Abstract
SMART (Simple Modular Architecture Research Tool, http://smart.embl-heidelberg.de) is a web-based resource used for the annotation of protein domains and the analysis of domain architectures, with particular emphasis on mobile eukaryotic domains. Extensive annotation for each domain family is available, providing information relating to function, subcellular localization, phyletic distribution and tertiary structure. The January 2002 release has added more than 200 hand-curated domain models. This brings the total to over 600 domain families that are widely represented among nuclear, signalling and extracellular proteins. Annotation now includes links to the Online Mendelian Inheritance in Man (OMIM) database in cases where a human disease is associated with one or more mutations in a particular domain. We have implemented new analysis methods and updated others. New advanced queries provide direct access to the SMART relational database using SQL. This database now contains information on intrinsic sequence features such as transmembrane regions, coiled-coils, signal peptides and internal repeats. SMART output can now be easily included in users’ documents. A SMART mirror has been created at http://smart.ox.ac.uk.
INTRODUCTION
The task of identifying homologous domains by sequence similarity is often made more difficult by differences in domain architectures and by substantial divergence in sequence. As the number of completely sequenced eukaryotic genomes increases, so does the need for accurate prediction of domain homologies. Accordingly, SMART (1) has been developed to identify and annotate protein domains, particularly those in eukaryotes that are mobile and difficult to detect.
SMART consists of a library of Hidden Markov models (HMMs) (2). These provide a robust statistical model of amino acid preferences and insertion/deletion probabilities at each position in a sequence alignment. The current database covers more than 600 protein domain families. These are linked to multiple sequence alignments, embodied within a web-based domain annotation tool. SMART provides facilities to query the underlying relational database for proteins with particular domain combinations (with the option of restricting these to any taxonomic group) and to alert users to sequences that contain particular domain combinations, after these are newly available in databases.
IMPROVED DOMAIN COVERAGE
The majority of domain alignments represented in SMART have been established using standard database searching methods (3,4). Over the past 2 years, in order to augment the SMART domain set, we have striven to develop semi-automatic search methods to identify new and biologically interesting domains. Of more than 200 domains added, many were identified in-house by investigating sequence regions that had no previous domain annotations.
IMPROVED ANNOTATION
Improvements in the annotation of domains, with respect to human disease and cellular localisation, have been implemented in the latest version of SMART.
SMART now provides information on known human heritable genetic disorders arising from missense mutations located within specified domains. Of the 10 121 missense mutations annotated in SWISS-PROT (http://ca.expasy.org/sprot/; 5), many of which are derived from OMIM; 3085 mutations could be mapped onto 170 different SMART domain types in 335 out of 734 human disease gene sequences (6).
For each domain family, SMART now provides estimated probabilities that each domain is part of a secreted, cytoplasmic and nuclear protein. These probabilities derive from observed patterns of domain co-occurrence and their correlations with protein localisations. The method for generating these probability values and an estimation of its accuracy will be presented elsewhere.
STRUCTURAL CHANGES AND THE SMART DATABASE
The core of SMART is a relational database management system (RDBMS) (3) powered by PostgreSQL (http://www.postgresql.org) that stores information on SMART domains. Each domain’s hit borders, raw bit score and Expect (E) value are recorded, together with protein accession code, description and species name.
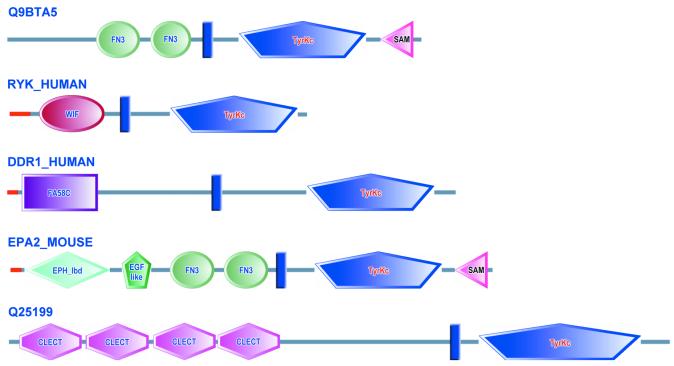
For each protein in the relational database, intrinsic features such as transmembrane regions (7), coiled-coils (8), signal peptides (9) and internal repeats (10) are now included. Users can now query the RDBMS for proteins containing not only particular domains, but also specified intrinsic features (‘TRANS’, transmembrane regions; ‘COIL’, coiled-coils; ‘SIGNAL’, signal peptides). For example, it is possible to identify receptor tyrosine kinases by searching for proteins that contain both a tyrosine kinase domain, and a predicted transmembrane region (Fig. 1).
Figure 1.
