Abstract
Sunflowers of the genus Helianthus are models for hybridization research and contain three of the best-studied examples of homoploid hybrid speciation. To understand a broader picture of hybridization within the annual sunflowers, we used whole-genome resequencing to conduct a phylogenomic analysis and test for gene flow between lineages. We find that all annual sunflower species tested have evidence of admixture, suggesting hybridization was common during the radiation of the genus. Support for the major species tree decreases with increasing recombination rate, consistent with hybridization and introgression contributing to discordant topologies. Admixture graphs found hybridization to be associated with the origins of the three putative hybrid species (Helianthus anomalus, Helianthus deserticola, and Helianthus paradoxus). However, the hybridization events are more ancient than suggested by previous work. Furthermore, H. anomalus and H. deserticola appear to have arisen from a single hybridization event involving an unexpected donor, rather than through multiple independent events as previously proposed. This means our results are consistent with, but not definitive proof of, two ancient independent homoploid hybrid speciation events in the genus. Using a broader data set that covers the whole Helianthus genus, including perennial species, we find that signals of introgression span the genus and beyond, suggesting highly divergent introgression and/or the sorting of ancient haplotypes. Thus, Helianthus can be viewed as a syngameon in which largely reproductively isolated species are linked together by occasional or frequent gene flow.
Keywords: hybrid speciation, introgression, Helianthus, genomics, gene flow, sunflower
Introduction
The evolutionary importance of hybridization and introgression has been explored since Darwin's time (Darwin 1859). Initially, although zoologists thought that hybridization was relatively rare and unimportant (Mayr 1963), botanists have long emphasized the role of hybridization and introgression in both adaptation and speciation (Anderson 1949; Heiser 1949a; Anderson and Stebbins 1954). Genomics eventually proved botanists correct in this regard; evidence of past hybridization is common in both animals and plants (Fontaine et al. 2015; Mallet et al. 2016; Figueiró et al. 2017; Calfee et al. 2021; Dagilis et al. 2021). In several cases, alleles responsible for ecologically important traits have been found to be acquired through introgression (Jones et al. 2018; Suarez-Gonzalez et al. 2018; Oziolor et al. 2019), supporting the notion that introgression not only occurs, but can be evolutionarily important.
Hybridization can also result in the formation of new species through homoploid hybrid speciation, in which an admixed lineage acquires reproductive isolation from its progenitors through a combination of alleles or chromosomal rearrangements from both parents (Stebbins 1957; Grant 1958; Schumer et al. 2014). One way this can occur is if two parental species differ at both habitat and mate choice. A hybrid population between these two may have alternate parental alleles for these two traits, and therefore, be separated from one parental species by habitat and the other by mate choice (Wang et al. 2021). Hybrid speciation can also occur when hybridization results in the ability to colonize a new ecological niche, reject parental species as potential mates, or have unique combinations of chromosomal rearrangements (Rieseberg et al. 1995; Gross and Rieseberg 2005; Melo et al. 2009). Schumer et al. (2014) argued that three criteria must be satisfied to demonstrate homoploid hybrid speciation: (1) reproductive isolation from parental species, (2) prior admixture, and (3) evidence that reproductive barriers arose via hybridization. Evaluation of the natural hybridization literature indicates that while admixed lineages are relatively common (criterion 2), few case studies satisfy criteria 1 and 3 (Schumer et al. 2014). Three examples of hybrid species thought to address all three criteria are within the sunflower genus, Helianthus.
The widespread species Helianthus annuus and Helianthus petiolaris are thought to be the parents of three independent homoploid hybrid species, Helianthus anomalus, Helianthus deserticola, and Helianthus paradoxus. These were initially identified as hybrid species through early genetic studies that found they had a mixture of chloroplast DNA, allozyme, and ribosomal DNA markers from both parental species (Rieseberg, Carter, et al. 1990; Rieseberg 1991). The hybrid species have strong ecological separation based on habitat choice; H. anomalus occurs on sand dunes, H. deserticola on the sandy soil of the desert floor, and H. paradoxus in brackish salt marshes (Heiser et al. 1969). Comparative QTL mapping of hybrids and parental species suggested that this ecological separation was achieved by combinations of parental alleles that allowed for extreme (transgressive) phenotypes (Rieseberg et al. 2003). Subsequent experiments found that such transgressive phenotypes were favored when segregating hybrids of H. annuus-H. petiolaris were transplanted into the natural habitats of the three ancient hybrid species, with some synthetic hybrids rivaling the ancient hybrids in fitness (Lexer et al. 2003; Gross et al. 2004; Ludwig et al. 2004).
In addition to ecological isolation, the three hybrid species are partially reproductively isolated by hybrid pollen sterility caused mainly by chromosomal rearrangements (Lai et al. 2005). This was hypothesized to be due to the hybrid species having a combination of parental chromosomal rearrangements as well as unique changes (Rieseberg et al. 1995; Lai et al. 2005). Synthetic hybrids of H. annuus–H. petiolaris not only recovered fertility in a small number of generations, but also were more compatible with the putative hybrid species than were the parental species (Rieseberg et al. 1996; Rieseberg 2000, 2006). Microsatellite, chloroplast, and karyotypic analyses suggested that all hybrid species originated between 63,000 and 208,000 generations before the present and a single (H. paradoxus) origin or multiple (H. anomalus and H. deserticola) origins depending on the species (Schwarzbach and Rieseberg 2002; Welch and Rieseberg 2002; Gross et al. 2003). The strong link between admixture and reproductive isolation, in terms of ecology and karyotype, comparisons with experimental hybrid lineages, and partial replication across three species have made Helianthus a model for understanding homoploid hybrid speciation.
Although the Helianthus hybrid species were probed with cutting-edge molecular markers when initially identified, they have been relatively unexamined in the genomics age. Studies using genotyping-by-sequencing grouped H. anomalus and H. deserticola together in a phylogenetic tree, but did not probe further (Baute et al. 2016). Moody and Rieseberg (2012) sequenced ten nuclear genes for multiple individuals of most annual sunflowers, including the hybrid species. They found extensive discordance between individual gene trees and notably that H. anomalus and H. deserticola grouped together and that H. paradoxus grouped with H. annuus, although the overall species tree did not agree with previous expectations. The most recent phylogeny of Helianthus did not include any hybrid species (Stephens et al. 2015). In each of these cases, modern approaches for identifying gene flow and hybrid ancestry were not performed. This is important because sunflower species are both young—Helianthus crown age is 3.6 Ma—and generally have a high effective population size (Strasburg and Rieseberg 2008). This means that incomplete lineage sorting (ILS) is likely high and discordant topologies that could be produced by hybridization may also be present without any interspecies gene flow. In addition, signals of hybridization can sometimes derive from unexpected sources (Owens et al. 2021).
Beyond the three well-characterized Helianthus hybrid species, numerous other cases of hybridization and introgression have been reported in the genus (Heiser et al. 1969; Rieseberg et al. 1991), leading Rieseberg (1991) to conclude that evolution in the group “must be considered reticulate, rather than exclusively dichotomous and branching.” Although many of these early cases—which were based on relatively limited evidence—have been subsequently confirmed with genomic data (Kane et al. 2009; Owens et al. 2016; Mondon et al. 2018; Lee-Yaw et al. 2019; Zhang et al. 2019), others have not (Owens et al. 2021), and several remain to be tested. This highlights that the combination of modern genomic data sets and phylogenetic techniques allows for hypotheses about hybridization to be rigorously tested (Hibbins and Hahn 2022).
Here, we use modern phylogenomic techniques to assess the extent of admixture during the evolution of the genus. To do this, we first use whole-genome sequencing data for most annual sunflowers to infer the genome-wide phylogeny of this group. We then apply tests of introgression and gene flow across the annual clade, including the three hybrid species, H. anomalus, H. deserticola, and H. paradoxus. This allows us to revisit criterion 2 of homoploid hybrid speciation for these three species, and to gain greater insight regarding their ancestry and relationship to the broader patterns of admixture within the annual sunflower clade. Lastly, using previously published phylogenomic data (Stephens et al. 2015), we extend our analyses across the entire genus.
Results
Data Filtering
We generated whole-genome sequence data for two H. paradoxus samples. We called variants against the H. annuus HA412-HOv2 reference genome for these samples as well as previously sequenced data for two samples of eight other annual species, or subspecies, and four outgroups (supplementary table S1, Supplementary Material online; Todesco et al. 2020). We retained 5,877,513 variables and 28,274,146 invariant sites after filtering for quality, mappability, and call rate (supplementary fig. S1, Supplementary Material online). We divided the genome into a total of 11,797 10 kbp non-overlapping windows and retained 3,267 that had ≥2,000 called bases.
Additionally, to understand the evolutionary history of the cytoplasmic genomes, we aligned all sequenced data to a reduced reference containing only the chloroplast and mitochondrial genomes. After calling variants, we found 744 and 1,477 SNPs and 166 and 90 indels in the cytoplasmic and mitochondrial genomes, respectively.
