Abstract
Background
Digital chest X-ray (dCXR) computer-aided detection (CAD) technology uses lung shape and texture analysis to determine the probability of tuberculosis (TB). However, many patients with previously treated TB have sequelae, which also distort lung shape and texture. We evaluated the diagnostic performance of 2 CAD systems for triage of active TB in patients with previously treated TB.
Methods
We conducted a retrospective analysis of data from a cross-sectional active TB case finding study. Participants ≥15 years, with ≥1 current TB symptom and complete data on history of previous TB, dCXR, and TB microbiological reference (Xpert MTB/RIF) were included. dCXRs were evaluated using CAD4TB (v.7.0) and qXR (v.3.0). We determined the diagnostic accuracy of both systems, overall and stratified by history of TB, using a single threshold for each system that achieved 90% sensitivity and maximized specificity in the overall population.
Results
Of 1884 participants, 452 (24.0%) had a history of previous TB. Prevalence of microbiologically confirmed TB among those with and without history of previous TB was 12.4% and 16.9%, respectively. Using CAD4TB, sensitivity and specificity were 89.3% (95% CI: 78.1–96.0%) and 24.0% (19.9–28.5%) and 90.5% (86.1–93.3%) and 60.3% (57.4–63.0%) among those with and without previous TB, respectively. Using qXR, sensitivity and specificity were 94.6% (95% CI: 85.1–98.9%) and 22.2% (18.2–26.6%) and 89.7% (85.1–93.2%) and 61.8% (58.9–64.5%) among those with and without previous TB, respectively.
Conclusions
The performance of CAD systems as a TB triage tool is decreased among persons previously treated for TB.
Keywords: tuberculosis, computer-aided detection, prior TB, CXR, triage
Among adults with presumptive tuberculosis in Zambia, at a fixed abnormality threshold that achieves 90% sensitivity, the specificity of 2 computer-aided detection systems for reading digital chest X-rays for active tuberculosis was substantially reduced in persons with previous tuberculosis.
Early tuberculosis (TB) case detection and treatment are global priorities to achieve the End TB strategy goals [1]. To facilitate early TB case detection, highly sensitive, easy-to-use, and affordable TB screening and triage tools should be widely accessible to the most high-risk groups and in high-prevalence settings. Chest X-ray (CXR) presents an opportunity for high-throughput screening and triaging of patients and has higher sensitivity and specificity than symptom-based screening [2, 3]. There is widespread use of CXR in well-resourced settings. However, its widespread scale-up and use in many high-TB-burden, resource-constrained settings have partly been limited by a lack of staff with sufficient training and experience to interpret CXRs [4]. Advances in digital radiography and computer-aided detection (CAD) for reading CXRs now make it easier to rapidly and reliably interpret CXRs; CAD has similar or higher performance to human readers [2, 4–6]. Additionally, digital CXR (dCXR) CAD has been demonstrated to be scalable and sustainable in high-burden settings [7–10].
World Health Organization (WHO) guidelines for systematic screening for TB disease now recommend CAD as an alternative to human readers [2, 11]. Computer-aided detection uses lung shape and texture analysis to determine the likelihood of TB [12], and CXRs are assigned a numerical score that ranges between 1 and 100 (or 0–1), with higher scores representing a greater likelihood of TB-associated abnormalities [4]. When CAD systems are used for screening/triage, it is recommended that the user sets a threshold—a dichotomous score for classifying which patients are at greater risk for TB; persons with a CAD score exceeding this threshold should be referred for definitive microbiological testing, which includes either culture or nucleic acid amplification testing [2]. The sensitivity and specificity of CAD systems at a fixed abnormality threshold may differ according to age, sex, history of previous TB, and sputum smear status [4, 11, 13–15]. Hence, there is a need to optimize the abnormality threshold for different subgroups. However, anecdotal evidence suggests that, in many real-world settings, 1 abnormality threshold is applied across different patient populations.
In sub-Saharan Africa, approximately 12% of TB notifications are among persons with a history of previous TB [16–18]. In some settings, over 80% of patients previously treated for TB, have post-pulmonary TB radiological changes, which most commonly include cavitation, volume loss, and fibrosis [19, 20]. When such individuals present to care, it may be difficult to distinguish whether radiological changes are due to previous TB sequelae or a new active TB episode, even for expert radiologists [21]. This can result in overtreatment of patients for TB, leading to unnecessary exposure to anti-TB therapy, failure to treat the true cause of illness, and wastage of limited health resources [18].
Given that post-TB radiographic sequelae distort the lung shape and texture, similar to active TB, the performance of CAD for TB screening/triaging may be affected. However, to date, there is little evidence on how previous TB affects the accuracy of CXR artificial intelligence (AI) systems and none on when these 2 subpopulations are further stratified by sex or human immunodeficiency virus (HIV) status. We, therefore, undertook this study to determine the comparative performance of 2 CXR CAD systems when used for TB triage among individuals with and without previously treated TB in Lusaka, Zambia.
