Abstract
Three-dimensional structures are now known within many protein families and it is quite likely, in searching a sequence database, that one will encounter a homolog with known structure. The goal of Entrez’s 3D-structure database is to make this information, and the functional annotation it can provide, easily accessible to molecular biologists. To this end Entrez’s search engine provides three powerful features. (i) Sequence and structure neighbors; one may select all sequences similar to one of interest, for example, and link to any known 3D structures. (ii) Links between databases; one may search by term matching in MEDLINE, for example, and link to 3D structures reported in these articles. (iii) Sequence and structure visualization; identifying a homolog with known structure, one may view molecular-graphic and alignment displays, to infer approximate 3D structure. In this article we focus on two features of Entrez’s Molecular Modeling Database (MMDB) not described previously: links from individual biopolymer chains within 3D structures to a systematic taxonomy of organisms represented in molecular databases, and links from individual chains (and compact 3D domains within them) to structure neighbors, other chains (and 3D domains) with similar 3D structure. MMDB may be accessed at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Structure.
MMDB CONTENTS
Data sources
Experimental 3D structure data for Entrez (1) are retrieved from the RCSB Protein Data Bank (PDB) (2). Theoretical models from PDB are omitted. Agreement of atomic coordinate and chemical-sequence data is checked and sequence data are automatically modified, if necessary, to achieve exact agreement with coordinates. Data are mapped into an easily-parsed form encoded in the ASN.1 language (3). This validation and encoding allows Entrez’s molecular-graphics viewer, Cn3D (4), to efficiently support integrated sequence, structure and alignment displays. Author-annotated features provided by PDB are fully recorded in MMDB (5). Uniformly defined secondary-structure and 3D-domain features are added, to support structure neighbor calculations. Coordinate subsets representing backbone-only and single-conformer models are also added, to support Cn3D visualization and structure neighbor calculations. MMDB currently contains ∼15 000 structure entries, corresponding to ∼35 000 chains and ∼50 000 3D domains.
Links, neighbors and visualization
Sequences derived from MMDB entries are entered into Entrez’s protein and nucleic acid sequence databases, preserving a link to the corresponding 3D structure. Links to the MEDLINE scientific literature database are generated by processing citation data within MMDB. These links allow Entrez to provide access to publications describing the original structure determination. Sequence neighbors of MMDB-derived sequences are identified automatically using the BLAST algorithm (6). Sequence-neighbor relationships are reciprocal, and MMDB-derived sequences also appear as neighbors of other sequences in Entrez. Structure neighbors are identified using the VAST algorithm, a structure–structure alignment method (7). While VAST uses a conservative significance threshold, the structural similarities it detects often represent remote relationships not detectable by sequence comparison. Some structural similarities may represent evolutionary convergence, however, and the Cn3D viewer provides 3D superpositions, so that users may examine and interpret structural similarities for themselves. Cn3D is available at http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml.
Taxonomy links
Links to NCBI’s taxonomy database (1) are generated by semi-automatic processing of ‘source’ and other descriptive text provided by PDB. Since PDB staff refer to the taxonomy database when creating ‘source’ descriptions (2), links normally follow the genus and species information provided. In some cases ‘source’ descriptions may omit genus and species, and refer only to the manner in which a sample was obtained or prepared. In these cases other descriptive information is examined manually, and sequence-similarity searches are sometimes conducted in an effort to determine an appropriate taxonomy link. The ‘source’ string for PDB entry 1FU2, for example, is ‘synthetic construct’. The primary citation provided by PDB indicates that the sample is human insulin, however, and a link to taxon Homo sapiens was therefore assigned within MMDB. We note that taxonomy is assigned at the level of individual chains and also recorded in MMDB-derived sequence records. Taxonomy assignments have been made for all MMDB entries, and new organisms represented only in MMDB have been added to the taxonomy database, in consultation with NCBI taxonomists.
