Abstract
Microplastics pollution is major threat to ecosystems and is impacting abiotic and biotic components. Microplastics are diverse and highly complex contaminants that transport other contaminants and microbes. Current methods to remove microplastics include biodegradation, incineration, landfilling, and recycling. Here we review microplastics with focus on sources, toxicity, and biodegradation. We discuss the role of algae, fungi, bacteria in the biodegradation, and we present biotechnological methods to enhance degradation, e.g., gene editing tools and bioinformatics.
Keywords: Microplastics, Incineration, Microplastic degrading microorganisms, Biodegradation, Synthetic biology, Biotechnological interventions, CRISPR
Introduction
Plastic pollution is considered to be one of the most significant threats to global ecosystems and is known to impact both the abiotic and biotic components (Ogonowski et al. 2018; Everaert et al. 2020; Lusher et al. 2021; Liu et al. 2022; Su et al. 2022; Federici et al. 2022). In 2018, global plastic production increased to more than 360 million tons, and it is expected to triple by the year 2050 (Gumel et al. 2013; Plastics Europe 2020). According to a survey made by Plastics Europe (the Association of Plastics Manufacturers in Europe) and the European Association of Plastics Recycling and Recovery Organisations, Asia is the largest producer and consumer of plastic goods, with China contributing the lion’s share (32%) to this “white pollution”, while the rest of Asia produces nearly 19%.
Whereas, Europe, Canada, Mexico, and the USA produce fewer plastics than Asia (Tiwari et al. 2020; https://plasticseurope.org/knowledge-hub/plastics-the-facts-2021/).
Moreover, the coronavirus disease 2019 pandemic has increased the use of one-time useable plastic wares, gloves, masks, tissues, and other personal protective equipment which along with the generation of municipal waste has exacerbated the plastic pollution crisis (De-la-Torre and Aragaw 2021; Morgana et al. 2021; Patrício Silva et al. 2021; Yang et al. 2022). Most single-use masks and personal protective equipment are made up of various polymeric substances such as polypropylene, polyurethane, polyacrylonitrile, polyethylene, and polystyrene. Also, the imposition of lockdown has led to the surge in the use of different types of plastic largely composed of high-density polyethylene, low-density polyethylene, polypropylene, and polyethylene terephthalate. The incorrect disposal and poor waste management of plastic items have led to their ubiquitous presence in all environments globally, and due to their persistent nature and low biodegradability, they will remain in these environments for prolonged periods of time (Andrady 2017; Aragaw 2020; Patrício Silva 2021).
It has been reported that around 4.8–12.8 million metric tons of plastic debris are disposed of in the ocean without a proper management strategy (Jambeck et al. 2015). This disposal of plastic debris into the ocean has led to a series of impacts on marine life (Galloway et al. 2017; Mendoza et al. 2018; Peng et al. 2020) and human health (Keswani et al. 2016). They result in problems like ingestion, entanglement, and suffocation to different marine species leading to reduced life quality, and impairment of feeding and reproductive ability (Staffieri et al. 2019; Wilcox et al. 2018). In addition, most plastics are positively buoyant and can be transported over a long distance, acting as carriers of non-native and invasive species (De-la-Torre et al. 2021).
Wasted plastics, exposed to ultraviolet (UV) irradiation and other environmental degradation processes, can degrade and fragment into smaller pieces: large microplastics (1 mm–5 mm), microplastics (1 µm–1 mm) and nanoplastics (1 nm–1 µm) (Waller et al. 2017; De-la-Torre 2020; Atugoda et al. 2022). However, there is no scientific consensus on the definition of microplastics. Here we define microplastic as plastics that are ≤ 5000 µm (or ≤ 5 mm) in diameter (ISO/TR 21960:2020), with "large microplastic" referring to the larger portion (> 1000 µm or > 1 mm) (Frias and Nash 2019; GESAMP 2019; Hartmann et al. 2019; Hale et al. 2020; Koelmans et al. 2020). Microplastics are the major contributors to plastic pollution in the marine ecosystem, freshwater ecosystem, soil ecosystem, and agroecosystem (Nizzetto et al. 2016; Mendoza et al. 2019; Wang et al. 2019b; Wong et al. 2020; Chia et al. 2021; Razeghi et al. 2021) and are known to be globally ubiquitous (Jambeck et al. 2015). They also contaminate soil through sewage sludge and wastewater which are widely used as fertilizers. Moreover, plastics are also used in agriculture mainly in a greenhouse, low tunnels and mulching and also as a coating for fertilizers, hormones, pesticides, and packaging material (Nizzetto et al. 2016).
Recently, several studies have investigated the distribution, uptake, fate, behavior, effects, and removal strategies of microplastics (Bahtt et al. 2021; Wong et al. 2020; Anik et al. 2021). Nevertheless, the effectiveness of the methods developed for microplastic remediation still remains unclear. Research in microplastic degradation has progressed focusing on biological and non-biological approaches. Microplastic treatments enabled by the action of microorganisms such as algae, fungi, and bacteria are considered attracting tools for cost-effective and eco-friendly degradation approaches. While research papers and reviews have recently been published on the microorganism-mediated degradation and remediation strategies (Chen et al. 2022; Bahtt et al. 2021; Qin et al. 2021; Kotova et al. 2021; Cholewinski et al. 2022), only few articles have addressed plastic degradation focusing on the use of modern biotechnological methods in the enhancement of microplastic degradation and there remains a lack of knowledge with respect to biotechnological interventions for microplastic removal (Danso et al. 2019; Patrício Silva et al. 2021). Therefore, it is crucial to summarize and analyze the current state of knowledge to determine microplastic degradation by microorganism, as well as to promote a better understanding of how modern biotechnological methods can be enabled to manage and degrade microplastics.
This review provides some background information on the impact of microplastic sources, their effects on marine life including microalgae, and their potential impact on humans. Then it categorizes the different types of microorganisms and enzymes associated with the degradation of microplastics. Finally, different biotechnological methods to increase the efficiency of bacterial cell degradation of microplastics and their possible application in field studies are discussed at length.
Sources of microplastics
Microplastics derive from two different origins. The primary microplastics are generated from cosmetics, household products, drug delivery systems (Patel et al. 2009), and polymeric raw materials (pellets, flakes, powders) composed of polyethylene, polystyrene, polyvinyl chloride, polyamide nylon 6 and polypropylene, among others. Personal care products such as toothpaste, scrubs, cleaning materials and cosmetics are known to contain irregularly shaped microplastics of 0.5 to < 0.1 mm in diameter, which are mainly marketed as “micro-beads” or “micro-exfoliates” and contribute to primary microplastics (Fendall and Sewell 2009). However, the existing wastewater treatment plants have shown that tertiary treatment of water is not a source of microplastic pollution, as these pollutants are effectively removed by the skimming and settling treatment processes (Carr et al. 2016).
The secondary microplastics are generated due to extensive fragmentation of large plastic items or particles in presence of environmental factors such as high temperature and exposure to UV radiation, stress, reactive ozone, oxidation, and atmospheric pressure (Tiwari et al. 2020; John et al. 2021). Polymeric materials can withstand oxidative-thermal degradation only when antioxidants and stabilizers are added. Physical abrasion also generates secondary microplastics. Moreover, biological agents like bacteria, fungi and algae are known to produce a plethora of enzymes which play a crucial role in microplastic degradation (Chia et al. 2020; John et al. 2021; Othman et al. 2021; Chen et al. 2022; Manzi et al. 2022; Miri et al. 2022; Zhu et al. 2022).
Secondary microplastics are mostly generated from a large plastic object made from the same polymers improperly disposed of in land and water systems, which undergoes physical abrasion leading to the weakening of the chemical bonds and subsequent oxidative-thermal degradation (Gerritse et al. 2020). Other secondary microplastic sources include the disintegration of synthetic fibers during the washing of clothes and commercial activities like thermal cutting of polystyrene. The increased use of single-use plastics has contributed to the overproduction of polyethylene, polypropylene, and polyethylene terephthalate products. Moreover, a wide range of electronic, automobile, textile, and paint industries also discharge microplastic products into the water bodies and river catchment areas which can lead to microplastic pollution (Kay et al. 2018; Wang et al. 2019b; Chia et al. 2021).
Impact of microplastics on marine organisms and humans
Microplastics could have a huge impact on the aquatic flora and fauna (Sathicq et al. 2021) (Fig. 1a) as they act as a vector for the transport of absorbed heavy metals, bacterial fish pathogens, multidrug resistant E. coli, persistent organic pollutants etc. (Enders et al. 2015; Viršek et al. 2017; Caruso, 2019; Song et al. 2020), and the possible leaching of chemical components added during their manufacturing process (Groh et al. 2019; Bacha et al. 2023). In addition, microplastics provide a novel habitat for the growth of microbial biofilms containing algae, bacteria and fungi and can potentially spread microbial pathogens and antimicrobial resistance (Zettler et al. 2013; Wu et al. 2019; Guo et al. 2020; Yuan et al. 2020).
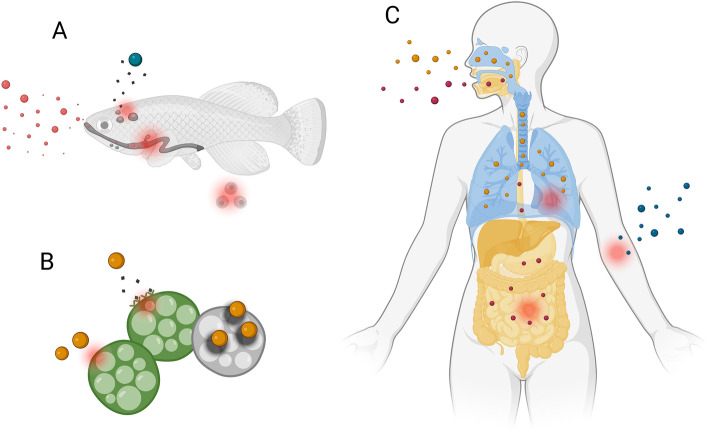
Fig. 1.
Microplastics impact on living organisms. A Microplastics can enter aquatic fauna food chain, causing intestinal blockage and alteration in nutrients absorption, endocrine disruption, immunological and neurological effects, and loss of reproductive functionalities. B Microplastics and toxic leachate can damage microalgal cell walls, cause metabolic dysfunctions, and cause impairment of photosynthesis due to shading effects. C The three main routes of human microplastics exposures are identified as ingestion, inhalation, and dermal contact, triggering inflammatory and immune reactions. Created with BioRender.com
As most microplastics have a size range similar to the foods that are normally consumed by the zooplankton, they can accidentally enter the food chain (Gregory 2009). This selection of microplastics instead of food particles can cause a loss of energy resources and also sublethal effects on the species reproductive pattern (Enders et al. 2015). Moreover, the ingested plastic can cause intestinal blockage, which subsequently reduces the absorption of nutrients and also causes a change in hormonal balance (Derraik 2002). The improper absorption of the nutrients may result in a decrease in the energy reserves and deficiency in food assimilation which in turn impacts growth and reproduction (Besseling et al. 2013; Wright et al. 2013a) and also decreases the ability of the organisms to survive in adverse environmental conditions (Bugoni et al. 2001).
Microplastic exposure can also cause other sublethal effects in marine organisms, including oxidative stress, altered gene expression, inflammation, and effects on the immune system and central nervous system (for review see Wright et al. 2013b; Vethaak and Martinez 2020). These adverse effects may be caused by residual monomers and additives release from plastic particles rather than by the particles themselves. Smaller microplastics below 10 µm were found to be more toxic to aquatic organisms than larger plastic particles.
The fishing and aquaculture industry is strongly impacted by microplastic pollution, and its viability and productivity are affected by the presence of plastic waste in water bodies (Rochman et al. 2015) and microplastic contaminated seafood products (Bråte et al. 2016; Smith et al. 2018; Curren et al. 2020; Li et al. 2021; Pan et al. 2022). The ubiquitous, persistent, and anticipated increase in microplastics pollution could in the long run also have a significant impact on marine biodiversity and ecological processes, such as primary producers at the basis of the food chain (Guzzetti et al. 2018; Vethaak and Martinez 2020).
The impact of microplastics on the growth and diversity of the microalgal population usually varied (Liu et al. 2019) (Fig. 1b). According to Sjollema et al. (2016), uncharged polystyrene particles negatively affected microalgae growth of Dunaliella tertiolecta at high concentrations (250 mg/l) and with decreasing particle size. The study made by Khoironi et al. (2019) reported that the growth of Spirulina sp. was severely impacted by the presence of high concentrations of microplastics, especially due to shading effects and reduced light intensity, with subsequent impairment of photosynthesis, and the damage of microalgal cell walls (Khoironi et al. 2019). Li et al. (2018) reported that both polyethylene and polypropylene gradually degrade and generate microsized plastics and release potentially toxic additives including plasticizers, polychlorinated biphenyls, dichlorodiphenyltrichloroethane, and heavy metals such as cadmium, chromium, bromium, copper, and titanium which cause cell membrane damage and growth inhibition. Capolupo et al. (2020) reported that in the case of Raphidocelis subcapitata and Skeletonema costatum the cell growth is inhibited due to the leaching of additives.
Raphidocelis subcapitata, on the other hand, showed a higher growth rate in presence of plastic microbeads (Canniff and Hoang 2018), and similar enhancement of cell growth and photosynthetic activity was evident in Dunaliella salina in presence of larger microplastics (Chae et al. 2019). This enhancement of growth seemed related to the trace concentration of additive chemicals such as stabilizers, phthalates, and endocrine disruptors, which are leached out of microplastics (Chae et al. 2019).
The potential effect of microplastics on humans is far from understood and requires further research (Vethaak and Legler 2021). Microplastics have been reported in different foods such as mussels, commercial fish, and table salt (Li et al. 2018) and three different pathways of exposure to microplastics are identified, including ingestion of food containing microplastics, inhalation of microplastics in the air, and dermal contact with these particles (Revel et al. 2018) (Fig. 1c). According to a study made by Cox et al. (2019), it was estimated that each person usually intakes around 39,000–52,000 microplastics each year. These microplastics cause intestinal blockage and result in the inflammatory response and changes in gut microbe composition and metabolism.
As stated earlier, microplastics can also be inhaled and the outdoor microplastic concentration is between 0.3 and 1.5 particles per m3, whereas the indoor concentration is 0.4–56.5 particles per m3 (Dris et al. 2017). The deposition of microplastics is largely dependent on the size and density of the particles. The less-dense smaller particles tend to deposit deepest in the lungs, causing the release of chemotactic factors and resulting in chronic inflammation (Oliveira et al. 2020a; b). The presence of microplastics in human lung tissue and human blood was very recently confirmed (Jenner et al. 2022; Leslie et al. 2022).
It was also speculated that nanoparticles can transverse the dermal barrier (Revel et al. 2018) causing low inflammatory reactions and fibrous encapsulation (Oliveira et al. 2020a; b). Once in contact with mucous membranes or absorbed by the body, microplastics generate oxidative stress and cytotoxicity, mainly due to their persistent nature in the body and the leaching of toxic additives, which may result in inflammation, immune reactions, neurological damage, metabolic disruptions, deoxyribonucleic acid (DNA) damage, and even cancer (Wright and Kelly 2017; Revel et al. 2018; Rahman et al. 2021; Vethaak and Legler 2021; Gruber et al. 2022).
Overall, microplastics and nanoplastics derived from several sources, in particular from the environmental degradation of waste plastics, can exert toxic effects on organisms in all trophic levels. They can enter aquatic fauna food chain, causing intestinal blockage and alteration in nutrients adsorption, endocrine disruption, immunological and neurological effects, and loss of reproductive functionalities. Micro- and nanoplastics and toxic leachate can damage microalgal cell walls and metabolic dysfunctions, and impairment of photosynthesis due to shading effects. Micro- and nanoplastics can enter the human body through ingestion, inhalation, and dermal contact, triggering inflammatory, and immune reactions.
Processes of microplastic degradation
Most of the conventional methods discussed for the reuse of microplastic degradation include a primary method where the plastic scrap is re-introduced in the heating cycle of the processing unit, followed by the conversion of waste to new plastic products by blending it with a virgin polymer which can considerably reduce the cost of production. Sometimes, plastic wastes are chemically or thermochemically altered to be recycled in the industrial loop. However, in most cases due to poor management strategies, these microplastic particles are not disposed of properly or segregated properly. Most of them get mixed up with the organic components in a landfill, which is used for composting or anaerobic digestion, leading to excessive pollution and to the production of toxic compounds such as dioxins, phthalates, tetrabromobisphenol A, polybrominated diphenyl ethers, and toxic metals such as cadmium and lead (Verma et al. 2016).