Species and Gene Trees
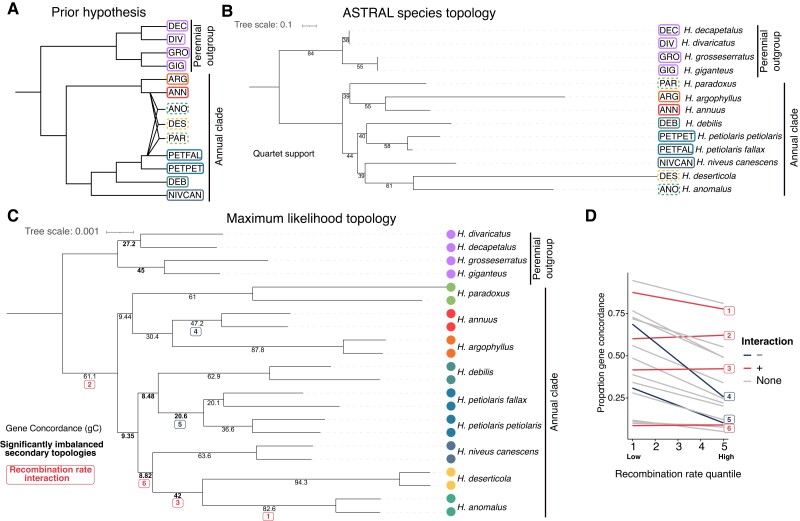
The species topology for non-hybrid taxa from ASTRAL (fig. 1A and B) is mostly consistent with previous phylogenies that used fewer markers (Stephens et al. 2015; Baute et al. 2016; Zhang et al. 2019). Quadripartition support at nodes ranged from 38% to 84%, suggesting substantial variation in topology at gene trees. The maximum likelihood concatenated tree also found the same topology as ASTRAL and monophyly for all samples of the same species (fig. 1B).
Fig. 1.
Phylogenetics of annual sunflowers. (A) The prior hypothesis of annual species relationships (Rieseberg 1991; Stephens et al. 2015). (B) Phylogenetic tree from combined 10 kbp gene trees used as input for ASTRAL species topology. Quartet support plotted on branches. (C) Concatenated maximum likelihood tree using IQ-TREE2. Gene concordance plotted on nodes. Bolded values have significantly imbalanced secondary topologies. (D) The relationship between gene concordance proportion and recombination quantile for each node with <90% overall gene concordance. Significant interactions with node ID are highlighted here and on panel C.
As outlined in the introduction, the three putative homoploid hybrid taxa, H. paradoxus, H. deserticola, and H. anomalus are thought to have originated from hybridization between H. annuus and H. petiolaris (Rieseberg 1991). Since ASTRAL does not account for hybridization, we would expect the potential hybrid species to group near the dominant parental species. We found that H. paradoxus grouped with H. annuus and its sister species Helianthus argophyllus, whereas H. anomalus and H. deserticola grouped with Helianthus niveus canescens (fig. 1B). Quadripartition support for these groupings is low (each 39%), as expected for hybrid lineages, although this was not unique to branches involving the previously identified hybrid species.
We further examined variation in gene tree topology by calculating the gene concordance factor for each branch on our concatenated tree using gene trees. We found relatively low support at branches separating species compared with branches separating samples within the species (fig. 1C). When including all samples, each tree was unique. When we subsampled tips down to a single individual per species we found higher agreement; the most common tree was found 6 times out of 3,242 trees.
For the cytoplasmic genomes, we found two separate clades for both mitochondrial and chloroplast genomes (supplementary fig. S2, Supplementary Material online). One clade contained H. annuus, H. argophyllus, H. petiolaris petiolaris, and H. anomalus, the other contained H. deserticola, H. petiolaris fallax, and H. niveus canescens. For H. paradoxus and Helianthus debilis, the two individuals fell into different clades for both genomes.
Gene Tree Discordance
To explore why gene concordance values were low, we compared the proportion of trees matching the two alternate topologies for every quadripartition using IQ-TREE2's gene concordance measure (Minh, Hahn, et al. 2020). Under purely ILS, we expect that both alternate topologies should be found at equal proportions, whereas introgression will lead to imbalances. We found that at five nodes within the annual species, there has been a significant (χ2, P < 0.05) imbalance in alternate topologies (fig. 1C).
Selection against introgressed ancestry can lead to reduced introgression in regions of low recombination (Schumer et al. 2018; Martin et al. 2019). We explored this by using a binomial regression to quantify the relationship between support for the species topology and the recombination rate (as identified from the H. annuus genetic map) at each node with >10% support for alternate topologies. In other words, are genomic regions with lower recombination more likely to show species topology? We found a significant negative effect of the recombination rate (β1 = −0.30, P = e−14) on support for the species topology (fig. 1C and D). Interestingly, we find a significant interaction between the node and recombination rate at six nodes. At four nodes, most of which include the H. anomalus/H. deserticola clade, the relationship is less negative or positive (fig. 1D). At the node separating the H. annuus samples, the relationship with recombination is most strongly negative.
Helianthus niveus canescens was previously considered a variety of H. petiolaris, with which it intergrades for part of its range (Heiser et al. 1969). Our species topology places H. niveus canescens with H. anomalus and H. deserticola instead of H. petiolaris, but with relatively low support and overall less support at low recombination regions (fig. 1D). We counted the number of trees where H. niveus canescens grouped with H. petiolaris petiolaris and H. petiolaris fallax to the exclusion of H. anomalus and H. deserticola, as well as the reverse (supplementary fig. S3, Supplementary Material online). We found more support for H. niveus canescens together with H. petiolaris.
Admixture in the Annual Sunflowers
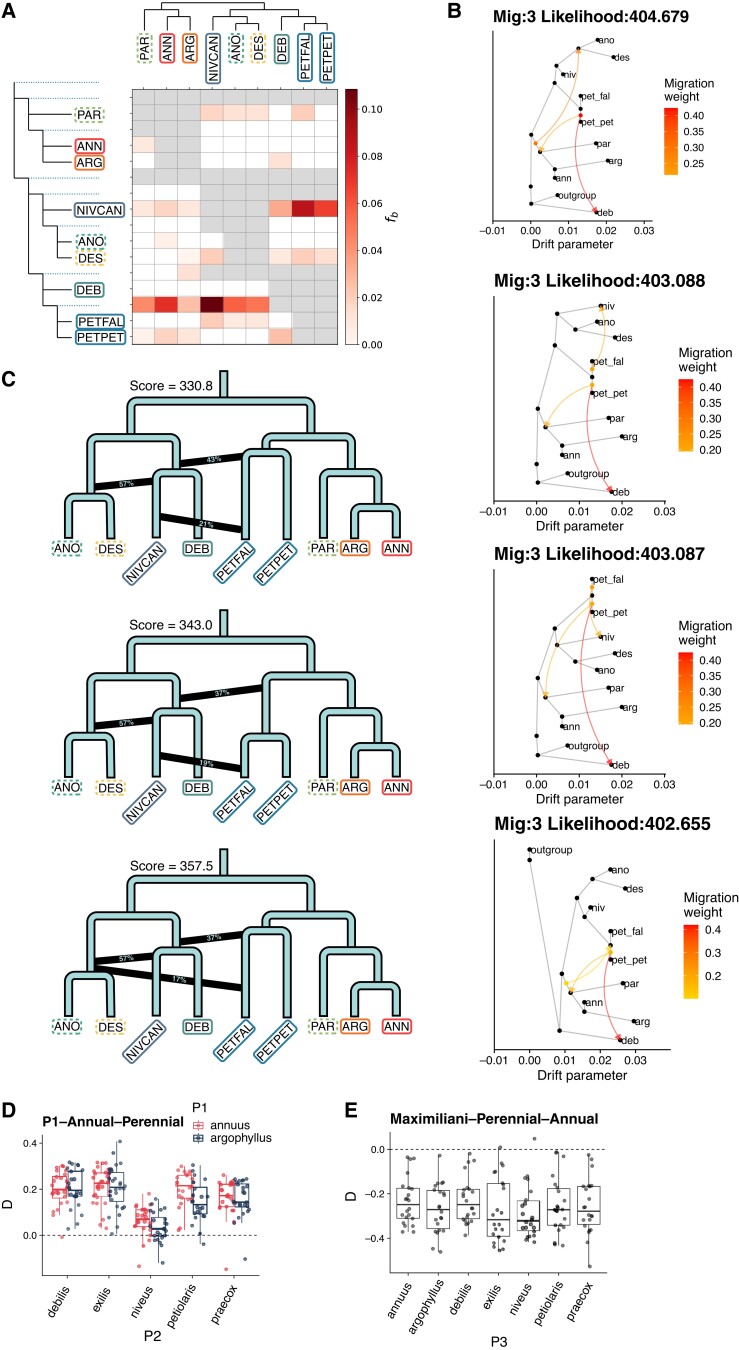
To assess the extent of hybridization during the evolution of the annual sunflowers, we used comprehensive approaches to test for admixture across all species. The fbranch analysis, which uses the f4-ratio test to identify introgression within a phylogenetic framework, found many significant signals (fig. 2A, supplementary table S2, Supplementary Material online). This approach tests all possible sets of four samples and synthesizes the results to show where introgression signals are shared among relatives, which can highlight admixture events that occurred in ancestors. The fbranch patterns are dependent on the underlying phylogeny, so to explore this effect, we repeated fbranch using the top nine most common species tree topologies (supplementary fig. S4, Supplementary Material online). Although no topology completely removes significant fbranch signal, this shows that introgression signals from fbranch can come from either close phylogenetic position or introgression. For example, in three of the top nine trees, H. paradoxus is placed within the H. petiolaris clade, but its fbranch signal shows it is much more similar to H. annuus and H. argophyllus than others in the H. petiolaris clade (supplementary fig. S4, Supplementary Material online). The three strongest patterns are the following:
Fig. 2.
Introgression across sunflowers. (A) Fbranch statistic for annual species using the ASTRAL species topology. (B) The four best-supported TreeMix graphs with three admixture edges. A higher likelihood score is better. (C) The best-supported admixture graph from ADMIXTOOLS2 with three admixture edges. Branch lengths are not to scale. A lower score is better. (D) ABBA-BABA with the groupings [H. annuus OR H. argophyllus], other annual species, perennial species, Phoebanthus. Positive values indicate more ABBA counts. E) ABBA-BABA with the groupings H. maximiliani, other perennials, annual species, Phoebanthus.
