morpholino (MO) on the left and an nSMase2-targeting MO on the right sides of gastrulating chicken embryos (schematized in Fig. 2A). The electroporated embryos were then allowed to develop to migratory neural crest stages, at which point neural crest cells were visualized in whole mount. While the control MO did not disrupt neural crest EMT or migration away from the midline, neural crest cells that received the nSMase2 MO showed a significant reduction in migration distance and these cells appeared to accumulate at the midline in the dorsal neural tube (Fig. 2B).
Fig. 2.
nSMase2-mediated ceramide production is required for neural crest EMT. A, Control (blue) and knockdown (red) reagents were delivered bilaterally into gastrulating chicken embryos (stage HH4), then embryos were incubated to neural crest migration stages for all in vivo electroporation experiments. Perturbations (Right side) were then compared to the contralateral internal control side (Left). B, Embryos were electroporated with plasmid encoding H2B-RFP as a lineage tracer together with a nonbinding control MO (Control MO, Left) and nSMase2-targeting MO (nSMase2 MO, Right). Pax7 immunolabeling shows failure of neural crest cells to migrate away from the midline following nSMase2 knockdown. Parallel coordinate plots display normalized Pax7+ neural crest migration area, demonstrating significantly reduced neural crest migration following nSMase2 MO or nSMase2 gRNA electroporations. Each point represents one embryo with lines connecting measurements from the same embryo (n = 27 MO-electroporated embryos and n = 8 gRNA-electroporated embryos). C, Control or nSMase2 MOs were electroporated together with H2B-RFP, then transverse sections were immunolabeled for the laminin-rich basement membrane. On the control side, neural crest cells have remodeled a channel permissive of migration (open arrow), while nSMase2-deficient neural crest cells fail to remodel the basement membrane (filled arrow). Stacked bar plots display basement membrane remodeling scores as percentages of sections (nSMase2 MO, n = 24 sections (3 each from eight independent embryos); nSMase2 gRNA, n = 18 sections (3 each from six independent embryos)). D, Pax7 and ceramide immunolabeling in transverse section reveals that ceramide is present on the membranes of control neural crest cells (filled yellow arrows), and this labeling is lost following nSMase2 knockdown. Parallel coordinate plot shows reduced ceramide immunolabeling within the cranial neural crest region (dashed yellow outlines) following nSMase2 knockdown, each point represents the mean of three sections from an individual embryo (n = 10 embryos). (Scale bars represent 100 µm (B) and 50 µm (C and D).) **P < 0.01, ****P < 0.0001, two-tailed paired t test.
To determine whether the effect of nSMase2 MO is specific to a loss of nSMase2 production, we performed two additional, independent methods of nSMase2 inhibition, both of which phenocopied nSMase2 MO. First, using a CRISPR/Cas9-mediated knockout strategy, we observed that electroporation of constructs encoding Cas9 and nSMase2-targetting gRNA induced a significant reduction in Smpd3, but not Tfap2b, transcripts (SI Appendix, Fig. S2A); this knockdown is consistent with mutation-induced nonsense-mediated decay. We then quantitated the migration area of cranial neural crest cells and observed a similar reduction in migration following CRISPR-mediated nSMase2 knockdown compared with nSMase2 MO (Fig. 2B and SI Appendix, Fig. S2B). Finally, treatment with the small-molecule inhibitor of neutral sphingomyelinases, GW4869 (27), similarly blocked neural crest EMT compared to DMSO control treatments (SI Appendix, Fig. S2C).
We next asked why nSMase2-deficient neural crest cells did not undergo migration and instead appeared to accumulate within the dorsal midline (Fig. 2B). To address this, we generated transverse sections and immunolabeled to detect laminin in the basement membrane. During EMT, the neural crest remodels the basement membrane in response to Wnt signaling to produce a channel through which neural crest cells delaminate from the neuroepithelium (28). While the basement membrane remodeled normally on the control side (Fig. 2C, open arrow), we observed a high frequency of incomplete or obstructed remodeling events following either nSMase2 MO or nSMase2 gRNA electroporations (Fig. 2C, closed arrow). Importantly, markers of mitosis and apoptosis were unaffected by nSMase2 knockdown within the neural crest (SI Appendix, Fig. S3), and thus aberrant proliferation or survival does not account for the observed reduction in migration. These results suggest that nSMase2-deficient neural crest cells fail to remodel the basement membrane as required to complete EMT, and therefore are unable to fully delaminate and migrate away from the dorsal neural tube.
Finally, we asked whether nSMase2 functions during neural crest development through its canonical role in metabolizing sphingomyelin to produce ceramide (22, 24). To this end, we compared ceramide immunoreactivity in neural crest cells in vivo and found that nSMase2 knockdown significantly decreased ceramide levels (Fig. 2D), demonstrating that nSMase2 is indeed necessary for ceramide production in the cranial neural crest. Together, these results demonstrate that nSMase2 function is critical for cranial neural crest delamination and suggest that ceramide production by nSMase2 is required to undergo EMT in vivo.
nSMase2 Is Required to Activate pro-EMT Gene Expression Changes.
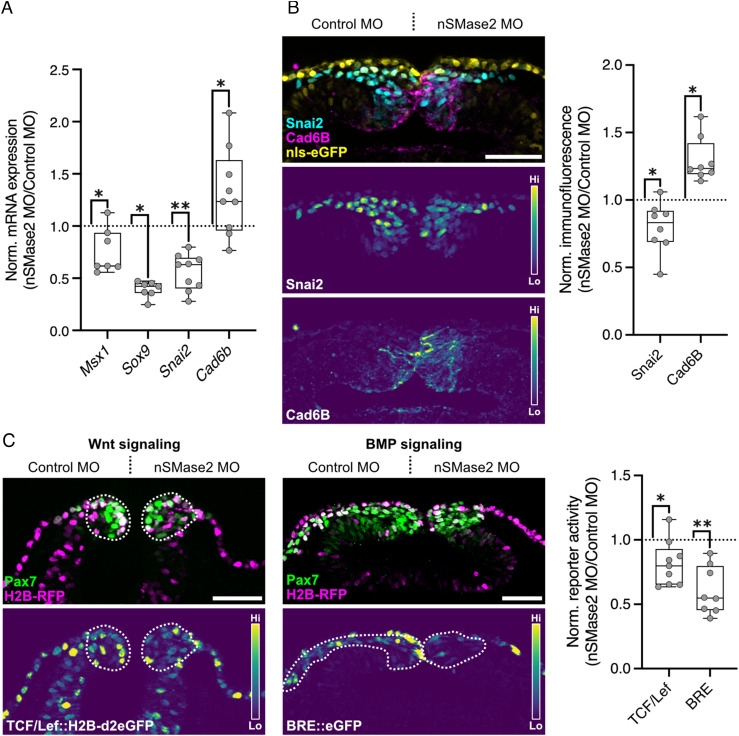
The observed loss of cranial neural crest delamination suggests that nSMase2 may act upstream of the EMT gene regulatory program. To examine this possibility, we performed HCR to visualize the expression of multiple markers that reflect neural crest specification (Msx1, Sox9, and Tfap2b) and EMT progression (Sox9 and Snai2) (15, 29). During neural crest EMT, Snai2 directly represses expression of the premigratory cadherin CAD6B (30), and thus we also examined the expression of Cad6b in nSMase2 morphants as a premigratory neural crest marker. nSMase2-deficient neural crest displayed significant downregulation of Msx1, Sox9, and Snai2 expression and elevated expression of Cad6b transcripts (Fig. 3A and SI Appendix, Fig. S4). Notably, we observed no significant effect on the expression of the neural crest specification marker Tfap2b following nSMase2 knockdown (SI Appendix, Fig. S2A). We next performed immunolabeling for Snai2 and Cad6B and, consistent with the HCR results, nSMase2 knockdown reduced Snai2 and elevated Cad6B protein levels in the cranial neural crest (Fig. 3B). As Smpd3 expression begins during neural crest specification (SI Appendix, Fig. S1) and its inhibition after specification blocks EMT (SI Appendix, Fig. S2C), the cumulative results suggest that nSMase2 activity is not required for the initiation of neural crest specification. Instead, its role is to maintain specified neural crest identity and facilitate the transition to a migratory gene expression signature during EMT.