METHODS
Study Design and Participants
This was an analysis of a previously reported prospective cross-sectional TB case finding study conducted at a public, primary healthcare facility in Lusaka, Zambia, between July 2017 and December 2018. The study setting and enrollment procedures of the cross-sectional study have been described previously [22]. Briefly, consecutive individuals presenting to the health facility and at the community TB screening points for any reason were screened for TB as described under test procedures. Because the performance assessment of CAD focused on a triage use case, analysis was limited to individuals with 1 or more current TB symptom(s) (current cough, chest pain, shortness of breath, night sweats, weight loss, or fevers). Individuals aged less than 15 years were excluded following WHO recommendations to restrict use of CAD systems as an alternative to human readers to individuals aged 15 years and older [2]. Additionally, individuals included in the present study were required to have a dCXR, TB microbiological reference result (Xpert MTB/RIF; Cepheid, Sunnyvale, CA, USA) and information on self-reported previous TB history.
Test Procedures
All participants underwent a TB symptom screen using a standardized screening register, which also captured information on self-reported history of previous TB [22]. Patients were asked if they had ever been treated for TB in the past; verification of a self-reported history of previous TB using medical records was not undertaken. When available, participants received a postero-anterior dCXR, using the DELFT CXR system (Delft Imaging Systems, Netherlands) mounted onto a mobile truck; this single CXR truck supported both community- and facility-based screening and thus not all participants received a CXR. All participants with either an abnormal CXR as defined by a CAD score of 60 or higher using CAD4TB version 5.0 (Delft Imaging Systems, Netherlands) and/or 1 or more TB symptoms, submitted a spot sputum sample for Xpert MTB/RIF (Cepheid, USA). Good-quality sputum samples [22] were processed for Xpert testing. Xpert tests were repeated if the initial test result was either invalid or an error.
All participants’ dCXRs were evaluated retrospectively using 2 CAD systems: CAD4TB version 7.0 (Delft Imaging Systems, Netherlands) and qXR version 3.0 (Qure.ai, India). The CAD systems scored anonymized images independently and blinded to all clinical information. Both CAD systems assigned each individual’s dCXR a numerical score from 0 to 100.
Definitions and Data Analysis
A positive sputum Xpert MTB/RIF was used as the reference standard to define TB cases; culture is not routinely available for patients in Zambia undergoing TB evaluation. Descriptive analyses were undertaken to describe the population that was included in this study, stratified by previous TB history and to compare the symptomatic individuals 15 years and older who were included in the analysis with those who were excluded—Fisher’s exact, chi-square, or Wilcoxon rank-sum tests were used as appropriate. Area under receiver operating curve (AUROC) values were determined to evaluate the performance of each CAD system for triage use against the reference standard and were undertaken overall and according to previous TB status and by sex and HIV status. Further analyses were performed to determine the diagnostic accuracy of both CAD systems for different patient subpopulations when a fixed abnormality threshold is used. A fixed threshold was selected that achieved an overall 90% sensitivity among all participants, while maximizing the specificity (15 for CAD4TB and 6 for qXR). The final analysis aimed to determine the optimal cutoff threshold for each CAD system in each patient subgroup that most approximated the WHO’s minimum target product profile (TPP) for screening and triage of 90% sensitivity and 70% specificity [2]. All data were analyzed using Stata version 17.0 (StataCorp).
Ethics Statement
This study was approved by the University of Zambia Bioethics Research Committee (UNZA BREC; no. 012-05-17). Verbal consent was obtained from the participants as per protocol; the study received a waiver for written consent as all study procedures aligned with standards of care. This study is reported in accordance with the Standards for Reporting of Diagnostic Accuracy Studies guidelines [23].
RESULTS
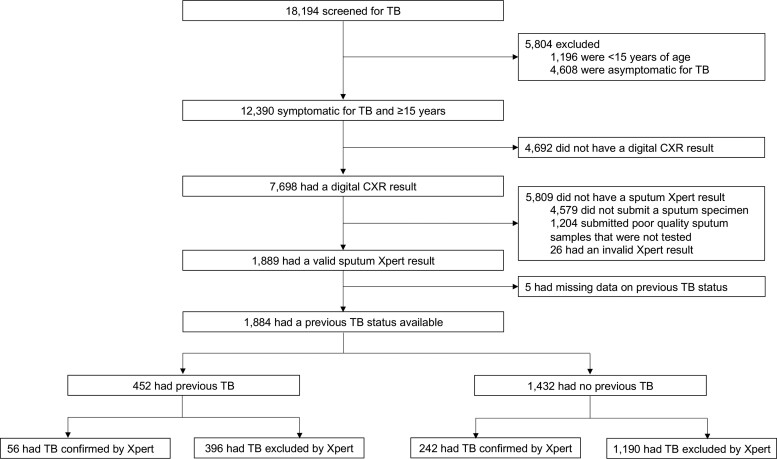
Overall, 18 194 individuals were screened for TB during the active case finding study, of whom 12 390 (68.1%) were 15 years or older and symptomatic for TB (Figure 1). Of these, 7698 (62.1%) had a dCXR, of whom 1884 (24.4%) had sputum Xpert results and data on history of previous TB and were included in the analysis. Of 1884 participants, 1184 (62.8%) were male, the median age was 38 (interquartile range: 29–48) years, and 702 (37.3%) were HIV positive (Table 1). There were 298 (prevalence: 15.8%; 95% confidence interval [CI]: 14.2–17.5) persons with microbiologically confirmed TB. Participants with complete data and included in the analysis were more likely to have been screened at the health facility (instead of the community) compared with those excluded from the analysis, and were more likely be older, male, HIV-positive, and have a previous TB history and multiple TB symptoms (Supplementary Table 1).