H. petiolaris has a signal of shared ancestry with the H. annuus clade.
H. niveus has a signal of shared ancestry with both H. petiolaris and the sister pair, H. anomalus and H. deserticola.
H. debilis has a signal of shared ancestry with an unsampled lineage basal to the H. petiolaris clade.
The three hybrid species do not stand out in terms of the extent of admixture.
A different picture comes from the TreeMix analysis, which uses the covariance of population allele frequency to estimate phylogeny and migration edges. We found that three was the optimal number of migration edges and explained 99.9% of the variance. Above four edges, there are diminishing returns in likelihood gain and variance explained (supplementary fig. S5, Supplementary Material online). The admixture is found to be associated with the origins of the three putative hybrid species in the most likely TreeMix graphs at both three and four edges (supplementary fig. S6, Supplementary Material online). The origin of H. paradoxus appears to involve ancient hybridization between the ancestor of the H. annuus clade and H. petiolaris, although this admixture is inherited by the other sampled members of the H. annuus clade. Likewise, there is an admixture event between a basal lineage of the H. annuus clade and the ancestor of H. deserticola and H. anomalus. The third signal in common between the top three admixture graphs was between H. debilis and H. petiolaris. However, this likely is an artifact of misplacing H. debilis as basal in the tree and may instead suggest that it has ancestry from a lineage at the base of the annual sunflower clade. When we look other graphs that are close in likelihood to the top graphs, we frequently find an admixture between H. niveus canescens and H. petiolaris fallax, although these results in the loss of the admixture event associated with H. anomalus and H. deserticola. One limitation of this method is that multiple topologies have very similar likelihood values, indicating that the program cannot easily distinguish between different admixture scenarios, in this case.
We also used ADMIXTOOLS2, which builds an admixture graph that explains f-statistics between populations. Admixture scenarios seen using ADMIXTOOLS2 have both similarities and differences with the TreeMix graphs. Out-of-sample scores decrease with increasing admixture edges from one to two, and from two to three, but not from three to four, suggesting that three admixture edges explain much of the f2 incongruence (supplementary fig. S7, Supplementary Material online). Although bootstrapping is unable to distinguish between the top three graphs with three admixture nodes (P > 0.1), they each present a similar pattern that distinguishes them from the species tree (fig. 2C). The strongest signal is associated with the origin of two of the putative hybrid species (H. anomalus and H. deserticola), but from admixture with the ancestor of H. niveus canescens and an unsampled or extinct basal lineage. Helianthus petiolaris is placed at the base of the annuus clade in the ADMIXTOOLS2 graph, implying that it shares ancestry with members of the annuus clade, including H. paradoxus. In addition, a recent admixture is seen between H. niveus canescens and H. petiolaris fallax, similar to that observed in the TreeMix analysis.
Triplet Tests of Hybrid Ancestry
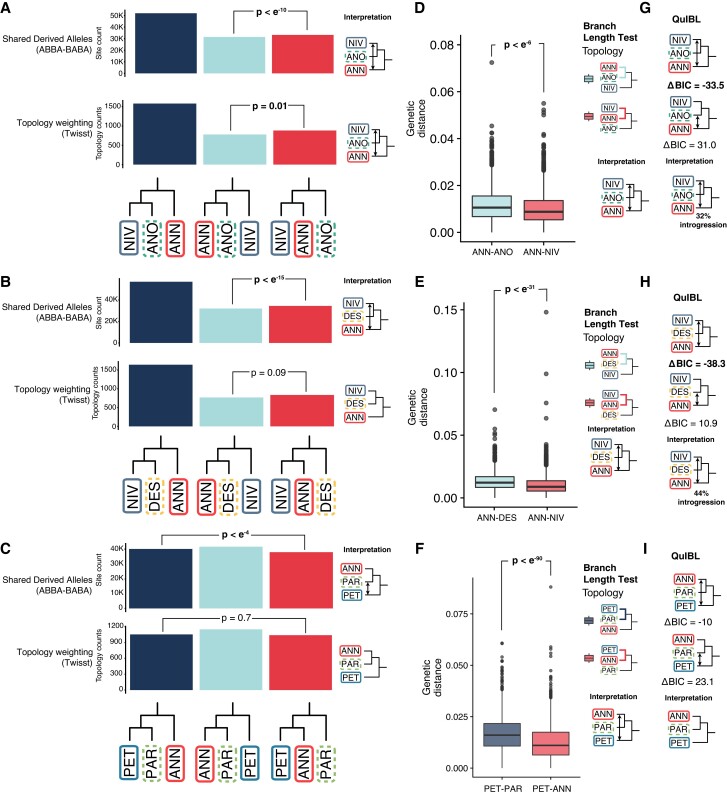
Although the admixture graphs (above) detected signals of hybridization associated with the origins of the three hybrid species, there were differences relative to previous hypotheses (Rieseberg 1991). Most notably, the origin of H. anomalus and H. deserticola appears to involve hybridization with the ancestor of H. niveus canescens rather than H. petiolaris fallax. Also, there was uncertainty as to whether the signal of admixture associated with the origin of H. paradoxus was independent from other members of the annuus clade. Therefore, we explicitly tested hypotheses of hybrid ancestry for H. anomalus, H. deserticola, and H. paradoxus against their putative parents. In the case of H. paradoxus, the testing is relative to H. annuus and H. petiolaris fallax, whereas for H. anomalus and H. deserticola, the putative parents are H. annuus and H. niveus canescens. For each test, we expect to see that the hybrid species is more closely related to both parental species, compared with the relationship between the parental species themselves, and that tests of introgression show that the dominant signal of introgression involves the hybrid species and the parental species, rather than between the parental species. We used four different approaches. Patterson's D compares the amounts of shared derived alleles in a trio (Green et al. 2010; Malinsky et al. 2021). In this case, we are using Twisst to compare the counts of different gene tree topologies (Martin and Van Belleghem 2017). The branch length test compares the branch lengths in alternate topology gene trees. Lastly, QuIBL models ILS and introgression based on branch lengths (Edelman et al. 2019; Suvorov et al. 2022). We note that the results of triplet tests should be interpreted with caution because of the long history of introgression between the putative parental clades, which can bias the results (Yatabe et al. 2007; Strasburg and Rieseberg 2008; Kane et al. 2009). For the topology-based tests (Patterson's D and Twisst), this means that the extent of introgression from the minor parent must be significantly greater than that between the parental lineages. For the distance-based tests (branch length and QuIBL), both the extent and the timing of introgression become critical, as the distance-based tests have little power for detecting ancient admixture.
For H. paradoxus, we consistently find that H. annuus is the closest species, but an inconsistent evidence of admixture with H. petiolaris fallax (fig. 3). Patterson's D finds significant H. paradoxus—H. petiolaris fallax admixture (fig. 3C). The results from Twisst are in the same direction, but are not significant (fig. 3C). In contrast, the branch length test supports greater H. annuus—H. petiolaris fallax introgression and QuIBL supports neither (fig. 3F and I). We find qualitatively similar results when using H. petiolaris petiolaris as a potential parental species. The branch length test is more sensitive to recent introgression, such as that ongoing between H. annuus and H. petiolaris (Yatabe et al. 2007), whereas D has greater power to detect ancient hybridization, perhaps accounting for the apparent discrepancy between the different tests. At the genomic window level, there is considerable variation in D with values ranging from −1 to 1 (supplementary fig. S8, Supplementary Material online).
Fig. 3.
Triplet tests of previous hypotheses of hybrid ancestry. (A–C) Counts of shared derived alleles and topological weighting for proposed hybrid species. P-values from block bootstrapping and chi-square tests for ABBA-BABA and Twisst, respectively. (D–F) Branch length test of introgression showing the genetic distance between taxa at gene trees with secondary topologies. (G–I) QuIBL delta BIC values and interpretation. Lower values indicate more support for introgression.
For H. anomalus and H. deserticola, we see a consistent pattern for all four tests; they are more closely related to H. niveus canescens and there is more admixture between the potential parental species, than between the hybrid species and H. annuus (fig. 3A, B, D, E, G, H). This could indicate that the hybrid species are ancient, and that the minor parent is at the base of the H. annuus clade (as suggested by TreeMix), and therefore, has a relatively small contribution from contemporary H. annuus or that the other parent of H. anomalus and H. deserticola is a now extinct basal lineage, as suggested in the ADMIXTOOLS2 graph.
Admixture Across the Sunflower Genus
To assess introgression across the rest of the genus, as well as to confirm our findings within the annual clade, we expanded our analysis of gene flow using a previously published sequence capture data set spanning the genus. With this data, we used Dsuite to calculate fbranch and D for all trios (supplementary fig. S9, Supplementary Material online, supplementary table S3, Supplementary Material online). We found evidence of introgression between perennial species, including Helianthus verticilliatus—H. giganteus, H. divaricatus—H. arizonensis, and H. atrorubens—Helianthus mollis. This data set also affirms the signal of introgression between H. petiolaris and both H. niveus and H. annuus. However, no introgression was found involving H. debilis. Surprisingly, we also find introgression-like signals between annual and perennial species. Helianthus maximilliani shares more derived alleles with annual species than other perennials (fig. 2E). Strangely, both H. annuus and H. argophyllus share fewer derived alleles with perennials than other annual species, which is consistent with widespread annual-perennial introgression excluding H. annuus/H. argophyllus, or introgression between the H. annuus/H. argophyllus clade and the outgroup Phoebanthus (fig. 2D).
