Abstract
Neurofilament and tau proteins are neuron-specific cytoskeletal proteins that are enriched in axons, regulated by many of the same protein kinases, interact physically, and are the principal constituents of neurofibrillary lesions in major adult-onset dementias. Both proteins share functions related to the modulation of stability and functions of the microtubule network in axons, axonal transport and scaffolding of organelles, long-term synaptic potentiation, and learning and memory. Expression of these proteins is regulated not only at the transcriptional level but also through posttranscriptional control of pre-mRNA splicing, mRNA stability, transport, localization, local translation and degradation. Current evidence suggests that posttranscriptional determinants of their levels are usually regulated by RNA-binding proteins and microRNAs primarily through 3’-untranslated regions of neurofilament and tau mRNAs. Dysregulations of neurofilament and tau expression caused by mutations or pathologies of RNA-binding proteins such as TDP43, FUS and microRNAs are increasingly recognized in association with varied neurological disorders. In this review, we summarize the current understanding of posttranscriptional control of neurofilament and tau by examining the posttranscriptional regulation of neurofilament and tau by RNA-binding proteins and microRNAs implicated in health and diseases.
Keywords: Neurofilament, Tau, mRNA, RNA-binding protein, miRNA, Posttranscriptional control
1. Introduction
Neurofilament (Nf) and tau proteins are not only abundant integral components of neurofibrillary tangles and but also hyperphosphorylated in Alzheimer disease (AD) (Grundke-Iqbal et al., 1986; Rudrabhatla et al., 2010, 2011). Tau protein may also be a constituent of Lewy bodies (Ishizawa et al., 2003) composed of alpha-synuclein and Nf proteins (Goldman et al., 1983; Kanazawa et al., 2008) commonly found in Parkinson disease (PD) and the Lewy body dementias. Previous studies also showed that neurofilament proteins colocalized with tau aggregates in transgenic mice overexpressing human tau protein (Ishihara et al., 1999). Moreover, deletion of neurofilament proteins resulted in a dramatic decrease in the total number of tau-positive spheroids and attenuated neurodegenerative disease phenotype in these mice, suggesting a role for neurofilament proteins in the pathogenesis of neurofibrillary tau lesions in neurodegenerative disorders (Ishihara et al., 2001). Recently, neurofilament (Yuan and Nixon, 2021) and tau proteins (Holper et al., 2022) have been used together as biomarkers of neuronal integrity reflecting the magnitude of neuronal injury and neurodegeneration.
Neurofilaments (Nfs) are almost exclusively expressed in neurons. Nfs are obligate 10 nm heteropolymers composed of four different subunits, each composed of a central alpha-helical rod region, a short variable amino-terminal head domain, and a highly variable length tail at the C-terminal end. CNS neurofilaments are composed of Nf heavy (NfH), Nf medium (NfM), Nf light (NfL) and alpha-internexin (INA) subunits exhibiting apparent molecular masses of 200 kDa, 145–160 kDa, 70 kDa, and 59–65 kDa respectively (Yuan et al., 2006). In the PNS, the 57–58 kDa intermediate filament protein peripherin (PRPH) replaces INA (Yuan et al., 2012). In mature neurons, these polymers, often many micrometers in length, bundle and extensively cross-link within a cytoskeletal protein network of exceptional stability (many months) and provide structural support to maintain the large calibers of myelinated axons crucial for nerve conduction velocity (Ohara et al., 1993). Nf proteins (NfPs) have also recently found to play unique roles in synapse function (Yuan et al., 2015). Specifically, Nf subunits can be transported from soma to synaptic terminals in oligomeric form, including heterodimers (Yuan et al., 2003) or portions of them could be synthesized locally (Yuan and Nixon, 2016). The Nf network comprising heterodimers, oligomers and short filaments could function as a regulatory scaffold for reversible organelle and protein complex docking (Rao et al., 2011; Yuan et al., 2015). Notably, NfL subunits were found to modulate the turnover, levels, and synaptic activity of the NMDA receptor and thereby influence LTP and NMDA-mediated behaviors (Yuan et al., 2018). By contrast, NfM selectively binds D1R and regulates sensitization / desensitization at the post-synaptic membrane by anchoring internalized D1 dopamine receptors on endosomes and modulating their recycling back to the membrane surface to increase D1-mediated activities (Yuan et al., 2015). Loss of NfPs reduces conduction velocity and cause neuropathy in quails (Ohara et al., 1993), mice (Zhu et al., 1997), and humans (Abe et al., 2009; Fu and Yuan, 2018; Yum et al., 2009). Mutations of NfPs can cause neurological diseases and the abnormal accumulation of NfPs in soma and axons is a common feature of many neurological conditions (Table 1). NfPs therefore play prominent roles in both physiology and pathology of the neuron, especially axons.
Table 1.
Mutations or pathologies of Nf & Tau proteins linked to neurological diseases.
| Genes | Mutations or pathologies of Nf or Tau | Linked diseases | References |
|---|---|---|---|
|
| |||
| NEFL | Both | Charcot-Marie-Tooth disease type 1 F | (Jordanova et al., 2003) |
| Charcot-Marie-Tooth disease type 2E | (Mersiyanova et al., 2000) | ||
| Charcot-Marie-Tooth disease, Dominant intermediate G | (Berciano et al., 2016) | ||
| Congenital myopathy | (Agrawal et al., 2014) | ||
| Hereditary spastic paraplegia | (Mul et al., 2020) | ||
| NEFH | Both | Charcot-Marie-Tooth disease, Axonal, Type 2CC | (Rebelo et al., 2016) |
| Amyotrophic lateral sclerosis | (Al-Chalabi et al., 1999) | ||
| PRPH | Both | Amyotrophic lateral sclerosis | (Leung et al., 2004) |
| Peripheral neuropathy | (Bjornsdottir et al., 2019) | ||
| INA | Pathology | Neuronal intermediate filament inclusion disease | (Cairns et al., 2004) |
| NEFM | Both | Parkinson disease | (Lavedan et al., 2002) |
| MAPT | Both | Frontotemporal dementia with or without | (Hutton et al., 1998) |
| parkinsonism Pick disease | (Murrell et al., 1999) | ||
| Progressive supranuclear palsy 1 | (Poorkaj et al., 2002) | ||
| Atypical progressive supranuclear palsy 1 | (Pastor et al., 2001) | ||
| Parkinson disease | (Martin et al., 2001) | ||
The tau proteins are a group of six highly abundant and soluble microtubule-associated protein isoforms produced by alternative splicing from the gene MAPT (Weingarten et al., 1975). They are believed to function primarily to facilitate assembly and stabilization of microtubules mainly in axons (Wang and Mandelkow, 2016). Although generally regarded as neuron-specific cytoskeleton proteins, tau isoforms are also expressed at very low levels in CNS astrocytes and oligodendrocytes. The presence of normal tau proteins in brains seems to be important for the physiological function of the nervous system since their deletion leads to muscle weakness, hyperactivity and impaired fear conditioning in mice (Ikegami et al., 2000). MAPT mutations can cause frontotemporal dementia and several other “tauopathies” (Table 1). Paired helical filaments of tau proteins are the main component of neurofibrillary tangles that are hallmark intraneuronal lesions in the brain in Alzheimer’s disease (AD) and several other neurodegenerative “tauopathies”. In summary, tau, like NfPs, plays multiple roles in the function and dysfunction of the nervous system, especially axons.
Both tau and NfPs undergo posttranslational modifications such as phosphorylation, glycosylation, nitration, oxidation and ubiquitination. Hyper-phosphorylation of tau (Karikari et al., 2020), NfH, and NfM (Rudrabhatla et al., 2010) are particularly implicated in neurodegenerative disorders including Alzheimer disease. It is also relevant that many of the kinases/phosphatases regulating these two classes of proteins are shared in common. Although posttranslational modifications of NfPs and tau are implicated in the pathogenesis of varied neurodegenerative diseases, new evidence has emerged for involvement of Nf / tau RNA-mediated mechanisms in a number of degenerative pathologies of the CNS (Chen et al., 2014; Kumar et al., 2021; Robertson et al., 2003; Santa-Maria et al., 2015). These RNA-mediated mechanisms include alternative splicing, a general phenomenon in eukaryotic genomes, which in humans impacts about 95% of multi-exonic genes (Pan et al., 2008; Wang et al., 2008). Given that alternative splicing is more common in the brain than in other tissues (Yeo et al., 2004), the posttranscriptional control of NfP and tau expression through RNA-binding proteins (RBPs) and microRNAs (miRNAs) is particularly active (Table 2). By transporting repressed mRNAs to distant compartments within the cell, posttranscriptional regulation of NfPs and tau in neurons is able to regulate local protein synthesis for immediate needs in synaptic terminals, thereby avoiding the delays in transporting new protein from the perikaryon. Moreover, RBPs play multiple roles in posttranscriptional control of RNAs, such as RNA biogenesis, alternative splicing, polyadenylation, maturation, stability, localization, transport and translation efficiency in neurons (Gebauer et al., 2021). Not surprisingly, mutations in RBPs are increasingly recognized to cause varied neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Table 3). Additional post-transcriptional control is exerted by miRNA, which is a small single-stranded non-coding RNA molecule (18–23 nucleotides long) that binds to and silences target mRNA at the posttranscriptional level (Gebert and MacRae, 2019). Each miRNA binds many target transcripts, providing additional broad control that complements canonical transcriptional pathways and posttranscriptional regulation by RBPs. Recently, miRNA defects have also been linked to neurodegeneration (Cheng et al., 2014; Hebert et al., 2010; Juzwik et al., 2019; Kim et al., 2007; Schaefer et al., 2007). Although posttranslational regulation of Nf and tau proteins have been previously extensively described, the importance of their posttranscriptional regulation has been under-appreciated. In this review, we discuss our current understanding of posttranscriptional control of Nf and tau proteins through RBPs and miRNAs and its influences on the brain in health and neurological diseases.
Table 2.