Figure 1.
Study flow diagram. Abbreviations: CXR, chest X-ray; TB, tuberculosis.
Table 1.
Characteristics of Study Participants
| Characteristic | All (N = 1884) |
Previous TB (n = 452) |
No Previous TB (n = 1432) |
P |
|---|---|---|---|---|
| Median (IQR) age, y | 38 (29–48) | 41 (34–50) | 37 (28–47) | <.001 |
| Male sex | 1184 (62.8) | 326 (71.1) | 858 (59.9) | <.001 |
| HIV statusa | ||||
| ȃPositive | 702 (37.3) | 250 (58.1) | 452 (33.6) | <.001 |
| ȃNegative | 1073 (56.9) | 180 (41.9) | 893 (66.4) | … |
| Current symptoms | ||||
| ȃCough | 1446 (76.7) | 355 (78.7) | 1091(76.2) | .28 |
| ȃFever | 634 (33.6) | 171(37.8) | 463 (32.3) | .033 |
| ȃNight sweats | 626 (33.2) | 153 (33.8) | 473 (35.2) | .76 |
| ȃWeight loss | 914 (48.5) | 234 (51.8) | 680 (47.5) | .11 |
| ȃChest pain | 1459(77.4) | 327 (72.3) | 1132 (79.0) | .003 |
| ȃShortness of breath | 909 (48.2) | 208 (46.0) | 701 (48.9) | .20 |
| Symptom classification | ||||
| ȃ1 Symptom | 350 (18.6) | 75 (16.6) | 275 (19.2) | .10 |
| ȃ2 Symptoms | 405 (21.2) | 96 (21.2) | 309 (21.6) | … |
| ȃ3 Symptoms | 369 (19.6) | 106 (23.4) | 263 (18.4) | … |
| ȃ≥4 Symptoms | 760 (37.0) | 175 (38.7) | 585 (40.8) | … |
| Sputum Xpert MTB/RIFb positive | 298 (15.8) | 56 (12.3) | 242 (16.9) | .022 |
All values represent n (%) except where explicitly noted. Abbreviations: HIV, human immunodeficiency virus; IQR, interquartile range; TB, tuberculosis.
There were 109 patients with an unknown HIV status (n = 22 persons with previous TB and n = 87 persons without previous TB.
Cepheid (Sunnyvale, CA).
Characteristics According to Previous Tuberculosis Status
Overall, 452 (23.9%) had a history of previous TB, while 1432 (76.1%) participants did not have a history of previous TB. Persons with previous TB were more likely to be male, older, and HIV positive (Table 1). The prevalence of microbiologically confirmed TB was higher among those without previous TB (16.9%; 95% CI: 15.0–18.9%) compared with those with previous TB (12.4%; 95% CI: 9.5–15.8%) (P = .022).
Discriminatory Value of CAD Software by Previous Tuberculosis Disease
Both CAD systems demonstrated an AUROC of at least 0.85 among all participants (Supplementary Table 2). The performance of the CAD4TB system was higher in individuals without previous TB compared with those with previous TB, while the performance of qXR was comparable in the 2 subpopulations (Supplementary Table 2). There was no difference in the AUROC values between HIV-positive and HIV-negative individuals and men and women, irrespective of previous TB history (Supplementary Table 2).
Diagnostic Accuracy of CAD Software at a Fixed Abnormality Threshold
Using a fixed abnormality threshold for each software system, the sensitivity of both CAD systems was high and did not differ by previous TB status (Table 2); however, the specificity of both systems was substantially lower among patients with a previous TB history. For both CAD systems, specificity was higher among women compared with men, irrespective of previous TB status. While the sensitivity and specificity were comparable among the HIV-negative and HIV-positive individuals with previous TB, specificity was higher among HIV-negative compared with HIV-positive individuals without previous TB (Table 2).
Table 2.