Currently, several physical, as well as chemical methods are popularly used for disposing of microplastic particles including incineration, landfilling, and recycling. Chemical recycling processes such as pyrolysis are extremely popular at the commercial level (Thiounn et al. 2020). In the slow pyrolysis methods, the plastic waste is converted to a mixture of char and tarry products which are treated at three different temperatures including 300, 425 and 550 °C (Dussud et al. 2018). There exist several pieces of research which focus on the pyrolysis of polypropylene, polystyrene, and polypropylene from where heat energy can be recovered. The contaminated, mixed, or degraded residues which are not suitable for recycling can be used as feedstocks for waste-to-energy strategies such as pyrolysis (an endothermic cracking process without oxidation), and incineration (oxidation of plastics) (Prata et al. 2020).
Degradation of microplastics can occur by physical, chemical, and biological methods and the biological degradation process is associated with a plethora of enzymes (Padervand et al. 2020; Bacha et al. 2021; Fig. 2). The basic process includes steps like the degradation of polymers to smaller particles, followed by the degradation of the smaller polymers to oligomers, dimers, and monomers. This degradation is followed by mineralization steps which are aided by microbes (see Fig. 3 for a representative scheme of polyethylene mineralization).
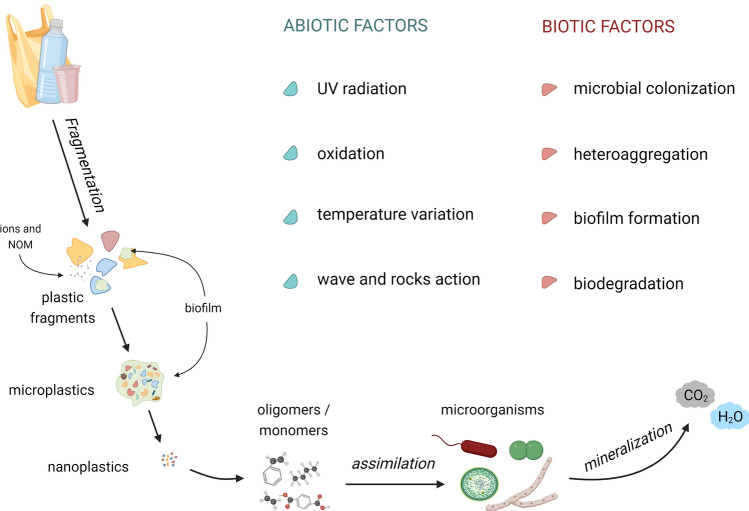
Fig. 2.
Processes of microplastic degradation through abiotic and biotic combined factors. Microplastics derive from the fragmentation of plastic debris and can further degrade into smaller plastic particles at the nanoscale (nanoplastics). Due to abiotic factors and extracellular enzymes, nanoplastics are degraded into oligomers and monomers, then internalized by microorganisms and used as carbon source, resulting in the complete mineralization of plastic. Created with BioRender.com
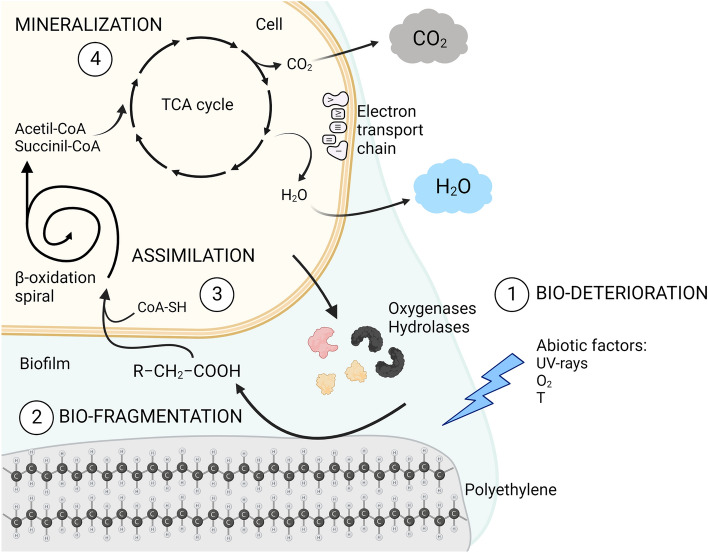
Fig. 3.
Mineralization process for polyethylene. Due to a combined effect of abiotic factors and extracellular enzymes, plastic undergoes bio-deterioration and bio-fragmentation processes, resulting in the release of oligomers and monomers. Thanks to specific cell transport mechanisms, monomers are internalized by microorganisms and enter the catabolic pathways as carbon source. The final products of cells’ aerobic metabolism, which results in the mineralization of plastic, are carbon dioxide and water. Created with BioRender.com
On complete mineralization, carbon dioxide is evolved along with the formation of several intermediate compounds which are used as a source of energy to promote the growth of microbes. The different extracellular enzymes that play a pivotal role in microplastics degradation include esterases, lipases, lignins peroxides, laccases, and manganese peroxides, which increase the hydrophilicity of microplastics and convert them to carbonyl or alcohol residues (Taniguchi et al. 2019). Hydrolases enzymes, such as lipases, esterase, and cutinase, act on plastic surfaces and degrade microplastics by enhancing the chain cleavage reactions. These enzymes fail to diffuse into the polymer, but they act on the surface resulting in the formation of cracks. The monomers generated are assimilated into the cytoplasm of microbes and finally enter into different metabolic pathways.
Although extensive research has already been done using extracellular enzymes in microplastics biodegradation, very little information is still now available on the role of intracellular enzymes in the degradation of microplastics; moreover, the pathways involved in the uptake of monomers are still not clear. Normally, after the fragmentation of the microplastics, the metabolic intermediates with carbonyl and hydroxyl groups are metabolized within the cell using the tricarboxylic acid cycle and β -oxidation pathway (Taniguchi et al. 2019). This process is followed by the complete mineralization of plastic debris into H2O, CO2, N2, and CH4 (Zettler et al. 2013). Researchers have made a thorough research on the process of surface colonization of the microplastics by degrading consortium forming a biofilm on the particles. The attachment process of the microbes occurs through several mechanisms including biofouling, and degradation of plasticizers followed by the attack on the backbone of the polymer which is subsequently associated with hydration and penetration of microbes in the polymer structure.
Moreover, for efficient biodegradation, several factors are required which include the availability of potential microbial degrading organisms which possess suitable enzymes and metabolic pathways and other environmental factors such as temperature, pH, salinity, and moisture content (Raddadi and Fava 2019; Syranidou et al. 2019; Matjašič et al. 2021; Miri et al. 2022; Lin et al. 2022). The biodegradation of microplastics is also influenced by the surface and the structure of the polymer, amorphous and crystalline regions, crystal size, and lamellar thickness of polymers. Shabbir et al. reported polyhydroxyalkanoates depolymerase enzymes to hydrolyze the chains structures in the amorphous state on the surface of fragmentation films followed by erosion of chains in the crystalline state (Shabbir et al. 2020).
In brief, microbial microplastic degradation involves abiotic and biotic combined factors. Microplastics derive from the fragmentation of plastic debris and can further degrade into smaller plastic particles at the nanoscale (nanoplastics). Due to a combined effect of abiotic factors and extracellular enzymes, plastic undergoes bio-deterioration and bio-fragmentation processes, resulting in the release of oligomers and monomers. Thanks to specific cell transport mechanisms, monomers are internalized by microorganisms and enter the catabolic pathways as carbon source. The final products of cells’ aerobic metabolism, which results in the mineralization of plastic, are carbon dioxide and water.
Techniques to monitor microplastic biodegradation
Different techniques have been applied to study microbial degradation of microplastics, which includes weight loss measurement due to leaching, CO2 production due to degradation of low molecular weight polymers and loss of additives which affect the strength of microplastics (Baldera-Moreno et al. 2022). To get direct proof of the degradation process, morphological, chemical, thermal, and structural properties are investigated using various techniques/methods such as scanning electron microscopy, laser diffraction particle, differential scanning calorimetry, dynamic light scattering, X-ray diffraction, etc. (Huang et al. 2022). Chemical changes are usually tracked by vibrational spectroscopy techniques, such as Fourier transform infrared spectroscopy, nuclear magnetic resonance, mass spectrometry, and gas chromatography (Donelli et al. 2009; Chamas et al. 2020; Ivleva, 2021; La Nasa et al. 2021; Miao et al. 2020; Du et al. 2021).
The gravimetric weight loss method is another widely used method for the determination of the biodegradation of microplastics. However, this method should be used carefully as biodegradation of microplastics is an extremely slow process (Raddadi and Fava 2019), and depends on the incubation time and the assay conditions. Spectroscopy techniques can be used to determine the microplastics biodegradation efficiency of microbes, by monitoring the changes in the chemical functional groups of the polymer due to microbial activity (Singh and Sharma 2008). These changes may occur in hydrogen bonding, end-group modification, cross-linking and copolymer compositions. Fourier-transform infrared spectroscopy is considered one of the most efficient methods for the detection of chemical changes in the polymer and reference spectra of a wide variety of polymers are available in libraries for comparison (e.g., Celina et al. 1997).
Scanning electron microscopy allows the detection of microbial biofilm formation and surface degradation by monitoring the physical aspects of the polymer surface. The evaluation of polymer biodegradation can be done by checking the formation of cracks and holes in the polymer (Raddadi and Fava 2019).
Modification of microplastics tensile strength and elongation at break are the signals of microbial biodegradation. Indeed, microbial degradation leads to significant changes in mechanical properties and biochemical modification of polymers due to the formation of cross-linking bonds or film disintegration and shortening of the polymer chain (Nowak et al. 2011; Othman et al. 2021).
Role of algae in the degradation of microplastics
Microalgae and their enzymes and toxins can be effectively used in the biological breakdown of polymeric material (Moog et al. 2019; Chia et al. 2020; Manzi et al. 2022). The main advantage is that they do not require a rich carbon source for growth when compared to the bacterial system and are adapted to a wide variety of habitats where most of the microplastics occur (Yan et al. 2016). Microalgae are known to colonize the plastic surfaces in wastewater streams and this adhesion initiates plastic degradation by the production of ligninolytic and exopolysaccharide enzymes. Mostly these polymers serve as a carbon source and increase the cellular proteins and carbohydrates and increase the growth rate. Very recently, surface degradation or breakdown of low-density polyethylene sheet through algal colonization has been identified using scanning electron microscopy (Sanniyasi et al. 2021).
Algal biodegradation occurs mainly in different processes such as corrosion, hydrolysis, penetration, fouling, etc. (Chia et al. 2020). Both Oscillatoria subbrevis and Phormidium lucidum were also found to be able to colonize the surface of low-density polyethylene and degrade it without any pro-oxidative additives or pretreatment (Sarmah and Rout 2018). Bisphenol A, an additive with estrogenic activity commonly found in the polymers, was degraded by a combination of bacteria and algae including Chlorella fusca var. vacuolate, Chlamydomonas mexicana, Stephanodiscus hantzschii, and Chlorella vulgaris (Hirooka et al. 2005; Li et al. 2009; Ji et al. 2014).
In most cases, the degradation of microplastics is associated with the formation of biofilms on the surface of polymers. Several cyanobacterial strains, including the genus Microcystis, Rivularia, Pleurocapsa, Synechococcus, Prochlorothrix, Leptolyngbya Calothrix, and Scytonema, were also able to form biofilms on the microplastic polymers (Bryant et al. 2016; Debroas et al. 2017; Dussud et al. 2018; Muthukrishnan et al. 2019). Besides cyanobacterial species, diatoms are also present in the biofilms which help in photosynthesis (AmaralZettler et al. 2020).
With the recent advances in different biotechnological processes, several genetically modified microalgal cell factories can be created which are capable of producing and secreting enzymes required for plastic degradation (Shen et al. 2019). Green microalgae Chlamydomonas reinhardtii was genetically modified to produce polyethylene terephthalate hydrolase, able to degrade polyethylene terephthalate films and terephthalic acid (Kim et al. 2020). A similar modification was also successfully done in P. tricornutum which produced polyethylene terephthalate hydrolase and showed catalytic activity against polyethylene terephthalate and the copolymer polyethylene terephthalate glycol (Moong et al. 2019).
In short, microalgae could serve as effective microplastic degraders, thanks to their capability of using plastic monomers as carbon source by producing degrading enzymes and the ease of culture. The possibility of genetically engineering algae strains to enhance degradation capability has provided a promising environmentally friendly solution to biologically degrade polyethylene terephthalate using microalgae via synthetic biology.
Fungal degradation of microplastics
The fungi largely consist of a diverse group of organisms which are largely saprotrophs, or opportunistic or obligate parasites. They have tremendous adaptivity and can grow in a wide range of habitats both aquatic and terrestrial ecosystems under diverse environmental conditions. As well as being able to tolerate several toxic chemicals and metals, they produce a diverse range of extracellular enzymes and natural biosurfactants such as hydrophobins that can degrade complex polymers into simple monomers, making them a source of electrons and carbons for microorganisms, thus facilitating the degradation and mineralization of complex pollutants (Olicón-Hernández et al. 2017).
The main genus associated with the degradation of different types of polymers such as polyethylene, polypropylene, and polyethylene terephthalate includes Zalerion maritimum, Aspergillus niger, Cladosporium, and Penicillium simplicissimum (Paço et al. 2017; de Oliveira et al., 2020a, b; Devi et al. 2015), which use microplastics as sole carbon source after degradation by extracellular enzymes. They promote the formation of different types of chemical bonds (having carbonyl, carboxyl, and ester functional groups) and decrease their hydrophobicity. Similar degradation of polyurethane was evident in fungal strains such as Aspergillus fumigatus, Aspergillus tubingensis, Cladosporium pseudocladosporioides, Fusarium solani, and Penicillium chrysogenum and strains of Pestalotiopsis microspora (Khan et al. 2017; Álvarez-Barragán et al. 2016; Magnin et al. 2020; 2015; Russell et al. 2011).
In most cases, serine hydrolase plays a pivotal role in polyurethane degradation. Degradation of high-density polyethylene from marine coastal areas by two fungal strains Aspergillus tubingensis VRKPT1 and Aspergillus flavus VRKPT2 was reported to be 6.02 ± 0.2 and 8.51 ± 0.1%, respectively (Devi et al. 2015). Recently, Kunlere et al. reported the promising degradation of low-density polyethylene by Mucor circinelloides and Aspergillus flavus isolated from a municipal landfill (Kunlere et al. 2019).
Pretreatment of the microplastics, for example, polyethylene, with chemicals such as nitric acid and sodium hydroxide is known to accelerate the rate of biodegradation of polyethylene by Aspergillus niger (Nwachukwu et al. 2010). Physical pretreatment processes including thermo-oxidization at 80 °C for 15 days were required to cause the degradation in low-density polyethylene mediated by Aspergillus niger and Penicillium pinophilum, showing 0.57 and 0.37% after incubation over 30 months (Volke-Sepúlveda et al. 2002). Similarly, Aspergillus spp. and Lysinibacillus spp. showed 29.5% of biodegradation of UV-irradiated and 15.8% of biodegradation of non-UV-irradiated polymer films (Esmaeili 2013).
Fungal enzymes associated with the degradation of microplastics
Fungi produce a diverse range of intra and extracellular enzymes which can catalyze diverse reactions and have the ability to degrade petroleum-based polymers. The intracellular enzymes perform a major role in fungal adaptation and detoxification processes (Schwartz et al. 2018). The enzyme systems associated with cytochrome P450 family epoxidases and transferases are associated with oxidation and conjugation reactions and help in the metabolism of aliphatic, alicyclic, and aromatic molecules. They perform a wide range of reactions such as epoxidation, sulfoxidation, desulfuration, dehalogenation, deamination, and epoxidation (Shin et al. 2018). The cytochrome P450 families of enzymes help in the preservation of hyphal wall integrity and formation of spore wall and utilized cofactors like heme, NADPH + H+, and FAD.