Fig. 3.
nSMase2 is required for the transcriptional activation of the EMT program and for Wnt and BMP signaling. A, Relative mRNA expression for neural crest markers in nSMase2 knockdown embryos at stage HH9- (representative images displayed in SI Appendix, Fig. S4, Msx1 n = 7 embryos, Sox9 n = 7 embryos, Snai2 n = 9 embryos, Cad6b n = 7 embryos). B, Immunolabeling for Snai2 and Cad6B proteins in nSMase2 knockdown sections at HH9. Box plot shows relative Snai2 and Cad6B protein levels as measured by fluorescence intensity (Snai2 n = 8 embryos, Cad6B n = 8 embryos). C, nSMase2 knockdown reagents were electroporated together with the Wnt-responsive TCF/Lef::H2B-d2EGFP (Left) or the BMP-responsive BRE::eGFP (Center) constructs and imaged in transverse sections at HH9− and HH10−, respectively. Relative eGFP fluorescence intensity within the Pax7+ neural crest domain shows a significant reduction in Wnt and BMP output in nSMase2-depleted neural crest cells (TCF/Lef n = 9 embryos, BRE n = 8 embryos). (Scale bars represent 50 µm.) *P < 0.05, **P < 0.01, two-tailed paired t test. Each point in A represents one embryo examined in whole mount, and in B and C represents the mean of three sections from individual embryos.
Wnt and BMP Signaling Pathways Require nSMase2 Function.
Activation of the EMT gene regulatory network in neural crest involves signaling by the canonical Wnt and BMP pathways (15, 29, 31). To test the possibility that sphingolipid metabolism at the plasma membrane may regulate the activity of these pathways, we turned to GFP reporter constructs which measure transcriptional output of canonical Wnt (T-cell factor (TCF)/Lef::H2B-d2eGFP) and BMP (BMP-responsive element (BRE)::eGFP) signaling in cranial neural crest cells (32–36). We electroporated these reporters together with control or nSMase2 MOs, and then compared GFP intensity within the cranial neural crest. At the onset of EMT and prior to migration (HH9−), we observed a significant reduction in Wnt/β-catenin signaling upon nSMase2 knockdown (Fig. 3C). Since SNAI2 and SOX9 are direct targets of Wnt signaling (37–39), this accounts for the observed loss of Snai2 and Sox9 expression (Fig. 3A). Similarly, we observed a significant reduction in BMP signaling activity (Fig. 3C), consistent with the reduced expression of the BMP target Msx1 (40) (Fig. 3A). Together, these results demonstrate that nSMase2 function is required for developmental signaling cascades, including those of the Wnt and BMP pathways.
nSMase2 Regulates Ceramide Production in Neural Crest Plasma Membranes.
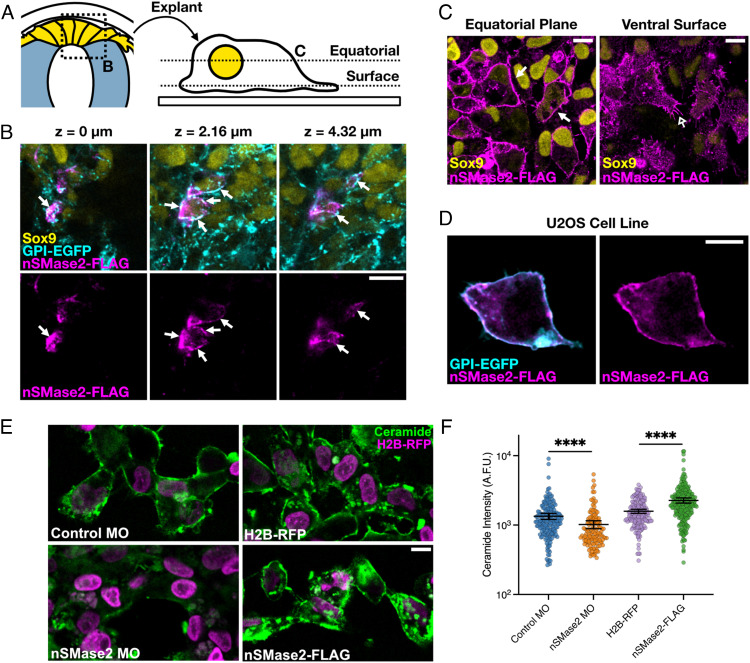
We next sought to determine the mechanism by which sphingolipid metabolism controls the activation of developmental signaling. Wnt and BMP/transforming growth factor β (TGF-β) receptor complexes can collect within sphingolipid- and cholesterol-rich membrane microdomains such as caveolae (19, 41–46), raising the possibility that nSMase2 may act at the plasma membrane to regulate cell signaling. To test this, we first examined the subcellular localization of FLAG-tagged chicken nSMase2 in vivo, ex vivo, and in vitro. The results showed that nSMase2 localizes predominantly to the plasma membrane in neural crest cells both in vivo (Fig. 4 A and B) and ex vivo in primary neural crest explant culture (Fig. 4C). Moreover, a similar localization is observed in vitro in the human U2OS cell line, which bears morphological similarity to premigratory neural crest (Fig. 4D). We next quantitated surface ceramide levels in neural crest explants in controls versus nSMase2 knockdown or overexpression (Fig. 4E). Whereas nSMase2 knockdown reduced ceramide levels, its overexpression elevated ceramide immunoreactivity (Fig. 4F). Together, these results demonstrate that nSMase2 functions in neural crest cells at the plasma membrane where it produces ceramide.
Fig. 4.
nSMase2 regulates ceramide production in the plasma membrane of neural crest cells. A. Schematic illustrating displays presented in panels B (high magnification section through premigratory neural crest in the dorsal neural tube) and C (equatorial and ventral surface planes of the same field of view of explanted neural crest cells). B. Three focal planes from a single z-stack show that nSMase2-FLAG localizes to the plasma membrane together with GPI-eGFP in the Sox9-positive premigratory neural crest cells in vivo (closed arrows). C-D. nSMase2-FLAG also localizes to the plasma membrane (closed arrow) and filopodia (open arrow) in explanted neural crest cells ex vivo (C), and in U2OS human osteosarcoma cells (D). E. Embryos were electroporated with the indicated reagents and neural crest cells were explanted and cultured ex vivo. After the start of migration, explants were stained for ceramide and single-cell ceramide levels were quantitated. F. Dot plot of ceramide intensity values, with each dot representing a single cell (n = 193 Control MO, n = 134 nSMase2 MO, n = 173 H2B-RFP, n = 235 nSMase2-FLAG cells) and overlayed bars displaying the mean and 95% CIs. ****P < 0.0001; Mann–Whitney U test. (All scale bars represent 10 µm.)
nSMase2 Expression Is Necessary for Endocytosis.
Ceramide production within sphingolipid- and cholesterol-rich membrane microdomains is sufficient to induce spontaneous membrane curvature and trigger membrane internalization (47–50), thus may potentiate the endocytosis of microdomain-localized proteins. Importantly, endocytosis of Wnt and BMP/TGF-β complexes into signalosomes facilitates the most potent signaling output (41, 42, 46, 51–61). These observations, together with our finding that nSMase2 is necessary for Wnt and BMP signaling, prompted us to ask whether nSMase2 function potentiates signaling activity by promoting endocytosis of signaling complexes.
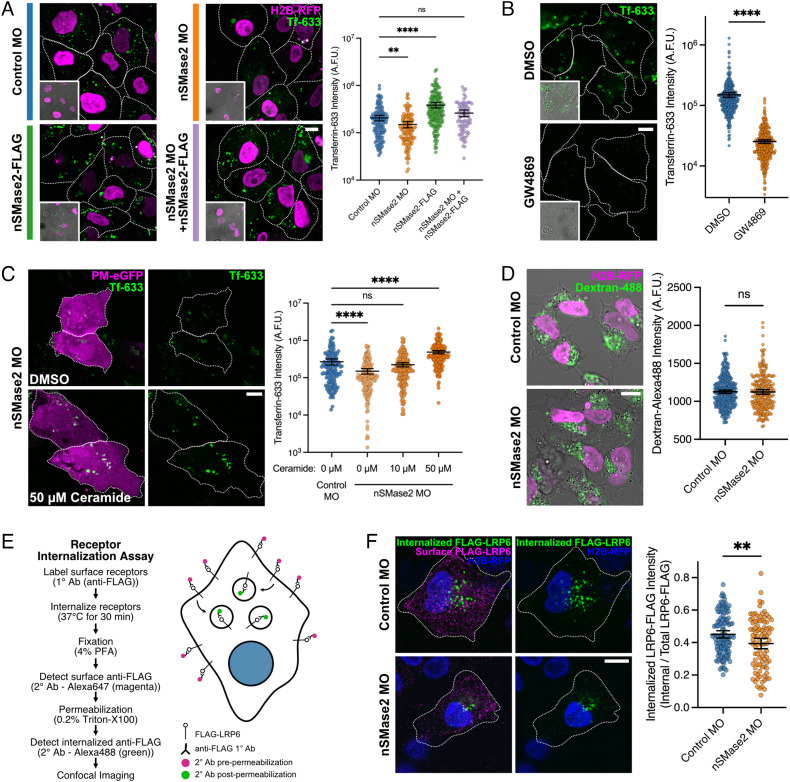
We first tested this hypothesis by assaying receptor-mediated endocytosis ex vivo in neural crest explants using fluorescently labeled transferrin (Tf-633) (62). Explanted neural crest cells were exposed to Tf-633 for 30 min, and internalized Tf-633 fluorescence intensity was measured (Fig. 5A). The results showed that electroporation with nSMase2 MO resulted in a significant reduction in Tf-633 internalization compared with that of control MO. Conversely, nSMase2-FLAG overexpression promoted increased Tf-633 internalization. Importantly, the effects of nSMase2 MO were rescued by coelectroporation of nSMase2-FLAG overexpressing constructs (Fig. 5A), thereby demonstrating that this phenotype is specific to nSMase2 knockdown. Further, neutral sphingomyelinase inhibition by GW4869 treatment also potently inhibited Tf-633 endocytosis (Fig. 5B). We next asked whether ceramide could rescue the loss of nSMase2 by supplying nSMase2-deficient neural crest explants with increasing concentrations of exogenous ceramide. Importantly, we found that ceramide addition showed a dose-dependent rescue of nSMase2 knockdown and even surpassed control explants in Tf-633 uptake (Fig. 5C). Finally, to determine whether nSMase2 functions specifically in receptor-mediated endocytosis mechanisms (e.g., transferrin), or if it also functions in pinocytosis, we performed engulfment assays using fluorescent dextran and found no difference between control and nSMase2-deficient neural crest cells (Fig. 5D). Together, these results demonstrate that ceramide production by nSMase2 is necessary for and sufficient to up-regulate endocytosis in neural crest cells and suggest that ceramide functions specifically during receptor-mediated endocytosis.