Performance of Computer-Aided Detection When Using a Fixed Abnormality Threshold According to Previous Tuberculosis Treatment Status
| Sensitivity (95% CI) |
Specificity (95% CI) |
Positive Likelihood Ratio (95% CI) |
Negative Likelihood Ratio (95% CI) |
|
|---|---|---|---|---|
| Overall | ||||
| ȃCAD4TBb (n = 1884) | 90.3 (86.3–93.4) | 51.2 (48.7–53.7) | 1.8 (1.7–2.0) | .2 (.1–.3) |
| ȃqXRc (n = 1884) | 90.6 (86.7–93.7) | 51.9 (49.4–54.4) | 1.9 (1.8–2.0) | .2 (.1–.3) |
| Previous TB | ||||
| ȃCAD4TB | ||||
| ȃȃAll (n = 452) | 89.3 (78.1–96.0) | 24.0 (19.9–28.5) | 1.2 (1.1–1.3) | .4 (.2–1.0) |
| ȃȃȃSex | ||||
| ȃȃȃȃMale (n = 326) | 91.1 (78.8–97.5) | 17.4 (13.2–22.4) | 1.1 (1.0–1.2) | .5 (.2–1.3) |
| ȃȃȃȃFemale (n = 126) | 81.8 (48.2–97.7) | 40.0 (31.0–49.6) | 1.4 (1.0–1.9) | .5 (.1–1.6) |
| ȃȃȃHIV statusa | ||||
| ȃȃȃȃHIV positive (n = 250) | 87.1 (70.2–96.4) | 28.3 (22.4–34.8) | 1.2 (1.0–1.4) | .5 (.2–1.2) |
| ȃȃȃȃHIV negative (n = 180) | 90.5 (69.6–98.8) | 18.9 (13.1–25.8) | 1.1 (1.0–1.3) | .5 (.1–2.0) |
| ȃqXR | ||||
| ȃȃAll (n = 452) | 94.6 (85.1–98.9) | 22.2 (18.2–26.6) | 1.2 (1.1–1.3) | .2 (.1–.7) |
| ȃȃȃSex | ||||
| ȃȃȃȃMale (n = 326) | 97.8 (88.2–99.2) | 16.4 (12.2–21.2) | 1.2 (1.1–1.25) | .3 (.0–1.0) |
| ȃȃȃȃFemale (n = 126) | 81.8 (48.2–97.7) | 36.5 (27.7–46.0) | 1.3 (.9–1.8) | .5 (.1–1.8) |
| ȃȃHIV statusa | ||||
| ȃȃȃHIV positive (n = 250) | 93.5 (78.6–99.2) | 26.5 (20.8–32.9) | 1.3 (1.1–1.4) | .2 (.1–.9) |
| ȃȃȃHIV negative (n = 180) | 95.2 (76.2–99.9) | 15.7 (10.4–22.3) | 1.1 (1.0–1.3) | .3 (.0–2.1) |
| No previous TB | ||||
| ȃCAD4TB | ||||
| ȃȃAll (n = 1432) | 90.5 (86.1–93.3) | 60.3 (57.4–63.0) | 2.3 (2.1–2.5) | .2 (.1–.2) |
| ȃȃȃSex | ||||
| ȃȃȃȃMale (n = 858) | 91.5 (86.4–95.2) | 52.6 (48.7–56.4) | 1.9 (1.8–2.1) | .2 (.1–.3) |
| ȃȃȃȃFemale (n = 574) | 87.7 (77.2–94.5) | 70.5 (66.4–74.5) | 3.0 (2.5–3.5) | .2 (.1–.3) |
| ȃȃȃHIV statusa | ||||
| ȃȃȃȃHIV positive (n = 452) | 89.5 (81.1–95.1) | 52.7 (47.5–57.9) | 1.9 (1.7–2.2) | .2 (.1–.4) |
| ȃȃȃȃHIV negative (n = 893) | 91.5 (85.7–95.6) | 64.4 (60.9–67.9) | 2.6 (2.3–2.9) | .1 (.1–.2) |
| ȃȃqXR | ||||
| ȃȃAll (n = 1432) | 89.7 (85.1–93.2) | 61.8 (58.9–64.5) | 2.4 (2.2–2.6) | .2 (.1–.2) |
| ȃȃȃSex | ||||
| ȃȃȃȃMale (n = 858) | 91.0 (85.7–94.7) | 53.7 (49.9–57.5) | 2.0 (1.8–2.2) | .2 (.1–.3) |
| ȃȃȃȃFemale (n = 574) | 86.2 (75.3–93.5) | 72.5 (68.4–76.3) | 3.1 (2.6–3.7) | .2 (.1–.4) |
| ȃȃȃHIV statusa | ||||
| ȃȃȃȃHIV positive (n = 452) | 89.5 (81.1–95.1) | 52.5 (47.2–57.7) | 1.9 (1.7–2.1) | .2 (.1–.4) |
| ȃȃȃȃHIV negative (n = 893) | 90.1 (84.0–94.5) | 66.8 (63.3–70.2) | 2.7 (2.4–3.1) | .1 (.1–.2) |
The fixed threshold for each software was determined on the basis of the CAD score that achieved at least 90% sensitivity while optimizing specificity among all study participants. These thresholds were 15 for CAD4TB and 6 for qXR, respectively. Abbreviations: CAD, computer-aided detection; CI, confidence interval; HIV, human immunodeficiency virus; TB, tuberculosis.
There were 109 patients with an unknown HIV status (n = 22 persons with previous TB and n = 87 persons without previous TB).
Delft Imaging Systems, Netherlands.
Qure.ai, India.
Optimum CAD Thresholds for Different Subpopulations
Among persons with previous TB, the optimum thresholds for achieving at least 90% sensitivity differed substantially by sex and by HIV status for both CAD software systems (Table 3); only qXR among HIV-negative persons approximated the minimum WHO-recommended criteria (90.5% sensitivity, 65.4% specificity). Among persons without a history of previous TB history, diagnostic accuracy was substantially higher and there was less variation in optimal threshold cutoffs by HIV status and sex for both software systems (Table 3); both CAD4TB and qXR among HIV-negative persons approximated but did not meet minimum WHO recommendations.
Table 3.