Functions of Nf & Tau mRNA binding proteins and miRNAs.
| Function | RNA binding proteins & miRNAs | Binding to Nf or Tau | Cis-acting RNA element | References |
|---|---|---|---|---|
|
| ||||
| mRNA stabilizers | TDP43 | NEFL | (UG)6–9 | (Strong et al., 2007) |
| ARHGEF28 | NEFL | 68 nucleotides spanning the end of the open reading frame & beginning of the 3’-UTR | (Canete-Soler et al., 2001) | |
| PABP | NEFL | Full-length instability determinant | (Stefanizzi and Canete-Soler, 2007) | |
| NGF | NEFL | Undetermined | (Ikenaka et al., 1990) | |
| miR-b1336 & miR-b2403 | NEFL | 3’-UTR | (Ishtiaq et al., 2014) | |
| ELAV4 | MAPT | U-rich segment of 3’-UTR | (Aranda-Abreu et al., 1999) | |
| SRSF2 | MAPT | Exon 10 | (Qian et al., 2011) | |
| mRNA destabilizers | SOD1 | NEFL | Undetermined | (Ge et al., 2005) |
| 14–3-3 proteins | NEFL | GCTTGC motif Ml | (Ge et al., 2007) | |
| Aldolase A | NEFL | UG sites | (Canete-Soler et al., 2005) | |
| Aldolase C | NEFL | UG sites | (Canete-Soler et al., 2005) | |
| NF-RBP1 | NEFL & NEFH | 45 nt at the proximal edge of the NfL 3’-UTR & 36 nt in the mid-distal region of the NfH 3’-UTR | (Canete-Soler and Schlaepfer, 2000) | |
| miR-219 | MAPT | 3’-UTR | (Santa-Maria et al., 2015) | |
| TDP43 | MAPT | 2 UG repeats of its 3’-UTR | (Gu et al., 2017) | |
| mRNA modulation | FUS | NEFL, NEFM, NEFH & MAPT | 3’-UTR | (Lagier-Tourenne et al., 2012) |
| miR-146a, miR624–5p & miR-582–3p | NEFL | 3’-UTR | (Campos-Melo et al., 2013) | |
| MiR-9–5p, miR-20b-5p, miR-92a-3p, miR-125b-5b & miR-233–3p | NEFM & NEFH | 3’-UTR | (Campos-Melo et al., 2018) | |
| Enhancer of mRNA transport | hnRNP K | NEFL, NEFM & NEFH | Prefer CU-rich elements | (Liu et al., 2008; Thyagarajan and Szaro, 2008) |
| Enhancer of mRNA translation | hnRNP K | NEFM, INA & MAPT | Prefer CU-rich elements | (Liu et al., 2008; Liu and Szaro, 2011) |
| ELAVL2 | NEFM | Non-canonical AREs in the 3’-UTR | (Antic et al., 1999) | |
| Translation inhibition | ELAV4, IGF2BP1 & G3BP1 | MAPT | 3’-UTR | (Atlas et al., 2007) |
| Structural effects on mRNA | 14–3-3 proteins | NEFL | GCTTGC motif M2 | (Ge et al., 2007) |
| Pre-mRNA splicing | Musashi-1 | MAPT | Exon 10 | (Cuadrado et al., 2002) |
| SWAP, hnRNP G | MAPT | Exon 10 | (Wang et al., 2004) | |
| miR-16, miR-132 | MAPT | Exon 10 | (Hebert et al., 2012) | |
| FUS | MAPT | Exons 3 & 10 | (Orozco et al., 2012) | |
| FMRP | MAPT | Exon 10 | (Zhou et al., 2017) | |
| Undetermined | hnRNP E1/E2 | NEFL, NEFM & NEFH | 3’-UTR | (Thyagarajan and Szaro, 2004, 2008) |
Table 3.
Mutations of RNA-binding proteins linked to ALS/FTD spectrum.
| Genes | RNA-binding proteins | Mutations | References |
|---|---|---|---|
|
| |||
| TARDBP | Yes | Missense | (Benajiba et al., 2009; Borroni et al., 2009) |
| FUS | Yes | Missense | (Broustal et al., 2010; Van Langenhove et al., 2010) |
| HNRNPA1 | Yes | Missense | (Kim et al., 2013) |
| HNRNPA2/B1 | Yes | Missense | (Kim et al., 2013) |
| TIA1 | Yes | Missense | (Mackenzie et al., 2017) |
| MATR3 | Yes | Missense | (Johnson et al., 2014) |
| TAF15 | Yes | Missense | (Ticozzi et al., 2011) |
| EWSR1 | Yes | Missense | (Couthouis et al., 2012) |
| ANG | Yes | Missense | (van Es et al., 2009) |
| GLE1 | Yes | Multiple | (Kaneb et al., 2015) |
| SETX | Yes | Missense | (Chen et al., 2004) |
| ARHGEF28 | Yes | Frameshift | (Droppelmann et al., 2013b) |
| SOD1 | Yes | Missense | (Katz et al., 2012) |
| ATXN2 | Yes | PolyQ expansions | (Elden et al., 2010) |
| C9orf72 | RNA toxicity | G4C2 repeat expansion | (DeJesus-Hernandez et al., 2011) |
2. Posttranscriptional regulation of NfL
2.1. NfL mRNA dynamics
Each NfP has a distinct structure that potentially could have differential functions. Among the five Nf subunits, NfL is the most abundant and forms the core of neurofilaments in both PNS and CNS. The stoichiometries of NfPs in neurons are temporally and spatially controlled. The levels of expression of NfL are tightly coupled with those of other Nf subunits and can influence their levels. When NfL is absent in mice, NfH levels decline most, followed by NfM and PRPH (Yuan et al., 2012) although INA levels are only marginally affected (Yuan et al., 2003). While transcriptional control determines the restriction of Nf expression to neurons, the abundance of Nf gene products in different neurons at different developmental stages is also regulated post-transcriptionally (Schwartz et al., 1995). Although there is additional evidence for translational control (Antic et al., 1999), a major degree of posttranscriptional control is exerted at the level of Nf mRNA stability (Schwartz et al., 1992). Transcription of the NfL gene yields NfL mRNA (Fig. 1A), which was originally believed to be translated exclusively in the perikaryon and proximal dendrites, has gradually been appreciated to occur in all compartments of neurons (Alami et al., 2014; Holt et al., 2019; Lee and Hollenbeck, 2003; Sotelo-Silveira et al., 2000). The impact of this regulation on NfPs is exemplified by studies in zebrafish. In contrast to the single mammalian Nfl gene, zebrafish have two NfL genes, called nefla and neflb (Wang et al., 2019). Both gene knockdown and rescue experiments show that neflb rather than nefla plays an indispensable role in nervous system development. Two isoforms of the latter gene, neflE3 and neflE4, are generated by alternative splicing of neflb mRNA (Fig. 1B) (Demy et al., 2020). Alternative use of two polyadenylation sites in exon 4 in the mouse NfL gene also generates 2.3-and 3.5-kb mRNAs (Nakahira et al., 1990). These two NfL mRNAs are much more stable in mature neurons than in cultured PC12 cells (Nakahira et al., 1990; Schwartz et al., 1992). When adult DRG neurons are placed in tissue culture, NfL mRNAs are rapidly destabilized. Therefore, NfL mRNA half-lives in the presence of actinomycin in dorsal root ganglion (DRG) neurons of adult rat are estimated to be at least 5 days (Moskowitz and Oblinger, 1994) whereas those in PC12 cells are only a few hours (Nakahira et al., 1990). Blockade of either transcription or protein synthesis using inhibitors in cultured adult DRG neurons prevents the destabilization of NfL mRNAs (Schwartz et al., 1992). NfL mRNAs in cultured neuronal cells can also be stabilized by nerve growth factors (Ikenaka et al., 1990) and destabilized by okadaic acid, which inhibits protein phosphatase 2 A (Sasahara et al., 1996). The stability of NfL mRNAs in vivo is also highly regulated. The levels of NfL mRNAs are dependent upon axonal- or target cell-derived signals (Schwartz et al., 1990). NfL mRNAs can also be stabilized during postnatal development (Schwartz et al., 1994). The observations that NfL showed maximal rates of gene transcription at postnatal day 5 followed by a decline at later times whereas the steady-state levels of NfL mRNAs changed in manner opposite to that of transcription and increased progressively during the postnatal intervals suggests mRNA stabilization is the main factor regulating the steady-state levels of NfL mRNAs in vivo (Moskowitz and Oblinger, 1995). Due to this posttranscriptional regulation, overexpression in transgenic mice of NfL mRNA from 3- to 5-fold in different regions of CNS resulted only in a mild increase of 10–50% in the levels of NfL proteins (Beaudet et al., 1993). Stability of NfL mRNA may also relate to other developmental phenomena that are characteristic of mature neurons such as the axotomy-induced 2- to 3-fold downregulation of NfL mRNA (Goldstein et al., 1988; Hoffman et al., 1987). In summary, stability of NfL mRNA may contribute to the high level of expression of NfL protein both In vitro and in vivo.
Fig. 1.