Fig. 5.
nSMase2 is necessary for receptor-mediated endocytosis but not for pinocytosis. A, Electroporated primary neural crest cells were explanted at premigratory stages. After 16 h, the explants were incubated with Transferrin-Alexa-633-conjugate (Tf-633) in the culture media for 30 min to allow for receptor-mediated endocytosis. The explants were then fixed and Tf-633 internalization was measured by confocal microscopy. The results show reduced endocytosis in nSMase2-deficient cells compared to controls, increased Tf-633 internalization by nSMase2-FLAG overexpression, and that nSMase2-FLAG overexpression rescues nSMase2 MO (n = 135 Control MO, n = 112 nSMase2 MO, n = 163 nSMase2 FLAG, and n = 74 nSMase2 MO + nSMase2-FLAG cells). ns not significant, **P < 0.01, ****P < 0.0001; Kruskal–Wallis test (P < 0.0001) with Dunn’s posthoc analysis. B, Neutral sphingomyelinase inhibition by GW4869 (50 µM) blocks Tf-633 endocytosis in neural crest explants compared to DMSO controls (n = 317 DMSO, n = 364 GW4869 cells). ****P < 0.0001; Mann–Whitney U test. C, Control and nSMase2-deficient neural crest explants were pretreated with DMSO or increasing concentrations of exogenous C12 ceramide, and Tf-633 endocytosis efficiency was analyzed revealing that ceramide rescues nSMase2 knockdown (n = 140 Control MO, n = 142 nSMase2 MO, n = 190 nSMase2 MO + 10 µM Ceramide, n = 182 nSMase2 MO + 50 µM Ceramide cells). ns not significant, ****P < 0.0001; Kruskal–Wallis test (P < 0.0001) with Dunn’s multiple comparison analysis. D, Dextran-488 uptake in control and nSMase2-deficient neural crest explants suggest that nSMase2 is dispensable for pinocytosis (n = 335 Control MO, n = 235 nSMase2 MO cells). ns not significant; two-tailed unpaired t test. E, Overview of receptor internalization assay designed to measure endocytosis of FLAG-LRP6 in control and nSMase2-deficient neural crest cells. F, Control and nSMase2-deficient neural crest explants were treated with anti-FLAG to track the endocytosis of FLAG-LRP6. Displayed are cell surface FLAG-LRP6 (magenta), internalized FLAG-LRP6 (green), and electroporation marker H2B-RFP (blue). The results show significantly reduced internalized FLAG-LRP6 following nSMase2 knockdown compared with controls (n = 124 Control MO, n = 94 nSMase2 MO cells pooled from five embryos). **P < 0.01; two-sample Kolmogorov–Smirnov test. Dot plots display fluorescent intensity measures for Transferrin-633 (A, B, and D), or Dextran-488 (C), or internalized FLAG-LRP6 normalized to total FLAG-LRP6 expression (F), with each dot representing a single neural crest cell. Overlayed bars display mean and 95% CIs. A.F.U. arbitrary fluorescence units. (All scale bars represent 10 µm.) White dotted lines indicate cell borders.
To determine whether this function is common to other cell types, we assayed receptor-mediated endocytosis of Tf-633 in the epithelial U2OS cell line. Compared with an RFP-only control, transfection of a dominant-negative Dynamin 1 mutant (Dyn1(K44A)) (63, 64) resulted in reduced Tf-633 internalization (SI Appendix, Fig. S5), as predicted. Next, we found that nSMase2 overexpression increased Tf-633 internalization (SI Appendix, Fig. S5), similar to our findings in neural crest cells (Fig. 5A). In contrast, overexpression of a mutant of nSMase2 (nSMase2(N130A)) which lacks all catalytic function (65) had no effect on endocytosis (SI Appendix, Fig. S5), further demonstrating that nSMase2-dependent hydrolysis of sphingomyelin into ceramide regulates endocytosis. These findings suggest that the role of nSMase2 in regulating endocytosis is generalizable across multiple species and cell types.
We next asked whether nSMase2 is required for the internalization of membrane receptors involved in Wnt signaling. Wnt signals activate the co-receptor protein LRP6 and trigger its aggregation into caveolae (46) where caveolin-mediated endocytosis of LRP6 is necessary for Wnt activity and nuclear β-catenin accumulation (54). If nSMase2 is required for Wnt signaling by promoting endocytosis of activated Wnt complexes, we predicted that nSMase2 knockdown would reduce LRP6 internalization. We tested this prediction by expressing extracellularly FLAG-tagged LRP6 (FLAG-LRP6), and then performing a receptor internalization assay on control and nSMase2-deficient neural crest explants (schematized in Fig. 5E) (66). FLAG-LRP6 was bound by anti-FLAG antibodies on live cells which were then allowed to endocytose this protein complex for 30 min. We then fixed the explants and performed secondary antibody detection before and after permeabilization to distinguish between surface-localized (magenta) and endocytosed (green) FLAG-LRP6. The results showed reduced FLAG-LRP6 internalization in nSMase2 knockdown neural crest explants compared with that of controls (Fig. 5F), thus demonstrating that nSMase2 function enhances the endocytosis of Wnt receptors in neural crest cells.
Inhibition of Endocytosis Phenocopies Loss of nSMase2 during Neural Crest EMT.
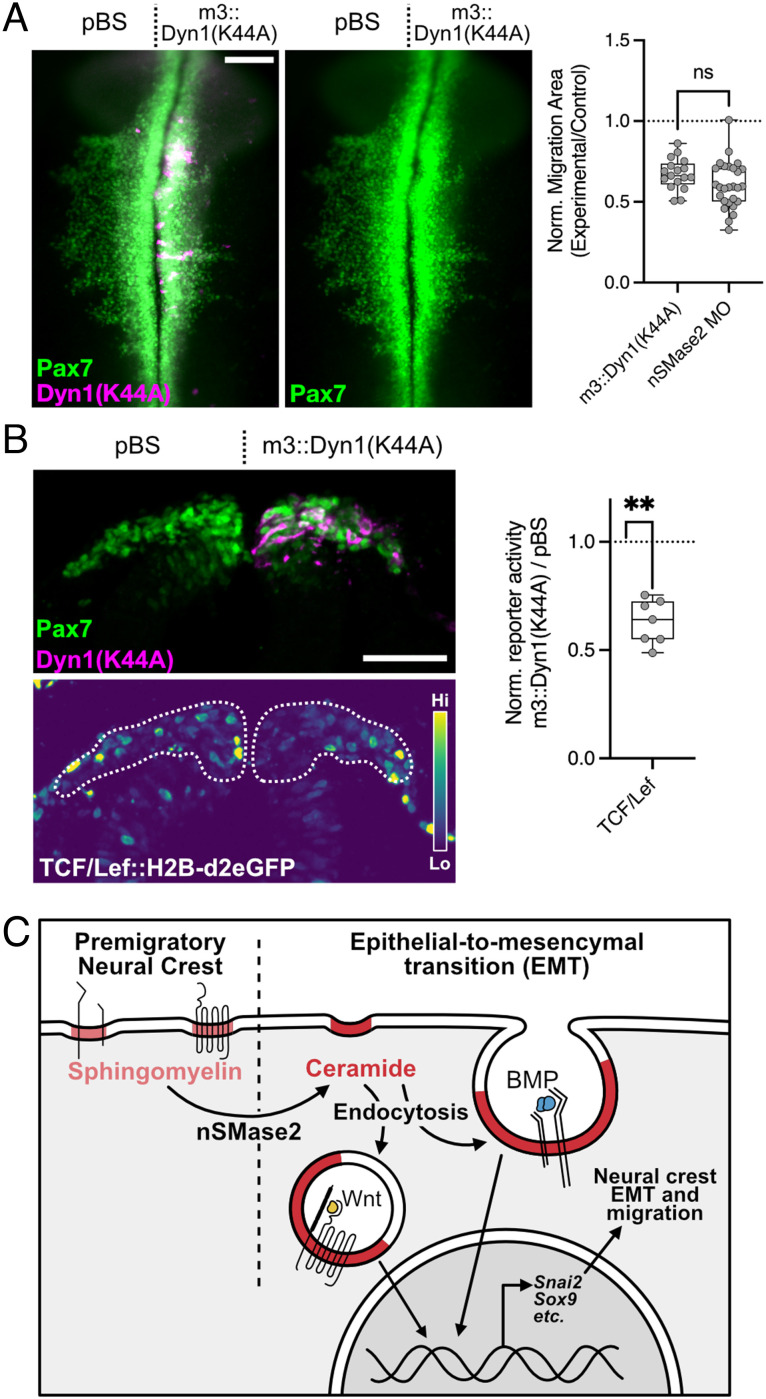
Finally, to test this mechanism in vivo, we asked whether inhibition of endocytosis within the neural crest during EMT reproduces the phenotypes observed following nSMase2 knockdown. We inhibited endocytosis specifically within cranial neural crest cells by expressing the dominant negative Dyn1(K44A) mutant under control of the neural crest-specific FoxD3 NC1.1m3 enhancer (m3::Dyn1(K44A)) (67). Following electroporation of m3::Dyn1(K44A), we observed a significant reduction in neural crest migration area that phenocopied nSMase2 knockdown (Fig. 6A). We also observed reduced Wnt/β-catenin signaling during EMT within the cranial neural crest (Fig. 6B). Taken together, these data indicate that endocytosis is required in the neural crest for Wnt/β-catenin signaling and EMT; by phenocopying nSMase2 knockdown, these findings are consistent with a model in which endocytosis acts downstream of nSMase2-dependent ceramide production to regulate cell signaling and neural crest EMT (Fig. 6C).