On the other hand, extracellular enzymes include hydrolases which are involved in the breakdown of complex polymers (Sánchez, 2009) and increase the solubility of the pollutants subsequently reducing bioaccumulation (Olicón-Hernández et al. 2017). Enzymes belonging to the class II peroxidases such as manganese peroxidase and lignin peroxidase, laccases, and dye-decolorizing peroxidases, which oxidize a wide range of substrates, can be used as efficient tools for environmental cleaning. Lignin degrading fungi produce laccase which catalyzes the oxidation of aromatic and non-aromatic substrates such as chlorophenolic or nonphenolic compounds (polymethylmethacrylate and polyhydroxybutyrate (Straub et al. 2017). The thermostability of these enzymes may promote their uses in large-scale reactors where the degradation of polypropylene can be carried out at a high temperature, facilitating high kinetics reactions. A detailed list of different fungi associated with microplastic degradation is reported in Table 1.
Table 1.
Microplastic degradation by fungi
| Source of microbes | Isolated fungal strains | Type of microplastic degraded | Pretreatment | Incubation period | % of degradation | Enzymes | References |
|---|---|---|---|---|---|---|---|
| Not reported | Aspergillus sp. Penicillium sp. | Polypropylene/butylene-adipate-co-terephthalate | 30 days | De Oliveira et al. ( 2020a, b) | |||
| Marine sediments | Zalerion maritimum | Polyethylene pellets | 28 days | Paço et al. (2017) | |||
| Not reported | Bjerkandera adusta | Polypropylene and biomass | Gamma irradiated | Ligninase | Butnaru et al. (2016) | ||
| Marine coastal area | Aspergillus flavus VRKPT2 | High-density polyethylene | 30 days | Devi et al. (2015) | |||
| Endophytes of Humboldtia brunonis, Psychotria flavida | Aspergillus sp. Paecilomyces Lilacinus, Lasiodiplodia theobromae | Polypropylene | Laccase | Sheik et al. (2015) | |||
| Waste dump | Aspergillus niger, Aspergillus terreus, Aureobasidium pullulans, Paecilomyces varioti, Penicillium funiculosum, Penicillium ochrochloron, Scopulariopsis brevicaulis, Trichoderma viride | Low-density polyethylene | UV-irradiated | 28 °C and relative humidity of > 90% for 84 days | 24%, 60% and 58% of its initial mass | Nowak et al. (2012) | |
| Soil, wall paint coated with polyurethane | Fusarium solani, Spicaria spp., Alternaria solani, and Aspergillus flavus | Polyester polyurethane | 100% | Ibrahim et al. (2011) | |||
| Culture collection | Penicillium pinophilum ATCC 11,797 | Low-density polyethylene powder | 31 months | Volke-Sepúlveda et al. (2002) | |||
| Not reported | Aspergillus niger, Penicillium funiculosum, Chaetomium globosum, Gliocladium virens and Pullularia pullulans | Low-density polyethylene | 28 days | Chandra and Rustgi (1997) |
Overall, a wide variety of fungal strains are capable of degrading plastics into more environmentally acceptable compounds, thanks to the production of a plethora of intracellular and extracellular enzymes, including oxidases and hydrolases, and natural biosurfactants such as hydrophobins.
Bacterial degradation of microplastics
Diverse studies have been conducted using bacteria for the degradation of microplastics. Bacteria capable of degrading microplastics have been isolated from a wide range of habitats including contaminated sediments, wastewater, sludge, compost, municipal landfills (Mehmood et al. 2016; Awasthi et al. 2020), and also from extreme climatic conditions like the Antarctic soils, mangrove, and marine sediments. Moreover, microplastic degrading microbes have also been isolated from the gut microflora of earthworms. It is generally reported that microbes living in polluted sites often develop an ability to activate the enzymatic system responsible for microplastic degradation.
Both pure cultures and bacterial consortiums can be used for microplastic degradation. However, pure cultures present several advantages in the degradation process, offering a convenient way to study metabolic pathways involved in the process. Moreover, the impact of environmental factors such as temperature, pH, substrate characteristics, and surfactants affecting the degradation process can be more easily monitored (Janssen et al. 2002). However, the main disadvantage is an extremely slow rate of degradation. Thus, more innovative methods are required to optimize conditions and improve the degrading bacterial isolates to shorten the degradation process. The use of a consortium of bacteria is usually preferred as it has been shown that biodegradation by a single bacterium often results in the generation of toxic end products (Dobretsov et al. 2013), which can be successfully eliminated in a stable microbial community (Singh and Wahid 2015).
The main process of degradation is represented by physicochemical degradation which reduces the polymer length and alters the functional groups of microplastics, making them more susceptible to microbial enzyme activity. Biodegradation using enzymes involves the action of lipases, esterases, laccases, amidases, cutinases, hydrolases, and carboxylesterases (Barth et al. 2016; Chen et al. 2020; Amobonye et al. 2021; Inderthal et al. 2021; Gómez-Méndez et al. 2018). Thus, in-depth knowledge of the metabolic pathways and associated enzymes is necessary to perform an efficient biodegradation process.
Physiochemical pre-treatment, including chemical oxidizing agents, thermooxidation, and UV irradiation of microplastics, is recommended to promote plastic biodegradation. These pre-treatments include UV irradiation, nitric acid treatment and blending with polymers like starch derivatives, cellulosic esters, polyhydroxybutyrate, poly(3-hydroxybutyrate-co-3-hydroxyvalerate), and polycaprolactone, which increase the biodegradability of polypropylene (Gironi and Piemonte 2011). The addition of prooxidative and biodegradable substances like starch to low-density polyethylene, high-density polyethylene, polyvinyl alcohol, and polystyrene has been reported to enhance their biodegradability, promoting amylase activity (Zadjelovic et al. 2020).
The earliest study of microplastics biodegrading microorganisms was conducted by Cacciari et al. (1993), using a consortium of Pseudomonas chlororaphis, Pseudomonas stutzeri, and Vibrio sp. to degrade polypropylene. In the same study, the addition of starch was also reported to increase the biodegradation ability.
Later on, both Arkatkar et al. and Fontanella et al. reported biodegradation of polypropylene using a consortium of Bacillus subtilis, B. flexus, Pseudomonas stutzeri, and Rhodococcus rhodochrous, respectively (Arkatkar et al. 2010; Fontanella et al. 2013). These microbial isolates were found to form a biofilm, as reported in the study of Kowalczyk et al. (2016), by isolating Achromobacter xylosoxidans.
In a study conducted by Auta et al. (2018), two bacterial strains, belonging to Bacillus and Rhodococcus, isolated from mangrove sediments, showed polypropylene degradation efficiency of 4.0 and 6.4% after 40 days of incubation, respectively. They also reported that Bacillus cereus and Bacillus gottheilii were able to degrade microplastics (Auta et al. 2017). B. gottheilii induced microplastics weight loss percentages of 6.2%, 3.0%, 3.6%, and 5.8% for polyethylene, polyethylene terephthalate, polypropylene, and polystyrene, respectively.
Few other bacteria associated with polypropylene degradation included Bacillus, Pseudomonas, Chelatococcus, and Lysinibacillus fusiformis, which were obtained from a wide variety of habitats including mangrove habitat, compost, cow dung, and land contaminated with plastic wastes.
Gut microflora of several arthropods like Tenebrio molitor (mealworms) (Yang et al. 2015), Plodia interpunctella (Indian meal moth) (Yang et al. 2014) and Galleria mellonella (wax moths) (Kong et al. 2019) have also been reported to harbor microbes having microplastics biodegradation properties. A study by Yang et al. (2014) isolated two bacteria, Enterobacter asburiae YT1 and Bacillus sp. YP1, from the gut of waxworms capable of degrading polyethylene by decreasing the hydrophobicity and damaging the surface of polypropylene. A later study conducted by Yang et al. (2015) isolated a bacterial strain Exiguobacterium sp. from the guts of mealworms able to form biofilm and degrade polystyrene.
Efficient biodegradation of low-density polyethylene was obtained using strains like Microbacterium paraoxydans and Pseudomonas aeruginosa, which showed nearly 61.0% and 50.5% degradation, respectively, within 2 months of incubation (Rajandas et al. 2012). Similarly, the biofilm of Pseudomonas sp. AKS2 has been reported to degrade low-density polyethylene up to 5 ± 1% within an incubation period of 45 days (Tribedi and Sil 2013) without any pretreatment. Likewise, degradation of polyethylene was also reported by isolating Rhodococcus ruber C208 at the rate of 0.86% per week (Sivan et al. 2006).
A consortium of bacteria consisting of Bacillus sp. and Paenibacillus sp. was able to reduce the dry weight of microplastics by 14.7% in 60 days (Park and Kim 2019). Moreover, Huerta Lwanga et al. (2018) investigated the earthworm (Lumbricus terrestris)-mediated degradation of low-density polyethylene. The isolates from the gut included genera Actinobacteria and Firmicutes which were also studied separately and observed to be able to degrade low-density polyethylene microplastics and release volatile compounds like eicosane, docosane, and tricosane. A consortium of Enterobacter and Pseudomonas from cow dung enhanced weight loss up to 15% within 120 days (Skariyachan et al. 2021).
Several marine hydrocarbonoclastic bacteria such as Alcanivorax borkumensis showed the ability to degrade alkyl cycloalkanes, isoprenoid hydrocarbons, alkanes, and branched aliphatic compounds (Davoodi et al. 2020). The research was done on the same isolate that previously showed biofilm formation on low-density polyethylene in the presence of pyruvate, hexadecane and yeast extract and the low-density polyethylene films (Delacuvellerie et al. 2019).
It was also stated that the presence of alkanes modifies the cell membrane hydrophilicity and produces biosurfactants to interact with the plastic surface and the formation of COOH/OH and C=O functional groups. Several actinomycetes including Rhodococcus ruber and Streptomyces were also involved in polyethylene biodegradation (Sivan 2011).
Overall, among the different genera of bacteria associated with microplastic degradation, 21% belonged to Pseudomonas, about 15% to Bacillus and 17% derived from mixtures of these two genera (Matjašič et al. 2021). Other bacteria associated with microplastic biodegradation included Enterobacter asburiae, Bacillus sp., Nocardia asteroids, Rhodococcus rhodochrous (Bonhomme et al. 2003), Streptomyces badius, Rhodococcus ruber, Comamonas acidovorans and Clostridium thermocellum (Paço et al. 2019), Exiguobacterium sp., Ideonella sakaiensis (Tanasupawat et al. 2016), Pseudomonas chlororaphis, Pseudomonas putida AJ, and Thermomonospora fusca (Ghosh et al. 2013). A detailed list of different bacteria and actinomycetes associated with microplastic degradation is reported in Tables 2 and 3.
Table 2.
Microplastic degradation by bacterial isolates
| Source of microbes | Isolated bacterial strains | Type of microplastic degraded | Incubation period | % of degradation | Biodegradation detection method/techniques | References |
|---|---|---|---|---|---|---|
| Polluted soil samples | Lysinibacillus sp. | Polypropylene, polyethylene | 26 days | 4 and 9% | Gas chromatography – mass spectrometry, Scanning electron microscopy | Jeon et al. (2021) |
| Cow dung sample | Enterobacter sp nov. bt DSCE01, Enterobacter cloacae nov. bt DSCE02, and Pseudomonas aeruginosa nov. bt DSCE-CD03 | Low-density polyethylene, polypropylene | 160 days | 64.25 ± 2% and 63.00 ± 2% | Weight loss | Skariyachan et al. (2021) |
| Compost | Bacillus cereus, Bacillus thuringiensis, Bacillus licheniformis | Polypropylene and poly-L-lactide | 6 months | Fourier-transform infrared spectroscopy; Thermogravimetric analysis | Jain et al. (2022) | |
| Municipal landfill sediment | Bacillus sp. and Paenibacillus sp. | Polyethylene | 60 days | 14.7 % | Field-emission scanning electron microscope, Fourier transform infrared spectrometer, Gas chromatography-mass spectrometer, Scanning electron microscopy, Thermogravimetric analyzer | Park and Kim (2019) |
| Biofilm composed by Pirellulaceae, Phycisphaerales, Cyclobacteriaceae, and Roseococcus | Polyethylene, polypropylene | NA | DNA extraction, amplification and sequencing (evaluation of the effects of substrate type on microbial communities) | Miao et al. (2019) | ||
| Earthworm gut | Bacillus simplex and Bacillus sp. | Low-density polyethylene | 21 days | Scanning electron microscopy | Huerta Lwanga et al. (2018) | |
| Mangrove sediments | Bacillus sp. strain 27 | Polypropylene | 40 days | 4.0% | Weight loss; Fourier-transform infrared spectroscopy; Scanning electron microscopy | Auta et al. (2018) |
| Mangrove sediment | Bacillus gottheilii | Polyethylene, polyethylene terephthalate, polypropylene, and polystyrene | 40 days | 6.2%, 3.0%, 3.6%, 5.8% | Weight loss; Fourier-transform infrared spectroscopy; Scanning electron microscopy | Auta et al. (2017) |
| Bacillus cereus | Polyethylene, polyethylene terephthalate, polystyrene | 40 days | 1.6%, 6.6%, and 7.4% | |||
| Compost | Bacillus thuringiensis | Polypropylene and poly-L-lactide | 15 days | 12% | Fourier-transform infrared spectroscopy; Scanning electron microscopy; Thermogravimetric analysis | Jain et al. (2018) |
| Compost | Bacillus licheniformis | Polypropylene and poly-L-lactide | 15 days | 10% | Fourier-transform infrared spectroscopy; Scanning electron microscopy; Thermogravimetric analysis | Jain et al. (2018) |
| Sewage treatment plants (STP) | Microbial consortia (including Aneurinibacillus sp. and Brevibacillus sp.) | Low density polyethylene, high density polyethylene and polypropylene | 140 days | 47%, 58% and 56% | Fourier-transform infrared spectroscopy; Scanning electron microscopy; Atomic force microscopy; Energy dispersive spectroscopy; Nuclear magnetic resonance; Gas chromatography-mass spectrometry | Skariyachan et al. (2018) |
| Mangrove sediments in Peninsular Malaysia | Bacillus cereus | Polypropylene | 40 days | 12% | Weight loss | Helen et al. (2017) |
| Mangrove sediments in Peninsular Malaysia | Sporosarcina globispora | Polypropylene | 40 days | 11% | Weight loss | Helen et al. (2017) |
| Sandy beaches in Northern Crete, Chania, Greece | Agios Onoufrios and Kalathas | Polystyrene films | 6 months | 0.19% | Weight loss; Fourier-transform infrared spectroscopy; Scanning electron microscopy | Syranidou et al.(2017) |
| Pseudomonas aeruginosa and Escherichia coli | Polyurethanes | 2.5% and 2.4 % | Weight loss; Scanning electron microscopy; Tensile strength and elongation at break | Uscátegui et al. (2016) | ||
| Municipal solid waste | Stenotrophomonas panacihumi PA3-2 | Polypropylene | 90 days | 20.3 ± 1.39% | Weight loss; Fourier-transform infrared spectroscopy | Jeon and Kim (2016) |
| Nitrosomonas sp., Nitrobacter sp., Burkholderia sp. and Pseudomonas sp. | High density polyethylene, low density polyethylene, polypropylene | 90 days | 15%-20% (High density polyethylene), 5%-9% (polypropylene), | Weight loss | Muenmee et al. (2016) | |
| Plastic-eating mealworms | Exiguobacterium sp. strain YT2 | Polystyrene | 60 days | 7.4% ± 0.4% | Cross polarization—magic angle spinning nuclear magnetic resonance; Thermogravimetric analysis coupled with fourier-transform infrared spectroscopy | Yang et al. (2015) |
| National Environmental Engineering Research Institute, Nagpur India | B. flexus + P. azotoformans | UV treated polymers | 12 months | 22.7% | Weight loss; Fourier-transform infrared spectroscopy | Aravinthan et al. (2016) |
| Plastic-eating waxworms gut | Bacillus sp. YP1 | Polyethylene films | 28 days | 10.7 ± 0.2% | Weight loss; Fourier-transform infrared spectroscopy | Yang et al. (2014) |
| Enterobacter asburiae YT1 | 6.1 ± 0.3% | |||||
| Compost | Chelatococcus sp. E1 | Low-molecular-weight polyethylene | 80 days | 44.5% | Gel permeation chromatography; Fourier-transform infrared spectroscopy; Nuclear magnetic resonance; Tensile strength | Jeon and Kim (2013) |
Table 3.