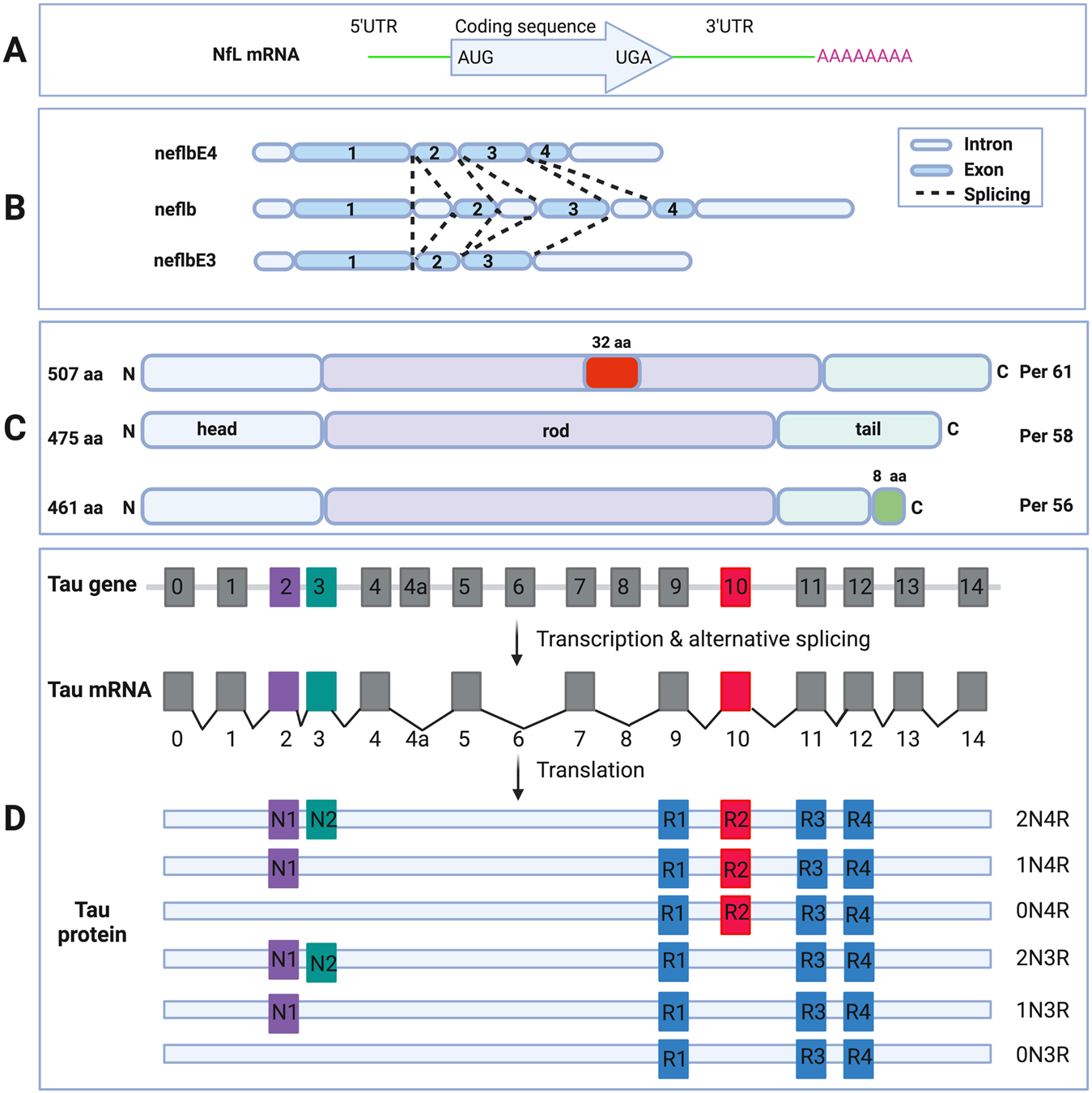
Schematic diagram of NfL mRNA (A). 5’UTR, protein coding region, 3’UTR and the poly-A tail are shown (upper box). Schematic diagram of the neflb isoforms, generated by alternative splicing of the same mRNA in zebrafish (B). neflb is expressed at all embryonic and larval stages in zebrafish, which a splicing shift from neflbE3 to neflbE4 occurring during CNS development. Schematic diagram of the PRPH isoforms, generated by alternative splicing of the same mRNA (C). Per 58 is the major constitutive form; Per 61 results from the insertion of 32 aa in the rod domain; Per 56 results from the replacing the C-terminal 21 aa with a different 8 aa sequence. Schematic diagram of MAPT gene, mRNA and splice isoforms of tau in the human brain (D). Human MAPT gene contains 16 exons. E1, E4, E5, E7, E9, E11, E12 and E13 are constitutive, whereas others are subject to alternative splicing. E0 and E1 encode the 5’UTR, whereas E14 is part of the 3’UTR. E4a, E6 and E8 are transcribed only in peripheral tissues. The alternative splicing of E2, E3 and E10 give rise to 6 different mRNAs, which are translated to 6 isoforms. These isoforms differ by the presence of 1 or 2 N-terminal inserts encoded by E2 (purple box) and E3 (green box), in combination with the presence of either 3 or 4 repeat regions coded by E9 (blue box), E10 (red box), E11 (blue box) and E12 (blue box) in the C-terminus.
2.2. 3’-UTR of NfL mRNA and regulation by RBPs
The involvement of 3’-untranslated region (3’-UTR) in mRNA stability (Hentze, 1991; Sachs, 1993) and the conservation of 3’-UTR in Nf mRNAs (Schwartz et al., 1994) suggest that the 3’-UTR in Nf genes may harbor determinants of Nf expression. In fact, deletion of 3’-UTR from mouse NfL leads to a 3-fold aberrant up-regulation of NfL mRNA between embryonic days 15 and 18, and loss of its susceptibility to axotomy-induced down-regulation, supporting this hypothesis (Schwartz et al., 1995). 3’-UTR in NfL mRNAs can also trigger aggregation of NfL protein in neuronal cells (Lin et al., 2004). Interestingly, overexpression of 3’-UTR of NfL mRNA or expression of an NfL transgene with a 36-base pair c-myc mutant mRNA stability determinants could also reproduce selective degeneration of motor neurons with aggregates of RNA-binding proteins in primary motor neurons in culture (Lin et al., 2003) or degenerating motor neurons of transgenic mice (Canete-Soler et al., 1999; Nie et al., 2002). Later studies demonstrated that a major stability determinant is located to a 68-nucleotide (nt) sequence that forms the junction between 3’-coding region and 3’-UTR of NfL mRNAs (Canete-Soler et al., 1998b), and also is the binding site of a unique ribonucleoprotein complex (Canete-Soler et al., 1998a). Further investigations showed that p190RhoGEF, a RhoA-specific guanine nucleotide exchange factor, can bind to this 68-nt sequence in mouse (Canete-Soler et al., 2001) and human (Volkening et al., 2010) and increases NfL mRNA stability by nearly 2-fold. This factor is also involved in aggregation of NfL protein (Lin et al., 2005) and co-aggregation of p190RhoGEF and NfL protein is associated with loss of NfL mRNAs in degenerating spinal cord motor neurons in ALS (Droppelmann et al., 2013a; Menzies et al., 2002; Wong et al., 2000). Correspondingly, p190RhoGEF gene mutation is also linked to familial ALS (Droppelmann et al., 2013b). Intriguingly, binding of p190RhoGEF to NfL mRNA is directly competed by BC1 RNA (Ge et al., 2002), a 152 nucleotide RNA polymerase III transcript that is highly expressed postnatally in neurons (Cao et al., 2006), indicating possible co-regulation of NfL mRNA by RBPs and non-coding RNAs. In addition to p190RhoGEF, aldolases A and C also bind to this sequence and negatively regulate the stability of NfL mRNAs (Canete-Soler et al., 2005) whereas poly (A)-binding protein positively regulates their stability (Stefanizzi and Canete-Soler, 2007). Still other proteins, such as wild-type and mutant superoxide dismutase 1 (SOD1) (Ge et al., 2005), TAR DNA-binding protein of 43 kDa (TDP43) (Strong et al., 2007), fused in sarcoma (FUS) (Lagier-Tourenne et al., 2012), 14–3–3 proteins (Ge et al., 2007), hnRNP K, E1 and E2 (Thyagarajan and Szaro, 2008), have also been identified as RNA-binding proteins that bind NfL mRNAs. It is also reported that mRNP granules containing TDP43 and NfL mRNA show predominantly anterograde movements and net anterograde displacement, whereas those that contain NfL mRNA without TDP43 show predominantly retrograde movements and net retrograde displacement (Alami et al., 2014). The functional significance of this observation is not yet understood. Recent studies showed that disruption of the balance of the two isoforms of NfL mRNAs through partial deletion of TDP43 affects Nf assembly and motor axon growth with resultant motor deficits in zebrafish (Demy et al., 2020). More recent studies demonstrated that TDP43 proteinopathy can cause translational suppression of Nf mRNAs resulting in 3-4-fold decrease in the levels of NfPs in mouse brain (Kumar et al., 2021). Thus, 3’-UTR of NfL mRNA may serve as a hub for posttranscriptional control as the target of many RBPs.
2.3. Regulation by miRNAs and interaction between miRNAs and RBPs
Beyond the interaction of RBPS with the 3’UTR of NfL mRNAs, miRNAs could also participate in NfL mRNA stability and translational regulation through mRNA decay or translational silencing (Du and Zamore, 2005; Gebert and MacRae, 2019). In fact, miRNA expression profile was shown to be broadly altered in the spinal cord of sporadic ALS and miRNAs including miR-146a, miR-424-5p and miR-582-3p (Campos-Melo et al., 2013), miR-b1336 and miR-b2403 (Ishtiaq et al., 2014) are capable of interacting with NfL mRNA 3’UTR and could play a role in the regulation of its mRNA levels. Notably, RNA is not surrounded by a lipid membrane, instead their fate is generally controlled by binding with many RBPs that form ribonucleoprotein (RNP) complexes. These membraneless compartments are called RNA granules, a category that includes transport granules, stress granules, storage granules, P-bodies and activity-dependent granules in the neuronal cytoplasm (Kiebler and Bassell, 2006; Wolozin and Ivanov, 2019). RNA granules form via liquid-liquid phase separation, which is driven by a dynamic network of multivalent interactions of protein containing intrinsically disordered regions and low complex domains such as TDP43 and FUS. On the one hand, both stress granules and P-bodies exist in a constant state of dynamic flux mediated by the nature of associated miRNAs (Anderson and Kedersha, 2006, 2008; Stoecklin and Kedersha, 2013). On the other hand, miRNAs biogenesis and metabolism are regulated by RBPs (Loffreda et al., 2015; Stroynowska-Czerwinska et al., 2014) and dysfunction of both miRNAs and RBPs is often observed in neurodegenerative diseases (Kinoshita et al., 2021). For example, TDP43 and FUS are involved in the regulation of miR-183/96/182 biogenesis and memory suppressor protein phosphatase 1, and their alterations results in age-related memory decline in the pathogenesis of ALS and FTD (Jawaid et al., 2019). Therefore, these miRNAs could interact with those RBPs binding NfL mRNAs and coordinate the posttranscription of NfL gene expression under physiological conditions and become dysfunctional in disease states (Jiang and Coller, 2012).
3. Posttranscriptional regulation of NfH
Although NfH expression is low, when compared to NfL, in developing neurons and only increases postnatally (Pachter and Liem, 1984; Shaw and Weber, 1982), the dissociation between rates of NfH gene transcription and the steady-state levels of NfH mRNAs during postnatal development are similar to those of the NfL gene, with maximal transcription at early times followed by a decline at later times whereas NfH mRNA levels increase substantially during the same interval (Moskowitz and Oblinger, 1995). Like NfL and NfM genes, NfH mRNA levels also decline following nerve transection (Goldstein et al., 1988). However, unlike NfL transgenic mice, the CNS of transgenic mice bearing multiple copies of the human NfH gene showed about 3-fold increase in the levels of both NfH mRNA and protein compared to wild-type mice, indicating NfH and NfL genes are still subject to different posttranscriptional regulation in the CNS (Beaudet et al., 1993). Although NfL mRNA is inhibited in degenerating spinal cord motor neurons in ALS, NfH mRNA seems to be not suppressed (Chen et al., 2014; Menzies et al., 2002). Poly (C)-sensitive complexes regulate the stability of NfH mRNA through binding a 36-nt sequence in the mid-distal region of the NfH 3’UTR (Canete-Soler and Schlaepfer, 2000). Other RNA-binding proteins like hnRNP K, E1 and E2 also bind NfH mRNAs for posttranscriptional regulation (Thyagarajan and Szaro, 2008). TDP43 was also shown to interact with human NfH mRNA from spinal cord homogenates but was not co-immunoprecipitated from HEK293T cells that had been transiently transfected with a partial sequence of hNfH lacking a component of the 5’ coding region of the hNfH construct (Strong et al., 2007), indicating that the 5’UTR or 5’ coding region of NfH has the potential to interact with TDP43. Like in NfL mRNAs, a small group of ALS-linked miRNAs that are expressed in human motor neurons also directly regulate NfH transcript levels (Campos-Melo et al., 2018).