Fig. 6.
Endocytosis is necessary for Wnt signaling and neural crest EMT in vivo. A, Embryos were electroporated with the dominant negative Dyn1(K44A) under control of the neural crest-specific FoxD3 NC1.1m3 enhancer (m3::Dyn1(K44A)) and examined in whole mount at HH9+ for neural crest migration by Pax7 immunolabeling. Neural crest migration was quantitated from endocytosis-inhibited embryos, revealing a reduction in migration area. Comparing relative migration area between m3::Dyn1(K44A) embryos with nSMase2 knockdown embryos shows that endocytosis inhibition phenocopies nSMase2 knockdown (n = 17 m3:Dyn1(K44A) and 27 nSMase2 MO embryos). ns not significant; two-tailed unpaired t test. Each point represents one embryo. nSMase2 MO data are reproduced here from Fig. 2B. B, The Wnt-responsive TCF/Lef::H2B-d2eGFP reporter was coelectroporated with m3::Dyn1(K44A). Embryos were collected and examined in transverse section at HH9+. Normalized eGFP fluorescence quantitation shows reduced Wnt signaling following Dyn1(K44A) expression compared to a nonexpressing pBluescript (pBS) control (n = 8 embryos). **P < 0.01; two-tailed paired t test. (Scale bars represent 100 µm (A) and 50 µm (B).) C, Our results support a model in which premigratory neural crest cells membranes are sphingomyelin rich. At the onset of EMT, nSMase2 expression converts sphingomyelin into ceramide to promote membrane curvature. This facilitates endocytosis of activated signaling complexes (Wnt and BMP). Internalization of these complexes enables activation of pro-EMT transcriptional targets (including SNAI2 and SOX9) which regulate neural crest EMT and subsequent cell migration.
Discussion
Here, we show that premigratory neural crest cells initiate the expression of nSMase2 to convert plasma membrane sphingomyelin into ceramide in preparation for EMT. Ceramide production has strong effects on lateral membrane organization by forming microdomains that drive receptor clustering and plasma membrane curvature (19, 47–48, 49–71). Accordingly, we observed that Wnt and BMP signaling in vivo requires the production of ceramide during neural crest EMT (Fig. 3C). Furthermore, we find that nSMase2 is necessary for and sufficient to upregulate endocytosis in neural crest cells (Fig. 5A), and that nSMase2 is required for internalization of LRP6 (Fig. 5F). This is consistent with evidence that many signaling pathways require endocytosis of activated signaling complexes (41, 42, 44–45, 46–51, 52–59). Finally, inhibition of endocytosis mimics the effects of nSMase2 knockdown in cranial neural crest EMT (Fig. 6 A and B). This model is further supported by the observation that nSMase2-deficient neural crest cells display elevated levels of the cell adhesion protein cadherin-6B (Fig. 3B), which is normally cleared from the membrane during EMT by dynamin-dependent endocytosis (72). Together, our results suggest that temporally regulated ceramide production by nSMase2 mechanistically activates endocytosis to facilitate neural crest EMT. Importantly, these results reveal that changes in lipid metabolism are a critical driver of developmental signaling that is required for the transition to a mesenchymal phenotype.
In many cancers, EMT and motility are enabled by the downregulation of ceramide or the upregulation of sphingomyelin production, whereas ceramide production shows pro-apoptotic and anti-EMT effects (6–8). These observations appear in contrast with our findings during developmental EMT, where ceramide production instead activates EMT gene expression and motility without affecting proliferation or apoptosis. In reconciling these observations, it is important to consider the subcellular localization of the sphingolipid metabolism. In the plasma membrane, sphingomyelin synthesis maintains epithelial properties in collecting duct cells (73), while mesenchymalization instead is achieved by the production of ceramide and reduction of sphingomyelin, by nSMase2 during neural crest development. Notably, chemotherapeutic agents also trigger acute ceramide production in the plasma membrane by nSMase2 to drive mesenchymal behaviors in HeLa cells including loss of adhesion and increased migratory capacity (74). In contrast, in endoplasmic reticulum membranes, ceramide production de novo is “anti-mesenchymal” and decreases cancer cells’ motility and metastatic potential (6, 8). Thus, similar bioactive lipids can have opposite effects based on their subcellular localization, and it will be of great interest to determine these context-specific mechanisms that regulate these opposing responses.
The observation that expression of Smpd3 and ceramide was present in multiple non-neural crest tissues (e.g., neural tube, Fig. 1E and SI Appendix, Fig. S1F) is not surprising given the abundance of many sphingolipids, the variety of metabolic pathways regulating their production, and the diverse physiological roles they play (75). Interestingly, the developing embryo regulates the expression of lipid metabolic pathways with specific spatiotemporal control, and that these expression domains activate context-dependent functions. For example, in addition to a neural crest EMT phenotype, we also note neural tube closure defects in a subset of nSMase2 knockdown embryos, consistent with the finding that exogenous ceramide induces neural tube closure defects (14) and suggests that precise levels of ceramide contribute to curvature of the neural tube epithelium.
We cannot exclude the possibility that loss of neural crest migration in our nSMase2- and endocytosis-inhibited embryos may partially reflect defective motility separate from EMT. However, several other phenotypes illustrate that nSMase2 functions to regulate EMT prior to the onset of neural crest motility, namely: 1) the loss of Snai2 upregulation and Cad6b downregulation, 2) the loss of Wnt signaling activity at stage HH9−, prior to migration, and 3) the basement membrane remodeling defects observed following nSMase2 knockdown. Further, we observe migration of nSMase2-deficient cells in our ex vivo culture system, suggesting that nSMase2 is not required for cell motility per se. We do find that migratory neural crest cells maintain Smpd3 expression after EMT; this suggests that nSMase2 may indeed play a second role in regulating certain aspects of neural crest migration, possibly through functioning in the production and release of extracellular vesicles (47, 76, 77).
Our results show that nSMase2 is necessary for transferrin endocytosis, and exogenous ceramide can rescue nSMase2 loss (Fig. 5). This is very intriguing given the lateral organization of transferrin receptors (TfR) with respect to ceramide—transferrin/TfR binding preferentially occurs within disordered plasma membrane microdomains and away from ceramide (68, 78, 79). In this context, ceramide displaces TfR away from ceramide-rich ordered domains and into disordered clathrin-coated pits for internalization (79). This may reflect one possible mechanism by which ceramide sorts specific cargo for endocytosis in vivo. We also observed that nSMase2 is required for endocytosis of the Wnt co-receptor LRP6 (Fig. 5F) and propose that loss of receptor endocytosis accounts for the reduced Wnt and BMP transcriptional responses following nSMase2 knockdown (Fig. 3C). Wnt and BMP signals can be potentiated by endocytosis by either clathrin-coated pits (55, 56, 80), caveolae (42, 45, 46, 53, 54), or macropinocytosis (59, 60). Since we found pinocytosis unaffected by nSMase2 perturbation (Fig. 5C), this suggests that nSMase2 function is specific to receptor-mediated pathways. One possibility in neural crest cells is that ceramide production organizes activated receptor complexes into clathrin-coated pits in a similar manner as seen by TfR (79). However, biochemical fractionation indicates that Wnt ligands preferentially bind and activate receptor complexes in caveolin-rich, detergent-resistant ordered membrane domains rather than detergent-soluble, clathrin-containing domains (43, 44), making clathrin-dependent endocytosis less likely and instead supporting a caveolar mechanism for Wnt cargo endocytosis.