Discussion
For three-fourths of a century, Helianthus sunflowers have served as a model for the study of hybridization and its evolutionary role (Heiser 1947; Rieseberg 1991; Todesco et al. 2020). Early studies documented hybrid swarms between different sunflower species (Heiser 1947; Heiser 1949a; Heiser 1951), demonstrating that chromosomal rearrangements contributed to hybrid sterility (Chandler et al. 1986; Lai et al. 2005), and speculated on possible ecotypes and species that might be the products of hybridization (Heiser 1949b; Heiser 1958; Rieseberg 1991). Later molecular phylogenetic work and early low-resolution genomic studies supported some of these hypotheses (Rieseberg, Beckstrom-Sternberg, et al. 1990; Yatabe et al. 2007), but not others (Rieseberg et al. 1988), and also suggested new hypotheses, including the putative origins of three homoploid hybrid species (Rieseberg 1991). Here, we have taken advantage of new high-resolution genomic data and computational methods to further explore the role of hybridization in sunflower evolution. We pay particular attention to the three homoploid hybrid species, but we also examine other well-studied cases of hybridization in the Helianthus (and potential new cases) with the goal of reconciling the new data and analyses with previous work. Lastly, we explore the effects of variation in recombination rate on tree topology and introgression.
Hybridization in Sunflowers
The whole-genome phylogeny for sunflowers is consistent with that found by previous multilocus studies (Stephens et al. 2015; Baute et al. 2016; Zhang et al. 2019). Leveraging this phylogeny, we employed several different methods to search for possible cases of admixture during the evolution of the genus. Remarkably, all species included in our study appear to be admixed, although the level of the admixture depends, in part, on the particular analysis performed (figs. 2 and 4). Overall, the strongest signal of hybridization occurs between two subspecies of H. petiolaris and H. annuus. Helianthus petiolaris is sympatric with H. annuus and contemporary hybridization is well-documented (Heiser 1947; Rieseberg et al. 1998). Previous studies have documented “rampant” introgression between the species (Yatabe et al. 2007) and have suggested that interspecific gene flow was common in the past as well (Strasburg and Rieseberg 2008).
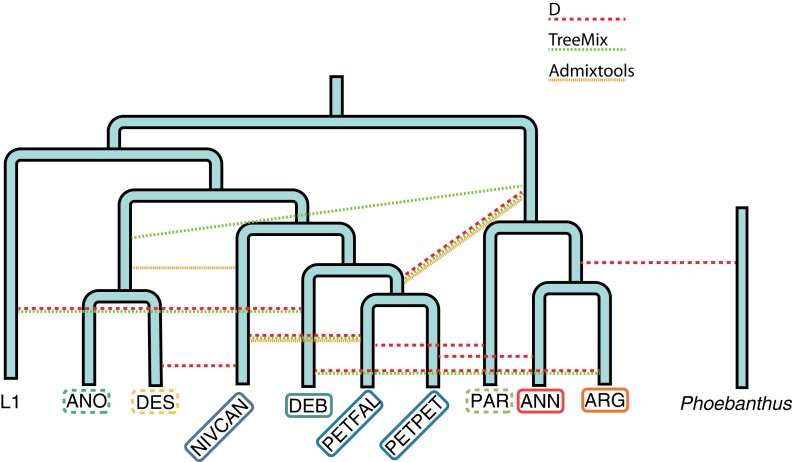
Fig. 4.
The proposed annual sunflower phylogeny with introgression events identified in the present study. L1 represents an unknown lineage seen through admixture analyses of H. debilis. Dotted lines indicate signatures of admixture seen in different analyses.
Another strong signal of admixture is found between H. petiolaris and H. niveus canescens, confirming a previous report (Zhang et al. 2019). Helianthus petiolaris intergrades with H. niveus canescens phenotypically, and Heiser et al. (1969) suggested that it might be due to hybridization. Although both types of phylogenetic analysis suggested that H. niveus canescens grouped with H. anomalus and H. deserticola rather than H. petiolaris, there were more gene trees that placed H. niveus canescens with H. petiolaris. Considering the weight of the evidence, we support that H. niveus canescens was involved with an admixture of the H. anomalus/deserticola ancestor, but was originally a member of the H. petiolaris clade. The beach sunflower, H. debilis, is sister to H. petiolaris in our tree, but fbranch shows that it is less similar to all other annual sunflower species (fig. 2A). This implies that it has ancestry from a lineage at the base of the annual sunflowers, as also suggested by TreeMix (fig. 4). Lastly, both the TreeMix and ADMIXTOOLS2 graphs suggest a shared hybrid origin for H. anomalus and H. deserticola, which we will discuss in more length below.
The analysis of cytoplasmic genomes supports an earlier work finding two clades of chloroplast haplotypes in annual sunflowers (Lee-Yaw et al. 2019). Previous phylogeographic studies identified both chloroplast clades in H. anomalus and H. deserticola (Schwarzbach and Rieseberg 2002; Gross et al. 2003), but only a single clade in H. paradoxus (Welch and Rieseberg 2002). In contrast, the present study found only a single cytoplasmic genome in the former two species, but both in the latter. This discrepancy appears to result from limited geographic sampling for H. anomalus and H. deserticola in the present study, as well as the inclusion of a recently discovered disjunct population of H. paradoxus from Mexico that was not analyzed previously. We find that H. petiolaris and H. debilis also have variations in the cytoplasmic clade, which was seen in several other species in a broader sampling of the annual sunflower species (Lee-Yaw et al. 2019). Simulations of chloroplast evolution by Lee-Yaw et al. (2019) suggested that cytoplasmic mismatch was mainly due to introgression rather than ILS.
We used an alternate data set generated by Stephens et al. (2015) to assess introgression across the entire Helianthus genus. We found evidence of hybridization between H. verticilliatus and H. giganteus. Helianthus verticilliatus was once thought to be a hybrid species but was later disproven by molecular data (Heiser et al. 1969; Ellis et al. 2006). Interestingly, H. verticilliatus was previously thought to be a subspecies of H. giganteus, and the two species overlap in range (Farwall 1916). Similarly, introgression between H. mollis and H. atrorubens is highly plausible considering they are known to hybridize (Beatley 1963). In contrast, the proposed introgression between H. divaricatus and H. arizonensis is difficult to explain because the species do not share any current range and there are no plausible proxy ancestry donors, but it is possible that their ranges overlapped in the past.
Surprisingly, there was identified introgression between the annual and perennial clades, despite strong post-pollination reproductive barriers between them. Most crosses between the annual and perennial sunflower species fail to set seed, and the few successful hybrids that have been reported typically have low pollen viability (Heiser et al. 1969). Crosses involving the hexaploid perennials, represent an exception to this rule (Atlagić and Terzić 2006), but we found the strongest signal of introgression with a diploid perennial Helianthus maximiliani, which is incompatible with members of the annual clade (Henn et al. 1998). Thus, we likely are seeing the outcome of an ancient hybridization, which occurred before reproductive isolation was complete. However, the most unexpected pattern is a signal of ancestral allele sharing between H. annuus/argophyllus and the outgroup, Pheobanthus (fig. 2E). These two lineages diverged 2.5–5.4 Ma, and are therefore, expected to be completely reproductively isolated (Owens and Rieseberg 2014; Mason 2018), again suggesting that the signal of admixture is from ancient hybridization with a now extinct species. Nonetheless, we recommend experimental crosses be undertaken between Pheobanthus and members of the annual clade to see if hybrids can be made. It is also possible that this signal represents a change in the evolutionary rate in H. annuus and H. argophyllus. This has been proposed to explain similar patterns of introgression signals in Papilio butterflies (Xiong et al. 2022). More broadly, our results suggest that admixture has been common in Helianthus throughout its evolutionary history, and that it has had a profound effect on phylogenomic relationships in the genus (fig. 4). Thus, Helianthus can be viewed as a syngameon, in which, gene flow connects the otherwise distinct species (Grant 1981). Our findings also address earlier concerns that hybridization in Helianthus might be a recent consequence of anthropogenic disturbance, and therefore, of little significance to its long-term evolution (Schemske 2000). Whether these ancient hybridization events triggered diversification, as suggested in other groups (e.g., Meier et al. 2017), remains unclear.
Ancestry of Putative Homoploid Hybrid Species
The TreeMix and ADMIXTOOLS2 graphs indicate that the three hybrid species are admixed, thereby fulfilling the second criterion for homoploid hybrid speciation. However, there are differences relative to the original scenario put forward by Rieseberg (1991). In particular, the hybridization events associated with the origins of the hybrid species appear to have occurred further back in time than previously believed. For H. paradoxus, a potential hybrid speciation event is older than previously anticipated (Welch and Rieseberg 2002), because individual gene trees place H. paradoxus most often at the base of the annuus clade, therefore, at least older than 1 ma. For H. anomalus and H. deserticola, our phylogenomic results suggest they are sister species, and share an admixture close in time to their origin. This would also place a potential hybrid speciation event further in the past than previous works suggested (Schwarzbach and Rieseberg 2002; Gross et al. 2003; see supplementary discussion for reconciliation with previous microsatellite dating of the hybrid species origin).