4. Posttranscriptional regulation of NfM
During postnatal development, some differences exist in the regulatory mechanisms that control NfM compared to NfL and NfH mRNA levels. Unlike NfL and NfH, NfM transcription rates were largely dissociated from steady-state mRNA level changes between postnatal days 2 and 10 (Moskowitz and Oblinger, 1995). However, the transcriptional activity of the NfM gene and the steady-state levels of NfM mRNA increased in a coordinate manner between postnatal days 2 and 10. Like NfL and NfH genes, NfM mRNA levels also decline following nerve transection (Goldstein et al., 1988). In addition to NfL mRNA involvement in neurodegeneration, NfM mRNA could also be associated with ALS since NSC34 cells expressing G37R or G93A SOD1 showed selective reductions of both NfM and NfL mRNA and proteins (Menzies et al., 2002). Similar to NfL and NfH, RNA-binding proteins hnRNP K, E1 and E2 can also bind 3’UTR of NfM mRNA and could regulate its stability (Thyagarajan and Szaro, 2004, 2008). Further studies showed that hnRNP K knockdown can compromise NfM mRNA nucleocytoplasmic export and disrupt their loading onto polysomes for translation in Xenopus laevis (Liu et al., 2008, 2012). This interaction of hnRNP K and NfM mRNA could be modulated through the phosphorylation by extracellular signal-regulated kinase (Hutchins et al., 2015). Other RBPs like ELAV-like RNA binding protein 2 also binds to 3’UTR of NfM mRNA and can cause an increase in translation initiation of NfM mRNA in transfected cells (Antic et al., 1999). Besides 3’UTR, its introns may also be involved in the posttranscriptional regulation of NfM expression in neurons (Wang and Szaro, 2016). Like in NfL and NfH mRNAs, a small group of ALS-linked miRNAs that are expressed in human motor neurons also directly regulate NfM transcript levels (Campos-Melo et al., 2018).
5. Posttranscriptional regulation of INA
Unlike NfL, MfM and NfH found in both the CNS and PNS, INA is expressed predominantly in the CNS in the adult (Pachter and Liem, 1985), indicating its special role in the normal functioning of neurons in the brain and spinal cord. In contrast to the downregulation of NfL, NfM and NfH mRNAs after peripheral nerve injury, INA mRNA increases after facial nerve lesions, suggesting its role is in the early stage of neuronal regeneration (McGraw et al., 2002). Interestingly, the level of INA must be tightly regulated since overexpression of INA at 2–3 times the normal level produces mice with motor coordination deficiencies and neurodegeneration in aged animals (Ching et al., 1999). Remarkably, more prominent aggregation of INA than other NfPs are the neuropathological hallmark of Nf inclusion disease, a form of FTD (Cairns et al., 2004). Like NfL mRNA, INA mRNA is also reduced in motor neurons in ALS (Wong et al., 2000). INA expression is also regulated post-transcriptionally by hnRNP K since hnRNP K knockdown yields defects in its nuclear export and translation but not transcription (Liu and Szaro, 2011).
6. Posttranscriptional regulation of PRPH
PRPH is the first NfP expressed by PC12 cells stimulated by nerve growth factors (Leonard et al., 1988; Portier et al., 1983a; Thompson et al., 1992) followed by expression of NfL, NfM and NfH (Lindenbaum et al., 1988). Unlike NfL, NfM and NfH, which are found in both the CNS and PNS, PRPH is expressed predominantly in the PNS and in neurons of the CNS that have projections to the periphery, including spinal motor neurons in the adult, indicating its special role in the normal functioning of neurons in peripheral nerves (Parysek and Goldman, 1988; Portier et al., 1983b). In contrast to the downregulation of NfL, NfM and NfH mRNAs after peripheral nerve injury, PRPH mRNA increases following peripheral axotomy in motor neurons and dorsal root ganglia (Oblinger et al., 1989; Troy et al., 1990; Wong and Oblinger, 1990) or some CNS neurons after stab lesion or cerebral ischemia (Beaulieu et al., 2002), implying its role in the early stage of neuronal regeneration. The proinflammatory cytokines, interleukin-6 and leukemia inhibitory factor, can also induce PRPH expression through the JAK-STAT signaling pathway (Djabali et al., 1993; Lecomte et al., 1998; Sterneck et al., 1996). In addition to the main species of 58 kDA PRPH (Per 58), two other forms have been identified in the mouse (Fig. 1 C). Per 56 is made by a receptor on exon 9 of the PRPH gene transcript which induces a frame shift and replacement of a 21 amino acid in the C-terminus found on the dominant Per 58 form with a new 8 amino acid sequence. Per 61 is generated by the in-frame retention of intron 4 leading to a 32 amino acid insertion within coil 2b of the alpha-helical domain of PRPH (Landon et al., 1989, 2000). Per 58 and Per 56 are normally co-expressed whereas Per 61 is unable to assemble properly and is not found in the normal PRPH expressed adult motor neurons but it is expressed in motor neurons of mutant SOD1 transgenic mice, suggesting that mutant SOD1 induces differential splicing of the PRPH pre-mRNA (Robertson et al., 2003). Per 61 is also toxic to primary motor neurons in culture and induces PRPH and Nf aggregate formation, indicating alternatively spliced variants of PRPH may contribute to neurodegeneration in ALS. In fact, the pathologies of PRPH along with other NfPs have been found in spheroids in patients with ALS and overexpression of PRPH causes late onset death of motor neurons in mice (Beaulieu et al., 1999). In addition to PRPH pathology, mutations of PRPH gene have also been linked to ALS (Gros-Louis et al., 2004; Leung et al., 2004). Like NfL and INA mRNAs, PRPH mRNA is also reduced in motor neurons in ALS (Wong et al., 2000). After nucleocytoplasmic export, PRPH mRNA form particles with RNPs (Chang et al., 2006). The messenger RNPs (mRNPs) of PRPH are translationally silent while moving along microtubules and initiate translation when they cease moving. Unlike NfM mRNA which binds hnRNP K, PRPH mRNA does not bind hnRNP K and knockdown of hnRNP K has no effect on PRPH mRNA nucleocytoplasmic export and translation (Liu et al., 2008).
Since current evidences suggest significant differences in the transcriptional regulation of different Nf proteins, these Nf subunits may show not only coordinate mRNA expression but also independent regulation. These findings are consistent with the different roles of NfPs in different neuronal compartments (Kriz et al., 2000; Yuan et al., 2015, 2018).
7. Posttranscriptional regulation of tau
7.1. tau mRNA dynamics
Human tau protein is encoded by a single gene located on chromosome 17q21 (Andreadis, 2005). By alternative splicing, six isoforms are produced in CNS whereas with apparent molecular masses between 48 and 67 kDa and additional isoforms of 110 kDa that include exons 4a and exon 6 are expressed in PNS (Fischer and Baas, 2020). The expression of tau mRNAs in the spinal cord appeared to precede that in the brain, and the expression in the brainstem appeared to precede that in cerebral cortex and cerebellum (Takemura et al., 1991). Interestingly, tau pre-mRNA is alternatively spliced to produce exon 10+ and exon 10− tau isoforms that are expressed in approximately equal amounts in adult human brain (Fig. 1D)(Avila et al., 2004). Some tau disease-causing mutations alter the expression levels of tau isoform in FTD (Hutton et al., 1998) and a recent study identified a new non-aggregative splicing form of human tau and showed that its level is decreased in AD brains (Garcia-Escudero et al., 2021). Like Nf mRNAs, transport, stability and local translation of tau mRNA may also be dependent on RBPs and miRNAs. Interestingly, it was suggested that motor protein KIF3 could anchor RBP HuD and transport the HuD-tau mRNA complex along axons (Aronov et al., 2002). Dysfunction of these process could lead to neurodegeneration. Unlike other mRNA targets, tau mRNA is a relatively stable molecule (Sadot et al., 1995) with a half-life of 16 h in neuronal cells. Its stability is likely determined by interaction of 3’UTR of tau mRNA, RBPs and miRNAs (Aronov et al., 1999).
7.2. 3’-UTR of tau mRNA and regulation by RBPs
Two RBPs of 43 and 38 kDa were first identified to bind a minimal sequence of 91 nucleotides within the 3’UTR of tau mRNA (Behar et al., 1995). These RBPs were later characterized as embryonic lethal abnormal vision (ELAV)-like RNA-binding proteins (HuD) (Aranda-Abreu et al., 1999). More RBPs, including the insulin-like growth factor mRNA binding-protein 1 (IMP-1) and RAS-GAP SH3 domain binding protein (G3BP), were later also found to bind tau mRNA and interact with HuD (Atlas et al., 2004). The posttranscriptional regulation of tau expression by thyroid hormone (T3) is also mediated by RBP called musashi-1 (Cuadrado et al., 2002). Splicing factor SC35 can also stabilize tau mRNA through binding to the SC35-like element of exon 10 and thus promotes 4-repeat tau expression (Qian et al., 2011). Other tau exon 10 regulators include suppressor-of white-apricot (SWAP) and hnRNP G (Wang et al., 2004). FUS also directly binds to tau pre-mRNA and loss of FUS promotes pathological tau splicing (Orozco et al., 2012), indicating a possible link between FUS and tau pathologies in FTD. Similarly, TDP43 can bind to the UG repeats of tau 3’UTR, promoting its mRNA instability and leading to suppressed tau protein expression (Gu et al., 2017). In fact, the presence of TDP43 pathologies in about 97% of ALS and 50% FTD and significant tau pathologies in about 50% of ALS-FTD spectrum also supports the intracellular interaction of TDP43 and tau (Behrouzi et al., 2016; Moszczynski et al., 2019). Like INA, tau is also regulated post-transcriptionally by hnRNP K as hnRNP K knockdown yielded defect in its nuclear export and translation but not transcription (Liu and Szaro, 2011). G3BP1and IMP1 have also been identified to bind to the 3’UTR of the tau mRNA (Atlas et al., 2007). Induction of RNA protein granules by G3BP1 or IMP1 could lead to more than 30-fold increase in the ratio of high molecular weight to low molecular weight tau mRNA and about a 12-fold increased ratio of high molecular weight to low molecular weight tau protein (Moschner et al., 2014). Serine and arginine rich splicing factor 2 (SRSF2) is another RBP that plays important roles in splicing of mRNA precursors and its levels are elevated in the brains of patients with progressive supranuclear palsy characterized by intracellular aggregation of 4 R tau isoforms (Bruch et al., 2014). SRSF2 upregulation through inhibition of mitochondrial complex I could also promote 4 R tau in human neurons. Tau mRNA is also a component of granules containing fragile X mental retardation protein (FMRP)-and Staufen 1-positive RNA granules within the dendritic component of primary hippocampal neurons where AMPA and NMDA receptor stimulation potentially enhance local translation of tau proteins (Kobayashi et al., 2017). On the one hand, posttranscriptional regulation of tau is controlled by RBPs. On the other hand, tau itself negatively regulates protein synthesis and alters synaptic function by interacting with ribosomal proteins (Koren et al., 2019; Meier et al., 2016; Papanikolopoulou et al., 2019).