Microplastic degradation by actinomycetes
| Source of microbes | Isolated actinomycetes strains | Type of microplastic degraded | Incubation period | % of degradation | Biodegradation detection Method/techniques | References |
|---|---|---|---|---|---|---|
| Antarctic soil | Pseudomonas sp. ADL15 and Rhodococcus sp. ADL36 | Polypropylene | 40 days | 17.3% and 7.3% | Weight loss; Fourier-transform infrared spectroscopy | Habib et al. (2020) |
| Not reported | Rhodococcus ruber strain C208 | Polyethylene | 2 months | 7.5% | Weight loss; Scanning electron microscopy | Sivan et al. (2006) |
| Soils from waste coal, a forest and an extinct volcano crater |
Bacterial consortia Arthrobacter viscosus, Micrococcus lylae, Micrococcus luteus, Bacillus mycoides, Bacillus cereus, Bacillus pumilus; Bacillus thuringiensis |
Low-density polyethylene | 225 days | 17.03% | Weight loss; Fourier-transform infrared spectroscopy; Scanning electron microscopy; elongation at brake | Nowak et al. (2011) |
| Not reported | Microbacterium paraoxydans | Polyethylene (pre-treated with nitric acid) | 2 months | 61.0% | Weight loss; Fourier-transform infrared spectroscopy | Rajandas et al. (2012) |
| Mangrove sediment | Rhodococcus | Polypropylene | 40 days | 6.4% | Weight loss; Fourier-transform infrared spectroscopy; Scanning electron microscopy | Auta et al. (2018) |
| Actinomadura sp. T16-1 (Enzyme production) | Polylactic acid (production of polylactic acid-degrading enzyme) | 96 h | Not available | Enzyme activity | Sukkhum et al. (2009) | |
| Rhodococcus ruber | Polystyrene | 2 months | 0.8% | Weight loss | Mor and Sivan (2008) | |
| Rhodococcus rhodochrous ATCC 29,672 | Two polypropylene films (Statistical copolymer and block copolymer) | 6 months | Not available | Fourier-transform infrared spectroscopy; Proton nuclear magnetic resonance; ADP/ATP ratio | Fontanella et al. (2013) |
To summarize, bacteria capable of degrading microplastics have been isolated from a wide range of habitats including contaminated sediments, wastewater, sludge, compost, municipal landfills, extreme environments, and microbiota. Bacteria have been tested for microplastics degradation, both using pure cultures and microbial consortium. Bacterial consortium, in particular, show greater efficiency and community stability.
Modern biotechnological methods to enhance microplastic degradation
Microplastics are gradually gaining attention due to their ubiquitous presence and negative impact on human health and the ecosystem. Microplastic exposure has increased many folds after the coronavirus disease 2019 pandemic due to the excessive use of single-use personal protective equipment products (Anand et al. 2022; De-la-Torre and Aragaw 2021; Yang et al. 2022). Microplastic exposure may occur through inhalation, digestion, and dermal absorption and might cause health issues like neurotoxicity, disrupt endocrine system, carcinogenicity, and metabolic disruptions (Naqash et al. 2020; Rahman et al. 2021; see also Section “Processes of microplastic degradation”). Thus, there is a need for efficient microplastic bioremediation.
Polyethylene terephthalate is a widely used thermoplastic polymer used for packaging which can degrade and generate microplastics. Isolates like, Ideonella sakaiensis 201-F6 are reported to produce polyethylene terephthalate-hydrolyzing enzymes able to degrade polyethylene terephthalate to terephthalic acid, and ethylene glycol which is non-hazardous monomers. Other bacterial strains can be genetically engineered by cloning the encoding genes of Ideonella sakaiensis 201-F6, promoting the generation of modified strains able to degrade polyethylene terephthalate in non-hazardous monomers. Moog et al. (2019) introduced polyethylene terephthalate-hydrolyzing enzymes into photosynthetic microalga, Phaeodactylum tricornutum which showed efficient polyethylene terephthalate hydrolyzing activity.
Genetic modifications have also been made to promote the capture of polyvinyl chloride within the bacterial biofilm (Liu et al. 2021). Pseudomonas aeruginosa was genetically engineered by deleting the wspF gene to increase the formation of sticky exopolymeric substances which enhance its capacity to accumulate microplastics in its biofilm. Moreover, yhjH gene was designed under the control of an arabinose-induced promoter and was introduced into the bacterium. Since the function of yhjH was to decrease cyclic dimeric guanosine monophosphate levels, induced expression of the gene reduced the biofilm formation suitable enough to release captured microplastics. The synthetic ‘capture and release’ system would enable the creation of efficient microplastics scavengers for the bioremediation of aquatic ecosystems.
The advent of different genetic engineering methods has enabled us to manipulate the genetic materials of microbes and enhance their biodegrading efficiency. Several procedures involving recombinant DNA technology, gene cloning, and genetic modification have been done to improve the bioremediation ability of the microbes in presence of different hydrocarbons and heavy metals (Kumar et al. 2020). However, till now very few works have been conducted on the application of genetic engineering for creating a better strain for degradation of plastics.
These techniques are used for the construction of novel pathways and can alter enzyme specificity and their affinity toward different microplastics. For successful gene editing, it is necessary to find suitable genes required for metabolizing and degrading microplastics and suitable host organisms like E. coli in which these genes are expressed. The main processes involved are polymerase chain reaction, antisense ribonucleic acid (RNA) technology, and site-directed mutagenesis. Antisense RNA technology has emerged as a new tool for genetic editing as artificially synthesized antisense RNA can effectively regulate the expression of genes in host cells. On the other hand, site-directed mutagenesis is also used to alter the activity of genes associated with microplastic degradation. In a recent study, Lameh et al. (2022) reported mutation of carboxylesterase by in silico site‐directed mutagenesis to produce BTA‐hydrolase in Archaeoglobus fulgidus to enhance its ability to degrade polyethylene terephthalate.
The main enzymes associated with microplastic degradation, such as manganese-dependent peroxidase, were produced by a genetically engineered strain of E. coli and S. cerevisiae BY 4741, similarly, laccase enzymes were produced by a modified genetic strain of E. coli BL21 and P. chrysosporium (Sharma et al. 2018; Paço et al. 2019). These genetically modified enzymes are capable of better degradation of polyethylene terephthalate.
Enzyme cutinase produced by microbes for the breakdown of polyester linkage can also be used in polyethylene terephthalate degradation which acts at an optimal temperature of 75 °C. Genetic engineered yeast produces bacterial cutinase which prevents formation in strategic positions with sugars which helps in the degradation of polyethylene terephthalate (Shirke et al. 2018). Islam et al. (2019) reported that genetically engineered cutinase enzyme reduces the degradation time from 41.8 to 6.2 h when compared with wild strain.
A similar enhanced biodegradation ability was observed in a consortium of marine microbes (Syranidou et al. 2019). However, despite their better ability in laboratory conditions, most of these genetically modified organisms have displayed unsatisfactory results in field studies.
Gene editing tools
Gene editing tools have been applied for genome engineering of plants, animals, and microorganisms for the expression of specific genes (Paço et al. 2019; Tang et al. 2020; Nidhi et al. 2021; Ozyigit et al. 2021; Bhattacharyya et al. 2022; Biswas et al. 2022; Mandal et al. 2022; Jiang et al. 2022). With the advent of different types of gene editing tools such as zinc finger proteins, transcription activator-like effector nucleases, and more recently, the clustered regularly interspaced palindromic repeats (CRISPR)/Cas9, the manipulation of organisms has become easier (Jiang et al. 2013; Gaj et al. 2013). Genome editing also helps in the manipulation of a gene of interest which can perform the loss and gain of function experiments which alter the expression of different genes.
This strategy can be efficiently used to incorporate genes encoding enzymes like polyethylene terephthalate hydrolase, dehalogenase, esterase, depolymerase, and laccase which are associated with microplastic degradation. Three different CRISPR sequences were identified in Streptomyces albogriseolus LBX-2 which makes it a suitable organism for genetic engineering, where the main enzyme associated with polyethylene degradation is oxygenase (Shao et al. 2019).
Bioinformatics
Bioinformatics has also become an effective tool for enhancing the biodegradation of plastic debris including microplastic particles (Purohit et al. 2020). Various types of databases such as The University of Minnesota Biocatalysis/Biodegradation Database, The Environmental Contaminant Biotransformation Pathway Resource, MetaCyc database, and BioCyc database related to biodegradation pathways have been established to evaluate the process of biodegradation by providing information on the metabolic pathways, the microbial enzymes and genes associated with the process (Gao et al. 2010; Wicker et al. 2016; Karp et al. 2019; Caspi et al. 2020). These databases and computational methods help to recognize enzymes involved in a metabolic pathway of interest and help in forecasting the biodegradation routes of toxic chemicals, providing a platform in which a novel approach for the biodegradation of plastic can be designed (Ali et al. 2021).
Despite all these advantages, the major disadvantage associated with bioinformatics is the lack of experimental data and its validation which is required for future research. Moreover, there is a wide knowledge gap between diverse groups of synthetic polymer degrader microorganisms and their responsible enzymes. Hence, an extensive investigation is required to identify suitable metabolic pathways for the degradation of polymers and their associated enzymes. In the near future, a combination of approaches, using bioinformatic tools, metabolic engineering, genetics, molecular, and system biology may help us to find a suitable and sustainable option for the biodegradation of microplastics.
Biosafety issues associated with genetically modified organisms
The advent of different genetic engineering techniques, and synthetic biological and genetic tools has allowed the development of genetically modified microbial scavengers for the mitigation of diverse types of pollutants (Mohamed et al. 2020; Wang et al. 2019a, b). However, several regulatory hurdles hamper the use of genetically modified microorganisms in an onsite experiment. Also, these genetically modified microbes have shown their efficiency in laboratory conditions, but onsite experiments are required to validate their effectiveness.
A diverse range of genetic tools has been developed only to prevent the negative impacts of genetically modified organisms on the field, including antibiotic gene-free genetic engineering tools (Ji et al. 2019) and suicide genetic systems (Honjo et al. 2019; Marguet et al. 2010; Scott et al. 2017). By using synthetic biology and metabolic engineering, an attempt has been already made to engineer microorganisms, which can be efficiently used as self-eliminate microbial scavengers for the bioremediation of various toxic environmental pollutants (Moog et al. 2019; French et al. 2020).
These techniques look promising and can be applied in the case of microplastics bioremediation. For biomedical applications of synthetically engineered microorganisms, programmed cell death circuits have been developed using synthetic biology (Marguet et al. 2010; Sedlmayer et al. 2018; Tran et al. 2021). After bioremediation is finished, these programmed cell death circuits can be employed to eradicate the microbial scavengers by themselves.
In short, the development of genetic engineering techniques has opened the possibility of modifying bacteria introducing exogenous genes for specific enzymes involved in the degradation of plastic. Moreover, modern gene editing tools, such as CRISPR/Cas 9, make the manipulation of organisms easier and more precise. However, some concerns of biosafety still limit the use of genetically modified microorganisms in an onsite experiment, limiting the real evaluation of gene editing effectiveness in microplastic bioremediation.
Conclusion
There are a lot of hurdles and limitations in the application of microbes for the biodegradation of microplastics which can be overcome by different genetic manipulations. However, most of genetically modified microbes have only been validated under laboratory conditions and reports on their efficiency in field conditions are largely lacking. Also, the knowledge associated with different metabolic pathways and enzymes is largely lacking. The recent advances in metagenomic analysis and engineering of uncultivated microbial communities, sampled from contaminated sites, can assist in the development of novel processes of bioremediation (Schloss and Handelsman 2005; Shilpa et al. 2022) and culture-independent techniques can open up new avenues for the discovery of novel metabolic pathways and enzymes.
Acknowledgements
Authors sincerely apologize to colleagues/researchers whose work could not be discussed and cited in this manuscript due to space limitations. The authors are thankful to their respective departments/institutes for providing space and other necessary facilities which helped to draft this manuscript.
Authors contributions
All authors contributed to drafting and critical revision of the article and approved the article before submission.
Funding
Not applicable.
Declarations
Conflicts of interest
The authors declared no potential conflicts of interest.
Consent for publication
All authors agreed on the publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Satarupa Dey, Email: dey1919@gmail.com.
Abhijit Dey, Email: abhijit.dbs@presiuniv.ac.in.
Stefania Federici, Email: stefania.federici@unibs.it.