Important questions remain about how endocytic routes are utilized by each signaling pathway in neural crest cells, and how ceramide production appropriately sorts these receptor complexes. It is possible that the biochemical structure of ceramide itself interacts with transmembrane proteins to sort cargo, induces membrane alterations such as curvature to cluster proteins, or may even be further modified by subsequent enzymes to produce another bioactive lipid necessary for receptor sorting. Finally, the experiments presented herein have been designed to measure cargo internalization in neural crest cells by nSMase2; thus, we cannot exclude the possibility that nSMase2/ceramide plays additional downstream roles in intracellular trafficking of the Wnt, BMP, and Tf receptors. Together, our findings illustrate how dynamic changes in lipid metabolism are utilized during development to alter plasma membrane organization and have broad reaching effects on critical processes such as EMT and cell migration.
Materials and Methods
RNA Sequencing Analysis.
Bulk RNA Sequencing (RNA Seq) reads from fluorescence-activated cell sorted FoxD3-positive premigratory (HH8+ to HH9−) and migratory (HH9+ to HH10) chicken cranial neural crest cells, and FoxD3-negative nonneural crest cells, were downloaded from NCBI (BioProject PRJNA497574) (20) and analyzed as previously described (36). Briefly, reads were trimmed using Cutadapt (81) and aligned to the chicken genome (GRCg6a) using BowTie2 (82). Transcripts were then counted using featureCounts (83), and differential expression analysis between premigratory and migratory gene expression was carried out using DESeq2 (84). Volcano plots (Fig. 1A) highlight genes with lipid-related GO terms (21) with an adjusted P-value < 0.05, base mean value >45, and log2-fold change value >0.7. Complete Euclidean distance clustering was performed using pheatmap (Fig. 1B) (85), and displayed are each premigratory, migratory, and nonneural crest cell biological replicate. Box plots (Fig. 1F) were used to display the expression levels of Smpd3 from this neural crest analysis, and from bulk RNA Seq analysis of dissected somites (BioProject PRJNA602335) (26).
Embryos and Perturbations.
Fertilized chicken eggs were obtained from commercial sources (Sunstate Ranch and AA Lab Eggs). Ex ovo electroporations were performed using five pulses of 5.6 V for 50 ms at 100 ms intervals and cultured in albumin with 1% penicillin/streptomycin, then incubated to the desired HH stage (86, 87). For transverse sections, fixed embryos were incubated in 5% sucrose for 30 min at room temperature, 15% sucrose overnight at 4°C, 7.5% gelatin at 39°C overnight, flash-frozen in liquid nitrogen, and cryosectioned at a thickness of 18 µm. All reagents electroporated either expressed a fluorescently tagged protein or were coelectroporated with nuclear GFP- or RFP-expressing constructs (88, 89); fluorescent protein expression was used to screen for electroporation efficiency and only embryos with high efficiency were included in the analysis. MOs were electroporated at 0.8 mM and synthesized with the following sequences (Gene Tools): control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′), translation-blocking nSMase2 MO (5′- GGTGTCACTGTGTCAAGCATCCATA-3′). pCI::nSMase2-FLAG, pTK-FoxD3-NC1.1m3::Dyn1(K44A), and pCI::FLAG-LRP6 overexpressing constructs were electroporated at 3 µg/µl, 1.5 µg/µl, and 1 µg/µl, respectively. CRISPR-mediated knockdown was performed using a plasmid-based strategy, and a nonbinding control gRNA (5′-GCACTGCTACGATCTACACC-3′) and a nSMase2-targeting gRNA designed within the first exon of the SMPD3 gene locus (5′-GCAATCTGCGCAGCCCGAGA-3′), each at 1.5 µg/µl concentrations, were coexpressed with a Cas9-expressing plasmid (1.5 µg/µl) as previously described (90). For chemical neutral sphingomyelinase inhibition experiments, embryos were incubated in 50 µM GW4869 (Sigma-Aldrich) in albumin starting at stage HH8 and incubated until stage HH9+.
Hybridization Chain Reaction and In Situ Hybridization.
Reagents for third-generation hybridization chain reaction (HCR) were purchased from Molecular Technologies, and hybridization experiments were performed following manufacturer’s instructions (91). Probe sets were designed against Smpd3 (B1 initiator), Snai2 (B4), Cad6b (B5), Sox9 (B5), Msx1 (B4), and Tfap2b (B7) and detected using appropriate amplifier hairpins labeled with Alexa488, Alexa546, and Alexa647. For chromogenic in situ hybridization, a 970 bp fragment of the chicken Smpd3 transcript, corresponding to 76 bp of the 5′ untranslated region and first 894 bp of open reading frame, was amplified by PCR from stage HH9 cDNA using primers “nSMase2 FWD -76” and “nSMase2 REV 894 T7” (SI Appendix, Table S1). RNA probes were synthesized and labeled with DIG (Roche) using the T7 RNA polymerase (Promega) and purified with Illustra Probe-Quant G-50 Micro Columns (GE Healthcare). Hybridization and probe detection were carried out as previously described (35).
Immunohistochemistry.
For immunofluorescence analysis, embryos were fixed for 20 min at room temperature in 4% PFA in phosphate buffer, and all subsequent washes and incubations were performed in TBST + Ca2+ (50 mM Tris-HCl, 150 mM NaCl, 1 mM CaCl2, 0.5% Triton X-100). Blocking was performed in 10% donkey serum for 2 h at room temperature, and primary and secondary antibody incubations were carried out in 10% donkey serum for two nights at 4°C. Primary antibodies employed in this study include mouse IgG1 anti-Pax7 (1:10; DSHB #PAX7), mouse IgM anti-HNK-1 (1:5; DSHB #3H5), mouse IgG1 anti-Cad6B (1:5; DSHB #CCD6B-1), mouse IgM anticeramide (1:200 or 1:50; Sigma #MID15B4), rabbit anti-phosphohistone H3 (1:500; Millipore #06-570), rabbit anticleaved caspase 3 (1:300; R&D Systems #AF835), rabbit anti-Snai2 (1:500; Cell Signaling Technologies #9585), rabbit anti-Sox9 (1:1000, Millipore #AB5535), goat anti-GFP (1:500; Rockland #600-101-215M), and rabbit anti-RFP (1:500; MBL #PM005). Alexa Fluor 350-, 488-, 568-, 633-, or 647-conjugated donkey secondary antibodies (1:500; Molecular Probes) were used to detect primary antibodies.
For membrane ceramide immunolabeling in neural crest ex vivo, we pretreated live explants with anticeramide (1:50) antibodies for 20 min at 4°C, washed with ice-cold phosphate-buffered saline (PBS), and then fixed explants for 10 min with 4% PFA + 0.25% glutaraldehyde at room temperature. After fixation, cells were permeabilized for 20 min at 4°C in PBS + 0.1% Tween-20, blocked with 1% bovine serum albumin (BSA) in PBS at 4°C for 1 h, and then incubated overnight in 1:50 dilution of anticeramide in 1% BSA. Following three PBS washes, the explants were incubated in secondary antibodies in 1% BSA for 1 h at room temperature and washed three more times with PBS before imaging.
Construct Design and Cloning.
Cloning was performed by a combination of PCR reactions with high-fidelity DNA polymerase AccuPrime (ThermoFisher) and restriction digestion and ligation (enzymes from NEB). A 5′ Kozak consensus sequence was introduced to promote efficient translation of each overexpression construct, and all constructs were verified sequencing before use. All primer sequences are presented in SI Appendix, Table S1. CMV::Dyn1(K44A)-mRFP (Addgene plasmid #55795) (63), pCAG::GPI-eGFP (Addgene plasmid #32601) (92), TCF/Lef::GFP (Addgene plasmid #32610) (32), M38-TOP::d2EGFP (Addgene plasmid #17114), BRE::GFP (34), PM-GFP (Addgene plasmid #21213) (93), and CMV::VSVG-LRP6 (94) constructs were generous gifts from Catherine Berlot, Anna-Katerina Hadjantonakis, Randall Moon, Elisa Martí, Tobias Meyer, and Xi He.