Another important difference is that H. anomalus and H. deserticola are sister species and their major parent appears to be H. niveus canescens rather than H. petiolaris fallax as originally proposed (Rieseberg 1991). Although this was unexpected, H. niveus canescens intergrades phenotypically with H. petiolaris fallax and the two taxa share chloroplast and nuclear ribosomal DNA haplotypes, which were the markers employed to hypothesize a hybrid origin in the first place (Heiser et al. 1969; Beckstrom-Sternberg et al. 1991). As a consequence, some taxonomic treatments have considered H. niveus canescens to be a variety of H. petiolaris (Kearney and Peebles 1942; Schilling 2020). The present study indicates that H. niveus canescens is genetically distinct from H. petiolaris fallax, although there is considerable admixture between them, which might account for their phenotypic resemblance (Zhang et al. 2019). Fortunately, recognition that the ancestor of H. niveus canescens was a parent of H. anomalus and H. deserticola has little impact on the interpretation of previous work. The population of H. petiolaris fallax that was employed for most of the experimental studies of hybrid speciation came from the zone of intergradation with H. niveus canescens (Lexer et al. 2003; Rieseberg et al. 2003; Gross et al. 2004; Ludwig et al. 2004). The taxa are indistinguishable morphologically and ecologically in this area, and may recapitulate the ancestor of H. niveus canescens.
Despite the power of the phylogenomic analyses, some uncertainties remain (see supplementary discussion about the challenges for interpreting admixture signals in sunflowers). For H. paradoxus, it is unclear whether the signal of admixture associated with its origin is independent from other members of the annuus clade. The triplet tests of introgression provide indirect support for independence. For D, there is a significant enrichment for H. petiolaris—H. paradoxus sharing as predicted under the hybrid origin scenario; gene tree topology counts show the same pattern, but are not significant. However, the branch length test, which has greater power to detect recent introgression, indicates more introgression between H. annuus—H. petiolaris. These results may be reconciled by the long history of introgression between the annuus and petiolaris clades (Strasburg and Rieseberg 2008), with the branch length test successfully detecting recent introgression between H. annuus and H. petiolaris, and D offering evidence of more ancient hybridization associated with the evolution of H. paradox. Likewise. H. paradoxus shares more derived alleles with H. anomalus and H. deserticola than do other members of the annuus clade, offering further indirect support that H. paradoxus is the product of an independent hybridization event (supplementary table S2, Supplementary Material online).
There is also uncertainty regarding the parentage of H. anomalus and H. deserticola. Although the role of H. niveus canescens as one parent is strongly supported, the other hybrid parent is less clear. Is it the H. annuus lineage? The most likely TreeMix graph suggests that the ancestor of the H. annuus clade is the other parent, but this pattern is not strongly supported by admixtools, the triplet testing, or D. The hybrid species share fewer derived alleles with H. annuus and H. argophyllus when compared with H. debilis, H. niveus canescens, or H. petiolaris (supplementary table S2, Supplementary Material online), but they share more derived alleles with H. paradoxus when compared with H. debilis. This may be indicative of more recent introgression between the H. annuus lineage and members of the petiolaris clade sampled in this study, all of which overlap in geographic distribution and hybridize with H. annuus. Alternatively, H. anomalus and H. deserticola are a combination of H. niveus canescens and an unsampled or extinct basal lineage, as is suggested in the admixtools tree. Future comparisons with an allopatric control from the H. petiolaris clade (e.g., H. niveus niveus) will be needed to distinguish between these hypotheses.
When examined together, the tests for hybrid ancestry of the putative homoploid species show mixed results between different test methods (table 1). In particular, targeted trio-based tests generally showed negative results, whereas genus-wide tests show evidence of some admixture. We suspect that the negative signals in trio-based tests are a result of confounding with other gene flow, as well as their low power for detecting ancient hybridization because the exact parents are not available for testing. That being said, the positive signals in genus-wide tests are due in part to the looser hypothesis of admixture versus admixture from specific sources.
Table 1.
Results for Tests for Hybrid Ancestry of Proposed Hybrid Species.
| Approach | Methodology | Trio-specified? | Anomalus hybrid ancestry | Deserticola hybrid ancestry | Paradoxus hybrid ancestry |
|---|---|---|---|---|---|
| Treemix | Allele frequency covariance in a tree | No | Positive | Positive | Positive |
| ADMIXTOOLS2 | f2 tests in a tree | No | Positive | Positive | Positive |
| fbranch | f4 tests for specific tree-based sets | No | Positive | Positive | Positive |
| Patterson's D | Shared derived alleles for trios | Yes | Negative | Negative | Positive |
| Twisst | Gene tree topology counts | Yes | Negative | Negative | Negative |
| QuIBL | Model based branch lengths for secondary topologies | Yes | Negative | Negative | Negative |
Note.—Methodology describes the statistic being considered in the test.
Perhaps, one surprising aspect of our results is that the amount of admixture in “hybrid species” is not exceptional compared with the non-hybrid species. This highlights that the amount of hybrid ancestry is not critical for hybrid speciation, only that admixture was involved in reproductive isolation (Rieseberg 1997; Schumer et al. 2014), which we do not directly address in this study. Nonetheless, our results have implications for the interpretation of previous experimental studies that have attempted to make this link (e.g., Rieseberg et al. 1995, 1996, 2003; Lexer et al. 2003; Gross et al. 2004; Ludwig et al. 2004). For example, evidence that the homoploid hybrid species are more ancient than previously believed makes it more difficult to determine which ecological, phenotypic, and genomic changes are associated with hybridization and which changes arose as a consequence of divergence after hybrid speciation (Ungerer et al. 2006). We suspect that the clusters of parental markers found to be linked to QTLs in segregating hybrids, and which were also found in the hybrid species genomes (Rieseberg et al. 2003), likely correspond to inversions. Such inversions are frequent in Helianthus (Ostevik et al. 2020) and often appear to be ancient, pre-dating the species they are found in (Todesco et al. 2020). Future studies attempting to connect reproductive barriers with admixture should be conducted within a phylogenetic framework to better assess when key differences arose.
Previous works attempted to dissect parental ancestry across the putative hybrid genomes, finding strings of linked parental markers suggestive of the rapid establishment of the hybrid genomes (Ungerer et al. 1998; Buerkle and Rieseberg 2008). However, ILS, multiple ancient admixture events, structural variation, and the auto-correlational effects of recombination rate variation may all affect ancestry assignments across the genome. Future attempts at identifying and interpreting ancestry tracks will have to contend with this complexity.
Are there other examples of homoploid hybrid speciation in Helianthus? Probably—although the relevant data come from earlier studies rather than the present analysis (see supplementary discussion about outcomes of hybridization in the annual sunflower clade). Independently derived dune ecotypes of the prairie sunflower are isolated from non-dune populations by multiple reproductive barriers (Heiser 1958; Ostevik et al. 2016). Recent population genomic studies indicate that most of the traits underlying reproductive isolation map to ancient chromosomal inversions appear to have originated via introgression from a basal lineage in the annual clade (or even earlier) (Huang et al. 2020; Todesco et al. 2020). In an example involving more recent hybridization, a 77-day difference in flowering between the coastal and barrier island ecotypes of H. argophyllus was found to result from a 30 Mb introgression from H. annuus containing a functional copy of HaFT1 (Todesco et al. 2020). The dune ecotypes arguably represent new species, and one of them is described as H. neglectus in most taxonomic treatments (Heiser et al. 1969; Schilling 2020). Ecotypic differentiation in H. argophyllus represents an earlier stage of the speciation process, but it illustrates how the introgression of a single gene can cause significant reproductive isolation. It is noteworthy that all three potential new cases of homoploid hybrid speciation involve the colonization of ecologically divergent habitats that provide ecogeographic isolation similar to that seen in the ancient hybrids.
Recombination Rate and Introgression
Local recombination rate is a critical population genetics parameter that can affect ILS, gene flow, and genetic diversity (Ortíz-Barrientos et al. 2002; Nachman and Payseur 2012; Haenel et al. 2018). For example, introgressed loci are more likely to persist in regions of high recombination because they more quickly unlink from deleterious neighboring loci, such as genetic incompatibilities (Brandvain et al. 2014; Schumer et al. 2018). In our data set, higher recombination regions are less likely to support the inferred species topology. This is likely a combination of reduced introgression in low recombination regions, as well as lower Ne, which reduces ILS (Pease and Hahn 2013; Li et al. 2019; Martin et al. 2019). The effect is strongest for the node separating H. annuus samples, where the recombination rate estimates are most accurate. Sunflowers have highly labile chromosome structure and there are numerous large-scale rearrangements between species in this tree, but this pattern is retained even between our outgroup perennial sunflower species, suggesting that recombination rate is relatively conserved even if genome structure changes (Ostevik et al. 2020). At several nodes, we find that the relationship is reversed, which may be because at those nodes we have the introgressed topology, rather than the true species topology. Exploring the relationship between recombination rate and tree topology is useful for identifying an accurate species topology.
Conclusions
Despite vast improvements in the quantity of genetic data available to current researchers, disentangling introgression patterns are often challenging (Hibbins and Hahn 2022). When multiple introgression events have occurred, this can lead to false positives or false negatives, which is especially true when the true donors are not sampled or, in some cases, no longer extant. A broad sampling of species can help to reduce these issues and a broad sampling of populations within species can identify when introgression is geographically localized. Thus, when evaluating the plausibility of introgression signals detected using genomic data, it is important to consider factors such as reproductive compatibility and the likelihood of geographic overlap over the course of evolutionary divergence. That being said, the reconstruction of historical ranges is an inexact science, and information on extinct congeners is completely unknown for most wild genera.
A phylogenomic analysis below the genus level often finds evidence of introgression, especially in plants (Dagilis et al. 2021). This suggests that a significant proportion of plant species have some admixed ancestry. Despite this, homoploid hybrid species are rare because most studies fail to link admixture with the evolution of reproductive barriers (Schumer et al. 2014). Although attempts have been made to identify admixture-derived reproductive barriers through purely bioinformatic analyses (e.g., Sun et al. 2020; Wang et al. 2022), links between phenotypes, reproductive isolation, and introgressed ancestry are required to confirm the homoploid hybrid speciation. A notable exception to this is the work by Wang et al. (2021), which identified an admixed species in Ostryopsis, quantified reproductive barriers, and transgenically tested candidate genes acquired through admixture. Widespread introgression across the sunflower species raises the possibility that admixture played a role in other speciation events. In particular, chromosome rearrangements are both common and known to cause reproductive isolation in sunflowers (Lai et al. 2005; Ostevik et al. 2020), so future works should leverage chromosome-resolved genomes and QTL mapping to identify how or whether the introgressed rearrangements cause speciation.