References
- Ali SS, Elsamahy T, Koutra E, Kornaros M, El-Sheekh M, Abdelkarim EA, Sun J. Degradation of conventional plastic wastes in the environment: a review on current status of knowledge and future perspectives of disposal. Sci Total Environ. 2021;771:144719. doi: 10.1016/j.scitotenv.2020.144719. [DOI] [PubMed] [Google Scholar]
- Álvarez-Barragán J, Domínguez-Malfavón L, Vargas-Suárez M, González-Hernández R, Aguilar-Osorio G, Loza-Tavera H, Kivisaar M. Biodegradative Activities of Selected Environmental Fungi on a Polyester Polyurethane Varnish and Polyether Polyurethane Foams. Appl Environ Microbiol. 2016;82(17):5225–5235. doi: 10.1128/AEM.01344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral-Zettler LA, Zettler ER, Mincer TJ. Ecology of the plastisphere. Nat Rev Microbiol. 2020;18(3):139–151. doi: 10.1038/s41579-019-0308-0. [DOI] [PubMed] [Google Scholar]
- Amobonye A, Bhagwat P, Singh S, Pillai S. Plastic biodegradation: frontline microbes and their enzymes. Sci Total Environ. 2021;759:143536. doi: 10.1016/j.scitotenv.2020.143536. [DOI] [PubMed] [Google Scholar]
- Anand U, Li X, Sunita K, Lokhandwala S, Gautam P, Suresh S, Sarma H, Vellingiri B, Dey A, Bontempi E, Jiang G. SARS-CoV-2 and other pathogens in municipal wastewater, landfill leachate, and solid waste: a review about virus surveillance, infectivity, and inactivation. Environ Res. 2022;203:111839. doi: 10.1016/j.envres.2021.111839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrady AL. The plastic in microplastics: a review. Mar Pollut Bull. 2017;119:12–22. doi: 10.1016/j.marpolbul.2017.01.082. [DOI] [PubMed] [Google Scholar]
- Anik AH, Hossain S, Alam M, Sultan MB, Hasnine MT, Rahman MM. Microplastics pollution: a comprehensive review on the sources, fates, effects, and potential remediation. Environ Nanotechnol, Monit Manag. 2021;16:100530. doi: 10.1016/j.enmm.2021.100530. [DOI] [Google Scholar]
- Aragaw TA. Surgical face masks as a potential source for microplastic pollution in the COVID-19 scenario. Mar Pollut Bull. 2020;159:111517. doi: 10.1016/j.marpolbul.2020.111517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravinthan A, Arkatkar A, Juwarkar AA, Doble M. Synergistic growth of Bacillus and Pseudomonas and its degradation potential on pretreated polypropylene. Prep Biochem Biotechnol. 2016;46(2):109–115. doi: 10.1080/10826068.2014.985836. [DOI] [PubMed] [Google Scholar]
- Arkatkar A, Juwarkar AA, Bhaduri S, Uppara PV, Doble M. Growth of Pseudomonas and Bacillus biofilms on pretreated polypropylene surface. Int Biodeterior Biodegrad. 2010;64(6):530–536. doi: 10.1016/j.ibiod.2010.06.002. [DOI] [Google Scholar]
- Atugoda T, Piyumali H, Wijesekara H, Sonne C, Lam SS, Mahatantila K, Vithanage M. Nanoplastic occurrence, transformation and toxicity: a review. Environ Chem Lett. 2022 doi: 10.1007/s10311-022-01479-w. [DOI] [Google Scholar]
- Auta HS, Emenike CU, Fauziah SH. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environ Pollut. 2017;231:1552–1559. doi: 10.1016/j.envpol.2017.09.043. [DOI] [PubMed] [Google Scholar]
- Auta HS, Emenike CU, Jayanthi B, Fauziah SH. Growth kinetics and biodeterioration of polypropylene microplastics by Bacillus sp. and Rhodococcus sp. isolated from mangrove sediment. Mar Pollut Bull. 2018;127:15–21. doi: 10.1016/j.marpolbul.2017.11.036. [DOI] [PubMed] [Google Scholar]
- Danso D, Chow J, Streit WR. Plastics: environmental and biotechnological perspectives on microbial degradation. Appl Environ Microbiol. 2019 doi: 10.1128/AEM.01095-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi AK, Tan Q, Li J. Biotechnological potential for microplastic waste. Trends Biotechnol. 2020;38(11):1196–1199. doi: 10.1016/j.tibtech.2020.03.002. [DOI] [PubMed] [Google Scholar]
- Bacha AUR, Nabi I, Zhang L. Mechanisms and the Engineering Approaches for the Degradation of Microplastics. ACS ES&T Eng. 2021;1(11):1481–1501. doi: 10.1021/acsestengg.1c00216. [DOI] [Google Scholar]
- Bacha AUR, Nabi I, Zaheer M, Jin W, Yang L. Biodegradation of macro- and micro-plastics in environment: A review on mechanism, toxicity, and future perspectives. Sci Total Environ. 2023;858:160108. doi: 10.1016/j.scitotenv.2022.160108. [DOI] [PubMed] [Google Scholar]
- Baldera-Moreno Y, Pino V, Farres A, Banerjee A, Gordillo F, Andler R. Biotechnological aspects and mathematical modeling of the biodegradation of plastics under controlled conditions. Polymers. 2022;14(3):375. doi: 10.3390/polym14030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth M, Honak A, Oeser T, Wei R, Belisário-Ferrari MR, Then J, Schmidt J, Zimmermann W. A dual enzyme system composed of a polyester hydrolase and a carboxylesterase enhances the biocatalytic degradation of polyethylene terephthalate films. Biotechnol J. 2016;11(8):1082–1087. doi: 10.1002/biot.201600008. [DOI] [PubMed] [Google Scholar]
- Basak N, Meena SS. Exploring the plastic degrading ability of microbial communities through metagenomic approach. Mater Today: Proc. 2022;57:1924–1932. doi: 10.1016/j.matpr.2022.02.308. [DOI] [Google Scholar]
- Besseling E, Wegner A, Foekema EM, Van Den Heuvel-Greve MJ, Koelmans AA. Effects of microplastic on fitness and PCB bioaccumulation by the lugworm Arenicola marina (L) Environ Sci Technol. 2013;47(1):593–600. doi: 10.1021/es302763x. [DOI] [PubMed] [Google Scholar]
- Bhatt P, Pathak VM, Bagheri AR, Bilal M. Microplastic contaminants in the aqueous environment, fate, toxicity consequences, and remediation strategies. Environ Res. 2021;200:111762. doi: 10.1016/j.envres.2021.111762. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya N, Anand U, Kumar R, Ghorai M, Aftab T, Jha NK, Rajapaksha AU, Bundschuh J, Bontempi E, Dey A. Phytoremediation and sequestration of soil metals using the CRISPR/Cas9 technology to modify plants: a review. Environ Chem Lett. 2022 doi: 10.1007/s10311-022-01474-1. [DOI] [Google Scholar]
- Biswas P, Anand U, Ghorai M, Pandey DK, Jha NK, Behl T, Kumar M, Chauhan R, Shekhawat MS, Dey A. Unraveling the promise and limitations of CRISPR/Cas system in natural product research: approaches and challenges. Biotechnol J. 2022;17(7):2100507. doi: 10.1002/biot.202100507. [DOI] [PubMed] [Google Scholar]
- Bonhomme S, Cuer A, Delort AM, Lemaire J, Sancelme M, Scott G. Environmental biodegradation of polyethylene. Polym Degrad Stab. 2003;81(3):441–452. doi: 10.1016/S0141-3910(03)00129-0. [DOI] [Google Scholar]
- Bråte ILN, Eidsvoll DP, Steindal CC, Thomas KV. Plastic ingestion by Atlantic cod (Gadus morhua) from the Norwegian coast. Mar Pollut Bull. 2016;112(1–2):105–110. doi: 10.1016/j.marpolbul.2016.08.034. [DOI] [PubMed] [Google Scholar]
- Bryant JA, Clemente TM, Viviani DA, Fong AA, Thomas KA, Kemp P, Karl DM, White AE, DeLong EF (2016) Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems 1, 1–19. 10.1128/mSystems.00024-16 [DOI] [PMC free article] [PubMed]
- Bugoni L, Krause L, Petry MV. Marine debris and human impacts on sea turtles in southern Brazil. Mar Pollut Bull. 2001;42(12):1330–1334. doi: 10.1016/S0025-326X(01)00147-3. [DOI] [PubMed] [Google Scholar]
- Butnaru E, Darie-Niţă RN, Zaharescu T, Balaeş T, Tănase C, Hitruc G, Vasile C. Gamma irradiation assisted fungal degradation of the polypropylene/biomass composites. Radiat Phys Chem. 2016;125:134–144. doi: 10.1016/j.radphyschem.2016.04.003. [DOI] [Google Scholar]
- Cacciari I, Quatrini P, Zirletta G, Mincione E, Vinciguerra V, Lupattelli P, Giovannozzi Sermanni G. Isotactic polypropylene biodegradation by a microbial community: physicochemical characterization of metabolites produced. Appl Environ Microbiol. 1993;59(11):3695–3700. doi: 10.1128/aem.59.11.3695-3700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canniff PM, Hoang TC. Microplastic ingestion by Daphnia magna and its enhancement on algal growth. Sci Total Environ. 2018;633:500–507. doi: 10.1016/j.scitotenv.2018.03.176. [DOI] [PubMed] [Google Scholar]
- Capolupo M, Sørensen L, Jayasena KDR, Booth AM, Fabbri E. Chemical composition and ecotoxicity of plastic and car tire rubber leachates to aquatic organisms. Water Res. 2020;169:115270. doi: 10.1016/j.watres.2019.115270. [DOI] [PubMed] [Google Scholar]
- Carr SA, Liu J, Tesoro AG. Transport and fate of microplastic particles in wastewater treatment plants. Water Res. 2016;91:174–182. doi: 10.1016/j.watres.2016.01.002. [DOI] [PubMed] [Google Scholar]
- Caruso G. Microplastics as vectors of contaminants. Mar Pollut Bull. 2019;146:921–924. doi: 10.1016/j.marpolbul.2019.07.052. [DOI] [PubMed] [Google Scholar]
- Caspi R, Billington R, Keseler IM, Kothari A, Krummenacker M, Midford PE, Ong WK, Paley S, Subhraveti P, Karp PD. The MetaCyc database of metabolic pathways and enzymes-a 2019 update. Nucleic Acids Res. 2020;48(D1):D445–D453. doi: 10.1093/nar/gkz862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celina MDKO, Ottesen DK, Gillen KT, Clough RL. FTIR emission spectroscopy applied to polymer degradation. Polym Degrad Stab. 1997;58(1–2):15–31. doi: 10.1016/S0141-3910(96)00218-2. [DOI] [Google Scholar]
- Chae Y, Kim D, An YJ. Effects of micro-sized polyethylene spheres on the marine microalga Dunaliella salina: focusing on the algal cell to plastic particle size ratio. Aquat Toxicol. 2019;216:105296. doi: 10.1016/j.aquatox.2019.105296. [DOI] [PubMed] [Google Scholar]
- Chamas A, Moon H, Zheng J, Qiu Y, Tabassum T, Jang JH, Abu-Omar M, Scott SL, Suh S. Degradation rates of plastics in the environment. ACS Sustain Chem Eng. 2020;8(9):3494–3511. doi: 10.1021/acssuschemeng.9b06635. [DOI] [Google Scholar]
- Chandra R, Rustgi R. Biodegradation of maleated linear low-density polyethylene and starch blends. Polym Degrad Stab. 1997;56(2):185–202. doi: 10.1016/S0141-3910(96)00212-1. [DOI] [Google Scholar]
- Chen CC, Dai L, Ma L, Guo RT. Enzymatic degradation of plant biomass and synthetic polymers. Nat Rev Chem. 2020;4(3):114–126. doi: 10.1038/s41570-020-0163-6. [DOI] [PubMed] [Google Scholar]
- Chen J, Wu J, Sherrell PC, Chen J, Wang H, Zhang WX, Yang J. How to build a microplastics-free environment: strategies for microplastics degradation and plastics recycling. Adv Sci. 2022;9(6):2103764. doi: 10.1002/advs.202103764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia WY, Tang DYY, Khoo KS, Lup ANK, Chew KW. Nature’s fight against plastic pollution: algae for plastic biodegradation and bioplastics production. Environ Sci Ecotechnol. 2020;4:100065. doi: 10.1016/j.ese.2020.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia RW, Lee JY, Kim H, Jang J. Microplastic pollution in soil and groundwater: a review. Environ Chem Lett. 2021;19(6):4211–4224. doi: 10.1007/s10311-021-01297-6. [DOI] [Google Scholar]
- Cholewinski A, Dadzie E, Sherlock C, Anderson WA, Charles TC, Habib K, Zhao B. A critical review of microplastic degradation and material flow analysis towards a circular economy. Environ Pollut. 2022 doi: 10.1016/j.envpol.2022.120334. [DOI] [PubMed] [Google Scholar]
- Cox KD, Covernton GA, Davies HL, Dower JF, Juanes F, Dudas SE. Human consumption of microplastics. Environ Sci Technol. 2019;53(12):7068–7074. doi: 10.1021/acs.est.0c04032. [DOI] [PubMed] [Google Scholar]
- Curren E, Leaw CP, Lim PT, Leong SCY. Evidence of marine microplastics in commercially harvested seafood. Front Bioeng Biotechnol. 2020;8:562760. doi: 10.3389/fbioe.2020.562760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi SM, Miri S, Taheran M, Brar SK, Galvez-Cloutier R, Martel R. Bioremediation of unconventional oil contaminated ecosystems under natural and assisted conditions: a review. Environ Sci Technol. 2020;54(4):2054–2067. doi: 10.1021/acs.est.9b00906. [DOI] [PubMed] [Google Scholar]
- Dobretsov S, Abed RM, Teplitski M. Mini-review: Inhibition of biofouling by marine microorganisms. Biofouling. 2013;29(4):423–441. doi: 10.1080/08927014.2013.776042. [DOI] [PubMed] [Google Scholar]
- de Oliveira TA, Barbosa R, Mesquita AB, Ferreira JH, de Carvalho LH, Alves TS. Fungal degradation of reprocessed PP/PBAT/thermoplastic starch blends. J Mater Res Technol. 2020;9(2):2338–2349. doi: 10.1016/j.jmrt.2019.12.065. [DOI] [Google Scholar]
- Debroas D, Mone A, Ter Halle A. Plastics in the North Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci Total Environ. 2017;599–600:1222–1232. doi: 10.1016/j.scitotenv.2017.05.059. [DOI] [PubMed] [Google Scholar]
- Debroy A, George N, Mukherjee G. Role of biofilms in the degradation of microplastics in aquatic environments. J Chem Technol Biotechnol. 2021 doi: 10.1002/jctb.6978. [DOI] [Google Scholar]
- Delacuvellerie A, Cyriaque V, Gobert S, Benali S, Wattiez R. The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J Hazard Mater. 2019;380:120899. doi: 10.1016/j.jhazmat.2019.120899. [DOI] [PubMed] [Google Scholar]
- De-la-Torre GE. Microplastics: an emerging threat to food security and human health. J Food Sci Technol. 2020;57:1601–1608. doi: 10.1007/s13197-019-04138-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Torre GE, Aragaw TA. What we need to know about PPE associated with the COVID-19 pandemic in the marine environment. Mar Pollut Bull. 2021;163:111879. doi: 10.1016/j.marpolbul.2020.111879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-la-Torre GE, Dioses-Salinas DC, Pérez-Baca BL, Millones Cumpa LA, Pizarro- Ortega CI, Torres FG, Gonzales KN, Santillán L. Marine macroinvertebrates inhabiting plastic litter in Peru. Mar Pollut Bull. 2021;167:112296. doi: 10.1016/j.marpolbul.2021.112296. [DOI] [PubMed] [Google Scholar]
- Derraik JGB. The pollution of the marine environment by plastic debris: a review. Mar Pollut Bull. 2002;44:842–852. doi: 10.1016/S0025-326X(02)00220-5. [DOI] [PubMed] [Google Scholar]
- Devi RS, Kannan VR, Nivas D, Kannan K, Chandru S, Antony AR. Biodegradation of HDPE by Aspergillus spp. from marine ecosystem of Gulf of Mannar, India. Mar Pollut Bull. 2015;96(1–2):32–40. doi: 10.1016/j.marpolbul.2015.05.050. [DOI] [PubMed] [Google Scholar]
- Donelli I, Taddei P, Smet PF, Poelman D, Nierstrasz VA, Freddi G. Enzymatic surface modification and functionalization of PET: a water contact angle, FTIR, and fluorescence spectroscopy study. Biotechnol Bioeng. 2009;103(5):845–856. doi: 10.1002/bit.22316. [DOI] [PubMed] [Google Scholar]
- Dris R, Gasperi J, Mirande C, Mandin C, Guerrouache M, Langlois V, Tassin B. A first overview of textile fibers, including microplastics, in indoor and outdoor environments. Environ Pollut. 2017;221:453–458. doi: 10.1016/j.envpol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Du H, Xie Y, Wang J. Microplastic degradation methods and corresponding degradation mechanism: research status and future perspectives. J Hazard Mater. 2021;418:126377. doi: 10.1016/j.jhazmat.2021.126377. [DOI] [PubMed] [Google Scholar]