Computer-Aided Detection Cutoff Thresholds and Associated Diagnostic Accuracy for Approximating World Health Organization Target Product Profile Specifications Using a 90% Sensitivity Threshold for Tuberculosis (TB) Screening Tests, According to Previous TB Treatment Status
| CAD Threshold |
Sensitivity (95% CI) |
Specificity (95% CI) |
Positive Likelihood Ratio (95% CI) |
Negative Likelihood Ratio (95% CI) |
|
|---|---|---|---|---|---|
| Previous TB | |||||
| ȃCAD4TBb | |||||
| ȃȃAll (n = 452) | 10 | 91.1 (80.4–97.0) | 19.7 (15.9–24.0) | 1.1 (1.0–1.2) | .5 (.2–1.1) |
| ȃȃȃSex | |||||
| ȃȃȃȃMale (n = 326) | 32 | 91.1 (78.8–97.5) | 31.0 (25.6–36.7) | 1.3 (1.2–1.5) | .3 (.1–.7) |
| ȃȃȃȃFemale (n = 126) | 4 | 90.9 (58.7–99.8) | 20.0 (13.1–28.5) | 1.1 (.9–1.4) | .5 (.1–3.1) |
| ȃȃȃHIV statusa | |||||
| ȃȃȃȃHIV positive (n = 250) | 9 | 93.5 (78.6–99.2) | 22.4 (17.0–28.5) | 1.2 (1.1–1.3) | .3 (.1–1.1) |
| ȃȃȃȃHIV negative (n = 180) | 45 | 90.5 (69.6–98.8) | 39.0 (31.4–47.0) | 1.5 (1.2–1.8) | .2 (.1–.9) |
| ȃqXRc | |||||
| ȃȃAll (n = 452) | 14 | 91.1 (80.4–97.0) | 29.0 (24.6–33.8) | 1.3 (1.2–1.4) | .3 (.1–.7) |
| ȃȃȃSex | |||||
| ȃȃȃȃMale (n = 326) | 54 | 91.1 (78.8–97.5) | 50.9 (44.9–56.9) | 1.9 (1.6–2.2) | .2 (.1–.4) |
| ȃȃȃȃFemale (n = 126) | 2 | 90.9 (58.7–99.8) | 17.4 (11.0–25.6) | 1.1 (.0–1.4) | .5 (.1–3.5) |
| ȃȃȃHIV statusa | |||||
| ȃȃȃȃHIV positive (n = 250) | 6 | 93.5 (78.6–99.2) | 26.5 (20.8–32.9) | 1.3 (1.2–1.4) | .2 (.1–.9) |
| ȃȃȃȃHIV negative (n = 180) | 71 | 90.5 (69.6–98.8) | 65.4 (57.5–72.8) | 2.6 (2.0–3.4) | .1 (.0–.5) |
| No previous TB | |||||
| ȃCAD4TB | |||||
| ȃȃAll (n = 1432) | 17 | 89.7 (85.1–93.2) | 61.9 (59.1–64.7) | 2.4 (2.2–2.6) | .2 (.1–.2) |
| ȃȃȃSex | |||||
| ȃȃȃȃMale (n = 858) | 20 | 89.8 (84.4–93.9) | 56.8 (53.0–60.6) | 2.1 (1.9–2.3) | .2 (.1–.3) |
| ȃȃȃȃFemale (n = 574) | 7 | 92.3 (83.0–97.5) | 59.7 (55.3–64.0) | 2.3 (2.0–2.6) | .1 (.1–.3) |
| ȃȃȃHIV statusa | |||||
| ȃȃȃȃHIV positive (n = 452) | 7 | 93.0 (85.4–97.4) | 40.4 (35.4–45.7) | 1.6 (1.4 -1.7) | .2 (.1–.4) |
| ȃȃȃȃHIV negative (n = 893) | 18 | 90.1 (84.0–94.5) | 66.2 (62.7–69.6) | 2.7 (2.4–3.0) | .1 (.1–.2) |
| ȃqXR | |||||
| ȃȃAll (n = 1432) | 6 | 89.7 (85.1–93.2) | 61.8 (58.9–64.5) | 2.4 (2.2–2.5) | .2 (.1–.2) |
| ȃȃȃSex | |||||
| ȃȃȃȃMale (n = 858) | 8 | 89.8 (84.4–93.9) | 56.4 (52.6–60.2) | 2.1 (1.9–2.3) | .2 (.1–.3) |
| ȃȃȃȃFemale (n = 574) | 2 | 95.4 (87.1–99.0) | 47.7 (43.3–52.2) | 1.8 (1.6–2.0) | .1 (.0–.3) |
| ȃȃȃHIV statusa | |||||
| ȃȃȃȃHIV positive (n = 452) | 5 | 91.9 (83.9–96.7) | 51.6 (46.4–56.9) | 1.9 (1.7–2.1) | .2 (.1–.3) |
| ȃȃȃȃHIV negative (n = 893) | 6 | 90.1 (84.0–94.5) | 66.8 (63.3–70.2) | 2.7 (2.4–3.0) | .1 (.1–.2) |
Abbreviations: CAD, computer-aided detection; CI, confidence interval; HIV, human immunodeficiency virus; TB, tuberculosis.
There were 109 patients with an unknown HIV status (n = 22 persons with previous TB and n = 87 persons without previous TB).
Delft Imaging Systems, Netherlands.
Qure.ai, India.