7.3. Regulation of tau by miRNAs
Dicer is a key enzyme in the biogenesis of miRNAs, small non-coding RNAs that function as part of the RNA-induced silencing complex (RISC) to repress gene expression at the posttranscriptional level. Mice lacking dicer in the forebrain displayed progressive neurodegeneration accompanied by tau hyperphosphorylation (Hebert et al., 2010), suggesting miRNAs may contribute to tau pathologies in tauopathy. Later studies found that miR-132, which is downregulated in PSP, inhibits 4 R tau expression in neuronal cells (Smith et al., 2011). Further investigations also demonstrated decrease of miR132-3p levels in neurons enriched with hyperphosphorylated tau in AD brains (Lau et al., 2013). Another non-coding miRNA miR-219 is not only downregulated in AD and primary age-related tauopathy but also can bind directly to the 3’UTR of the tau mRNA and therefore represses tau synthesis at the posttranscriptional level (Santa-Maria et al., 2015).
8. Distinctions and inter-related functions of Nf and tau in neurons
Nf and tau proteins clearly belong to two distinct class of protein family with different structures and different, and in various contexts inter-related, functions in the neuron. The former is classified as highly insoluble proteins of intermediate filaments found in the cytoplasm of neurons whereas the latter is a highly soluble microtubule-associated protein. Their newly synthesized proteins are also transported at different rates along mouse optic axons (Mercken et al., 1995; Yuan et al., 2006). However, they are directly linked physically both under physiological and pathological conditions (Miyata et al., 1986; Rudrabhatla et al., 2011). Both Nf and tau proteins are especially enriched in axons and share functions related to microtubule polymerization and stability (Bocquet et al., 2009; Yadav et al., 2016), axonal transport and scaffolding of organelles in axons (Mueller et al., 2021; Perrot and Julien, 2009; Rao et al., 2011). Recent evidence also demonstrates their presence at the synapse (Ittner et al., 2010; Kimura et al., 2014; Yuan et al., 2015) and roles in long-term synaptic potentiation, learning and memory (Ahmed et al., 2014; Biundo et al., 2018; Yuan et al., 2015). Both NfPs and tau undergo posttranslational modifications such as phosphorylation, and hyperphosphorylation of tau (Karikari et al., 2020), NfH, and NfM (Rudrabhatla et al., 2010) are particularly implicated in Alzheimer disease. Both proteins also share many of the kinases / phosphatases for their regulation and are used as biomarkers of neurodegeneration often associated with their hyperphosphorylation or abnormal degradation. Deletion of NfP can also diminish tau pathological lesions and attenuate disease phenotype caused by tau abnormality (Ishihara et al., 2001).
9. Potential common mechanisms of posttranscriptional dysregulation in both Nf-associated diseases of the CNS and tauopathies
Both the mRNAs and proteins of Nf and tau can be transported into axons (Alami et al., 2014; Morita and Sobue, 2009), dendrite (Balaji et al., 2018) and synapses (Crispino et al., 1993; Kobayashi et al., 2017) (Fig. 2). Since Nf and tau proteins are especially enriched in axons, any damage to neuronal cytoskeleton in this domain could potentially lead to pathologies of both proteins. In fact, tau and Nf proteins are both components of neurofibrillary tangles in AD (Rudrabhatla et al., 2011). In addition to these tangles, approximately 57% of AD cases also have TDP43 deposits in the brain (Meneses et al., 2021). It is also known that Nf aggregates and TDP43 pathology are hallmarks of ALS (Hardiman et al., 2017). Moreover, a 17 kDa neurotoxic tau fragment (tau45–230) has been recently observed in extracts of both brain and ventral spinal cord of ALS patients, but not in controls (Vintilescu et al., 2016). More recently, elevated levels of tau and a phosphorylated form of the tau protein were reported in the brain of ALS cases who carried a mutation in the C9orf72 gene (Petrozziello et al., 2022). Similarly, PD was not initially considered to be a typical tauopathy. However, a genome-wide association study indicated a potential association between tauopathy and sporadic PD (Nalls et al., 2014). The aggregation and deposition of tau were also observed in about 50% of PD brains (Zhang et al., 2018). Since TDP43 has opposite effects on the stability of NfL (Strong et al., 2007) and tau mRNAs (Gu et al., 2017) through direct interaction with their 3’-UTR, this RBP could be involved in the posttranscriptional dysregulation of both Nf and tau proteins in these neurodegenerative diseases. Intriguingly, TDP43 may also modulate anterograde transport of messenger ribonucleoprotein granules (Chu et al., 2019) that could contain mRNAs of NfL (Alami et al., 2014) and tau (Aronov et al., 2002; Villace et al., 2004). In addition to TDP43, two other RBPs, FUS and Pabpc1, could also be involved in the dysregulation of Nf and tau mRNAs in diseases since the former can modulate mRNAs of Nefl, Nefm, Nefh and Mapt while the latter shields the NfL mRNA from aldolase attack (Stefanizzi and Canete-Soler, 2007) and co-localizes with small tau inclusions in tauopathy (Maziuk et al., 2018). Besides RBPs, some miRNAs may also have common targets involving both Nf and tau proteins since these two classes of proteins are both involved in disease pathologies of AD and PD (Goldman et al., 1983; Rudrabhatla et al., 2011; Zhang et al., 2018). However, the specific miRNAs targeting both Nf and tau have not been reported. Nevertheless, miRNA125b was shown to be involved in the modulation of NfM and NfH mRNAs (Campos-Melo et al., 2018) and this same miRNA also induces tau hyperphosphorylation and cognitive deficits in AD (Banzhaf--Strathmann et al., 2014). Therefore, regulation of tau protein by miRNA 125b needs to be further investigated. Another miRNA, miRNA 30b was also shown to suppress NfL expression through interaction with its 3’-UTR. This miRNA is also upregulated in the brains of both AD patients and APP transgenic mice (Song et al., 2019). Future studies will be needed to examine the posttranscriptional regulation of tau protein by this miRNA.
Fig. 2.
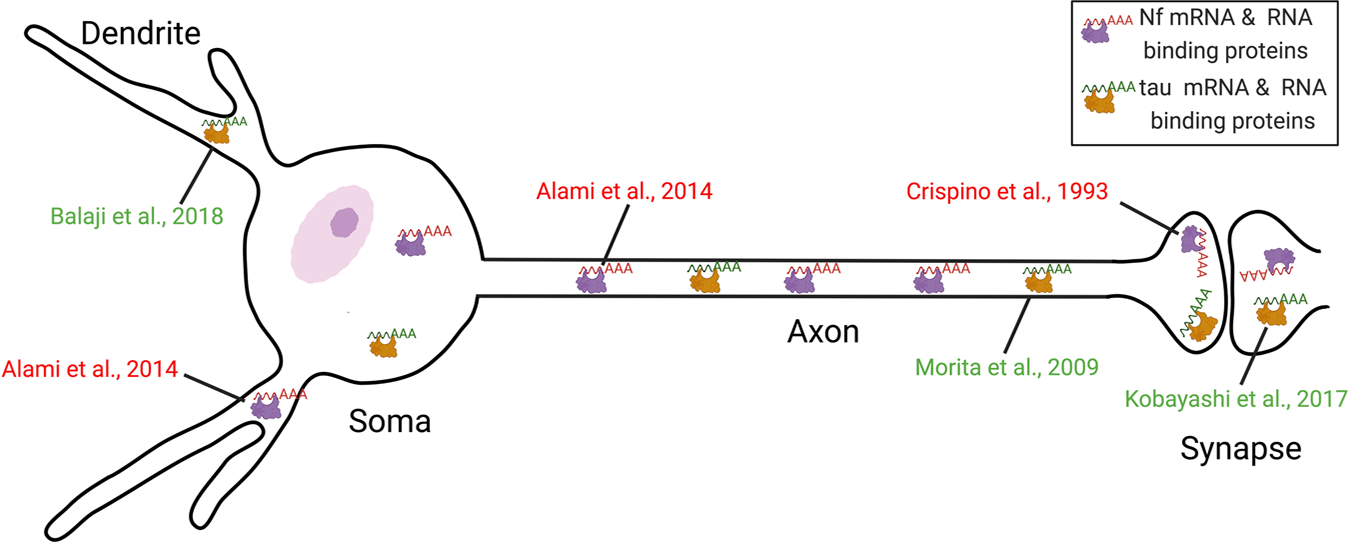
Distribution of the mRNA pools of Nf and tau in the different subcellular compartments of a neuron with references to the respective studies. mRNAs of Nf and tau can be transported into axons, dendrites and synapses in the form of transport granules composed of RNA binding proteins and RNAs.
10. Summary and future research
Protein synthesis is an essential mechanism to ensure proper function of neurons. For years, it was assumed that protein synthesis of Nf and tau occurs exclusively in the neuronal cell body (Fig. 3A–3B), from where the synthesized proteins are then transported through kinesins or dynein associated with microtubules (Fig. 3 C) to local compartments where they carry out their functions (Fig. 3D). In recent years, however, evidence has accumulated that transport of Nf and tau proteins from the soma to peripheral sub-compartments is only one way to supply them to distal neuronal processes. Another way to provide the proteins for immediate need in peripheral subcellular domains is the transport of their mRNAs in the form of transport granules associated with molecular motors or vesicles (Cioni et al., 2019) from the soma to axons, dendrites and their synaptic terminals for subsequent translation into proteins (Fig. 3E–3 F). During their transport, these transcripts are usually translationally repressed and once they reach their target, these mRNAs are translated into proteins to fulfill rapid local demand for new proteins and to provide subcellular functions such as survival of axons, synaptogenesis and experience-dependent plasticity. Despite the importance of posttranscriptional regulation of Nf and tau proteins, our current knowledge of trans-acting factors and cis-acting elements that affect Nf and tau pre-mRNA splicing, mature mRNA stability, transport, localization, and local translation in health and disease remains rather limited. Nevertheless, available evidence suggests that RBPs, including TDP43 and FUS, and miRNAs play crucial roles in posttranscriptional control of Nf and tau expression in both physiological and pathophysiological contexts. Control of Nf and tau mRNA stability, transport and local translation facilitates the temporal and spatial expression of varied NfP assemblies and tau variants in different neuronal compartments. Dysfunction or altered abundance of RBPs, such as TDP43, FUS and also miRNAs, and consequently changes in the relative levels of NfPs and tau may be involved in pathogenesis of neurodegenerative disorders. It will be of interest in the future to directly assess the half-lives of Nf and tau mRNAs and to identify additional RNA-binding proteins and miRNAs that stabilize or destabilize these mRNAs and their impact on aging and aging-related disease. It is also important to know if different Nf mRNAs are transported alone or together along axons and dendrites and how different RBPs and miRNA work together to co-ordinate the dynamic co-translation of NfPs and their assembly and interaction with tau proteins in different neuronal compartments.
