Abstract
TRIPLES is a web-accessible database of TRansposon-Insertion Phenotypes, Localization and Expression in Saccharomyces cerevisiae—a relational database housing nearly half a million data points generated from an ongoing study using large-scale transposon mutagenesis to characterize gene function in yeast. At present, TRIPLES contains three principal data sets (i.e. phenotypic data, protein localization data and expression data) for over 3500 annotated yeast genes as well as several hundred non-annotated open reading frames. In addition, the TRIPLES web site provides online order forms linked to each data set so that users may request any strain or reagent generated from this project free of charge. In response to user requests, the TRIPLES web site has undergone several recent modifications. Our localization data have been supplemented with approximately 500 fluorescent micrographs depicting actual staining patterns observed upon indirect immunofluorescence analysis of indicated epitope-tagged proteins. These localization data, as well as all other data sets within TRIPLES, are now available in full as tab-delimited text. To accommodate increased reagent requests, all orders are now cataloged in a separate database, and users are notified immediately of order receipt and shipment. Also, TRIPLES is one of five sites incorporated into the new functional analysis tool Function Junction provided by the Saccharomyces Genome Database. TRIPLES may be accessed from the Yale Genome Analysis Center (YGAC) homepage at http://ygac.med.yale.edu.
INTRODUCTION
Since its inception, the TRIPLES web site has provided convenient access to data from our transposon-based study of gene function in yeast. Described in detail elsewhere (1,2), this study utilizes a multifunctional transposon to generate random insertions throughout the yeast genome. These insertions may be used to derive a variety of informative alleles including reporter gene fusions, gene disruptions and epitope-tagged alleles (1). Gene fusions to transposon-encoded lacZ provide a means of generating expression profiles identifying sequences translated under given growth conditions. As this lacZ reporter is terminated by a series of stop codons, transposon insertion also results in truncation of its host gene, thereby potentially generating disruption alleles for subsequent phenotypic analysis. Finally, by means of Cre-lox recombination, an integrated transposon insertion may be modified such that the bulk of the transposon is excised, leaving behind a short stretch of epitope-coding sequence. These epitope-tagged alleles can be used to generate corresponding tagged proteins for immunolocalization. By this approach, a single insertion is sufficient to yield expression, phenotypic and localization data—a cumulatively unique data set maintained and updated in TRIPLES.
In addition to the value in this collected data, individual insertion alleles and transposon-tagged strains are useful laboratory reagents. As such, we make all strains from this project available free of charge to any interested researcher through the TRIPLES web site. Order forms are available both from the YGAC homepage as well as from links accompanying each data set within TRIPLES. All requests are typically processed and shipped within a week of receipt.
DESIGN AND IMPLEMENTATION
TRIPLES was implemented using the ORACLE database system, version 8i. Our web front-end was mainly implemented using Active Server Page (ASP), an integral part of the Microsoft IIS web server running on Windows NT. The ASP mechanism has enabled us to embed server-side code written in VBScript and JavaScript within HTML documents; we have also incorporated some PERL/CGI programs. To ensure code compatibility with different database platforms, we have used ODBC (Open Database Connectivity) to implement database access.
DATA SEARCHING AND RETRIEVAL
Users may access data within TRIPLES through either composite or category-specific searches. Composite searches can be used to retrieve records from multiple data sets (phenotypic, expression and localization data) for any given gene/insertion. As the name suggests, category-specific searches are helpful in querying a single type of data. In either case, searches may be initiated by supplying a gene name in either systematic (e.g. YIL046W) or standard form (e.g. MET30). Alternatively, data regarding a given insertion may be accessed through its clone ID, a unique designation (e.g. V66A9) assigned to each transposon-mutagenized strain in our collection based upon its position in a 96-well storage plate. Category-specific searches may also be initiated by selecting from a list of controlled vocabulary terms descriptive of that particular data set. For example, TRIPLES may be queried for all tagged proteins localizing to the nucleus by initiating a category-specific search of localization data with ‘nucleus’ chosen as the localization. To facilitate multi-level searching, the results of a given search may be used to initiate further category-specific searches. These and additional search options are demonstrated on the TRIPLES homepage at http://ygac.med.yale.edu/triples/triples.htm.
To ease data retrieval, all category-specific output reports are presented in tabular format and may be conveniently downloaded as tab-delimited text. If desired, these reports may be custom-formatted to display only those fields of greatest interest. In addition, category-specific reports may be sorted by data fields in order to group results in a logical manner. Each complete data set is available for downloading as a flat file, which we periodically update. To further enhance the utility of TRIPLES as an information resource, all composite reports also provide access to supplemental background literature through direct external links to corresponding entries within SGD (3), YPD (4) and GenBank (5).
NEW DATA/FEATURES
TRIPLES has grown significantly as a resource to the yeast scientific community over the last 2 years. Approximately doubling in total data content, TRIPLES now encompasses functional data for nearly 60% of the yeast genome (Table 1). At present, the TRIPLES database catalogs a collection of over 28 000 transposon insertion alleles, with each allele serving as a potentially useful laboratory reagent. More than 27 000 transposon-mutagenized yeast strains have been used in our studies of gene expression, disruption phenotypes and protein localization. This data is of value both to molecular biologists and computational biologists; to facilitate its easy dissemination, we now offer a ‘data download’ page accessible from our basic search form. Users may download complete data sets describing transposon insertions sites, gene expression, disruption phenotypes and protein localization. Each data set is available as tab-delimited text. As our phenotypic data set is very large, we provide users the opportunity to download results from each growth assay individually, thereby generating files more amenable to standard spreadsheet analysis.