The nSMase2 open reading frame was amplified from cDNA using “nSMase2 ATG XhoI” and “nSMase2 stop ClaI” or “nSMase2 FLAG ClaI” primers and ligated into pCI::H2B-RFP (88) between the XhoI and ClaI sites to produce pCI::nSMase2 and pCI::nSMase2-FLAG, respectively. The nSMase2(N130A) mutation was inserted by amplifying the 5′ and 3′ fragments of nSMase2 from cDNA using “nSMase2 FWD-76” and “nSMase2 N130A REV” and “nSMase2 N130A FWD” and “nSMase2 2462 REV,” respectively. These PCR fragments were then fused by amplification with “nSMase2 ATG XhoI” and “nSMase2 stop ClaI” and ligated into pCI::H2B-RFP between the XhoI and ClaI sites, producing pCI::nSMase2(N130A). pCAG::nSMase2-RFP was produced by amplifying the nSMase2 open reading frame without the stop codon from pCI::nSMase2 using “nSMase2 ATG SphI” and “nSMase2 nostop ClaI” primers and amplifying RFP from pCI::nSMase2 using “RFP ATG ClaI” and “RFP stop NotI” primers. These fragments were digested and ligated together between the SphI and NotI sites of pCI::H2B-RFP.
pTK-FoxD3-NC1.1m3::Dyn1(K44A)-RFP was produced by digesting pTK-FoxD3-NC1.1m3::GFP (67) with NheI and XbaI. CMV::Dyn1(K44A)-RFP was grown in One Shot INV110 dam-Escherichia coli (ThermoFisher) to prevent XbaI methylation and was similarly digested with NheI and XbaI. The resulting products were ligated together and sequenced to confirm orientation. A FLAG tag was inserted at the N terminus of HsLRP6, downstream the signal peptide, by amplifying CMV::LRP6 94 with “LRP6 Signal Peptide FWD” and “LRP6 Signal Peptide FLAG REV” to generate the signal peptide fragment and with “LRP6 FLAG FWD” and “LRP6 REV” to generate the C-terminal fragment. Both amplicons were assembled into pCI::H2B-RFP linearized by XhoI and ClaI using Gibson assembly. GFP was replaced in TCF/Lef::H2B-GFP (32) with d2EGFP from M38-TOP::d2EGFP by digestion and ligation between the AgeI and NotI cut sites. Finally, membrane-localized PM-GFP 93 was assembled in place of the H2B-RFP in pCI::H2B-RFP using Gibson assembly to produce pCAG::PM-GFP.
Tissue Culture, Transfection, and Neural Crest Explants.
Human osteosarcoma U2OS cells (ATCC #HTB-96) were cultured at 37°C in 5% CO2 in McCoy’s 5A Modified Media (ThermoFisher) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Corning). Cells were seeded for transfection in eight-well glass bottom chamber slides (ibidi # 80827) and transfected using lipofectamine 3000 (Invitrogen) at 80% confluency and then incubated for 24 h. After incubation, the cells were then analyzed for endocytosis efficiency.
Neural crest explants were prepared from control or electroporated chicken embryos as previously described (36). Briefly, polymer coverslip bottom imaging chambers (ibidi) were incubated for 1 h at 37°C with 25 µg/mL fibronectin in PBS; fibronectin was then aspirated and replaced with explant culture media (DMEM supplemented with 10% fetal bovine serum, 10% chick embryo extract, and 1% penicillin/streptomycin). Dorsal neural tube explants were cut from the midbrains of HH8+/HH9− embryos using fine scissors or polished tungsten needles. The explants were then cultured overnight at 37°C with 5% CO2 before endocytosis efficiency was analyzed.
Endocytosis and Receptor Internalization Assays.
To analyze transferrin or dextran uptake in cell culture and neural crest explant experiments, explanted neural crest cultures were washed in serum-free DMEM (ThermoFisher) and were serum-starved for 30 min at 37°C. Media was then replaced with 25 µg/mL Transferrin-Alexa-633-conjugate (Tf-633, ThermoFisher #T23362), or 20 µg/mL 3kDa Dextran-Alexa-488-conjugate (Dextran-488, ThermoFisher #D34682), in serum-free DMEM and incubated at 37°C for 30 or 10 min, respectively. For GW4869 neutral sphingomyelinase inhibition assays, explants were pretreated in serum-free media with GW4869 (50 µM) or DMSO for 30 min at 37°C, 25 µg/mL Tf-633 was added, and the explants were incubated for another 15 min at 37°C. For ceramide rescue experiments (Fig. 5C), explants were pretreated with DMSO or the indicated concentration of C12 ceramide (d18:1/12:0, Avanti Polar Lipids #860512) in serum-free DMEM at 37°C for 30 min after which 25 µg/mL Tf-633 was added for an additional 30 min at 37°C. After each endocytosis assay, cells were then washed 2× with Dulbecco's PBS (ThermoFisher) and fixed with 4% PFA in PBS for 10 min at room temperature before imaging.
Receptor internalization assays (66) were performed to determine the rate of FLAG-LRP6 internalization in neural crest explants and is outlined in Fig. 4E. Gastrulating embryos were electroporated with pCI::FLAG-LRP6 together with either control MO or nSMase2 MO and were cultured to stage HH8+/HH9−. Dorsal neural tube explants were cut and incubated as described above. To analyze FLAG-LRP6 internalization, explants were first incubated for 30 min at 37°C in serum-starving media (DMEM supplemented with 0.3% BSA). The explants were then blocked with 2% BSA in PBS for 10 min at 37°C, and then placed on ice to stop endocytosis. Cell surface FLAG-LRP6 was labeled with primary antibody solution (20 µg/mL anti-FLAG M2 in PBS with 2% BSA) for 20 min on ice after which unbound antibody was removed by three ice-cold PBS washes. The explants were then incubated at 37°C in serum-starving media for 30 min to allow internalization of anti-FLAG-bound receptors. After incubation, the explants were immediately transferred to ice and fixed with 4% PFA at room temperature for 10 min. Surface-bound primary antibody was then detected by incubation with donkey anti-mouse IgG AlexaFluor 647-conjugate in PBS/2% BSA (1:300) for 1 h on ice. After three PBS washes, the explants were permeabilized and reblocked with PBS/2% BSA/0.2% Triton X-100 for 30 min on ice. Internalized primary antibody was then detected by incubation with donkey anti-mouse IgG AlexaFluor 488-conjugate in PBS/2% BSA (1:300) for 30 min on ice. Unbound antibodies were removed by three PBS washes and then explants were imaged by confocal microscopy. Both the surface (magenta) and internalized (green) FLAG-LRP6 fractions were measured per cell, and Fig. 5B reports the internalized fraction normalized to the total FLAG-LRP6 expression (surface + internalized).
Microscopy and Image Analysis.
Whole mount and transverse section imaging was performed using a Zeiss Imager.M2 with an ApoTome.2 module or with a Zeiss LSM 880 upright confocal microscope. Fluorescent whole mount embryo images were collected as wide-field views (Figs. 2B and 6A and SI Appendix, Fig. S2 B and C), or displayed as a maximum intensity projection of Z-stacks (Fig. 1 C and D and SI Appendix, Figs. S2A and S4). Fluorescent transverse section images were displayed as a single optical plane (Figs. 2 C and D and 4B) or as maximum intensity projections of Z-stacks (Figs. 1E, 3 B and C, and 6B and SI Appendix, Figs. S1F and S3 A and C). Cell culture and explant experiments were imaged using a Zeiss LSM 800 or LSM 980 inverted confocal microscope and images were displayed as single equatorial focal plane images (Figs. 4 C–E and 5 and SI Appendix, Fig. S5).
Images were analyzed and prepared for display using Fiji (95). Chick embryo electroporations are heterogeneous and mosaic; as such, we performed analysis over large regions of interests (ROIs) that encompass many neural crest cells. This analysis approach has been used routinely to identify changes in fluorescence intensity and minimize noise from mosaic perturbations (28, 33, 35, 36, 96, 97). ROIs were manually drawn to measure fluorescence intensity and neural crest migration area, and to define boundaries for single-cell fluorescence intensity measurements or particle counting. For fluorescence measurements, corrected total cellular fluorescence values were calculated and normalized as previously described (35). For all particle counting analyses, thresholding parameters were optimized for each image dataset. For PH3-positive cell counting, median filtered images were thresholded using the Bernsen method with the Auto Local Threshold tool and the Analyze Particles feature was then used to quantitate particles larger than 20.0 µm2. For cleaved-Casp3 quantitation, median filtered images were subjected to uniform manual thresholding, and Analyze Particles was used to count puncta larger than 3 µm2. For Tf-633 particle counting in U2OS cells, images were subjected to median filtering, then thresholding using the Phansalkar method in the Auto Local Threshold tool. Analyze Particles was then used to count puncta with areas between 0.25 and 15.0 µm2.
Statistical Analysis.