- Dussud C, Meistertzheim AL, Conan P, Pujo-Pay M, George M, Fabre P, Coudane J, Higgs P, Elineau A, Pedrotti ML, Gorsky G, Ghiglione JF. Evidence of niche partitioning among bacteria living on plastics, organic particles and surrounding seawaters. Environ Pollut. 2018;236:807–816. doi: 10.1016/j.envpol.2017.12.027. [DOI] [PubMed] [Google Scholar]
- Enders K, Lenz R, Stedmon CA, Nielsen TG. Abundance, size and polymer composition of marine microplastics≥ 10 μm in the Atlantic Ocean and their modelled vertical distribution. Mar Pollut Bull. 2015;100(1):70–81. doi: 10.1016/j.marpolbul.2015.09.027. [DOI] [PubMed] [Google Scholar]
- Esmaeili A, Pourbabaee AA, Alikhani HA, Shabani F, Esmaeili E. Biodegradation of low-density polyethylene (LDPE) by mixed culture of Lysinibacillus xylanilyticus and Aspergillus niger in soil. PLoS ONE. 2013;8(9):e71720. doi: 10.1371/journal.pone.0071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everaert G, De Rijcke M, Lonneville B, Janssen CR, Backhaus T, Mees J, van Sebille E, Koelmans AA, Catarino AI, Vandegehuchte MB. Risks of floating microplastic in the global ocean. Environ Pollut. 2020;267:115499. doi: 10.1016/j.envpol.2020.115499. [DOI] [PubMed] [Google Scholar]
- Federici S, Ademovic Z, Amorim MJB, Bigalke M, Cocca M, Depero LE, Dutta J, Fritzsche W, Hartmann NB, Kalčikova G, Keller N, Meisel TC, Mitrano DM, Morrison L, Raquez J-M, Tubić A, Velimirovic M. COST action priority: an EU perspective on micro- and nanoplastics as global issues. Microplastics. 2022;1:282–290. doi: 10.3390/microplastics1020020. [DOI] [Google Scholar]
- Fendall LS, Sewell MA. Contributing to marine pollution by washing your face: microplastics in facial cleansers. Mar Pollut Bull. 2009;58(8):1225–1228. doi: 10.1016/j.marpolbul.2009.04.025. [DOI] [PubMed] [Google Scholar]
- Fontanella S, Bonhomme S, Brusson J, Pitteri S, Samuel G, Pichon G, et al. Comparison of biodegradability of various polypropylene films containing pro-oxidant additives based on Mn, Mn/Fe or Co. Polym Degrad Stab. 2013;98:875–884. doi: 10.1016/j.polymdegradstab.2013.01.002. [DOI] [Google Scholar]
- French KE, Zhou Z, Terry N. Horizontal ‘gene drives’ harness indigenous bacteria for bioremediation. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-72138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias JP, Nash R. Microplastics: finding a consensus on the definition. Mar Pollut Bull. 2019;138:145–147. doi: 10.1016/j.marpolbul.2018.11.022. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway TS, Cole M, Lewis C. Interactions of microplastic debris throughout the marine ecosystem. Nat Ecol Evol. 2017;1(5):1–8. doi: 10.1038/s41559-017-0116. [DOI] [PubMed] [Google Scholar]
- Gao J, Ellis LB and Wackett LP (2010) The university of Minnesota biocatalysis/biodegradation database: improving public access. Nucleic Acids Res, 38(suppl_1), D488-D491. 10.1093/nar/gkp771 [DOI] [PMC free article] [PubMed]
- Gerritse J, Leslie HA, de Tender CA, Devriese LI, Vethaak AD. Fragmentation of plastic objects in a laboratory seawater microcosm. Sci Rep. 2020;10(1):1–16. doi: 10.1038/s41598-020-67927-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GESAMP (2019) Guidelines for the monitoring and assessment of plastic litter and microplastics in the ocean. 130. In: Kershaw PJ, Turra A, Galgani F (Eds.), (IMO/FAO/UNESCO-IOC/UNIDO/WMO/IAEA/UN/UNEP/UNDP/ISA Joint group of experts on the scientific aspects of marine environmental protection). GESAMP Rep. Stud. No. 99 (2019). http://www.gesamp.org/publications/guidelines-for-the-monitoring-and-assessment-of-plastic-litter-in-the-ocean
- Ghosh SK, Pal S, Ray S. Study of microbes having potentiality for biodegradation of plastics. Environ Sci Pollut Res. 2013;20(7):4339–4355. doi: 10.1007/s11356-013-1706-x. [DOI] [PubMed] [Google Scholar]
- Gironi F, Piemonte V. Bioplastics and petroleum-based plastics: strengths and weaknesses. Energy Sources, Part a: Recovery, Utilization, Environ Eff. 2011;33(21):1949–1959. doi: 10.1080/15567030903436830. [DOI] [Google Scholar]
- Gómez-Méndez LD, Moreno-Bayona DA, Poutou-Pinales RA, Salcedo-Reyes JC, Pedroza-Rodríguez AM, Vargas A, Bogoya JM. Biodeterioration of plasma pretreated LDPE sheets by Pleurotus ostreatus. PLoS ONE. 2018;13(9):e0203786. doi: 10.1371/journal.pone.0203786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MR. Environmental implications of plastic debris in marine settings–entanglement, ingestion, smothering, hangers-on, hitch-hiking and alien invasions. Philos Trans R Soc Lond B Biol Sci. 2009;364(1526):2013–2025. doi: 10.1098/rstb.2008.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh KJ, Backhaus T, Carney-Almroth B, Geueke B, Inostroza PA, Lennquist A, Leslie HA, Maffini M, Slunge D, Trasande L, Warhurst AM. Overview of known plastic packaging-associated chemicals and their hazards. Sci Total Environ. 2019;651:3253–3268. doi: 10.1016/j.scitotenv.2018.10.015. [DOI] [PubMed] [Google Scholar]
- Gruber ES, Stadlbauer V, Pichler V, Resch-Fauster K, Todorovic A, Meisel TC, Kenner L. To waste or not to waste: questioning potential health risks of micro-and nanoplastics with a focus on their ingestion and potential carcinogenicity. Expos Health. 2022 doi: 10.1007/s12403-022-00470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumel AM, Annuar MSM, Chisti Y. Recent advances in the production, recovery and applications of polyhydroxyalkanoates. J Polym Environ. 2013;21(2):580–605. doi: 10.1007/s10924-012-0527-1. [DOI] [Google Scholar]
- Guo XP, Sun XL, Chen YR, Hou L, Liu M, Yang Y. Antibiotic resistance genes in biofilms on plastic wastes in an estuarine environment. Sci Total Environ. 2020;745:140916. doi: 10.1016/j.scitotenv.2020.140916. [DOI] [PubMed] [Google Scholar]
- Guzzetti E, Sureda A, Tejada S, Faggio C. Microplastic in marine organism: environmental and toxicological effects. Environ Toxicol Pharmacol. 2018;64:164–171. doi: 10.1016/j.etap.2018.10.009. [DOI] [PubMed] [Google Scholar]
- Habib S, Iruthayam A, Abd Shukor MY, Alias SA, Smykla J, Yasid NA. Biodeterioration of untreated polypropylene microplastic particles by antarctic bacteria. Polymers. 2020;12(11):2616. doi: 10.3390/polym12112616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale RC, Seeley ME, La Guardia MJ, Mai L, Zeng EY. A global perspective on microplastics. J Geophys Res: Oceans. 2020;125(1):e2018JC01479. doi: 10.1029/2018JC014719. [DOI] [Google Scholar]
- Hartmann NB, Huffer T, Thompson RC, Hassellöv M, Verschoor A, Daugaard AE, Rist S, Karlsson T, Brennholt N, Cole M and Herrling MP (2019) Are we speaking the same language? Recommendations for a definition and categorization framework for plastic debris. 10.1021/acs.est.8b05297 [DOI] [PubMed]
- Helen AS, Uche EC, Hamid FS. Screening for polypropylene degradation potential of bacteria isolated from mangrove ecosystems in Peninsular Malaysia. Int J Biosci Biochem Bioinform. 2017;7:245–251. [Google Scholar]
- Hirooka T, Nagase H, Uchida K, Hiroshige Y, Ehara Y, Nishikawa JI, Nishihara T, Miyamoto K, Hirata Z. Biodegradation of bisphenol A and disappearance of its estrogenic activity by the green alga Chlorella fusca var. vacuolata. Environ Toxicol Chem: Int J. 2005;24(8):1896–1901. doi: 10.1897/04-259R.1. [DOI] [PubMed] [Google Scholar]
- Honjo H, Iwasaki K, Soma Y, Tsuruno K, Hamada H, Hanai T. Synthetic microbial consortium with specific roles designated by genetic circuits for cooperative chemical production. Metab Eng. 2019;55:268–275. doi: 10.1016/j.ymben.2019.08.007. [DOI] [PubMed] [Google Scholar]
- Huang Z, Hu B, Wang H. Analytical methods for microplastics in the environment: a review. Environ Chem Lett. 2022 doi: 10.1007/s10311-022-01525-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta Lwanga E, Thapa B, Yang X, Gertsen H, Salánki T, Geissen V, Garbeva P. Decay of low-density polyethylene by bacteria extracted from earthworm's guts: a potential for soil restoration. Sci Total Environ. 2018;624:753–757. doi: 10.1016/j.scitotenv.2017.12.144. [DOI] [PubMed] [Google Scholar]
- Ibrahim IN, Maraqa A, Hameed KM, Saadoun IM, Maswadeh HM. Assessment of potential plastic-degrading fungi in Jordanian habitats. Turk J Bio. 2011;35(5):551–557. doi: 10.3906/biy-0901-9. [DOI] [Google Scholar]
- Inderthal H, Tai SL, Harrison ST. Non-hydrolyzable plastics–an interdisciplinary look at plastic bio-oxidation. Trends Biotechnol. 2021;39(1):12–23. doi: 10.1016/j.tibtech.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Islam S, Apitius L, Jakob F, Schwaneberg U. Targeting microplastic particles in the void of diluted suspensions. Environ Int. 2019;123:428–435. doi: 10.1016/j.envint.2018.12.029. [DOI] [PubMed] [Google Scholar]
- Ivleva NP. Chemical analysis of microplastics and nanoplastics: challenges, advanced methods, and perspectives. Chem Rev. 2021;121(19):11886–11936. doi: 10.1021/acs.chemrev.1c00178. [DOI] [PubMed] [Google Scholar]
- Jain K, Bhunia H, Reddy MS. Degradation of polypropylene-poly-L-lactide blend by bacteria isolated from compost. Bioremed J. 2018;22(3–4):73–90. doi: 10.1080/10889868.2018.1516620. [DOI] [Google Scholar]
- Jain K, Bhunia H, Reddy MS. Degradation of polypropylene-poly-L-lactide blends by Bacillus isolates: a microcosm and field evaluation. Bioremediation J. 2022;26(1):64–75. doi: 10.1080/10889868.2021.1886037. [DOI] [Google Scholar]
- Jambeck JR, Geyer R, Wilcox C, Siegler TR, Perryman M, Andrady A, Narayan R, Law KL. Plastic waste inputs from land into the ocean. Science. 2015;347(6223):768–771. doi: 10.1126/science.1260352. [DOI] [PubMed] [Google Scholar]
- Janssen PH, Yates PS, Grinton BE, Taylor PM, Sait M. Acidobacteria Actinobacteria Proteobacteria Verrucomicrobia. Appl Environ Microbiol. 2002;68(5):2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner LC, Rotchell JM, Bennett RT, Cowen M, Tentzeris V, Sadofsky LR. Detection of microplastics in human lung tissue using μFTIR spectroscopy. Sci Total Environ. 2022;831:154907. doi: 10.1016/j.scitotenv.2022.154907. [DOI] [PubMed] [Google Scholar]
- Jeon HJ, Kim MN. Isolation of a thermophilic bacterium capable of low molecular–weight polyethylene degradation. Biodegradation. 2013;24:89–98. doi: 10.1007/s10532-012-9560-y. [DOI] [PubMed] [Google Scholar]
- Jeon HJ, Kim MN. Isolation of mesophilic bacterium for biodegradation of polypropylene. Int Biodeter Biodegrad. 2016;115:244–249. doi: 10.1016/j.ibiod.2016.08.025. [DOI] [Google Scholar]
- Jeon J-M, Park S-J, Choi T-R, Park J-H, Yang Y-H, Yoon J-J. Biodegradation of polyethylene and polypropylene by Lysinibacillus species JJY0216 isolated from soil grove. Poly Degrad Stab. 2021;191:109662. doi: 10.1016/j.polymdegradstab.2021.109662. [DOI] [Google Scholar]
- Ji MK, Kabra AN, Choi J, Hwang JH, Kim JR, Abou-Shanab RA, Oh YK, Jeon BH. Biodegradation of bisphenol A by the freshwater microalgae Chlamydomonas mexicana and Chlorella vulgaris. Ecol Eng. 2014;73:260–269. doi: 10.1016/j.ecoleng.2014.09.070. [DOI] [Google Scholar]
- Ji X, Lu P, van der Veen S. Development of a dual-antimicrobial counterselection method for markerless genetic engineering of bacterial genomes. Appl Microbiol Biotechnol. 2019;103(3):1465–1474. doi: 10.1007/s00253-018-9565-5. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Yan W, Tan Q, Xiao Y, Shi Y, Lei J, Li Z, Hou Y, Liu T, Li Y. The toxic differentiation of micro-and nanoplastics verified by gene-edited fluorescent Caenorhabditis elegans. Sci Total Environ. 2022 doi: 10.1016/j.scitotenv.2022.159058. [DOI] [PubMed] [Google Scholar]
- John J, Nandhini AR, Velayudhaperumal Chellam P, Sillanpää M. Microplastics in mangroves and coral reef ecosystems: a review. Environ Chem Lett. 2021 doi: 10.1007/s10311-021-01326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Billington R, Caspi R, Fulcher CA, Latendresse M, Kothari A, Keseler IM, Krummenacker M, Midford PE, Ong Q, Ong WK. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2019;20(4):1085–1093. doi: 10.1093/bib/bbx085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay P, Hiscoe R, Moberley I, Bajic L, McKenna N. Wastewater treatment plants as a source of microplastics in river catchments. Environ Sci Pollut Res. 2018;25(20):20264–20267. doi: 10.1007/s11356-018-2070-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswani A, Oliver DM, Gutierrez T, Quilliam RS. Microbial hitchhikers on marine plastic debris: human exposure risks at bathing waters and beach environments. Mar Environ Res. 2016;118:10–19. doi: 10.1016/j.marenvres.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Khan S, Nadir S, Shah ZU, Shah AA, Karunarathna SC, Xu J, Hasan F. Biodegradation of polyester polyurethane by Aspergillus tubingensis. Environ Pollut. 2017;225:469–480. doi: 10.1016/j.envpol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- Khoironi A, Anggoro S. Evaluation of the interaction among microalgae Spirulina sp, plastics polyethylene terephthalate and polypropylene in freshwater environment. J. Ecol. Eng. 2019;20:161–173. doi: 10.12911/22998993/108637. [DOI] [Google Scholar]
- Kim JW, Park SB, Tran QG, Cho DH, Choi DY, Lee YJ, Kim HS. Functional expression of polyethylene terephthalate-degrading enzyme (PETase) in green microalgae. Microb Cell Fact. 2020;19(1):1–9. doi: 10.1186/s12934-020-01355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto D, Yanagishita H, Endo A, Nakaiwa M, Nakane T, Akiya T. Remarkable antiagglomeration effect of a yeast biosurfactant, diacylmannosylerythritol, on ice-water slurry for cold thermal storage. Biotechnol Prog. 2001;17(2):362–365. doi: 10.1021/bp000159f. [DOI] [PubMed] [Google Scholar]
- Koelmans AA, Redondo-Hasselerharm PE, Mohamed Nor NH, Kooi M. Solving the nonalignment of methods and approaches used in microplastic research to consistently characterize risk. Environ Sci Technol. 2020;54(19):12307–12315. doi: 10.1021/acs.est.0c02982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HG, Kim HH, Chung JH, Jun J, Lee S, Kim HM, Ryu CM. The Galleria mellonella hologenome supports microbiota-independent metabolism of long-chain hydrocarbon beeswax. Cell Rep. 2019;26(9):2451–2464. doi: 10.1016/j.celrep.2019.02.018. [DOI] [PubMed] [Google Scholar]
- Kotova IB, Taktarova YV, Tsavkelova EA, Egorova MA, Bubnov IA, Malakhova DV, Bonch-Osmolovskaya EA. Microbial degradation of plastics and approaches to make it more efficient. Microbiology. 2021;90(6):671–701. doi: 10.1134/S0026261721060084. [DOI] [Google Scholar]
- Kowalczyk A, Chyc M, Ryszka P, Latowski D. Achromobacter xylosoxidans as a new microorganism strain colonizing high-density polyethylene as a key step to its biodegradation. Environ Sci Pollut Res. 2016;23(11):11349–11356. doi: 10.1007/s11356-016-6563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Xiong X, He M, Tsang DC, Gupta J, Khan E, Bolan NS. Microplastics as pollutants in agricultural soils. Environ Pollut. 2020;265:114980. doi: 10.1016/j.envpol.2020.114980. [DOI] [PubMed] [Google Scholar]