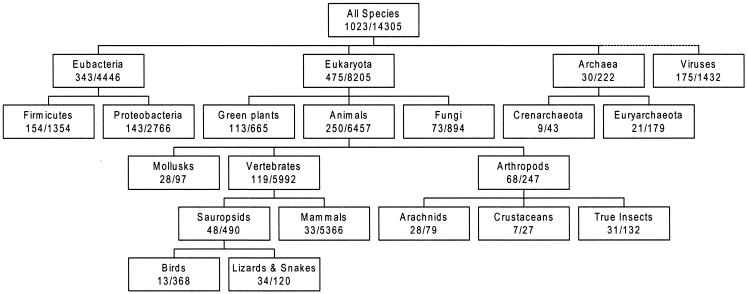
We emphasize that taxonomy links in MMDB provide more than a means to search for structures from a particular genus or species. In each case a complete lineage, or location in the ‘tree of life’, has been recorded via the link to the NCBI taxonomy database. This means that one can search in Entrez for all 3D structures from mammals, for example, or from other taxonomic groups above the level of genus and species. This type of search is not possible using PDB files, which do not contain lineage information. To illustrate this capability, we survey in Figure 1 how some major taxonomic groups are populated by the 3D-structure database. The figure shows, for selected taxa, the numbers of species for which one or more structures are known and the total number of structures by taxon. We also list in Table 1 the 10 species for which the most structures have been determined. Further information on MMDB taxonomy assignments is available at http://www.ncbi.nlm.nih.gov/Structure/PDBEAST/pdbeast.shtml. A browser for the NCBI taxonomy database, useful for identifying the scientific names of different taxa, is accessible at http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/index.cgi.
Figure 1.
Numbers of different species represented in MMDB and numbers of known structures, for selected taxa from the ‘tree of life’. The hierarchy of taxa is taken from the NCBI taxonomy database, with each box corresponding to a particular node in the hierarchy, as indicated by its scientific or common name. Also shown is the number of individual species within each taxon from which one or more 3D structures are known, followed by the number of structures in MMDB derived from these species. Figure 1 and Table 1 are based on structures available in MMDB as of July 2001. The total number of structures for ‘all species’ omits structures for which no taxonomy link is assigned, for example short oligonucleotides.
Table 1. Top 10 organisms in MMDBa.
| Organism | Chain count |
|---|---|
| Homo sapiens | 537 |
| Escherichia coli | 384 |
| Saccharomyces cerevisiae | 153 |
| Mus musculus | 127 |
| Bos taurus | 126 |
| Rattus norvegicus | 117 |
| Thermus thermophilus | 90 |
| Sus scrofa | 57 |
| Gallus gallus | 55 |
| Bacillus subtilis | 48 |
aGroups of sequence-similar chains (with BLAST e-value < 10–7) are counted only once, so as to illustrate the number of unrelated protein structures known from each species. The total number of chains with known 3D structure is larger, since structures of many sequence-similar chains have been determined more than once, with and without bound ligands, for example.
Related 3D domains
Calculations for MMDB’s structure neighbor database and visualization of VAST superpositions have always employed comparisons at the level of individual chains and compact 3D domains. In earlier versions of Entrez’s search engine, however, these were recorded only as ‘related structures’, a list containing the structure neighbors for all chains and 3D domains of a given structure. The current Entrez version links each structure to its ‘3D domains’, a list of all polypeptide chains and any compact domains within them. Each ‘3D domain’ is in turn linked to ‘related 3D domains’, that is, the structure neighbors of that particular chain or domain. In earlier versions, for example, structure neighbors of an antibody–lysozyme complex included both antibody and lysozyme structures. In the current version the structure neighbors of the ‘3D domain’ representing lysozyme list other lysozyme chains, while those of the antibody chains (and their compact domains) list other immunoglobulin family structures.
3D domains within individual polypeptide chains in MMDB are identified automatically, using an algorithm that searches for one or more breakpoints, falling between major secondary structure elements, such that the ratio of intra- to inter-domain contacts falls above a set threshold (8). This method is very similar to others proposed for identification of autonomously folding domains from 3D structure data, such as that of Holm and Sander (9). We emphasize that 3D domains identified in this way provide means to increase the sensitivity of structure neighbor calculations (7), and to present 3D superpositions based on compact domains as well as complete polypeptide chains. They are not intended to represent domains identified by comparative sequence and structure analysis, as modules that recur in related proteins, though there is often good agreement between domain boundaries identified by these methods (10). NCBI’s Conserved Domain Database (CDD) provides information on domains identified by comparative analysis (11) and is available at http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. We note that structure neighbors for domains with boundaries chosen by the user are available through VAST-Search at http://www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html.