Fig. 3.
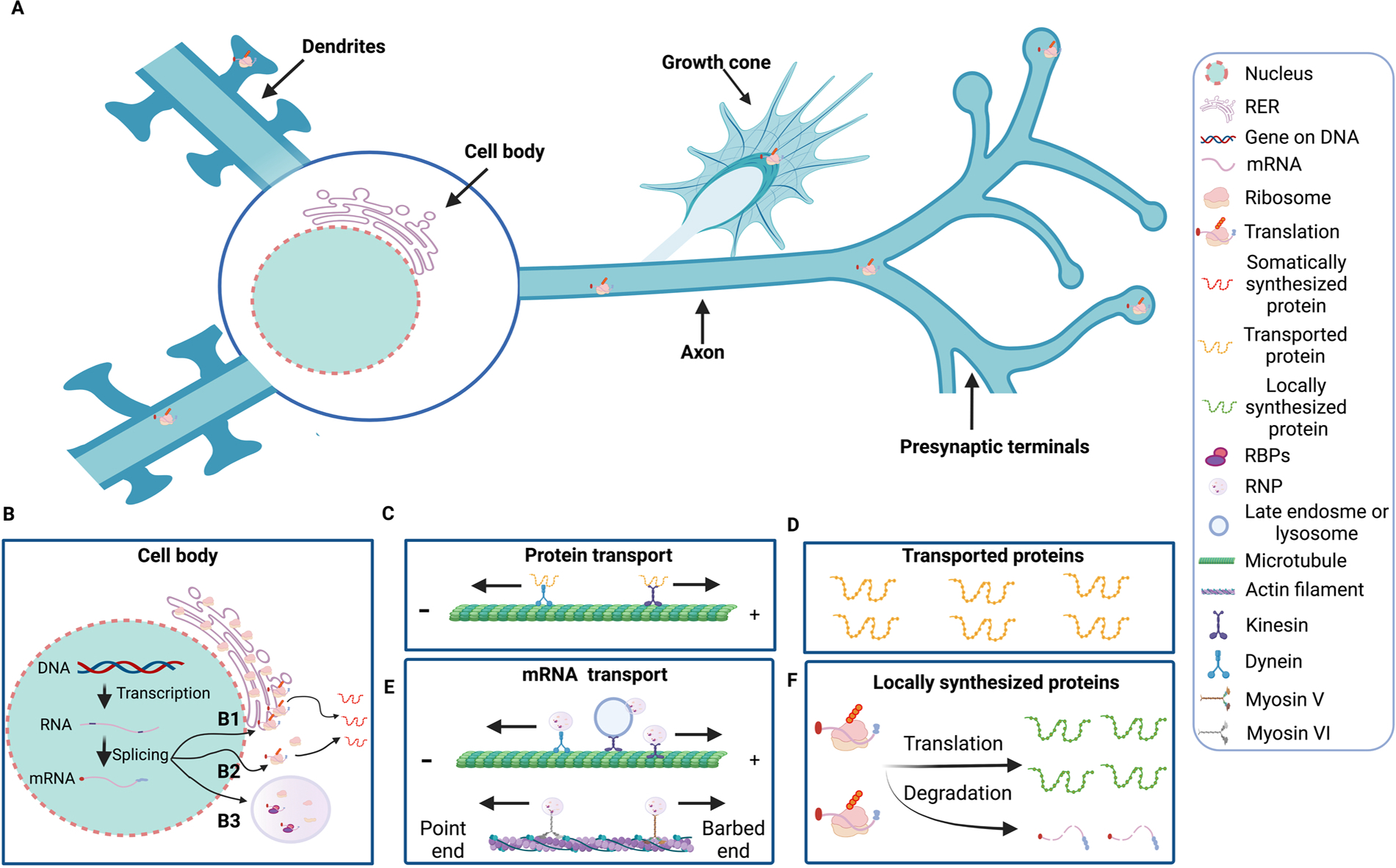
Control points of gene expression in neurons. Genes encoded in chromosomal DNA are transcribed into heterogeneous pre-mRNA, which undergo splicing, capping and addition of a poly-A tail to an RNA molecule (A). The nuclear mRNA forms complexes with ribonucleoproteins (RNPs), some of which exit from the nucleus (B). Once in the cytosol, mRNA can be translated by ribosomes associated to rough endoplasmic reticulum (RER) (B1) and free cytosolic ribosomes (B2). The lifetime of an mRNA molecule in the cytosol affects how many proteins can be made from it. Small regulatory RNAs called miRNAs can bind to target mRNAs and cause them to be cleaved. Translation starts with the binding of 40 S ribosomal subunit to cap, scanning of the 40 S subunit for start codon recognition, and then binding of the 60 S ribosomal subunit to 40 S subunit. Following these events, translation elongation, co-initiation of new rounds of translation, translation termination of each active ribosome, and lastly complete translation termination, which releases the mRNA to degradation. Synthesized proteins are transported through kinesins or dynein associated with microtubules (C) to local compartments where they carry out their functions (D). Alternatively, mRNA transcripts are associated to RNA-binding proteins (RBPs) and assembled as RNP granules (B3), which can be transported with kinesins and dynein or myosin linked to actin filaments. Late endosome or lysosomes can also act as vesicular transporters for RNA granules (E). During their transport, these transcripts are usually translationally repressed and once reach their target, these mRNAs are translated into proteins for immediate local needs (F).
Acknowledgements
Research by AY and RAN is supported by NIH P01AG017617 and R01AG062376 and The Leonard Litwin Scholar Award, New Vision Research (NVR2022-001-1).
Footnotes
Declaration of interest
The authors declare that there are no conflicts of interest.
Data Availability
No data was used for the research described in the article.
References
- Abe A, Numakura C, Saito K, Koide H, Oka N, Honma A, Kishikawa Y, Hayasaka K, 2009. Neurofilament light chain polypeptide gene mutations in Charcot-Marie-Tooth disease: nonsense mutation probably causes a recessive phenotype. J. Hum. Genet 54 (2), 94–97. 10.1038/jhg.2008.13. [DOI] [PubMed] [Google Scholar]
- Agrawal PB, Joshi M, Marinakis NS, Schmitz-Abe K, Ciarlini PD, Sargent JC, Markianos K, De Girolami U, Chad DA, Beggs AH, 2014. Expanding the phenotype associated with the NEFL mutation: neuromuscular disease in a family with overlapping myopathic and neurogenic findings. JAMA Neurol. 71 (11), 1413–1420. 10.1001/jamaneurol.2014.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, Van der Jeugd A, Blum D, Galas MC, D’Hooge R, Buee L, Balschun D, 2014. Cognition and hippocampal synaptic plasticity in mice with a homozygous tau deletion. Neurobiol. Aging 35 (11), 2474–2478. 10.1016/j.neurobiolaging.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, et al. , 2014. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 81 (3), 536–543. 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, Shaw CE, Powell JF, Leigh PN, 1999. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum. Mol. Genet 8 (2), 157–164. 10.1093/hmg/8.2.157. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2006. RNA granules. J. Cell Biol. 172 (6), 803–808. 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2008. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 33 (3), 141–150. 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Andreadis A, 2005. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys. Acta 1739 (2–3), 91–103. 10.1016/j.bbadis.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Antic D, Lu N, Keene JD, 1999. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13 (4), 449–461. 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Abreu GE, Behar L, Chung S, Furneaux H, Ginzburg I, 1999. Embryonic lethal abnormal vision-like RNA-binding proteins regulate neurite outgrowth and tau expression in PC12 cells. J. Neurosci. 19 (16), 6907–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronov S, Marx R, Ginzburg I, 1999. Identification of 3’UTR region implicated in tau mRNA stabilization in neuronal cells. J. Mol. Neurosci. 12 (2), 131–145. 10.1007/BF02736927. [DOI] [PubMed] [Google Scholar]
- Aronov S, Aranda G, Behar L, Ginzburg I, 2002. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J. Cell Sci. 115 (Pt 19), 3817–3827. 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- Atlas R, Behar L, Elliott E, Ginzburg I, 2004. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 89 (3), 613–626. 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- Atlas R, Behar L, Sapoznik S, Ginzburg I, 2007. Dynamic association with polysomes during P19 neuronal differentiation and an untranslated-region-dependent translation regulation of the tau mRNA by the tau mRNA-associated proteins IMP1, HuD, and G3BP1. J. Neurosci. Res. 85 (1), 173–183. 10.1002/jnr.21099. [DOI] [PubMed] [Google Scholar]
- Avila J, Lucas JJ, Perez M, Hernandez F, 2004. Role of tau protein in both physiological and pathological conditions. Physiol. Rev. 84 (2), 361–384. 10.1152/physrev.00024.2003. [DOI] [PubMed] [Google Scholar]
- Balaji V, Kaniyappan S, Mandelkow E, Wang Y, Mandelkow EM, 2018. Pathological missorting of endogenous MAPT/Tau in neurons caused by failure of protein degradation systems. Autophagy 14 (12), 2139–2154. 10.1080/15548627.2018.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf-Strathmann J, Benito E, May S, Arzberger T, Tahirovic S, Kretzschmar H, Fischer A, Edbauer D, 2014. MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 33 (15), 1667–1680. 10.15252/embj.201387576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudet L, Cote F, Houle D, Julien JP, 1993. Different posttranscriptional controls for the human neurofilament light and heavy genes in transgenic mice. Brain Res. Mol. Brain Res. 18 (1–2), 23–31. 10.1016/0169-328x(93)90170-t. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Nguyen MD, Julien JP, 1999. Late onset of motor neurons in mice overexpressing wild-type peripherin. J. Cell Biol. 147 (3), 531–544. 10.1083/jcb.147.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Kriz J, Julien JP, 2002. Induction of peripherin expression in subsets of brain neurons after lesion injury or cerebral ischemia. Brain Res. 946 (2), 153–161. 10.1016/s0006-8993(02)02830-5. [DOI] [PubMed] [Google Scholar]
- Behar L, Marx R, Sadot E, Barg J, Ginzburg I, 1995. cis-acting signals and trans-acting proteins are involved in tau mRNA targeting into neurites of differentiating neuronal cells. Int. J. Dev. Neurosci. 13 (2), 113–127. 10.1016/0736-5748(95)00001-w. [DOI] [PubMed] [Google Scholar]
- Behrouzi R, Liu X, Wu D, Robinson AC, Tanaguchi-Watanabe S, Rollinson S, Shi J, Tian J, Hamdalla HH, Ealing J, et al. , 2016. Pathological tau deposition in Motor Neurone Disease and frontotemporal lobar degeneration associated with TDP-43 proteinopathy. Acta Neuropathol. Commun. 4, 33. 10.1186/s40478-016-0301-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benajiba L, Le Ber I, Camuzat A, Lacoste M, Thomas-Anterion C, Couratier P, Legallic S, Salachas F, Hannequin D, Decousus M, et al. , 2009. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann. Neurol. 65 (4), 470–473. 10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- Berciano J, Peeters K, Garcia A, Lopez-Alburquerque T, Gallardo E, Hernandez-Fabian A, Pelayo-Negro AL, De Vriendt E, Infante J, Jordanova A, 2016. NEFL N98S mutation: another cause of dominant intermediate Charcot-Marie-Tooth disease with heterogeneous early-onset phenotype. J. Neurol. 263 (2), 361–369. 10.1007/s00415-015-7985-z. [DOI] [PubMed] [Google Scholar]
- Biundo F, Del Prete D, Zhang H, Arancio O, D’Adamio L, 2018. A role for tau in learning, memory and synaptic plasticity. Sci. Rep. 8 (1), 3184. 10.1038/s41598-018-21596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsdottir G, Ivarsdottir EV, Bjarnadottir K, Benonisdottir S, Gylfadottir SS, Arnadottir GA, Benediktsson R, Halldorsson GH, Helgadottir A, Jonasdottir A, et al. , 2019. A PRPH splice-donor variant associates with reduced sural nerve amplitude and risk of peripheral neuropathy. Nat. Commun. 10 (1), 1777. 10.1038/s41467-019-09719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet A, Berges R, Frank R, Robert P, Peterson AC, Eyer J, 2009. Neurofilaments bind tubulin and modulate its polymerization. J. Neurosci. 29 (35), 11043–11054. 10.1523/JNEUROSCI.1924-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni B, Bonvicini C, Alberici A, Buratti E, Agosti C, Archetti S, Papetti A, Stuani C, Di Luca M, Gennarelli M, et al. , 2009. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 30 (11), E974–E983. 10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- Broustal O, Camuzat A, Guillot-Noel L, Guy N, Millecamps S, Deffond D, Lacomblez L, Golfier V, Hannequin D, Salachas F, et al. , 2010. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J. Alzheimers Dis. 22 (3), 765–769. [PubMed] [Google Scholar]