Materials and Methods
Data Generation
We generated whole-genome resequencing data for two H. paradoxus samples. DNA was extracted from individual seedlings or dried leaf tissues using a modified CTAB extraction protocol (Murray and Thompson 1980; Zeng et al. 2002), and an indexed Illumina sequencing library was created, which included a duplex specific nuclease repeat depletion step (Todesco et al. 2020). This data were added to previously published whole-genome resequencing data to create a data set of nine species or subspecies of annual sunflowers (Hübner et al. 2019; Todesco et al. 2020; Owens et al. 2021). For each, we randomly selected two samples to be used in the analysis. We only included two per species because we were interested in understanding between-species relationships, rather than within-species diversity. Additionally, using fewer samples substantially reduced the computational bottleneck in both variant calling and subsequent phylogenetic analyses. To act as outgroups, we also included one sample each for four diploid perennial sunflower species (See supplementary table S1, Supplementary Material online).
For each sample, Illumina sequence data were aligned to the Helianthus annuus HA412-HOv2 genome using NextGenMap (v0.5.3) (Sedlazeck et al. 2013), PCR duplicates were marked with samtools v0.1.19 (Danecek et al. 2021), and variants were called using GATK (v4.1.4.1) (McKenna et al. 2010). We specifically included invariant sites in GATK genotypeGVCFs to facilitate downstream analyses. The sunflower genome contains a large proportion of transposable element repeats that hamper read alignment. To avoid those regions, we used GenMap (-K 50 -E 2) (v1.3) to calculate k-mer uniqueness or mappability for all positions in the genome (Pockrandt et al. 2020). We then removed all contiguous regions with mappability <1 that are >100 bp. This removed 2.43 Gbp out of 3.23 Gbp, but highly enriched retained regions for single-copy sequences. This set also excluded the cytoplasmic genomes.
For our data set, we filtered the variant and invariant sites separately. For the variant sites, we removed all indels and visualized the distribution of quality metrics of SNPs using bcftools (v1.14) to extract values and ggplot (v3.3.5) in R (v4.1.2) to plot (Wickham 2011; R core team 2021). We visually selected thresholds to remove outliers based on the distribution (bcftools view -e “INFO/DP > 1000’ -e “INFO/FS > 50’ -e “INFO/QD < 2’ -e “INFO/SOR > 4’ -e “INFO/MQ < 30') (supplementary fig. S1, Supplementary Material online). Both the variant and invariant sites were combined and then filtered to require ≥4 reads per genotype and ≥80% of samples genotyped using VCFtools (v0.1.16) (Danecek et al. 2011).
To explore relatedness in the cytoplasmic genomes, we aligned all samples sequence data to a reduced reference containing only the chloroplast and mitochondrial genomes, using BWA mem2 (Md et al. 2019). We then used bcftools to call bases across the entire reference (including invariant sites) using reads with a mapping quality >20 (Danecek et al. 2021). Resulting base calls were converted into fasta format using a custom perl script for the chloroplast and mitochondrial genomes separately. To retain alignment when outputting all sequences, necessary gaps were inserted at the ends of the alleles to ensure that all indel alleles had equal length for any position.
Code used to conduct analyses and make figures is deposited at https://github.com/owensgl/helianthus_hybrid_species_2021.
Gene and Species Tree Estimation
We took two approaches to explore genome-wide gene trees. In the first, we equalized information across windows by requiring each genomic window to have 10,000 called bp, leading to variable physical size. In the second approach, we equalized physical size but not information by dividing the entire genome into non-overlapping 10,000 bp windows and retained windows with ≥2,000 called bases. In both cases, the heterozygous sites were retained and coded using IUPAC coding.
For each of our genomic windows, we calculated a maximum likelihood phylogeny using IQ-Tree (v2.0.6) (Minh, Hahn, et al. 2020) with H. giganteus, a perennial species, as the outgroup. Additionally, a single concatenated species tree using all retained bases was created using maximum likelihood and model selection in IQ-Tree2. We used a coalescent approach in ASTRAL (v5.7.3) to estimate the species tree using both sets of gene trees separately (Zhang et al. 2018).
We explored the diversity of gene tree topologies by using the tool findCommonTrees.py (Edelman et al. 2019) to identify and count the occurrence of each topology. We found that every tree was unique; so we subsampled trees down to a single sample per species and a single perennial outgroup species to reduce the possible tree space. We repeated the counts of trees and visualized the most common gene trees.
Using the concatenated phylogeny as a backbone, we measured gene concordance using IQ-Tree2 (Minh, Schmidt, et al. 2020). This measures the percent of gene trees that support the topology and the two possible alternate topologies for each branch quartet. We matched the H. annuus recombination rate for each genomic window and calculated the percent support for each topology for each recombination quintile. To determine if the recombination rate affected support for the species topology, we used a binomial regression with the formula:
For each cytoplasmic genome, we created a single maximum likelihood phylogeny using IQ-Tree2, including model selection and 1,000 ultra-fast bootstraps (Minh, Schmidt, et al. 2020).
Broad Tests of Introgression
We used several methods that integrate signals of introgression between all samples. Using Dsuite, we calculated the fbranch statistic for the species phylogeny (Malinsky et al. 2021). This statistic incorporates all valid f4 ratios (similar to Patterson's D) as well as the phylogeny to identify branches that share allelic imbalance. This can help to identify introgression events that predate speciation times, and therefore, have a signal in multiple species.
We also used TreeMix to identify introgression events (Pickrell and Pritchard 2012). This program builds a graph model of population splits based on the covariance of allele frequencies. It allows for migration edges to be added to the graph that explain the remaining covariance not accounted for by the initial graph topology. Since TreeMix relies on covariance, we pruned our data set for SNPs in LD by only including sites with <0.8 r2 in 100,000 bp using ‘bcftools prune’ (Danecek et al. 2021), used a block size of 1,000 SNPs (-k 1000) and did not include sample size correction. We ran TreeMix with 0 to 10 migration edges, with 99 replicates using different starting seeds, and selected the optimal number of migration edges using the Evanno method in OptM (Fitak 2021). We visualized the replicates with the likelihood values within 10 of the highest likelihood for each number of migration edges (i.e., the models that were not the best, but were close).
Lastly, we built an admixture graph for the phylogeny using ADMIXTOOLS 2 (Patterson et al. 2012). We first converted our VCF file to eigenvector format using a custom Perl script, and then calculated all f2 statistics using the extract_f2 and f2_from_precomp functions. We used a 5 Mbp window for block bootstrapping. Admixture graphs were found using the find_graphs command with stop_gen = 200, stop_gen2 = 20, and perennials set as the consistent outgroup. This method finds graphs that fit the observed f-statistics in the data. The graph search can get stuck at local optima, so we repeated each search 100 times for each number of admixture events, from one to four, and retained the best-fitting graph in each iteration. From this, we picked the top three scoring graphs for each number of admixture events. We then used bootstrap-resampling of the out-of-sample scores for each graph using the function qpgraph_resample_multi to ask whether adding additional nodes was significantly better supported, by comparing the best graph with n admixture events with the best graph with n + 1 admixture events, as well as comparing graphs.
Trio-based Tests of Introgression
Given the high levels of discordant gene trees, and conflicting signals from broad tests of introgression, we explored trio-based tests of introgression using Dsuite to calculate Patterson's D (Green et al. 2010; Malinsky et al. 2021). This test looks at quartets with the relationship (((A, B), C), O) and asks if there is a greater derived allele sharing between A and C or B and C. Under ILS, both should share equal counts of derived alleles, but introgression will lead to imbalance. Significance was tested using a block bootstrap algorithm. We used the perennial species as an outgroup and tested all trio sets consistent with the species phylogeny.
For the putative hybrid species and their parents, we used three additional approaches: the branch length test (BLT), QuIBL, and Twisst. For these tests, we used genomic windows with equal physical size, as described above. The BLT test compares the tip-to-tip distance between inferred sister clades in the minor topologies using a Mann–Whitney test (Suvorov et al. 2022). Under ILS, both minor topologies should have equal distance, but if introgression is occurring, then the topology it produces should have reduced distance due to its more recent coalescence. These tests were done in R using TreeTools (Smith 2019), using the subsampled trees with only a single sample per species. QuIBL examines trios in gene trees and asks whether the branch length distribution fits better to a model that includes both ILS and later introgression, or purely ILS (Edelman et al. 2019). As before, we used our perennial samples as outgroups. We considered models with Bayesian information criterion score difference of ≥30 to be the evidence of introgression. We used Twisst to calculate topological weighting for each possible tree using equal size gene trees (Martin and Van Belleghem 2017). This takes into account phylogenetic position variation among individuals within a species and calculates a weight for how common a particular topology is at each gene tree. We normalized weights such that each genomic window had a total weight of one and counted the total weight for each topology. Similar to the Discordant Count Test from Suvorov et al. (2022), we compared the counts of the two minor topologies using a chi-square test, under a null hypothesis that ILS should produce equal counts of both minor topologies. We also calculated D across the genome using Dsuite, with a window size of 50 SNPs and a step size of 25 for each possible candidate trio.