USING MMDB
A simple query
MMDB is an integrated part of Entrez and can be accessed by querying Entrez’s ‘3D structure’ database for particular terms or keywords. This allows one to identify structures based on protein names, author names, publication dates, species names or other terms. A query such as this will produce a list of MMDB entries, and one may browse this list, following links to other databases, for example those to MEDLINE abstracts. At the time of writing, MMDB’s servers receive approximately 25 000 3D structure queries per day.
As an example, we consider a search with the terms ‘aminocyclopropane synthase’. This identifies several 3D structures available for this enzyme, including the structure with PDB identifier ‘1B8G’ (12), the protein from Malus x domestica, the apple tree. Following the link to ‘3D domains’, one sees that structure neighbors are available for eight different substructures, the complete chains A and B plus three compact domains identified within each. Following the link to ‘related 3D domains’ for domain ‘1B8G A 3’ (the third domain in chain A, as numbered from the N-terminus of the chain), one sees that 3D superpositions are available for over 1000 structure neighbors of this domain.
A more advanced query
Entrez provides a query refinement feature that allows one to combine the results of simple queries involving term-match hits, links or neighbors. To continue with the example above, suppose one wishes to identify some of the most evolutionarily distant structure neighbors of domain ‘1BG8 A 3’, as a means to identify conserved residues that may be associated with its binding and/or catalytic function. One option is to examine the tabular listing of VAST superposition statistics, available by following the link from the domain identifier ‘1BG8 A 3’, to choose structure neighbors with a low percentage of identical residues in the structural alignment. Another powerful method, however, is to choose structure neighbors from phylogenetically distant organisms. For this search it is necessary to combine results of an MMDB search by taxonomy with structure neighboring results.
As may be seen by following the taxonomy links from domain ‘1BG8 A 3’, this protein is derived from an organism (apple tree) in the superkingdom Eukaryota. The most distantly related organisms will be those from the two other superkingdom taxa, Eubacteria and Archaea. Searching Entrez’s ‘3D Domain’ database for ‘Archaea’ (with ‘limits’ set to ‘organism’), one finds that there are approximately 1000 3D domain structures known for this taxon. To select those that are also structure neighbors of 3D domain ‘1BG8 A 3’, one uses Entrez’s ‘history’ window to request the Boolean ‘AND’ of the 3D domains identified by each simple query: <1> AND <2>, where <1> and <2> represent query numbers as recorded in Entrez’s history list. Performing this search, one finds approximately 20 structures which are both structure neighbors of ‘1BG8 A 3’ and derived from Archaea, among them domain ‘1DJU A 3’, a domain from an aromatic aminotransferase from Pyrococcus horikoshii (13). Proceeding similarly for ‘Eubacteria’, one finds that several hundred structure neighbors of ‘1BG8 A 3’ derive from this taxon, including ‘1AMQ 2’, an aspartate aminotransferase from Escherichia coli (14).
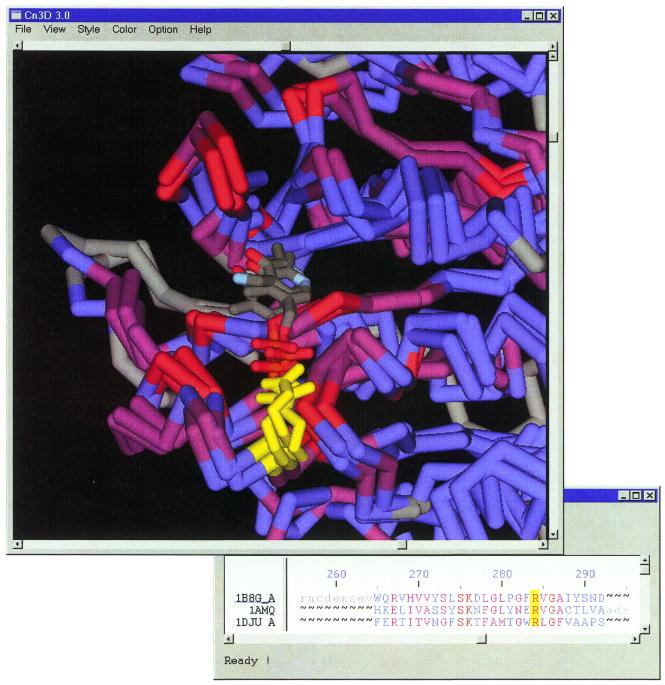
Visualization of structure neighbors is available from the ‘View’ link provided with tabular listings of VAST superposition statistics. Choosing the structure neighbors ‘1DJU A 3’ and ‘1AMQ 2’ from among the other neighbors of ‘1BG8 A 3’, and pressing the ‘View’ button, one may launch a Cn3D display as shown in Figure 2. Setting Cn3D to color aligned residues by variability, one can immediately see that conserved residues are concentrated in a single region of these domains. Furthermore, since each structure contains a bound pyridoxal phosphate cofactor (or related compound), one can verify that these conserved residues line the binding pocket, and are presumably necessary for cofactor binding and aminotransferase activity. We note that tabular listings of VAST superposition statistics provide several controls for sorting and subset selection, as an aid to browsing. To reproduce the superposition in Figure 2 it is helpful to select subset ‘all of MMDB’ and sort by ‘aligned residues’. This allows one to identify structure neighbors having extensive similarity (many aligned residues) and (in this example) with bound cofactors.