- Bruch J, Xu H, De Andrade A, Hoglinger G, 2014. Mitochondrial complex 1 inhibition increases 4-repeat isoform tau by SRSF2 upregulation. PLoS One 9 (11), e113070. 10.1371/journal.pone.0113070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Zhukareva V, Uryu K, Zhang B, Bigio E, Mackenzie IR, Gearing M, Duyckaerts C, Yokoo H, Nakazato Y, et al. , 2004. alpha-internexin is present in the pathological inclusions of neuronal intermediate filament inclusion disease. Am. J. Pathol. 164 (6), 2153–2161. 10.1016/s0002-9440(10)63773-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Melo D, Hawley ZCE, Strong MJ, 2018. Dysregulation of human NEFM and NEFH mRNA stability by ALS-linked miRNAs. Mol. Brain 11 (1), 43. 10.1186/s13041-018-0386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Melo D, Droppelmann CA, He Z, Volkening K, Strong MJ, 2013. Altered microRNA expression profile in Amyotrophic Lateral Sclerosis: a role in the regulation of NFL mRNA levels. Mol. Brain 6, 26. 10.1186/1756-6606-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canete-Soler R, Schlaepfer WW, 2000. Similar poly(C)-sensitive RNA-binding complexes regulate the stability of the heavy and light neurofilament mRNAs. Brain Res. 867 (1–2), 265–279. 10.1016/s0006-8993(00)02389-1. [DOI] [PubMed] [Google Scholar]
- Canete-Soler R, Schwartz ML, Hua Y, Schlaepfer WW, 1998a. Characterization of ribonucleoprotein complexes and their binding sites on the neurofilament light subunit mRNA. J. Biol. Chem. 273 (20), 12655–12661. 10.1074/jbc.273.20.12655. [DOI] [PubMed] [Google Scholar]
- Canete-Soler R, Schwartz ML, Hua Y, Schlaepfer WW, 1998b. Stability determinants are localized to the 3’-untranslated region and 3’-coding region of the neurofilament light subunit mRNA using a tetracycline-inducible promoter. J. Biol. Chem. 273 (20), 12650–12654. 10.1074/jbc.273.20.12650. [DOI] [PubMed] [Google Scholar]
- Canete-Soler R, Silberg DG, Gershon MD, Schlaepfer WW, 1999. Mutation in neurofilament transgene implicates RNA processing in the pathogenesis of neurodegenerative disease. J. Neurosci. 19 (4), 1273–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canete-Soler R, Reddy KS, Tolan DR, Zhai J, 2005. Aldolases a and C are ribonucleolytic components of a neuronal complex that regulates the stability of the light-neurofilament mRNA. J. Neurosci. 25 (17), 4353–4364. 10.1523/JNEUROSCI.0885-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canete-Soler R, Wu J, Zhai J, Shamim M, Schlaepfer WW, 2001. p190RhoGEF Binds to a destabilizing element in the 3’ untranslated region of light neurofilament subunit mRNA and alters the stability of the transcript. J. Biol. Chem. 276 (34), 32046–32050. 10.1074/jbc.M104104200. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH, 2006. Noncoding RNAs in the mammalian central nervous system. Ann. Rev. Neurosci. 29, 77–103. 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Chang L, Shav-Tal Y, Trcek T, Singer RH, Goldman RD, 2006. Assembling an intermediate filament network by dynamic cotranslation. J. Cell Biol. 172 (5), 747–758. 10.1083/jcb.200511033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Qian K, Du Z, Cao J, Petersen A, Liu H, Blackbourn L.Wt, Huang CL, Errigo A, Yin Y, et al. , 2014. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 14 (6), 796–809. 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, Dierick I, Abel A, Kennerson ML, Rabin BA, et al. , 2004. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am. J. Hum. Genet. 74 (6), 1128–1135. 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Zhang C, Xu C, Wang L, Zou X, Chen G, 2014. Age-dependent neuron loss is associated with impaired adult neurogenesis in forebrain neuron-specific Dicer conditional knockout mice. Int J. Biochem Cell Biol. 57, 186–196. 10.1016/j.biocel.2014.10.029. [DOI] [PubMed] [Google Scholar]
- Ching GY, Chien CL, Flores R, Liem RK, 1999. Overexpression of alpha-internexin causes abnormal neurofilamentous accumulations and motor coordination deficits in transgenic mice. J. Neurosci. 19 (8), 2974–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JF, Majumder P, Chatterjee B, Huang SL, Shen CJ, 2019. TDP-43 regulates coupled dendritic mRNA transport-translation processes in co-operation with FMRP and staufen1. Cell Rep. 29 (10), 3118–3133. 10.1016/j.celrep.2019.10.061. [DOI] [PubMed] [Google Scholar]
- Cioni JM, Lin JQ, Holtermann AV, Koppers M, Jakobs MAH, Azizi A, Turner-Bridger B, Shigeoka T, Franze K, Harris WA, et al. , 2019. Late endosomes Act as mRNA translation platforms and sustain mitochondria in axons. Cell 176 (1–2), 56–72. 10.1016/j.cell.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. , 2012. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum. Mol. Genet 21 (13), 2899–2911. 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino M, Capano CP, Kaplan BB, Giuditta A, 1993. Neurofilament proteins are synthesized in nerve endings from squid brain. J. Neurochem 61 (3), 1144–1146. 10.1111/j.1471-4159.1993.tb03632.x. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Garcia-Fernandez LF, Imai T, Okano H, Munoz A, 2002. Regulation of tau RNA maturation by thyroid hormone is mediated by the neural RNA-binding protein musashi-1. Mol. Cell Neurosci. 20 (2), 198–210. 10.1006/mcne.2002.1131. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. , 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72 (2), 245–256. 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demy DL, Campanari ML, Munoz-Ruiz R, Durham HD, Gentil BJ, Kabashi E, 2020. Functional characterization of neurofilament light splicing and misbalance in zebrafish. Cells 9 (5). 10.3390/cells9051238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djabali K, Zissopoulou A, de Hoop MJ, Georgatos SD, Dotti CG, 1993. Peripherin expression in hippocampal neurons induced by muscle soluble factor(s). J. Cell Biol. 123 (5), 1197–1206. 10.1083/jcb.123.5.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droppelmann CA, Keller BA, Campos-Melo D, Volkening K, Strong MJ, 2013a. Rho guanine nucleotide exchange factor is an NFL mRNA destabilizing factor that forms cytoplasmic inclusions in amyotrophic lateral sclerosis. Neurobiol. Aging 34 (1), 248–262. 10.1016/j.neurobiolaging.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Droppelmann CA, Wang J, Campos-Melo D, Keller B, Volkening K, Hegele RA, Strong MJ, 2013b. Detection of a novel frameshift mutation and regions with homozygosis within ARHGEF28 gene in familial amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Front. Degener. 14 (5–6), 444–451. 10.3109/21678421.2012.758288. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD, 2005. microPrimer: the biogenesis and function of microRNA. Development 132 (21), 4645–4652. 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. , 2010. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466 (7310), 1069–1075. 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es MA, Diekstra FP, Veldink JH, Baas F, Bourque PR, Schelhaas HJ, Strengman E, Hennekam EA, Lindhout D, Ophoff RA, et al. , 2009. A case of ALS-FTD in a large FALS pedigree with a K17I ANG mutation. Neurology 72 (3), 287–288. 10.1212/01.wnl.0000339487.84908.00. [DOI] [PubMed] [Google Scholar]
- Fischer I, Baas PW, 2020. Resurrecting the mysteries of Big Tau. Trends Neurosci. 43 (7), 493–504. 10.1016/j.tins.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Yuan Y, 2018. A novel homozygous nonsense mutation in NEFL causes autosomal recessive Charcot-Marie-Tooth disease. Neuromuscul. Disord. 28 (1), 44–47. 10.1016/j.nmd.2017.09.018. [DOI] [PubMed] [Google Scholar]
- Garcia-Escudero V, Ruiz-Gabarre D, Gargini R, Perez M, Garcia E, Cuadros R, Hernandez IH, Cabrera JR, Garcia-Escudero R, Lucas JJ, et al. , 2021. A new non-aggregative splicing isoform of human Tau is decreased in Alzheimer’s disease. Acta Neuropathol. 142 (1), 159–177. 10.1007/s00401-021-02317-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Wu J, Zhai J, Nie Z, Lin H, Schlaepfer WW, Canete-Soler R, 2002. Binding of p190RhoGEF to a destabilizing element on the light neurofilament mRNA is competed by BC1 RNA. J. Biol. Chem. 277 (45), 42701–42705. 10.1074/jbc.M206635200. [DOI] [PubMed] [Google Scholar]
- Ge WW, Wen W, Strong W, Leystra-Lantz C, Strong MJ, 2005. Mutant copper-zinc superoxide dismutase binds to and destabilizes human low molecular weight neurofilament mRNA. J. Biol. Chem. 280 (1), 118–124. 10.1074/jbc.M405065200. [DOI] [PubMed] [Google Scholar]
- Ge WW, Volkening K, Leystra-Lantz C, Jaffe H, Strong MJ, 2007. 14–3-3 protein binds to the low molecular weight neurofilament (NFL) mRNA 3’ UTR. Mol. Cell Neurosci. 10.1016/j.mcn.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Schwarzl T, Valcarcel J, Hentze MW, 2021. RNA-binding proteins in human genetic disease. Nat. Rev. Genet 22 (3), 185–198. 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- Gebert LFR, MacRae IJ, 2019. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 20 (1), 21–37. 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JE, Yen SH, Chiu FC, Peress NS, 1983. Lewy bodies of Parkinson’s disease contain neurofilament antigens. Science 221 (4615), 1082–1084. 10.1126/science.6308771. [DOI] [PubMed] [Google Scholar]