Quantifying Introgression Across the Genus
Given the amount of gene flow identified within annual sunflowers, we expanded our analysis of gene flow by using a previously published sequence capture set encompassing a majority of diploid species in the genus (Stephens et al. 2015). This data set included 170 aligned sequences totaling 106,862 bp and 11,407 parsimony informative sites. Since these markers were anonymous and genome positions are required for block bootstrapping, we aligned them to the H. annuus HA412-HOv2 reference genome using BLAST and selected the location hit with the highest e-value. Sequence capture gene alignments include numerous indels, so we were not able to combine this data set with our previous WGS data set. As before, we used Dsuite to calculate D, f4, and fbranch. For the species topology, we used the MP-EST tree from Stephens et al. (2015), with polytomies randomly resolved.
Supplementary Material
Acknowledgments
The authors thank Sarah Yakimowski and Robert Sivinski for providing tissues used for DNA extractions. This work was funded by a Banting Fellowship to GLO, as well as Discovery Grants from the Natural Sciences and Engineering Research Council of Canada to LHR and GLO. The authors thank two anonymous reviewers and the editor for helpful suggestions.
Contributor Information
Gregory L Owens, Department of Biology, University of Victoria, Victoria, BC, Canada.
Kaichi Huang, Department of Botany and Beaty Biodiversity Center, University of British Columbia, Vancouver, BC, Canada.
Marco Todesco, Department of Botany and Beaty Biodiversity Center, University of British Columbia, Vancouver, BC, Canada.
Loren H Rieseberg, Department of Botany and Beaty Biodiversity Center, University of British Columbia, Vancouver, BC, Canada.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online.
References
- Anderson E. 1949. Introgressive hybridization. New York: J. Wiley. [Google Scholar]
- Anderson E, Stebbins Jr GL. 1954. Hybridization as an evolutionary stimulus. Evolution. 378:88. [Google Scholar]
- Atlagić J, Terzić S. 2006. Cytogenetic study of hexaploid species Helianthus tuberosus and its F1 and BC1F1 hybrids with cultivated sunflower, H. annuus. Genetika. 38:203–213. [Google Scholar]
- Baute GJ, Owens GL, Bock DG, Rieseberg LH. 2016. Genome-wide GBS data provide a high-resolution view of wild Helianthus diversity, genetic structure and interspecies gene flow. Am J Bot. 103:2170–2177. [DOI] [PubMed] [Google Scholar]
- Beatley JC. 1963. The sunflowers (genus Helianthus) in Tennessee. J Tennessee Acad Sci. 38:135–154. [Google Scholar]
- Beckstrom-Sternberg S, Rieseberg LH, Doan K. 1991. Gene lineage analysis of populations of Helianthus niveus and H. petiolaris. Plant Syst Evol. 175:125–138. [Google Scholar]
- Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL. 2014. Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS genet. 10:e1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerkle CA, Rieseberg LH. 2008. The rate of genome stabilization in homoploid hybrid species. Evolution. 62:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfee E, Gates D, Lorant A, Perkins MT, Coop G, Ross-Ibarra J. 2021. Selective sorting of ancestral introgression in maize and teosinte along an elevational cline. PLoS genet. 17:e1009810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JM, Jan CC, Beard BH. 1986. Chromosomal differentiation among the annual Helianthus Species. Syst Bot. 11:354–371. [Google Scholar]
- Dagilis AJ, Peede D, Coughlan JM, Jofre GI, D’Agostino ER, Mavengere H, Tate AD, Matute DR. 2021. 15 Years of introgression studies: quantifying gene flow across eukaryotes. bioRxiv. [DOI] [PMC free article] [PubMed]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics. 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience. 10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. 1859. On the origin of species. London: John Murray. [Google Scholar]
- Edelman NB, Frandsen PB, Miyagi M, Clavijo B, Davey J, Dikow RB, García-Accinelli G, Van Belleghem SM, Patterson N, Neafsey DE, et al. 2019. Genomic architecture and introgression shape a butterfly radiation. Science. 366:594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JR, Pashley CH, Burke JM, McCauley DE. 2006. High genetic diversity in a rare and endangered sunflower as compared to a common congener. Mol Ecol. 15:2345–2355. [DOI] [PubMed] [Google Scholar]
- Farwall OA. 1916. Contribution to the botany of Michigan. No. 14. Rep Mich Acad Sci. 17:180. [Google Scholar]
- Figueiró HV, Li G, Trindade FJ, Assis J, Pais F, Fernandes G, Santos SH, Hughes GM, Komissarov A, Antunes A, et al. 2017. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci Adv. 3:e1700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitak RR. 2021. Optm: estimating the optimal number of migration edges on population trees using treemix. Biol Methods Protoc. 6:bpab017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, et al. 2015. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science. 347:1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. 1958. The regulation of recombination in plants. Cold Spring Harb Symp Quant. 23:337–363. [DOI] [PubMed] [Google Scholar]
- Grant V. 1981. Plant speciation. New York Chichester, West Sussex: Columbia University Press. [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the neandertal genome. Science. 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Kane NC, Lexer C, Ludwig F, Rosenthal DM, Donovan LA, Rieseberg LH. 2004. Reconstructing the origin(s) of Helianthus deserticola: survivorship and selection on the desert floor. Am Nat. 164:145–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Rieseberg LH. 2005. The ecological genetics of homoploid hybrid speciation. J Hered. 96:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BL, Schwarzbach AE, Rieseberg LH. 2003. Origin(s) of the diploid hybrid species Helianthus deserticola (Asteraceae). Am J Bot. 90:1708–1719. [DOI] [PubMed] [Google Scholar]
- Haenel Q, Laurentino TG, Roesti M, Berner D. 2018. Meta-analysis of chromosome-scale crossover rate variation in eukaryotes and its significance to evolutionary genomics. Mol Ecol. 27:2477–2497. [DOI] [PubMed] [Google Scholar]
- Heiser CB. 1947. Hybridization between the sunflower species Helianthus annuus and H. petiolaris. Evolution. 1:249–262. [Google Scholar]
- Heiser CB. 1949a. Natural hybridization with particular reference to introgression. Bot Rev. 15:645–687. [Google Scholar]
- Heiser CB. 1949b. Study in the evolution of the sunflower species Helianthus annuus and H. bolanderi. UC Pub Bot. 12:157–196. [Google Scholar]
- Heiser CB. 1951. Hybridization in the annual sunflowers: helianthus annuus x H. debilis var. cucumerifolius. Evolution. 5:42–51. [Google Scholar]
- Heiser CB. 1958. Three new annual sunflowers from the southwestern United States. Rhodora. 60:275–279. [Google Scholar]
- Heiser CB, Smith DM, Clevenger SB, Martin WC. 1969. The North American sunflowers (Helianthus). Mem Torrey Bot Club 22:1–218. [Google Scholar]
- Henn HJ, Wingender R, Schnabl H. 1998. Regeneration of fertile interspecific hybrids from protoplast fusions between Helianthus annuus L. And wild Helianthus species. Plant Cell Rep. 18:220–224. [DOI] [PubMed] [Google Scholar]
- Hibbins MS, Hahn MW. 2022. Phylogenomic approaches to detecting and characterizing introgression. Genetics. 220:iyab173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Andrew RL, Owens GL, Ostevik KL, Rieseberg LH. 2020. Multiple chromosomal inversions contribute to adaptive divergence of a dune sunflower ecotype. Mol Ecol. 29:2535–2549. [DOI] [PubMed] [Google Scholar]
- Hübner S, Bercovich N, Todesco M, Mandel JR, Odenheimer J, Ziegler JE, Lee JS, Baute GJ, Owens GL, Grassa CJ, et al. 2019. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants. 5:54–62. [DOI] [PubMed] [Google Scholar]
- Jones MR, Mills LS, Alves PC, Callahan CM, Alves JM, Lafferty DJ, Jiggins FM, Jensen JD, Melo-Ferreira J, Good JM. 2018. Adaptive introgression underlies polymorphic seasonal camouflage in snowshoe hares. Science. 360:1355–1358. [DOI] [PubMed] [Google Scholar]
- Kane NC, King M, Barker MS, Raduski A, Karrenberg S, Yatabe Y, Knapp SJ, Rieseberg LH. 2009. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution. 63:2061–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney TH, Peebles RH. 1942. Flowering plants and ferns of Arizona. Vol. 423. Washington (DC): U.S.D.A. Misc. Publ. [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH. 2005. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 171:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Yaw JA, Grassa CJ, Joly S, Andrew RL, Rieseberg LH. 2019. An evaluation of alternative explanations for widespread cytonuclear discordance in annual sunflowers (Helianthus). New Phyt. 221:515–526. [DOI] [PubMed] [Google Scholar]
- Lexer C, Welch M, Raymond O, Rieseberg LH. 2003. The origins of ecological divergence in Helianthus paradoxus (Asteraceae): selection on transgressive characters in a novel hybrid habitat. Evolution. 57:1989–2000. [DOI] [PubMed] [Google Scholar]
- Li G, Figueiró HV, Eizirik E, Murphy WJ. 2019. Recombination-aware phylogenomics reveals the structured genomic landscape of hybridizing cat species. Mol Biol Evol. 36:2111–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig F, Rosenthal D, Johnston JA, Kane N, Gross BL, Lexer C, Dudley SA, Rieseberg LH, Donovan LA. 2004. Selection on leaf ecophysiological traits in a hybrid Helianthus species and early generation hybrids in a desert dune habitat. Evolution. 58:2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinsky M, Matschiner M, Svardal H. 2021. Dsuite-fast D-statistics and related admixture evidence from VCF files. Mol Ecol Resour. 21:584–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Besansky N, Hahn MW. 2016. How reticulated are species?. BioEssays. 38:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Salazar C, Jiggins CD. 2019. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 17:e2006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Van Belleghem SM. 2017. Exploring evolutionary relationships across the genome using topology weighting. Genetics. 206:429–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason CM. 2018. How old are sunflowers? A molecular clock analysis of key divergences in the origin and diversification of Helianthus (Asteraceae). Int J Plant Sci. 179:182–191. [Google Scholar]