Figure 2.
Structural alignment of aminotransferase domains from the three superkingdoms of the NCBI taxonomy, Archaea, Eukaryota and Eubacteria. Conserved residues are shown in red, partially conserved residues in magenta, and non-conserved residues in blue. Grey regions are structurally dissimilar, and not structurally aligned. The figure was constructed using Cn3D. A conserved arginine residue in the pyridoxal phosphate-binding pocket is highlighted in yellow, to indicate the corresponding residue in Cn3D’s structure and sequence windows.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the NIH Intramural Research Program for support. We thank Scott Federhen, Detlef Leipe and other members of the NCBI taxonomy team for assistance with taxonomy assignments. Comments, suggestions and questions are welcome and should be addressed to info@ncbi.nlm.nih.gov.
REFERENCES
- 1.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D. Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westbrook J., Feng,Z., Jain,S., Bhat,T.N., Thanki,N., Ravichandran,V., Gilliland,G.L., Bluhm,W., Weissig,H., Greer,D.S. et al. (2002) The Protein Data Bank: unifying the archive. Nucleic Acids Res., 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohkawa H., Ostell,J. and Bryant,S. (1995) MMDB: an ASN.1 specification for macromolecular structure. Proc. Int. Conf. Intell. Syst. Mol. Biol., 3, 259–267. [PubMed] [Google Scholar]
- 4.Wang Y., Geer,L.Y., Chappey,C., Kans,J.A. and Bryant,S.H. (2000) Cn3D: sequence and structure views for Entrez. Trends Biochem. Sci., 25, 300–302. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Addess,K.J., Geer,L., Madej,T., Marchler-Bauer,A., Zimmerman,D. and Bryant,S.H. (2000) MMDB: 3D structure data in Entrez. Nucleic Acids Res., 28, 243–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 8.Madej T., Gibrat,J.F. and Bryant,S.H. (1995) Threading a database of protein cores. Proteins, 23, 356–369. [DOI] [PubMed] [Google Scholar]
- 9.Holm L. and Sander,C. (1994) Parser for protein folding units. Proteins, 19, 256–268. [DOI] [PubMed] [Google Scholar]
- 10.Matsuo Y. and Bryant,S.H. (1999) Identification of homologous core structures. Proteins, 35, 70–79. [PubMed] [Google Scholar]
- 11.Marchler-Bauer A., Panchenko,A.R., Shoemaker,B.A., Thiessen,P.A., Geer,L.Y. and Bryant,S.H. (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res., 30, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capitani G., Hohenester,E., Feng,L., Storici,P., Kirsch,J.F. and Jansonius,J.N. (1999) Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J. Mol. Biol., 294, 745–756. [DOI] [PubMed] [Google Scholar]
- 13.Matsui I., Matsui,E., Sakai,Y., Kikuchi,H., Kawarabayasi,Y., Ura,H., Kawaguchi,S., Kuramitsu,S. and Harata,K. (2000) The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii. J. Biol. Chem., 275, 4871–4879. [DOI] [PubMed] [Google Scholar]
- 14.Miyahara I., Hirotsu,K., Hayashi,H. and Kagamiyama,H. (1994) X-ray crystallographic study of pyridoxamine 5′-phosphate-type aspartate aminotransferases from Escherichia coli in three forms. J. Biochem. (Tokyo), 116, 1001–1012. [DOI] [PubMed] [Google Scholar]