DISCUSSION
In this study, we demonstrated that the overall discriminatory value (ie, AUROC) of CAD in reading dCXRs for the presence of active TB disease differs substantially according to a patient’s previous TB status when using CAD4TB but was comparable when using qXR. However, when using a fixed abnormality threshold defined by the optimal performance in the overall study population, the sensitivity of the 2 CAD systems was similarly high regardless of previous TB status, but their specificity was substantially lower among individuals with a history of previous TB.
Our study adds to and extends the scarce existing evidence base on how CAD system performance as a triage tool for TB is affected by an individual’s previous TB status [4, 6, 15, 24]. Similar to a study from Bangladesh that evaluated 5 AI algorithms, our study was conducted in a high-TB-burden setting, Xpert MTB/RIF was used as the reference standard, and we observed a decline in discriminatory power for CAD4TB among patients with previous TB [4]. In contrast to the same study, there was no statistically significant decline in the discriminatory power of qXR among those with previous TB compared with those without previous TB. This likely reflects differences in the demographic and clinical characteristics of participants [25]. Notably, we observed a significant reduction in the specificity of CAD among patients with previous TB when using a fixed abnormality threshold, which has also been reported in other studies [6, 15, 24].
The findings on CAD’s reduced triage performance among patients with prior TB are especially notable as the clinical presentation of patients with previously treated TB (eg, post-TB lung disease) but without current, active disease may be similar to that of previously treated patients with TB with recurrent, active TB. In such clinical scenarios, the discriminatory value of dCXR CAD is even more important in guiding clinicians in the appropriate evaluation of symptomatic patients with previous TB [21, 26]. Explicitly accounting for post-TB scarring during further CAD software development and adding a module that compares individuals’ previous and current CXR to identify new lesions [27] could potentially improve the performance of the CAD software among patients with previously treated TB.
Substantial differences in specificity between men and women (irrespective of previous TB history) were observed for both CAD software systems when a fixed threshold that approximated 90% sensitivity in the overall study population was applied. This likely, in large part, reflects the tradeoffs between the sensitivity and specificity at a fixed abnormality threshold as the AUROCs for these subgroups were similar. However, prior studies from Bangladesh and Kenya found lower accuracy of CAD for TB among women compared with men [4, 15]; when we applied an optimal cutoff, we found that qXR but not CAD4TB had lower specificity among women compared with men. Reduced CAD accuracy among women could, in part, be due to breast attenuation in some women, which reduces visualization of pertinent anatomy in the lower lung zones, especially if post-TB scarring is also present. Notably, when applying a fixed abnormality threshold, only small differences in CAD’s performance were observed according to HIV status, which was limited to among those without prior TB, where the specificity was lower in HIV-positive persons. These findings largely are in concordance with those from a South African study that reported comparable accuracy of CAD among HIV-positive and -negative individuals [28].
Irrespective of the CAD abnormality threshold (fixed or optimized), CAD systems generally fell short of achieving the minimum TPP specifications for TB screening and triage tools (90% sensitivity and 70% specificity) among different subpopulations [2]. At CAD thresholds required to achieve a 90% sensitivity, extremely low specificity (<30%) was observed for both CAD software systems among all persons with a previous TB history. This could be associated with substantially increased programmatic costs when this subgroup of patients is being evaluated for TB, due to a large proportion of patients requiring subsequent microbiological testing. However, patients with previous TB are at increased risk of drug-resistant TB (DR-TB), and thus it is important that triage tests in this subgroup retain high sensitivity—potentially at the cost of reduced specificity—in order to not delay the diagnosis of DR-TB. When applying optimized CAD cutoffs, diagnostic accuracy was significantly better in men compared with women for qXR and for HIV-negative compared with HIV-positive persons for both CAD systems regardless of prior TB status. Nonetheless, both software systems’ performance only approximated minimum TPP specifications among HIV-negative persons with (qXR only) and without previous TB (qXR and CAD4TB) [2]. While optimal CAD thresholds differed substantially according to both sex and HIV status among participants with previous TB, there tended to be smaller differences in optimal CAD thresholds among participants without previous TB. Additionally, the optimal abnormality thresholds in our study were significantly lower than those reported in the Bangladesh study [4]. Collectively, this suggests that defining and implementing optimal CAD cutoffs for triaging of TB among persons with previous TB—and potentially accounting for additional individual characteristics such as sex and HIV status—will be extremely important as access to this technology is scaled-up across different high-TB-burden settings [6, 14, 29]. It will also be important for future studies to evaluate the feasibility of different strategies for implementing differential CAD cutoffs to account for patient characteristics.
Strengths of this study were the inclusion of a large number of consecutively enrolled individuals from both community and health facility contexts in a high-HIV/TB-prevalence setting. Second, no participant CXR images in the present study were used for training of the CAD products, which mitigated the risk of overestimation of their accuracy. Third, our analysis of CAD performance among those with and without previous TB was further stratified by sex and HIV status, which has not been previously reported.