- Kunlere IO, Fagade OE, Nwadike BI. Biodegradation of low density polyethylene (LDPE) by certain indigenous bacteria and fungi. Int J Environ Stud. 2019;76(3):428–440. doi: 10.1080/00207233.2019.1579586. [DOI] [Google Scholar]
- Lin Z, Jin T, Zou T, Xu L, Xi B, Xu D, Fei J. Current progress on plastic/microplastic degradation: Fact influences and mechanism. Environ Pollut. 2022;304:119159. doi: 10.1016/j.envpol.2022.119159. [DOI] [PubMed] [Google Scholar]
- La Nasa J, Lomonaco T, Manco E, Ceccarini A, Fuoco R, Corti A, Modugno F, Castelvetro V, Degano I. Plastic breeze: Volatile organic compounds (VOCs) emitted by degrading macro-and microplastics analyzed by selected ion flow-tube mass spectrometry. Chemosphere. 2021;270:128612. doi: 10.1016/j.chemosphere.2020.128612. [DOI] [PubMed] [Google Scholar]
- Lameh F, Baseer AQ, Ashiru AG. Comparative molecular docking and molecular-dynamic simulation of wild-type-and mutant carboxylesterase with BTA-hydrolase for enhanced binding to plastic. Eng Life Sci. 2022;22(1):13–29. doi: 10.1002/elsc.202100083. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leslie HA, Van Velzen MJ, Brandsma SH, Vethaak AD, Garcia-Vallejo JJ, Lamoree MH. Discovery and quantification of plastic particle pollution in human blood. Environ Int. 2022;163:107199. doi: 10.1016/j.envint.2022.107199. [DOI] [PubMed] [Google Scholar]
- Li R, Chen GZ, Tam NFY, Luan TG, Shin PK, Cheung SG, Liu Y. Toxicity of bisphenol A and its bioaccumulation and removal by a marine microalga Stephanodiscus hantzschii. Ecotoxicol Environ Saf. 2009;72(2):321–328. doi: 10.1016/j.ecoenv.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Li J, Liu H, Chen JP. Microplastics in freshwater systems: a review on occurrence, environmental effects, and methods for microplastics detection. Water Res. 2018;137:362–374. doi: 10.1016/j.watres.2017.12.056. [DOI] [PubMed] [Google Scholar]
- Li Q, Ma C, Zhang Q, Shi H. Microplastics in shellfish and implications for food safety. Curr Opin Food Sci. 2021;40:192–197. doi: 10.1016/j.cofs.2021.04.017. [DOI] [Google Scholar]
- Liu G, Jiang R, You J, Muir DC, Zeng EY. Microplastic impacts on microalgae growth: effects of size and humic acid. Environ Sci Technol. 2019;54(3):1782–1789. doi: 10.1021/acs.est.9b06187. [DOI] [PubMed] [Google Scholar]
- Liu SY, Leung MML, Fang JKH, Chua SL. Engineering a microbial ‘trap and release’ mechanism for microplastics removal. Chem Eng J. 2021;404:127079. doi: 10.1016/j.cej.2020.127079. [DOI] [Google Scholar]
- Liu Q, Chen Y, Chen Z, Yang F, Xie Y, Yao W. Current status of microplastics and nanoplastics removal methods: Summary, comparison and prospect. Sci Total Environ. 2022 doi: 10.1016/j.scitotenv.2022.157991. [DOI] [PubMed] [Google Scholar]
- Lusher AL, Hurley R, Arp HPH, Booth AM, Bråte ILN, Gabrielsen GW, Gomiero A, Gomes T, Grøsvik BE, Green N, Haave M. Moving forward in microplastic research: a Norwegian perspective. Environ Int. 2021;157:106794. doi: 10.1016/j.envint.2021.106794. [DOI] [PubMed] [Google Scholar]
- Magnin A, Pollet E, Phalip V, Avérous L. Evaluation of biological degradation of polyurethanes. Biotechnol Adv. 2020;39:107457. doi: 10.1016/j.biotechadv.2019.107457. [DOI] [PubMed] [Google Scholar]
- Mandal S, Ghorai M, Anand U, Roy D, Kant N, Mishra T, Mane AB, Jha NK, Lal MK, Tiwari RK, Kumar M. Cytokinins: a genetic target for increasing yield potential in the CRISPR era. Front Genet. 2022 doi: 10.3389/fgene.2022.883930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzi HP, Abou-Shanab RA, Jeon BH, Wang J, Salama ES. Algae: a frontline photosynthetic organism in the microplastic catastrophe. Trends Plant Sci. 2022;S1360–1385(22):00157–161. doi: 10.1016/j.tplants.2022.06.005. [DOI] [PubMed] [Google Scholar]
- Marguet P, Tanouchi Y, Spitz E, Smith C, You L. Oscillations by minimal bacterial suicide circuits reveal hidden facets of host-circuit physiology. PLoS ONE. 2010;5(7):e11909. doi: 10.1371/journal.pone.0011909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matjašič T, Simčič T, Medvešček N, Bajt O, Dreo T, Mori N. Critical evaluation of biodegradation studies on synthetic plastics through a systematic literature review. Sci Total Environ. 2021;752:141959. doi: 10.1016/j.scitotenv.2020.141959. [DOI] [PubMed] [Google Scholar]
- Mehmood CT, Qazi IA, Hashmi I, Bhargava S, Deepa S. Biodegradation of low density polyethylene (LDPE) modified with dye sensitized titania and starch blend using Stenotrophomonas pavanii. Int Biodeterior Biodegrad. 2016;113:276–286. doi: 10.1016/j.ibiod.2016.01.025. [DOI] [Google Scholar]
- Mendoza LMR, Balcer M. Microplastics in freshwater environments: a review of quantification assessment. TrAC Trends Anal Chem. 2019;113:402–408. doi: 10.1016/j.trac.2018.10.020. [DOI] [Google Scholar]
- Mendoza LMR, Karapanagioti H, Álvarez NR. Micro (nanoplastics) in the marine environment: current knowledge and gaps. Curr Opin Environ Sci Health. 2018;1:47–51. doi: 10.1016/j.coesh.2017.11.004. [DOI] [Google Scholar]
- Miao L, Wang P, Hou J, Yao Y, Liu Z, Liu S, Li T. Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci Total Environ. 2019;650:2395–2402. doi: 10.1016/j.scitotenv.2018.09.378. [DOI] [PubMed] [Google Scholar]
- Miao F, Liu Y, Gao M, Yu X, Xiao P, Wang M, Wang S, Wang X. Degradation of polyvinyl chloride microplastics via an electro-Fenton-like system with a TiO2/graphite cathode. J Hazard Mater. 2020;399:123023. doi: 10.1016/j.jhazmat.2020.123023. [DOI] [PubMed] [Google Scholar]
- Miri S, Saini R, Davoodi SM, Pulicharla R, Brar SK, Magdouli S. Biodegradation of microplastics: better late than never. Chemosphere. 2022;286:131670. doi: 10.1016/j.chemosphere.2021.131670. [DOI] [PubMed] [Google Scholar]
- Mohamed MSM, El-Arabi NI, El-Hussein A, El-Maaty SA, Abdelhadi AA. Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol. 2020;65(4):687–696. doi: 10.1007/s12223-020-00771-y. [DOI] [PubMed] [Google Scholar]
- Moog D, Schmitt J, Senger J, Zarzycki J, Rexer KH, Linne U, Erb T, Maier UG. Using a marine microalga as a chassis for polyethylene terephthalate (PET) degradation. Microb Cell Fact. 2019;18(1):1–15. doi: 10.1186/s12934-019-1220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgana S, Casentini B, Amalfitano S. Uncovering the release of micro/nanoplastics from disposable face masks at times of COVID-19. J Hazard Mater. 2021;419:126507. doi: 10.1016/j.jhazmat.2021.126507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor R, Sivan A. Biofilm formation and partial biodegradation of polystyrene by the actinomycete Rhodococcus ruber: biodegradation of polystyrene. Biodegrad. 2008;19:851–858. doi: 10.1007/s10532-008-9188-0. [DOI] [PubMed] [Google Scholar]
- Muenmee S, Chiemchaisri W, Chiemchaisri C. Enhancement of biodegradation of plastic wastes via methane oxidation in semi-aerobic landfill. Int J Biodeterior Biodegrad. 2016;113:244–255. doi: 10.1016/j.ibiod.2016.03.016. [DOI] [Google Scholar]
- Muthukrishnan T, Al Khaburi M, Abed RMM. Fouling microbial communities on plastics compared with wood and steel: are they substrate- or location-specific? Microb Ecol. 2019;78:361–374. doi: 10.1007/s00248-018-1303-0. [DOI] [PubMed] [Google Scholar]
- Naqash N, Prakash S, Kapoor D, Singh R. Interaction of freshwater microplastics with biota and heavy metals: a review. Environ Chem Lett. 2020;18(6):1813–1824. doi: 10.1007/s10311-020-01044-3. [DOI] [Google Scholar]
- Nidhi S, Anand U, Oleksak P, Tripathi P, Lal JA, Thomas G, Kuca K, Tripathi V. Novel CRISPR–Cas systems: an updated review of the current achievements, applications, and future research perspectives. Int J Mol Sci. 2021;22(7):3327. doi: 10.3390/ijms22073327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzetto L, Futter M, Langaas S. Are agricultural soils dumps for microplastics of urban origin? Environ Sci Technol. 2016;50(20):10777–10779. doi: 10.1021/acs.est.6b04140. [DOI] [PubMed] [Google Scholar]
- Nowak B, Pająk J, Drozd-Bratkowicz M, Rymarz G. Microorganisms participating in the biodegradation of modified polyethylene films in different soils under laboratory conditions. Int Biodeterior Biodegradation. 2011;65(6):757–767. doi: 10.1016/j.ibiod.2011.04.007. [DOI] [Google Scholar]
- Nowak B, Pajk J, Karcz J (2012) Biodegradation of pre-aged modified polyethylene films. In: Kazmiruk, V. (Ed.), Scanning Electron Microscopy. InTech. 10.5772/35128
- Nwachukwu S, Obidi O, Odocha C. Occurrence and recalcitrance of polyethylene bag waste in Nigerian soils. Afr J Biotechnol. 2010;9(37):6096–6104. [Google Scholar]
- Ogonowski M, Gerdes Z, Gorokhova E. What we know and what we think we know about microplastic effects–a critical perspective. Curr Opin Environ Sci Health. 2018;1:41–46. doi: 10.1016/j.coesh.2017.09.001. [DOI] [Google Scholar]
- Olicón-Hernández DR, González-López J, Aranda E. Overview on the biochemical potential of filamentous fungi to degrade pharmaceutical compounds. Front Microbiol. 2017;8:1792. doi: 10.3389/fmicb.2017.01792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira J, Belchior A, da Silva VD, Rotter A, Petrovski Ž, Almeida PL, Gaudêncio SP. Marine environmental plastic pollution: mitigation by microorganism degradation and recycling valorization. Front Mar Sci. 2020;7:567126. doi: 10.3389/fmars.2020.567126. [DOI] [Google Scholar]
- Othman AR, Hasan HA, Muhamad MH, Ismail NI, Abdullah SRS. Microbial degradation of microplastics by enzymatic processes: a review. Environ Chem Lett. 2021;19(4):3057–3073. doi: 10.1007/s10311-021-01197-9. [DOI] [Google Scholar]
- Ozyigit II, Can H, Dogan I. Phytoremediation using genetically engineered plants to remove metals: a review. Environ Chem Lett. 2021;19(1):669–698. doi: 10.1007/s10311-020-01095-6. [DOI] [Google Scholar]
- Paço A, Duarte K, da Costa JP, Santos PS, Pereira R, Pereira ME, Freitas AC, Duarte AC, Rocha-Santos TA. Biodegradation of polyethylene microplastics by the marine fungus Zalerion maritimum. Sci Total Environ. 2017;586:10–15. doi: 10.1016/j.scitotenv.2017.02.017. [DOI] [PubMed] [Google Scholar]
- Paço A, Jacinto J, da Costa JP, Santos PS, Vitorino R, Duarte AC, Rocha-Santos T. Biotechnological tools for the effective management of plastics in the environment. Crit Rev Environ Sci Technol. 2019;49(5):410–441. doi: 10.1080/10643389.2018.1548862. [DOI] [Google Scholar]
- Padervand M, Lichtfouse E, Robert D, Wang C. Removal of microplastics from the environment. A review. Environ Chem Lett. 2020;18(3):807–828. doi: 10.1007/s10311-020-00983-1. [DOI] [Google Scholar]
- Pan Z, Liu Q, Xu J, Li W, Lin H. Microplastic contamination in seafood from Dongshan Bay in southeastern China and its health risk implication for human consumption. Environ Pollut. 2022;303:119163. doi: 10.1016/j.envpol.2022.119163. [DOI] [PubMed] [Google Scholar]
- Park SY, Kim CG. Biodegradation of micro-polyethylene particles by bacterial colonization of a mixed microbial consortium isolated from a landfill site. Chemosphere. 2019;222:527–533. doi: 10.1016/j.chemosphere.2019.01.159. [DOI] [PubMed] [Google Scholar]
- Patel MM, Goyal BR, Bhadada SV, Bhatt JS, Amin AF. Getting into the Brain. CNS Drugs. 2009;23(1):35–58. doi: 10.2165/0023210-200923010-00003. [DOI] [PubMed] [Google Scholar]
- Peng L, Fu D, Qi H, Lan CQ, Yu H, Ge C. Micro-and nano-plastics in marine environment: source, distribution and threats—a review. Sci Total Environ. 2020;698:134254. doi: 10.1016/j.scitotenv.2019.134254. [DOI] [PubMed] [Google Scholar]
- PlasticsEurope. Plastics–the Facts (2020) Avenue E. van Nieuwenhuyse 4/3, 1160 Brussels. Belgium: PlasticsEurope. https://www.plasticseurope.org/de/resources/publications/4312-plastics-facts-2020
- Prata JC, Silva ALP, Walker TR, Duarte AC, Rocha-Santos T. COVID-19 pandemic repercussions on the use and management of plastics. Environ Sci Technol. 2020;54:7760–7765. doi: 10.1021/acs.est.0c02178. [DOI] [PubMed] [Google Scholar]
- Purohit J, Chattopadhyay A, Teli B. Metagenomic exploration of plastic degrading microbes for biotechnological application. Curr Genomics. 2020;21(4):253–270. doi: 10.2174/1389202921999200525155711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZH, Mou JH, Chao CYH, Chopra SS, Daoud W, Leu SY, Lin CSK. Biotechnology of plastic waste degradation, recycling, and valorization: current advances and future perspectives. Chemsuschem. 2021;14(19):4103–4114. doi: 10.1002/cssc.202100752. [DOI] [PubMed] [Google Scholar]
- Raddadi N, Fava F. Biodegradation of oil-based plastics in the environment: existing knowledge and needs of research and innovation. Sci Total Environ. 2019;679:148–158. doi: 10.1016/j.scitotenv.2019.04.419. [DOI] [PubMed] [Google Scholar]
- Rahman A, Sarkar A, Yadav OP, Achari G, Slobodnik J. Potential human health risks due to environmental exposure to nano-and microplastics and knowledge gaps: a scoping review. Sci Total Environ. 2021;757:143872. doi: 10.1016/j.scitotenv.2020.143872. [DOI] [PubMed] [Google Scholar]
- Rajandas H, Parimannan S, Sathasivam K, Ravichandran M, Yin LS. A novel FTIR-ATR spectroscopy based technique for the estimation of low-density polyethylene biodegradation. Polym Test. 2012;31:1094–1099. doi: 10.1016/j.polymertesting.2012.07.015. [DOI] [Google Scholar]
- Razeghi N, Hamidian AH, Wu C, Zhang Y, Yang M. Microplastic sampling techniques in freshwaters and sediments: a review. Environ Chem Lett. 2021;19(6):4225–4252. doi: 10.1007/s10311-021-01227-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel M, Châtel A, Mouneyrac C. Micro (nano) plastics: a threat to human health? Curr Opin Environ Sci Health. 2018;1:17–23. doi: 10.1016/j.coesh.2017.10.003. [DOI] [Google Scholar]
- Rochman CM, Tahir A, Williams SL, Baxa DV, Lam R, Miller JT, Teh FC, Werorilangi S, Teh SJ. Anthropogenic debris in seafood: Plastic debris and fibers from textiles in fish and bivalves sold for human consumption. Sci Rep. 2015;5(1):1–10. doi: 10.1038/srep14340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JR, Huang J, Anand P, Kucera K, Sandoval AG, Dantzler KW, Hickman DS, Jee J, Kimovec FM, Koppstein D, Marks DH, Mittermiller PA, Nunez SJ, Santiago M, Townes MA, Vishnevetsky M, Williams NE, Nunez Vargas MP, Boulanger LA, Slack CB, Strobel SA. Biodegradation of polyester polyurethane by endophytic fungi. Appl Environ Microbiol. 2011;77(17):6076–6084. doi: 10.1128/AEM.00521-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol Adv. 2009;27(2):185–194. doi: 10.1016/j.biotechadv.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Sanniyasi E, Gopal RK, Gunasekar DK, Raj PP. Biodegradation of low-density polyethylene (LDPE) sheet by microalga, Uronema Africanum Borge. Sci Rep. 2021;11(1):1–33. doi: 10.1038/s41598-021-96315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah P, Rout J. Efficient biodegradation of low-density polyethylene by cyanobacteria isolated from submerged polyethylene surface in domestic sewage water. Environ Sci Pollut Res. 2018;25(33):33508–33520. doi: 10.1007/s11356-018-3079-7. [DOI] [PubMed] [Google Scholar]