Parallel coordinate plots (Fig. 2 B and D) show the control and experimental measurements, normalized to the mean of the control group, with solid lines connecting values from the same embryo. Box plots (Figs. 3 and 6 A and B and SI Appendix, Fig. S2A) display the ratio of experimental divided by corresponding paired control measurements, with the following box plot elements: center line, median; box limits, upper and lower quartiles; whiskers, minimum and maximum values; and points, individual embryo measurements. Dot plots (Figs. 4F and 5 and SI Appendix, Fig. S5) display individual cell values with the mean and 95% CIs overlaid. Statistical analyses were performed using GraphPad Prism 9 software. All datasets were tested for normal distribution with a Kolmogorov–Smirnov test—normally distributed data were analyzed with parametric tests (t test or one-way ANOVA), while nonnormally distributed data were analyzed with nonparametric tests (Mann–Whitney U or Kruskal–Wallis test). The specific tests used are reported in the corresponding figure legends. All embryo analyses show pooled biological replicates from at least three independent experiments, and all t tests passed power analysis in G*Power 3.1 (98) at a power cutoff of 0.80. Dot plots for Tf-633, Dextran-488, or FLAG-LRP6+ endocytosis in neural crest explants display pooled results from at least three independent biological replicates and dot plots for Tf-633 endocytosis in U2OS cells display pooled results from two independent experiments.
Code Availability.
All data analysis codes used in this study are available on GitHub at https://github.com/PiacentinoLab/2022_PNAS_nSMase2_Endocytosis.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We would like to thank Megan Martik, Erdinç Sezgin, Justin Bois, and Steven Wilbert for valuable discussion on experiment design and analysis and Alexis Camacho-Avila and Gabriel da Silva Pescador for technical support. We thank Catherine Berlot, Anna-Katerina Hadjantonakis, Randall Moon, Elisa Martí, Tobias Meyer, and Xi He for sharing reagents. Confocal imaging was supported by the Caltech Beckman Institute and the Arnold and Mabel Beckman Foundation to the Biological Imaging Facility. Funding for this work comes from the NIH grants K99DE029240 to M.L.P., K99DE028592 to E.J.H., R01DE027538 and R01DE027568 to M.E.B., and from the Caltech Summer Undergraduate Research Fellowship (SURF) to C.J.A.
Author contributions
M.L.P. and M.E.B. designed research; M.L.P., E.J.H., and C.J.A. performed research; M.L.P. contributed new reagents/analytic tools; M.L.P., E.J.H., and C.J.A. analyzed data; M.L.P. wrote the manuscript; and E.J.H. and M.E.B. edited the manuscript.
Competing interest
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the main text and/or supporting information. Source data can also be found with data analysis codes on GitHub at https://github.com/PiacentinoLab/2022_PNAS_nSMase2_Endocytosis. The Smpd3 mRNA sequence has been submitted to GenBank (Accession #MW142015).
Supporting Information
References
- 1.Nieto M. A., Huang R.Y.-J., Jackson R. A., Thiery J. P., EMT: 2016. Cell 166, 21–45 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Weinberg R. A., Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Front. Med. 12, 361–373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang J., Weinberg R. A., Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Meshcheryakova A., et al. , Exploring the role of sphingolipid machinery during the epithelial to mesenchymal transition program using an integrative approach. Oncotarget 7, 22295–22323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sampaio J. L., et al. , Membrane lipidome of an epithelial cell line. Proc. Natl. Acad. Sci. U. S. A. 108, 1903–1907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu P., White-Gilbertson S., Nganga R., Kester M., Voelkel-Johnson C., Expression of the SNAI2 transcriptional repressor is regulated by C16-ceramide. Cancer Biol. Ther. 20, 922–930 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng K., et al. , Sphingomyelin synthase 2 promotes an aggressive breast cancer phenotype by disrupting the homoeostasis of ceramide and sphingomyelin. Cell Death Dis. 10, 157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmond V., et al. , Downregulation of ceramide synthase-6 during epithelial-to-mesenchymal transition reduces plasma membrane fluidity and cancer cell motility. Oncogene 34, 996–1005 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y., et al. , Sphingosine-1-phosphate induced epithelial-mesenchymal transition of hepatocellular carcinoma via an MMP-7/ syndecan-1/TGF-β autocrine loop. Oncotarget 7, 63324–63337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W., Hind T., Lam B. W. S., Herr D. R., Sphingosine 1-phosphate signaling induces SNAI2 expression to promote cell invasion in breast cancer cells. FASEB J. 33, 7180–7191 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Kuriyama S., et al. , In vivo collective cell migration requires an LPAR2-dependent increase in tissue fluidity. J. Cell Biol. 206, 113–127 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sano H., Renault A. D., Lehmann R., Control of lateral migration and germ cell elimination by the Drosophila melanogaster lipid phosphate phosphatases Wunen and Wunen 2. J. Cell Biol. 171, 675–683 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kupperman E., An S., Osborne N., Waldron S., Stainier D. Y., A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature 406, 192–195 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Ross M. M., et al. , Ceramide: A novel inducer for neural tube defects. Dev. Dyn. 248, 979–996 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Martik M. L., Bronner M. E., Regulatory logic underlying diversification of the neural crest. Trends Genet. 33, 715–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piacentino M. L., Li Y., Bronner M. E., Epithelial-to-mesenchymal transition and different migration strategies as viewed from the neural crest. Curr. Opin. Cell Biol. 66, 43–50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palacios-Moreno J., et al. , Neuroblastoma tyrosine kinase signaling networks involve FYN and LYN in endosomes and lipid rafts. PLoS Comput. Biol. 11, e1004130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahmaniyan M., Qudeimat A., J. M., Kraveka, “Bioactive sphingolipids in neuroblastoma” in Neuroblastoma-Present and Future (InTech, 2012). [Google Scholar]
- 19.Sezgin E., Levental I., Mayor S., Eggeling C., The mystery of membrane organization: Composition, regulation and roles of lipid rafts. Nat. Rev. Mol. Cell Biol. 18, 361–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams R. M., et al. , Reconstruction of the global neural crest gene regulatory network in vivo. Dev. Cell 51, 255–276.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Gene Ontology Consortium , The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 47, D330–D338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tani M., Hannun Y. A., Analysis of membrane topology of neutral sphingomyelinase 2. FEBS Lett. 581, 1323–1328 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu B. X., Clarke C. J., Hannun Y. A., Mammalian neutral sphingomyelinases: Regulation and roles in cell signaling responses. Neuromolecular Med. 12, 320–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shamseddine A. A., Airola M. V., Hannun Y. A., Roles and regulation of neutral sphingomyelinase-2 in cellular and pathological processes. Adv. Biol. Regul. 57, 24–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levade T., Andrieu-Abadie N., Micheau O., Legembre P., Ségui B., Sphingolipids modulate the epithelial-mesenchymal transition in cancer. Cell Death Discov. 1, 15001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mok G. F., et al. , Characterising open chromatin in chick embryos identifies cis-regulatory elements important for paraxial mesoderm formation and axis extension. Nat. Commun. 12, 1157 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luberto C., et al. , Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J. Biol. Chem. 277, 41128–41139 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Hutchins E. J., Bronner M. E., Draxin alters laminin organization during basement membrane remodeling to control cranial neural crest EMT. Dev. Biol. 446, 151–158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers C. D., Nie S., Specifying neural crest cells: From chromatin to morphogens and factors in between. Wiley Interdiscip. Rev. Dev. Biol. 7, e322 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taneyhill L. A., Coles E. G., Bronner-Fraser M., Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development 134, 1481–1490 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pla P., Monsoro-Burq A. H., The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev. Biol. 444, S36–S46 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Ferrer-Vaquer A., et al. , A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev. Biol. 10, 121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutchins E. J., Bronner M. E., Draxin acts as a molecular rheostat of canonical Wnt signaling to control cranial neural crest EMT. J. Cell Biol. 217, 3683–3697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Dréau G., et al. , Canonical BMP7 activity is required for the generation of discrete neuronal populations in the dorsal spinal cord. Development 139, 259–268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piacentino M. L., Bronner M. E., Intracellular attenuation of BMP signaling via CKIP-1/Smurf1 is essential during neural crest induction. PLoS Biol. 16, e2004425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piacentino M. L., Hutchins E. J., Bronner M. E., Essential function and targets of BMP signaling during midbrain neural crest delamination. Dev. Biol. 477, 251–261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaBonne C., Bronner-Fraser M., Neural crest induction in Xenopus: Evidence for a two-signal model. Development 125, 2403–2414 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Luo T., Lee Y.-H., Saint-Jeannet J.-P., Sargent T. D., Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc. Natl. Acad. Sci. U. S. A. 100, 532–537 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bagheri-Fam S., et al. , Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 291, 382–397 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Tribulo C., Aybar M. J., Nguyen V. H., Mullins M. C., Mayor R., Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development 130, 6441–6452 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Ehrlich M., Endocytosis and trafficking of BMP receptors: Regulatory mechanisms for fine-tuning the signaling response in different cellular contexts. Cytokine Growth Factor Rev. 27, 35–42 (2016). [DOI] [PubMed] [Google Scholar]