There were, however, some limitations. First, we were unable to compare CAD’s performance with manually read CXRs. However, in a study in which we compared expert human readers’ performance with a reference standard, we also found that there was a limitation in discriminating radiographic changes due to scarring from old TB compared with changes due to prevalent, active TB [21]. Second, previous TB disease was based on self-report rather than programmatic records. Due to TB-related stigma, a small number of participants may not have reported their previous TB history, resulting in misclassification. Third, Xpert MTB/RIF was used as the reference standard. It is less sensitive than culture, which could have led to some patients with culture-positive, Xpert-negative TB being incorrectly classified as not having TB. Further, due to persistently positive Xpert MTB/RIF results among patients with previously treated TB [30], a small number of false-positive results may have contributed to the high prevalence of TB within this subpopulation. Also, participants in our analysis were predominantly drawn from a health facility setting, where a large proportion were HIV-positive, had previous TB, and were highly symptomatic for TB; this may limit the generalizability of our findings to community- and lower-level health settings where lower risk and less symptomatic persons—earlier in their disease course—may first be evaluated [22]. Finally, this study excluded asymptomatic persons (given the small number of such participants) and did not have data to perform analyses by smear status, both of which should be assessed as part of future evaluations of the performance of CAD for TB screening and triage.
In conclusion, CAD4TB and qXR had excellent overall discriminatory value as triage tools for TB; however, accuracy for both software systems was substantially decreased among those previously treated for TB when applying a fixed abnormality threshold. The optimal CAD threshold for detecting active TB disease differed substantially by previous TB history and further stratified by sex and HIV status. This suggests that different CAD threshold cutoffs are needed when triaging individuals with a previous TB history to optimize diagnostic performance in this population. Further evidence is needed regarding the performance of CAD software stratified according to the characteristics of the individual being evaluated, to allow for improved patient-specific and setting-specific calibration of CAD within local TB-control programs.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Mary Kagujje, Tuberculosis Department, Centre for Infectious Disease Research in Zambia, Lusaka, Zambia.
Andrew D Kerkhoff, Division of HIV, Infectious Diseases and Global Medicine Zuckerberg, San Francisco General Hospital and Trauma Center, University of California, San Francisco, San Francisco, California, USA.
Mutinta Nteeni, Department of Radiology, Levy Mwanawasa University Teaching Hospital, Lusaka, Zambia.
Ian Dunn, Department of Radiology, University of British Columbia, Vancouver, Canada.
Kondwelani Mateyo, Department of Internal Medicine, University Teaching Hospital, Lusaka, Zambia.
Monde Muyoyeta, Tuberculosis Department, Centre for Infectious Disease Research in Zambia, Lusaka, Zambia.
Notes
Author Contributions. Conceived and designed the study: M. M. Data curation and analysis: M. M., A. D. K. Implemented the study: M. K., M. M. Wrote the manuscript: M. K., A. D. K., and M. M. Critically reviewed the manuscript: M. N., I. D., K. M. Approved the final version to be published: All authors.
Acknowledgments. The authors thank DELFT and Qure.ai for allowing us to use their AI algorithms at no cost. Both companies had no influence over any aspect of this research and its findings. M. K. and M. M. report a grant from Qure.ai under one of its research studies and an ongoing project under which they procured qXR licenses.
Data sharing. The anonymized individual participant data and a data dictionary defining each field in the dataset used in this study can be available upon reasonable request to the senior author (mondemuy@gmail.com). Chest X-ray images will not be provided as these are withheld by the corresponding author’s organization to reserve their use for product evaluations. Additional related documents will be provided to those whose requests are approved, but secondary analysis must be done with investigator support.
Financial support. This work was supported by the Stop TB partnership’s TB REACH initiative with funding from the Government of Canada (grant number STBP/TBREACH/GSA/W5-26). The authors report that DELFT and Qure.ai both provided lisences for use of their products at no cost.
References
- 1. World Health Organization . The End TB strategy. Available from: https://www.who.int/tb/post2015_TBstrategy.pdf?ua=1. Accessed 19 June 2022.
- 2. World Health Organization . Module 2: screening—systematic screening for tuberculosis disease, in WHO consolidated guidelines on tuberculosis. Geneva, Switzerland: WHO; 2021. [PubMed] [Google Scholar]
- 3. Van’t Hoog A, Viney K, Biermann O, et al. Symptom-and chest-radiography screening for active pulmonary tuberculosis in HIV-negative adults and adults with unknown HIV status. Cochrane Database Syst Rev 2022; 3:CD010890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qin ZZ, Ahmed S, Sarker MS, et al. Tuberculosis detection from chest x-rays for triaging in a high tuberculosis-burden setting: an evaluation of five artificial intelligence algorithms. Lancet Digit Health 2021; 3:e543–54. [DOI] [PubMed] [Google Scholar]
- 5. Pande T, Cohen C, Pai M, Ahmad Khan F. Computer-aided detection of pulmonary tuberculosis on digital chest radiographs: a systematic review. Int J Tuberc Lung Dis 2016; 20:1226–30. [DOI] [PubMed] [Google Scholar]
- 6. Tavaziva G, Harris M, Abidi SK, et al. Chest X-ray analysis with deep learning-based software as a triage test for pulmonary tuberculosis: an individual patient data meta-analysis of diagnostic accuracy. Clin Infect Dis 2022; 74:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Basheer S, Jayakrishna V, Kamal AG. Computer assisted X-ray analysis system for detection of onset of tuberculosis. Int J Sci Eng Res 2013; 4:606–13. [Google Scholar]
- 8. Jaeger S, Karargyris A, Candemir S, et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imaging 2014; 33:233–45. [DOI] [PubMed] [Google Scholar]
- 9. World Health Organization . World Health Organization (WHO) information note: tuberculosis and COVID-19. 2020. Available from: https://www.who.int/docs/default-source/documents/tuberculosis/infonote-tb-covid-19.pdf. Accessed 2 July 2021.