Using intrinsic features in Architecture SMART queries. The SMART database was queried for all proteins containing a tyrosine kinase domain and a transmembrane region (TyrKc and TRANS). 387 proteins were found, including the five displayed here. Note that the text colour of domain names has been designed to correlate both with its subcellular localisation (blue, secreted; black, intracellular) and its catalytic activity (red, catalytic activity).
For the latest release of SMART, two new analytical methods have been employed. TMHMM2 is now being used to predict transmembrane sequences, since this method demonstrates 97–98% accuracy for transmembrane prediction (7). Internal sequence repeats are detected using Prospero (10), with a significance threshold probability of 10–4, after first filtering the sequence for low complexity and coiled-coil regions.
IMPROVED WEB INTERFACE
SMART provides a World Wide Web-based interface to its underlying relational database and HMMER-based search engine (3). In response to rapidly increasing demand, we have taken steps to dramatically improve the efficiency and response times of our server. Underlying code has been modified to use persistent database connections. Many speed optimisations have been made thereby providing users with a much faster and more productive environment.
Schematic representations of proteins are now generated dynamically and displayed as a single PNG (Portable Network Graphics) image. This enables easy ‘copy–paste’ inclusion of SMART output in users’ publications. SMART multiple sequence alignments may now be coloured by consensus using CHROMA (11). This highlights patterns of residue conservation, which can assist in clarifying questions of homology, and can draw attention to functional positions such as binding and active sites.
SMART database querying capabilities were recently greatly extended allowing users to build up more complex queries of the underlying relational database using SQL commands. The latest release of SMART also introduced options to retrieve FASTA-formatted sequences of domains or proteins that have been viewed using Architecture SMART. Thus, it is easier for users to generate full alignments for all the occurrences of a particular domain that occur in a given species.
APPLICATION OF SMART
Apart from its use as a web tool, SMART has been applied to large-scale annotation projects such as the annotation of the human genome draft sequence (12,13), the investigation of single domain families in model organisms (14), the study of sequence conservation in multiple alignments (15) and, in conjunction with genomic data, for the study of conservation of gene (i.e. intron/exon) structure (16). SMART will continue to be a valuable resource for large-scale sequence analysis studies.
SMART has also been incorporated into other domain and protein family resources that are used for the primary annotation of sequence databases. It is a component database of InterPro (17), which contributes to the annotation of SWISS-PROT sequences (5), and of the Conserved Domain Database (CDD) (http://web.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) which contributes to the annotation of RefSeq sequences (18).
CONCLUSIONS
Over the past years, SMART has developed and matured into an important and widely used biological web tool characterised by stability and fast response times. Our main goal has been to continue to provide improvements and feature expansion together with the highest quality of data. We are committed to maintain, improve and extend SMART to accommodate the rising needs of genome and proteome annotation and analysis.
REFERENCES
- 1.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Durbin R., Eddy,S., Krogh,A. and Mitchison,G. (1998) Biological Sequence Analysis. Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge, UK.
- 3.Schultz J., Copley,R.R., Doerks,T., Ponting,C.P. and Bork,P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res., 28, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponting C.P., Schultz,J., Copley,R.R., Andrade,M.A. and Bork,P. (2000) Evolution of domain families. Adv. Protein Chem., 54, 185–244. [DOI] [PubMed] [Google Scholar]
- 5.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodstadt L. and Ponting,C.P. (2001) Sequence variation and disease in the wake of the draft human genome. Hum. Mol. Genet., 10, 2209–2214. [DOI] [PubMed] [Google Scholar]
- 7.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 8.Lupas A., Van Dyke,M. and Stock,J. (1991) Predicting coiled coils from protein sequences. Science, 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 9.von Heijne G. (1987) Sequence Analysis in Molecular Biology: Treasure Trove or Trivial Pursuit. Academic Press, San Diego, CA, 429–436. [Google Scholar]
- 10.Mott R. (2000) Accurate formula for P-values of gapped local sequence and profile alignments. J. Mol. Biol., 300, 649–659. [DOI] [PubMed] [Google Scholar]
- 11.Goodstadt L. and Ponting,C.P. (2001) CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics, 17, 845–846. [DOI] [PubMed] [Google Scholar]
- 12.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 13.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 14.Hill E., Broadbent,I.D., Chothia,C. and Pettitt,J. (2001) Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J. Mol. Biol., 305, 1011–1024. [DOI] [PubMed] [Google Scholar]
- 15.Pei J. and Grishin,N.V. (2001) AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics, 17, 700–712. [DOI] [PubMed] [Google Scholar]
- 16.Betts M.J., Guigo,R., Agarwal,P. and Russell,R.B. (2001) Exon structure conservation despite low sequence similarity: a relic of dramatic events in evolution? EMBO J., 20, 5354–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]