- 42.He K., et al. , Internalization of the TGF-β type I receptor into caveolin-1 and EEA1 double-positive early endosomes. Cell Res. 25, 738–752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Özhan G., et al. , Lypd6 enhances Wnt/β-catenin signaling by promoting Lrp6 phosphorylation in raft plasma membrane domains. Dev. Cell 26, 331–345 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Sezgin E., et al. , Binding of canonical Wnt ligands to their receptor complexes occurs in ordered plasma membrane environments. FEBS J. 284, 2513–2526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramos M., Lamé M. W., Segall H. J., Wilson D. W., The BMP type II receptor is located in lipid rafts, including caveolae, of pulmonary endothelium in vivo and in vitro. Vascul. Pharmacol. 44, 50–59 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Bilic J., et al. , Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316, 1619–1622 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Trajkovic K., et al. , Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Holopainen J. M., Angelova M. I., Kinnunen P. K., Vectorial budding of vesicles by asymmetrical enzymatic formation of ceramide in giant liposomes. Biophys. J. 78, 830–838 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alonso A., Goñi F. M., The physical properties of ceramides in membranes. Annu. Rev. Biophys. 47, 633–654 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Shen H., et al. , Coupling between endocytosis and sphingosine kinase 1 recruitment. Nat. Cell Biol. 16, 652–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blitzer J. T., Nusse R., A critical role for endocytosis in Wnt signaling. BMC Cell Biol. 7, 28 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobrowolski R., De Robertis E. M., Endocytic control of growth factor signalling: Multivesicular bodies as signalling organelles. Nat. Rev. Mol. Cell Biol. 13, 53–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayes S., Chawla A., Corvera S., TGF beta receptor internalization into EEA1-enriched early endosomes: Role in signaling to Smad2. J. Cell Biol. 158, 1239–1249 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamamoto H., Komekado H., Kikuchi A., Caveolin is necessary for Wnt-3a-dependent internalization of LRP6 and accumulation of beta-catenin. Dev. Cell 11, 213–223 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Gammons M. V., Renko M., Johnson C. M., Rutherford T. J., Bienz M., Wnt Signalosome Assembly by DEP Domain Swapping of Dishevelled. Mol. Cell 64, 92–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hagemann A. I. H., et al. , In vivo analysis of formation and endocytosis of the Wnt/β-catenin signaling complex in zebrafish embryos. J. Cell Sci. 127, 3970–3982 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taelman V. F., et al. , Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143, 1136–1148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding Y., et al. , Bighead is a Wnt antagonist secreted by the Xenopus Spemann organizer that promotes Lrp6 endocytosis. Proc. Natl. Acad. Sci. U.S.A. 115, E9135–E9144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tejeda-Muñoz N., Albrecht L. V., Bui M. H., De Robertis E. M., Wnt canonical pathway activates macropinocytosis and lysosomal degradation of extracellular proteins. Proc. Natl. Acad. Sci. U.S.A. 116, 10402–10411 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albrecht L. V., Tejeda-Muñoz N., De Robertis E. M., Cell biology of canonical Wnt signaling. Annu. Rev. Cell Dev. Biol. 37, 369–389 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Tejeda-Muñoz N., et al. , Canonical Wnt signaling induces focal adhesion and Integrin beta-1 endocytosis. iScience 25, 104123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barysch S. V., Jahn R., Rizzoli S. O., A fluorescence-based in vitro assay for investigating early endosome dynamics. Nat. Protoc. 5, 1127–1137 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Hynes T. R., Mervine S. M., Yost E. A., Sabo J. L., Berlot C. H., Live cell imaging of Gs and the beta2-adrenergic receptor demonstrates that both alphas and beta1gamma7 internalize upon stimulation and exhibit similar trafficking patterns that differ from that of the beta2-adrenergic receptor. J. Biol. Chem. 279, 44101–44112 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Ochoa G. C., et al. , A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 150, 377–389 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Airola M. V., et al. , Structure of human nSMase2 reveals an interdomain allosteric activation mechanism for ceramide generation. Proc. Natl. Acad. Sci. U.S.A. 114, E5549–E5558 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rizzolio S., Tamagnone L., Antibody-feeding assay: A method to track the internalization of neuropilin-1 and other cell surface receptors. Methods Mol. Biol. 1493, 311–319 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Simões-Costa M. S., McKeown S. J., Tan-Cabugao J., Sauka-Spengler T., Bronner M. E., Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 8, e1003142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sot J., Bagatolli L. A., Goñi F. M., Alonso A., Detergent-resistant, ceramide-enriched domains in sphingomyelin/ceramide bilayers. Biophys. J. 90, 903–914 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.López-Montero I., Vélez M., Devaux P. F., Surface tension induced by sphingomyelin to ceramide conversion in lipid membranes. Biochim. Biophys. Acta 1768, 553–561 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Cremesti A. E., Goni F. M., Kolesnick R., Role of sphingomyelinase and ceramide in modulating rafts: Do biophysical properties determine biologic outcome? FEBS Lett. 531, 47–53 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Holopainen J. M., Subramanian M., Kinnunen P. K., Sphingomyelinase induces lipid microdomain formation in a fluid phosphatidylcholine/sphingomyelin membrane. Biochemistry 37, 17562–17570 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Padmanabhan R., Taneyhill L. A., Cadherin-6B undergoes macropinocytosis and clathrin-mediated endocytosis during cranial neural crest cell EMT. J. Cell Sci. 128, 1773–1786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandán Y. R., et al. , The inhibition of sphingomyelin synthase 1 activity induces collecting duct cells to lose their epithelial phenotype. Biochim. Biophys. Acta Mol. Cell Res. 1865, 309–322 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Canals D., et al. , Ceramide launches an acute anti-adhesion pro-migration cell signaling program in response to chemotherapy. FASEB J. 34, 7610–7630 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hannun Y. A., Obeid L. M., Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 175–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gustafson C. M., Roffers-Agarwal J., Gammill L. S., Chick cranial neural crest cells release extracellular vesicles that are critical for their migration. J. Cell Sci. 135, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sung B. H., Parent C. A., Weaver A. M., Extracellular vesicles: Critical players during cell migration. Dev. Cell 56, 1861–1874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Macdonald J. L., Pike L. J., A simplified method for the preparation of detergent-free lipid rafts. J. Lipid Res. 46, 1061–1067 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Abdel Shakor A. B., Atia M. M., Kwiatkowska K., Sobota A., Cell surface ceramide controls translocation of transferrin receptor to clathrin-coated pits. Cell. Signal. 24, 677–684 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Kim I., et al. , Clathrin and AP2 are required for PtdIns(4,5)P2-mediated formation of LRP6 signalosomes. J. Cell Biol. 200, 419–428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 82.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 84.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolde R., pheatmap: Pretty Heatmaps R package version 1.0.12 (2015).
- 86.Hamburger V., Hamilton H. L., A series of normal stages in the development of the chick embryo. J. Morphol. 88, 49–92 (1951). [PubMed] [Google Scholar]
- 87.Williams R. M., Sauka-Spengler T., Ex ovo electroporation of early chicken embryos. STAR Protoc. 2, 100424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Betancur P., Bronner-Fraser M., Sauka-Spengler T., Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. U.S.A. 107, 3570–3575 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Megason S. G., McMahon A. P., A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129, 2087–2098 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Gandhi S., Piacentino M. L., Vieceli F. M., Bronner M. E., Optimization of CRISPR/Cas9 genome editing for loss-of-function in the early chick embryo. Dev. Biol. 432, 86–97 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choi H. M. T., et al. , Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rhee J. M., et al. , In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis 44, 202–218 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teruel M. N., Blanpied T. A., Shen K., Augustine G. J., Meyer T., A versatile microporation technique for the transfection of cultured CNS neurons. J. Neurosci. Methods 93, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- 94.MacDonald B. T., Semenov M. V., Huang H., He X., Dissecting molecular differences between Wnt coreceptors LRP5 and LRP6. PLoS One 6, e23537 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schiffmacher A. T., Adomako-Ankomah A., Xie V., Taneyhill L. A., Cadherin-6B proteolytic N-terminal fragments promote chick cranial neural crest cell delamination by regulating extracellular matrix degradation. Dev. Biol. 444, S237–S251 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gandhi S., et al. , Bimodal function of chromatin remodeler Hmga1 in neural crest induction and Wnt-dependent emigration. Elife 9, 1–62 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Faul F., Erdfelder E., Lang A.-G., Buchner A., G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the main text and/or supporting information. Source data can also be found with data analysis codes on GitHub at https://github.com/PiacentinoLab/2022_PNAS_nSMase2_Endocytosis. The Smpd3 mRNA sequence has been submitted to GenBank (Accession #MW142015).