- 10. Muyoyeta M, Moyo M, Kasese N, et al. Implementation research to inform the use of Xpert MTB/RIF in primary health care facilities in high TB and HIV settings in resource constrained settings. PLoS One 2015; 10:e0126376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. World Health Organization . WHO consolidated guidelines on tuberculosis: module 2: screening: systematic screening for tuberculosis disease. Web Annex C: GRADE evidence to decision tables. 2021. Available from: https://apps.who.int/iris/bitstream/handle/10665/340243/9789240022713-eng.pdf. Accessed 2 July 2021.
- 12. Qin C, Yao D, Shi Y, Song Z. Computer-aided detection in chest radiography based on artificial intelligence: a survey. Biomed Eng Online 2018; 17:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khan FA, Majidulla A, Tavaziva G, et al. Chest X-ray analysis with deep learning-based software as a triage test for pulmonary tuberculosis: a prospective study of diagnostic accuracy for culture-confirmed disease. Lancet Digit Health 2020; 2:e573–81. [DOI] [PubMed] [Google Scholar]
- 14. Qin ZZ, Sander MS, Rai B, et al. Using artificial intelligence to read chest radiographs for tuberculosis detection: a multi-site evaluation of the diagnostic accuracy of three deep learning systems. Sci Rep 2019; 9:15000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mungai B, Ong'angò J, Ku CC, et al. Accuracy of computer-aided chest X-ray screening in the Kenya national Tuberculosis prevalence survey. medRxiv, doi: 10.1101/2021.10.21.21265321, 2021, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nabukenya-Mudiope MG, Kawuma HJ, Brouwer M, Mudiope P, Vassall A. Tuberculosis retreatment ‘others’ in comparison with classical retreatment cases; a retrospective cohort review. BMC Public Health 2015; 15:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dedefo MG, Sirata MT, Ejeta BM, et al. Treatment outcomes of tuberculosis retreatment case and its determinants in west Ethiopia. Open Respir Med J 2019; 13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metcalfe JZ, Mason P, Mungofa S, Sandy C, Hopewell PC. Empiric tuberculosis treatment in retreatment patients in high HIV/tuberculosis-burden settings. Lancet Infect Dis 2014; 14:794–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meghji J, Simpson H, Squire SB, Mortimer K. A systematic review of the prevalence and pattern of imaging defined post-TB lung disease. PLoS One 2016; 11:e0161176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ali MG, Muhammad ZS, Shahzad T, Yaseen A, Irfan M. Post tuberculosis sequelae in patients treated for tuberculosis: an observational study at a tertiary care center of a high TB burden country. Eur Respir Soc 2018; 52:PA2745. [Google Scholar]
- 21. Mateyo K, Kerkhoff AD, Dunn I, Nteeni MS, Muyoyeta M. Clinical and radiographic characteristics of presumptive tuberculosis patients previously treated for tuberculosis in Zambia. PLoS One 2022; 17:e0263116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kagujje M, Chilukutu L, Somwe P, et al. Active TB case finding in a high burden setting; comparison of community and facility-based strategies in Lusaka, Zambia. PLoS One 2020; 15:e0237931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Clin Chem 2015; 61:1446–52. [DOI] [PubMed] [Google Scholar]
- 24. Kik SV, Gelaw SM, Ruhwald M, et al. Diagnostic accuracy of chest X-ray interpretation for tuberculosis by three artificial intelligence-based software in a screening use-case: an individual patient meta-analysis of global data. medRxiv, doi: 10.1101/2022.01.24.22269730,2022, preprint: not peer reviewed. [DOI] [Google Scholar]
- 25. Zaidi SMA, Habib SS, Van Ginneken B, et al. Evaluation of the diagnostic accuracy of computer-aided detection of tuberculosis on chest radiography among private sector patients in Pakistan. Sci Rep 2018; 8:12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ayari A, Smadhi H, Mejri I, et al. Management of pulmonary tuberculosis sequelae. Eur Respir Soc 2015;46(Suppl 59):PA2762. [Google Scholar]
- 27. Bhalla AS, Goyal A, Guleria R, Gupta AK. Chest tuberculosis: radiological review and imaging recommendations. Indian J Radiol Imaging 2015; 25:213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fehr J, Konigorski S, Olivier S, et al. Computer-aided interpretation of chest radiography reveals the spectrum of tuberculosis in rural South Africa. NPJ Digit Med 2021; 4:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rahman MT, Codlin AJ, Rahman MM, et al. An evaluation of automated chest radiography reading software for tuberculosis screening among public- and private-sector patients. Eur Respir J 2017; 49:1602159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theron G, Venter R, Calligaro G, et al. Xpert MTB/RIF results in patients with previous tuberculosis: can we distinguish true from false positive results? Clin Infect Dis 2016; 62:995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.