Table 1. TRIPLES data sets (as of September 2001).
| Data sets | Entries | Numbers |
|---|---|---|
| Transposon insertions | Total | 28 545 |
| Sequenced/defined site of insertion | 22 587 | |
| Affected genes (annotated) | 3504 | |
| Expression data | Total records | 27 270 |
| Induced during vegetative growth | 26 878 | |
| Phenotypic data | Total records | 16 288 |
| Strains with mutant phenotypes | 7240 | |
| Localization data | Total records | 11 426 |
| Localizations (not background) | 5504 |
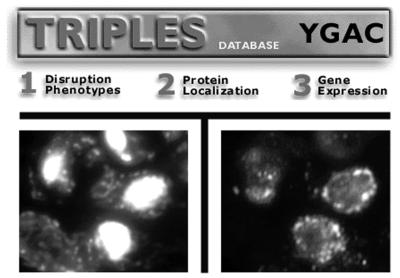
In response to user requests, protein localization data within TRIPLES is now supplemented with fluorescent micrographs illustrating actual staining patterns obtained upon indirect immmunofluorescence analysis of given epitope-tagged proteins (Fig. 1). Available images (JPEG files) may be viewed by clicking on any underlined localization within a composite or category-specific report. At present, approximately 500 such images are accessible through TRIPLES, establishing it as one of the largest web-accessible visual libraries of yeast protein localization. Additional text describing these images is available as online help within TRIPLES.
Figure 1.
Example micrographs linked to protein localization data in TRIPLES. Transposon-tagged proteins were visualized by indirect immunofluorescence with monoclonal antibodies directed against the transposon-encoded HA epitope. Approximately 500 images of subcellular staining patterns observed using this approach may now be viewed through TRIPLES. Two images are available per entry: in each case, the left-hand micrograph shows a cluster of transposon-tagged yeast cells stained with the DNA-binding dye DAPI; the right-hand micrograph is of the same cells stained with anti-HA antibody. Shown here is a set of micrographs from immunofluorescence analysis of strain V50A5, which contains an integrated epitope tag in-frame with the nucleoporin-encoding gene NUP159. Note the distinct staining pattern observed around the nuclear rim in the right-hand image.
The TRIPLES web site has continued to serve as a popular source of yeast strains and reagents: since its last release (6), over 400 requests for reagents have been placed through TRIPLES. In order to better service these requests, all orders placed online are now stored in an ACCESS database. Requests are assigned an order number; automatic email confirmation of order receipt is shipped to each user, and users are also notified by email of actual sample shipment. The efficiency of this system has allowed us to accommodate larger requests from researchers both here and abroad, without increasing ‘turn-around’ time.
SIGNIFICANCE
With an expanding repertoire of tools and technology facilitating large-scale research, fundamental advances in our understanding of biology lie within reach—provided that a spirit of cooperation continues to prevail in science. Free exchange of data and resources is central to our immediate and future progress; in that light, the TRIPLES database represents an important medium by which information and reagents can be shared among the scientific community. TRIPLES is also featured in the new SGD tool Function Junction (3), another helpful resource providing convenient access to data from a variety of functional genomic projects. Collectively, these types of resources exemplify the collaborative effort necessary to foster rapid advancement in molecular biology and, as such, represent a promising blueprint for scientific endeavor.
SUPPLEMENTARY MATERIAL
A summary of TRIPLES data sets (i.e. transposon insertion point data, gene expression data, phenotypic data, protein localization data) is available as Supplementary Material at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work is supported by NIH grant R01-CA77808 (to M.S.). A.K. is supported by a post-doctoral fellowship from the American Cancer Society.
REFERENCES
- 1.Ross-MacDonald P., Coelho,P.S.R., Roemer,T., Agarwal,S., Kumar,A., Jansen,R., Cheung,K.-H., Sheehan,A., Symoniatis,D., Umansky,L. et al. (1999) Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature, 402, 413–418. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A., des Etages,S.A., Coelho,P.S.R., Roeder,G.S. and Snyder,M. (2000) High-throughput methods for the large-scale analysis of gene function by transposon tagging. Methods Enzymol., 328, 550–574. [DOI] [PubMed] [Google Scholar]
- 3.Ball C., Jin,H., Sherlock,G., Weng,S., Matese,J.C., Andrada,R., Binkley,G., Dolinski,K., Dwight,S.S., Harris,M.A. et al. (2001) Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res., 29, 80–81. Updated article in this issue: Nucleic Acids Res. (2002), 30, 69–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costanzo M.C., Crawford,M.E., Hirschman,J.E., Kranz,J.E., Olsen,P., Robertson,L.S., Skrzypek,M.S., Braun,B.R., Lennon-Hopkins,K., Kondu,P. et al. (2001) YPD™, PombePD™ and WormPD™: model organism volumes of the BioKnowledge™ Library, an integrated resource for protein information. Nucleic Acids Res., 29, 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schrimi,L.M., Tatusova,T.A., Wagner,L. et al. (2001) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 29, 11–16. Updated article in this issue: Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar A., Cheung,K.-H., Ross-Macdonald,P., Coelho,P.S.R., Miller,P. and Snyder,M. (2000) TRIPLES: a database of gene function in Saccharomyces cerevisiae. Nucleic Acids Res., 28, 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.