- Mayr E. 1963. Animal species and evolution. Oxford: Harvard University Press. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Md V, Misra S, Li H, Aluru S. 2019. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. IEEE Parallel and Distributed Processing Symposium (IPDPS)
- Meier J, Marques D, Mwaiko S, Wagner CE, Excoffier L, Seehausen O. 2017. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat Commun. 8:14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo MC, Salazar C, Jiggins CD, Linares M. 2009. Assortative mating preferences among hybrids offers a route to hybrid speciation. Evolution. 63:1660–1665. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Hahn MW, Lanfear R. 2020. New methods to calculate concordance factors for phylogenomic datasets. Mol Biol Evol. 37:2727–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37:1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondon A, Owens GL, Poverene M, Cantamutto M, Rieseberg LH. 2018. Gene flow in argentinian sunflowers as revealed by genotyping by sequencing data. Evol Appl. 11:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody ML, Rieseberg LH. 2012. Sorting through the chaff, nDNA gene trees for phylogenetic inference and hybrid identification of annual sunflowers (Helianthus sect. Helianthus). Mol Phylogenet Evol. 64:145–155. [DOI] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8:4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Payseur BA. 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos Trans R Soc Lond, B, Biol Sci. 367:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz-Barrientos D, Reiland J, Hey J, Noor MA. 2002. Recombination and the divergence of hybridizing species. In: Etges WJ, Noor MAF, editors. Genetics of Mate choice: from sexual selection to sexual isolation. Boston: Kluwer Academic Publishers. p. 167–178. [PubMed] [Google Scholar]
- Ostevik KL, Andrew RL, Otto SP, Rieseberg LH. 2016. Multiple reproductive barriers separate recently diverged sunflower ecotypes. Evolution. 70:2322–2335. [DOI] [PubMed] [Google Scholar]
- Ostevik KL, Samuk K, Rieseberg LH. 2020. Ancestral reconstruction of karyotypes reveals an exceptional rate of nonrandom chromosomal evolution in sunflower. Genetics. 214:1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens GL, Baute GJ, Rieseberg LH. 2016. Revisiting a classic case of introgression: hybridization and gene flow in californian sunflowers. Mol Ecol. 25:2630–2643. [DOI] [PubMed] [Google Scholar]
- Owens GL, Rieseberg LH. 2014. Hybrid incompatibility is acquired faster in annual than in perennial species of sunflower and tarweed. Evolution. 68:893–900. [DOI] [PubMed] [Google Scholar]
- Owens GL, Todesco M, Bercovich N, Légaré JS, Mitchell N, Whitney KD, Rieseberg LH. 2021. Standing variation rather than recent adaptive introgression probably underlies differentiation of the texanus subspecies of Helianthus annuus. Mol Ecol. 30:6229–6245. [DOI] [PubMed] [Google Scholar]
- Oziolor EM, Reid NM, Yair S, Lee KM, Guberman VerPloeg S, Bruns PC, Shaw JR, Whitehead A, Matson CW. 2019. Adaptive introgression enables evolutionary rescue from extreme environmental pollution. Science. 364:455–457. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. 2012. Ancient admixture in human history. Genetics. 192:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. 2013. More accurate phylogenies inferred from low-recombination regions in the presence of incomplete lineage sorting. Evolution. 67:2376–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J, Pritchard J. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pockrandt C, Alzamel M, Iliopoulos CS, Reinert K. 2020. Genmap: ultra-fast computation of genome mappability. Bioinform. 36:3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2021. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rieseberg LH. 1991. Homoploid reticulate evolution in Helianthus: evidence from ribosomal genes. Am J Bot. 78:1218–1237. [Google Scholar]
- Rieseberg LH. 1997. Hybrid origins of plant species. Ann Rev Ecol Syst. 28:359–389. [Google Scholar]
- Rieseberg LH. 2000. Crossing relationships among ancient and experimental sunflower hybrid lineages. Evolution. 54:859–865. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. 2006. Hybrid speciation in wild sunflowers. Ann Mo Bot Gard. 93:34–48. [Google Scholar]
- Rieseberg LH, Baird S, Desrochers A. 1998. Patterns of mating in wild sunflower hybrid zones. Evolution. 52:713–726. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Beckstrom-Sternberg S, Doan K. 1990. Helianthus annuus ssp. texanus has chloroplast DNA and nuclear ribosomal RNA genes of Helianthus debilis ssp. cucumerifolius. PNAS. 87:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Beckstrom-Sternberg S, Liston A, Arias D. 1991. Phylogenetic and systematic inferences from chloroplast DNA and isozyme variation in Helianthus sect. Helianthus. Syst Bot. 16:50–76. [Google Scholar]
- Rieseberg LH, Carter R, Zona S. 1990. Molecular tests of the hypothesized hybrid origin of two diploid Helianthus species (Asteraceae). Evolution. 44:1498–1511. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C. 2003. Major ecological transitions in annual sunflowers facilitated by hybridization. Science. 301:1211–1216. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Sinervo B, Linder CR, Ungerer M, Arias DM. 1996. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science. 272:741–745. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Soltis DE, Palmer JD. 1988. A molecular reexamination of introgression between Helianthus annuus and H. bolanderi. Evolution. 42:227–238. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Van Fossen C, Desrochers A. 1995. Hybrid speciation accompanied by genomic reorganization in wild sunflowers. Nature. 375:313–316. [Google Scholar]
- Schemske DW. 2000. Understanding the origin of species. Evolution. 54:1069–1073. [Google Scholar]
- Schilling EE. 2020. Helianthus. In: Flora of North America editorial committee, eds. 1993. Vol 19. New York and Oxford: Flora of North America North of Mexico. Vol. 21, p. 141. [Google Scholar]
- Schumer M, Rosenthal GG, Andolfatto P. 2014. How common is hybrid speciation? Evolution. 68:1553–1560. [DOI] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science. 360:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbach AE, Rieseberg LH. 2002. Likely multiple origins of a diploid hybrid sunflower species. Mol Ecol. 11:1703–1717. [DOI] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Von Haeseler A. 2013. Nextgenmap: fast and accurate read mapping in highly polymorphic genomes. Bioinform. 29:2790–2791. [DOI] [PubMed] [Google Scholar]
- Smith MR. 2019. TreeTools: create, modify and analyse phylogenetic trees. Comprehensive R Archive Network.
- Stebbins GL. 1957. The hybrid origin of microspecies in the Elymus glaucus complex. Cytologia (Tokyo). 36:336–340. [Google Scholar]
- Stephens JD, Rogers WL, Mason CM, Donovan LA, Malmberg RL. 2015. Species tree estimation of diploid Helianthus (Asteraceae) using target enrichment. Am J Bot. 102:910–920. [DOI] [PubMed] [Google Scholar]
- Strasburg JL, Rieseberg LH. 2008. Molecular demographic history of the annual sunflowers Helianthus annuus and H. petiolaris—large effective population sizes and rates of long-term gene flow. Evolution. 62:1936–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Gonzalez A, Lexer C, Cronk QC. 2018. Adaptive introgression: a plant perspective. Biol Lett. 14:20170688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Lu Z, Zhu X, Ma H. 2020. Genomic basis of homoploid hybrid speciation within chestnut trees. Nat Commun. 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D’agostino ER, Price DK, Waddell PJ, Lang M, Courtier-Orgogozo V, et al. 2022. Widespread introgression across a phylogeny of 155 Drosophila genomes. Curr Biol. 32:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Owens GL, Bercovich N, Légaré JS, Soudi S, Burge DO, Huang K, Ostevik KL, Drummond EBM, et al. 2020. Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature. 584:602–607. [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Baird S, Pan J, Rieseberg LH. 1998. Rapid hybrid speciation in wild sunflowers. PNAS. 95:11757–11762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Strakosh SC, Zhen Y. 2006. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr Biol. 16:R872–R873. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang Y, Bi H, Lu Z, Ma Y, Yang X, Chen N, Tian B, Liu B, Mao X, et al. 2021. Hybrid speciation via inheritance of alternate alleles of parental isolating genes. Mol Plant. 14:208–222. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kang M, Li J, Zhang Z, Wang Y, Chen C, Yang Y, Liu J. 2022. Genomic evidence for homoploid hybrid speciation between ancestors of two different genera. Nat commun. 13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch ME, Rieseberg LH. 2002. Habitat divergence between a homoploid hybrid sunflower species, Helianthus paradoxus (Asteraceae), and its progenitors. Am J Bot. 89:472–478. [DOI] [PubMed] [Google Scholar]
- Wickham H. 2011. Ggplot2. Wiley Interdiscip Rev Comput Stat. 3:180–185. [Google Scholar]
- Xiong T, Li X, Yago M, Mallet J. 2022. Admixture of evolutionary rates across a butterfly hybrid zone. eLife. 11:e78135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. 2007. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 175:1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Zou Y, Bai J, Zheng H. 2002. Preparation of total DNA from recalcitrant plant taxa. Acta Bot Sin. 44:694–697. [Google Scholar]
- Zhang JQ, Imerovski I, Borkowski K, Huang K, Burge D, Rieseberg LH. 2019. Intraspecific genetic divergence within Helianthus niveus and the status of two new morphotypes from Mexico. Am J Bot. 106:1229–1239. [DOI] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 19:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.