- Sathicq MB, Sabatino R, Corno G, Di Cesare A. Are microplastic particles a hotspot for the spread and the persistence of antibiotic resistance in aquatic systems? Environ Pollut. 2021;279:116896. doi: 10.1016/j.envpol.2021.116896. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J (2005) Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome biol 6(8):1–4. 10.1186/gb-2005-6-8-229 [DOI] [PMC free article] [PubMed]
- Schwartz M, Perrot T, Aubert E, Dumarçay S, Favier F, Gérardin P, Gelhaye E. Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-26601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SR, Din MO, Bittihn P, Xiong L, Tsimring LS, Hasty J. A stabilized microbial ecosystem of self-limiting bacteria using synthetic quorum-regulated lysis. Nat Microbiol. 2017;2(8):1–9. doi: 10.1038/nmicrobiol.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlmayer F, Aubel D, Fussenegger M. Synthetic gene circuits for the detection, elimination and prevention of disease. Nat Biomed Eng. 2018;2(6):399–415. doi: 10.1038/s41551-018-0215-0. [DOI] [PubMed] [Google Scholar]
- Shabbir S, Faheem M, Ali N, Kerr PG, Wang LF, Kuppusamy S, Li Y. Periphytic biofilm: an innovative approach for biodegradation of microplastics. Sci Total Environ. 2020;717:137064. doi: 10.1016/j.scitotenv.2020.137064. [DOI] [PubMed] [Google Scholar]
- Shao H, Chen M, Fei X, Zhang R, Zhong Y, Ni W, Tan X. Complete genome sequence and characterization of a polyethylene biodegradation strain, Streptomyces albogriseolus LBX-2. Microorganisms. 2019;7(10):379. doi: 10.3390/microorganisms7100379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B, Dangi AK, Shukla P. Contemporary enzyme based technologies for bioremediation: a review. J Environ Manage. 2018;210:10–22. doi: 10.1016/j.jenvman.2017.12.075. [DOI] [PubMed] [Google Scholar]
- Sheik S, Chandrashekar KR, Swaroop K, Somashekarappa HM. Biodegradation of gamma irradiated low density polyethylene and polypropylene by endophytic fungi. Int Biodeterior Biodegrad. 2015;105:21–29. doi: 10.1016/j.ibiod.2015.08.006. [DOI] [Google Scholar]
- Shen M, Zeng G, Zhang Y, Wen X, Song B, Tang W. Can biotechnology strategies effectively manage environmental (micro) plastics? Sci Total Environ. 2019;697:134200. doi: 10.1016/j.scitotenv.2019.134200. [DOI] [PubMed] [Google Scholar]
- Shin J, Kim JE, Lee YW, Son H. Fungal cytochrome P450s and the P450 complement (CYPome) of Fusarium graminearum. Toxins. 2018;10(3):112. doi: 10.3390/toxins10030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirke AN, White C, Englaender JA, Zwarycz A, Butterfoss GL, Linhardt RJ, Gross RA. Stabilizing leaf and branch compost cutinase (LCC) with glycosylation: mechanism and effect on PET hydrolysis. Biochemistry. 2018;57:1190–1200. doi: 10.1021/acs.biochem.7b01189. [DOI] [PubMed] [Google Scholar]
- Silva ALP. New frontiers in remediation of (micro) plastics. Curr Opin Green Sustain Chem. 2021;28:100443. doi: 10.1016/j.cogsc.2020.100443. [DOI] [Google Scholar]
- Silva ALP, Prata JC, Duarte AC, Barcelò D, Rocha-Santos T. An urgent call to think globally and act locally on landfill disposable plastics under and after covid-19 pandemic: pollution prevention and technological (Bio) remediation solutions. Chem Eng J. 2021;426:131201. doi: 10.1016/j.cej.2021.131201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B, Sharma N. Mechanistic implications of plastic degradation. Polym Degrad Stab. 2008;93:561–584. doi: 10.1016/j.polymdegradstab.2007.11.008. [DOI] [Google Scholar]
- Singh L, Wahid ZA. Methods for enhancing bio-hydrogen production from biological process: a review. J Ind Eng Chem. 2015;21:70–80. doi: 10.1016/j.jiec.2014.05.035. [DOI] [Google Scholar]
- Sivan A. New perspectives in plastic biodegradation. Curr Opin Biotechnol. 2011;22:422–426. doi: 10.1016/j.copbio.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Sivan A, Szanto M, Pavlov V. Biofilm development of the polyethylene-degrading bacterium Rhodococcus ruber. Appl Microbiol Biotechnol. 2006;72:346–352. doi: 10.1007/s00253-005-0259-4. [DOI] [PubMed] [Google Scholar]
- Sjollema SB, Redondo-Hasselerharm P, Leslie HA, Kraak MH, Vethaak AD. Do plastic particles affect microalgal photosynthesis and growth? Aquat Toxicol. 2016;170:259–261. doi: 10.1016/j.aquatox.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Skariyachan S, Patil AA, Shankar A, Manjunath M, Bachappanavar N, Kiran S. Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polym Degrad Stab. 2018;149:52–68. doi: 10.1016/j.polymdegradstab.2018.01.018. [DOI] [Google Scholar]
- Skariyachan S, Taskeen N, Kishore AP, Krishna BV, Naidu G. Novel consortia of Enterobacter and Pseudomonas formulated from cow dung exhibited enhanced biodegradation of polyethylene and polypropylene. J Environ Manage. 2021;284:112030. doi: 10.1016/j.jenvman.2021.112030. [DOI] [PubMed] [Google Scholar]
- Smith M, Love DC, Rochman CM, Neff RA. Microplastics in seafood and the implications for human health. Curr Environ Health Rep. 2018;5(3):375–386. doi: 10.1007/s40572-018-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Jongmans-Hochschulz E, Mauder N, Imirzalioglu C, Wichels A, Gerdts G. The Travelling Particles: Investigating microplastics as possible transport vectors for multidrug resistant E. coli in the Weser estuary (Germany) Sci Total Environ. 2020;720:137603. doi: 10.1016/j.scitotenv.2020.137603. [DOI] [PubMed] [Google Scholar]
- Staffieri E, de Lucia GA, Camedda A, Poeta G, Battisti C. Pressure and impact of anthropogenic litter on marine and estuarine reptiles: an updated “blacklist” highlighting gaps of evidence. Environ Sci Pollut Res. 2019;26:1238–1249. doi: 10.1007/s11356-018-3616-4. [DOI] [PubMed] [Google Scholar]
- Straub S, Hirsch PE, Burkhardt-Holm P. Biodegradable and petroleum-based microplastics do not differ in their ingestion and excretion but in their biological effects in a freshwater invertebrate Gammarus fossarum. Int J Environ Res Public Health. 2017;14(7):774. doi: 10.3390/ijerph14070774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Xiong X, Zhang Y, Wu C, Xu X, Sun C, Shi H. Global transportation of plastics and microplastics: a critical review of pathways and influences. Sci Total Environ. 2022 doi: 10.1016/j.scitotenv.2022.154884. [DOI] [PubMed] [Google Scholar]
- Sukkhum S, Tokuyama S, Kitpreechavanich V. Development of fermentation process for PLA-degrading enzyme production by a new thermophilic Actinomadura sp. T16–1. Biotechnol Bioprocess Eng. 2009;14(3):302–306. doi: 10.1007/s12257-008-0207-0. [DOI] [Google Scholar]
- Syranidou E, Karkanorachaki K, Amorotti F, Franchini M, Repouskou E, Kaliva M, Vamvakaki M, Kolvenbach B, Fava F, Corvini PFX, Kalogerakis N. Biodegradation of weathered polystyrene films in seawater microcosms. Sci Rep. 2017;7(1):1–12. doi: 10.1038/s41598-017-18366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syranidou E, Karkanorachaki K, Amorotti F, Avgeropoulos A, Kolvenbach B, Zhou NY, Kalogerakis N. Biodegradation of mixture of plastic films by tailored marine consortia. J Hazard Mater. 2019;375:33–42. doi: 10.1016/j.jhazmat.2019.04.078. [DOI] [PubMed] [Google Scholar]
- Tanasupawat S, Takehana T, Yoshida S, Hiraga K, Oda K. Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly (ethylene terephthalate) Int J Syst Evol Microbiol. 2016;66(8):2813–2818. doi: 10.1099/ijsem.0.001058. [DOI] [PubMed] [Google Scholar]
- Tang DYY, Yew GY, Koyande AK, Chew KW, Vo DVN, Show PL. Green technology for the industrial production of biofuels and bioproducts from microalgae: a review. Environ Chem Lett. 2020;18(6):1967–1985. doi: 10.1007/s10311-020-01052-3. [DOI] [Google Scholar]
- Taniguchi I, Yoshida S, Hiraga K, Miyamoto K, Kimura Y, Oda K. Biodegradation of PET: current status and application aspects. ACS Catal. 2019;9(5):4089–4105. doi: 10.1021/acscatal.8b05171. [DOI] [Google Scholar]
- Thiounn T, Smith RC. Advances and approaches for chemical recycling of plastic waste. J Polym Sci. 2020;58(10):1347–1364. doi: 10.1002/pol.20190261. [DOI] [Google Scholar]
- Tiwari N, Santhiya D, Sharma JG. Microbial remediation of micro-nano plastics: current knowledge and future trends. Environ Pollut. 2020;265:115044. doi: 10.1016/j.envpol.2020.115044. [DOI] [PubMed] [Google Scholar]
- Tran KM, Lee HM, Thai TD, Shen J, Eyun SI, Na D. Synthetically engineered microbial scavengers for enhanced bioremediation. J Hazard Mater. 2021;419:126516. doi: 10.1016/j.jhazmat.2021.126516. [DOI] [PubMed] [Google Scholar]
- Tribedi P, Sil AK. Low-density polyethylene degradation by Pseudomonas sp. AKS2 biofilm. Environ Sci Pollut Res. 2013;20(6):4146–4153. doi: 10.1007/s11356-012-1378-y. [DOI] [PubMed] [Google Scholar]
- Uscátegui YL, Arévalo FR, Díaz LE, Cobo MI, Valero MF. Microbial degradation, cytotoxicity and antibacterial activity of polyurethanes based on modified castor oil and polycaprolactone. J Biomater Sci, Polym Ed. 2016;27(18):1860–1879. doi: 10.1080/09205063.2016.1239948. [DOI] [PubMed] [Google Scholar]
- Verma R, Vinoda KS, Papireddy M, Gowda ANS. Toxic pollutants from plastic waste-a review. Procedia Environ Sci. 2016;35:701–708. doi: 10.1016/j.proenv.2016.07.069. [DOI] [Google Scholar]
- Vethaak AD and Martínez-Gómez C (2020) Micro and nanoplastics in the aquatic environment with special reference to synthetic fibers. In: Wagterveld RM et al. (eds.), Synthetic nano-and microfibers, Chapter 9. Wetsus, European centre of excellence for sustainable water technology, Leeuwarden. http://hdl.handle.net/10508/11552
- Vethaak AD, Legler J. Microplastics and human health. Science. 2021;371(6530):672–674. doi: 10.1126/science.abe5041. [DOI] [PubMed] [Google Scholar]
- Viršek MK, Lovšin MN, Koren Š, Kržan A, Peterlin M. Microplastics as a vector for the transport of the bacterial fish pathogen species Aeromonas salmonicida. Mar Pollut Bull. 2017;125(1-2):301–309. doi: 10.1016/j.marpolbul.2017.08.024. [DOI] [PubMed] [Google Scholar]
- Volke-Sepúlveda T, Saucedo-Castañeda G, Gutiérrez-Rojas M, Manzur A, Favela-Torres E. Thermally treated low density polyethylene biodegradation by Penicillium pinophilum and Aspergillus niger. J Appl Polym Sci. 2002;83(2):305–314. doi: 10.1002/app.2245. [DOI] [Google Scholar]
- Waller CL, Griffiths HJ, Waluda CM, Thorpe SE, Loaiza I, Moreno B, Hughes KA. Microplastics in the Antarctic marine system: an emerging area of research. Sci Total Environ. 2017;598:220–227. doi: 10.1016/j.scitotenv.2017.03.283. [DOI] [PubMed] [Google Scholar]
- Wang B, Xu J, Gao J, Fu X, Han H, Li Z, Yao Q. Construction of an Escherichia coli strain to degrade phenol completely with two modified metabolic modules. J Hazard Mater. 2019;373:29–38. doi: 10.1016/j.jhazmat.2019.03.055. [DOI] [PubMed] [Google Scholar]
- Wang Z, Qin Y, Li W, Yang W, Meng Q, Yang J. Microplastic contamination in freshwater: first observation in lake ulansuhai, yellow river basin. China Environ Chem Lett. 2019;17(4):1821–1830. doi: 10.1007/s10311-019-00888-8. [DOI] [Google Scholar]
- Wicker J, Lorsbach T, Gütlein M, Schmid E, Latino D, Kramer S, Fenner K. enviPath–The environmental contaminant biotransformation pathway resource. Nucleic Acids Res. 2016;44(D1):D502–D508. doi: 10.1093/nar/gkv1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox C, Puckridge M, Schuyler QA, Townsend K, Hardesty BD. A quantitative analysis linking sea turtle mortality and plastic debris ingestion. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-30038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JKH, Lee KK, Tang KHD, Yap PS. Microplastics in the freshwater and terrestrial environments: prevalence, fates, impacts and sustainable solutions. Sci Total Environ. 2020;719:137512. doi: 10.1016/j.scitotenv.2020.137512. [DOI] [PubMed] [Google Scholar]
- Wright SL, Kelly FJ. Plastic and human health: a micro issue? Environ Sci Technol. 2017;51(12):6634–6647. doi: 10.1021/acs.est.7b00423. [DOI] [PubMed] [Google Scholar]
- Wright SL, Rowe D, Thompson RC, Galloway TS. Microplastic ingestion decreases energy reserves in marine worms. Curr Biol. 2013;23(23):R1031–R1033. doi: 10.1016/j.cub.2013.10.068. [DOI] [PubMed] [Google Scholar]
- Wright SL, Thompson RC, Galloway TS. The physical impacts of microplastics on marine organisms: a review. Environ Pollut. 2013;178:483–492. doi: 10.1016/j.envpol.2013.02.031. [DOI] [PubMed] [Google Scholar]
- Wu X, Pan J, Li M, Li Y, Bartlam M, Wang Y. Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 2019;165:114979. doi: 10.1016/j.watres.2019.114979. [DOI] [PubMed] [Google Scholar]
- Yan N, Fan C, Chen Y, Hu Z. The potential for microalgae as bioreactors to produce pharmaceuticals. Int J Mol Sci. 2016;17(6):962. doi: 10.3390/ijms17060962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yang Y, Wu WM, Zhao J, Jiang L. Evidence of polyethylene biodegradation by bacterial strains from the guts of plastic-eating waxworms. Environ Sci Technol. 2014;48:13776–13784. doi: 10.1021/es504038a. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yang J, Wu WM, Zhao J, Song Y, Gao L, Yang R, Jiang L. Biodegradation and mineralization of polystyrene by plastic-eating mealworms: part 2. Role of gut microorganisms. Environ Sci Technol. 2015;49(20):12087–12093. doi: 10.1021/acs.est.5b02663. [DOI] [PubMed] [Google Scholar]
- Yang S, Cheng Y, Liu T, Huang S, Yin L, Pu Y, Liang G. Impact of waste of COVID-19 protective equipment on the environment, animals and human health: a review. Environ Chem Lett. 2022 doi: 10.1007/s10311-022-01462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Ma J, Sun Y, Zhou T, Zhao Y, Yu F. Microbial degradation and other environmental aspects of microplastics/plastics. Sci Total Environ. 2020;715:136968. doi: 10.1016/j.scitotenv.2020.136968. [DOI] [PubMed] [Google Scholar]
- Zadjelovic V, Chhun A, Quareshy M, Silvano E, Hernandez-Fernaud JR, Aguilo-Ferretjans MM, Bosch R, Dorador C, Gibson MI, Christie-Oleza JA. Beyond oil degradation: enzymatic potential of Alcanivorax to degrade natural and synthetic polyesters. Environ Microbiol. 2020;22:1356–1369. doi: 10.1111/1462-2920.14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettler ER, Mincer TJ, Amaral-Zettler LA. Life in the "plastisphere": microbial communities on plastic marine debris. Environ Sci Technol. 2013;47(13):7137–7146. doi: 10.1021/es401288x. [DOI] [PubMed] [Google Scholar]
- Zhu B, Wang D, Wei N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022;40(1):22–37. doi: 10.1016/j.tibtech.2021.02.008. [DOI] [PubMed] [Google Scholar]