- Goldstein ME, Weiss SR, Lazzarini RA, Shneidman PS, Lees JF, Schlaepfer WW, 1988. mRNA levels of all three neurofilament proteins decline following nerve transection. Brain Res. 427 (3), 287–291. 10.1016/0169-328x(88)90051-4. [DOI] [PubMed] [Google Scholar]
- Gros-Louis F, Lariviere R, Gowing G, Laurent S, Camu W, Bouchard JP, Meininger V, Rouleau GA, Julien JP, 2004. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J. Biol. Chem. 279 (44), 45951–45956. 10.1074/jbc.M408139200. [DOI] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI, 1986. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA 83 (13), 4913–4917. 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Wu F, Xu W, Shi J, Hu W, Jin N, Qian W, Wang X, Iqbal K, Gong CX, et al. , 2017. TDP-43 suppresses tau expression via promoting its mRNA instability. Nucleic Acids Res 45 (10), 6177–6193. 10.1093/nar/gkx175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardiman O, Al-Chalabi A, Chio A, Corr EM, Logroscino G, Robberecht W, Shaw PJ, Simmons Z, van den Berg LH, 2017. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Prim. 3, 17085. 10.1038/nrdp.2017.85. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Sergeant N, Buee L, 2012. MicroRNAs and the regulation of tau metabolism. Int. J. Alzheimers Dis. 2012, 406561 10.1155/2012/406561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buee L, De Strooper B, 2010. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 19 (20), 3959–3969. 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- Hentze MW, 1991. Determinants and regulation of cytoplasmic mRNA stability in eukaryotic cells. Biochim Biophys. Acta 1090 (3), 281–292. 10.1016/0167-4781(91)90191-n. [DOI] [PubMed] [Google Scholar]
- Hoffman PN, Cleveland DW, Griffin JW, Landes PW, Cowan NJ, Price DL, 1987. Neurofilament gene expression: a major determinant of axonal caliber. Proc. Natl. Acad. Sci. USA 84 (10), 3472–3476. 10.1073/pnas.84.10.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holper S, Watson R, Yassi N, 2022. Tau as a Biomarker of Neurodegeneration. Int. J. Mol. Sci. 23 (13) 10.3390/ijms23137307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Martin KC, Schuman EM, 2019. Local translation in neurons: visualization and function. Nat. Struct. Mol. Biol. 26 (7), 557–566. 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- Hutchins EJ, Belrose JL, Szaro BG, 2015. Phosphorylation of heterogeneous nuclear ribonucleoprotein K at an extracellular signal-regulated kinase phosphorylation site promotes neurofilament-medium protein expression and axon outgrowth in Xenopus. Neurosci. Lett. 607, 59–65. 10.1016/j.neulet.2015.09.027. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, et al. , 1998. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature 393 (6686), 702–705. 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Ikegami S, Harada A, Hirokawa N, 2000. Muscle weakness, hyperactivity, and impairment in fear conditioning in tau-deficient mice. Neurosci. Lett. 279 (3), 129–132. 10.1016/s0304-3940(99)00964-7. [DOI] [PubMed] [Google Scholar]
- Ikenaka K, Nakahira K, Takayama C, Wada K, Hatanaka H, Mikoshiba K, 1990. Nerve growth factor rapidly induces expression of the 68-kDa neurofilament gene by posttranscriptional modification in PC12h-R cells. J. Biol. Chem. 265 (32), 19782–19785. [PubMed] [Google Scholar]
- Ishihara T, Hong M, Zhang B, Nakagawa Y, Lee MK, Trojanowski JQ, Lee VM, 1999. Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron 24 (3), 751–762. 10.1016/s0896-6273(00)81127-7. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Higuchi M, Zhang B, Yoshiyama Y, Hong M, Trojanowski JQ, Lee VM, 2001. Attenuated neurodegenerative disease phenotype in tau transgenic mouse lacking neurofilaments. J. Neurosci. 21 (16), 6026–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizawa T, Mattila P, Davies P, Wang D, Dickson DW, 2003. Colocalization of tau and alpha-synuclein epitopes in Lewy bodies. J. Neuropathol. Exp. Neurol. 62 (4), 389–397. 10.1093/jnen/62.4.389. [DOI] [PubMed] [Google Scholar]
- Ishtiaq M, Campos-Melo D, Volkening K, Strong MJ, 2014. Analysis of novel NEFL mRNA targeting microRNAs in amyotrophic lateral sclerosis. PLoS One 9 (1), e85653. 10.1371/journal.pone.0085653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, Wolfing H, Chieng BC, Christie MJ, Napier IA, et al. , 2010. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer’s disease mouse models. Cell 142 (3), 387–397. 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Jawaid A, Woldemichael BT, Kremer EA, Laferriere F, Gaur N, Afroz T, Polymenidou M, Mansuy IM, 2019. Memory Decline and Its Reversal in Aging and Neurodegeneration Involve miR-183/96/182 Biogenesis. Mol. Neurobiol. 56 (5), 3451–3462. 10.1007/s12035-018-1314-3. [DOI] [PubMed] [Google Scholar]
- Jiang P, Coller H, 2012. Functional interactions between microRNAs and RNA binding proteins. Microrna 1 (1), 70–79. 10.2174/2211536611201010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, Pliner HA, Abramzon Y, Marangi G, Winborn BJ, et al. , 2014. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat. Neurosci. 17 (5), 664–666. 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova A, De Jonghe P, Boerkoel CF, Takashima H, De Vriendt E, Ceuterick C, Martin JJ, Butler IJ, Mancias P, Papasozomenos S, et al. , 2003. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain 126 (Pt 3), 590–597. 10.1093/brain/awg059. [DOI] [PubMed] [Google Scholar]
- Juzwik CA, S SD, Zhang Y, Paradis-Isler N, Sylvester A, Amar-Zifkin A, Douglas C, Morquette B, Moore CS, Fournier AE, 2019. microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog. Neurobiol. 182, 101664 10.1016/j.pneurobio.2019.101664. [DOI] [PubMed] [Google Scholar]
- Kanazawa T, Uchihara T, Takahashi A, Nakamura A, Orimo S, Mizusawa H, 2008. Three-layered structure shared between Lewy bodies and lewy neurites-three-dimensional reconstruction of triple-labeled sections. Brain Pathol. 18 (3), 415–422. 10.1111/j.1750-3639.2008.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann AW, Belzil VV, Jao LE, Leblond CS, Girard SL, Daoud H, Noreau A, Rochefort D, Hince P, et al. , 2015. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum. Mol. Genet 24 (5), 1363–1373. 10.1093/hmg/ddu545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikari TK, Pascoal TA, Ashton NJ, Janelidze S, Benedet AL, Rodriguez JL, Chamoun M, Savard M, Kang MS, Therriault J, et al. , 2020. Blood phosphorylated tau 181 as a biomarker for Alzheimer’s disease: a diagnostic performance and prediction modelling study using data from four prospective cohorts. Lancet Neurol. 19 (5), 422–433. 10.1016/S1474-4422(20)30071-5. [DOI] [PubMed] [Google Scholar]
- Katz JS, Katzberg HD, Woolley SC, Marklund SL, Andersen PM, 2012. Combined fulminant frontotemporal dementia and amyotrophic lateral sclerosis associated with an I113T SOD1 mutation. Amyotroph. Lateral Scler. 13 (6), 567–569. 10.3109/17482968.2012.678365. [DOI] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ, 2006. Neuronal RNA granules: movers and makers. Neuron 51 (6), 685–690. 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. , 2013. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495 (7442), 467–473. 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A, 2007. A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317 (5842), 1220–1224. 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Whitcomb DJ, Jo J, Regan P, Piers T, Heo S, Brown C, Hashikawa T, Murayama M, Seok H, et al. , 2014. Microtubule-associated protein tau is essential for long-term depression in the hippocampus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369 (1633), 20130144. 10.1098/rstb.2013.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita C, Kubota N, Aoyama K, 2021. Interplay of RNA-binding proteins and microRNAs in neurodegenerative diseases. Int J. Mol. Sci. 22 (10) 10.3390/ijms22105292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Tanaka T, Soeda Y, Almeida OFX, Takashima A, 2017. Local somatodendritic translation and hyperphosphorylation of tau protein triggered by AMPA and NMDA receptor stimulation. EBioMedicine 20, 120–126. 10.1016/j.ebiom.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren SA, Hamm MJ, Meier SE, Weiss BE, Nation GK, Chishti EA, Arango JP, Chen J, Zhu H, Blalock EM, et al. , 2019. Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 137 (4), 571–583. 10.1007/s00401-019-01970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriz J, Zhu Q, Julien JP, Padjen AL, 2000. Electrophysiological properties of axons in mice lacking neurofilament subunit genes: disparity between conduction velocity and axon diameter in absence of NF-H. Brain Res. 885 (1), 32–44. 10.1016/s0006-8993(00)02899-7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Phaneuf D, Cordeau P Jr., Boutej H, Kriz J, Julien JP, 2021. Induction of autophagy mitigates TDP-43 pathology and translational repression of neurofilament mRNAs in mouse models of ALS/FTD. Mol. Neurodegener. 16 (1), 1. 10.1186/s13024-020-00420-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. , 2012. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15 (11), 1488–1497. 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landon F, Wolff A, de Nechaud B, 2000. Mouse peripherin isoforms. Biol. Cell 92 (6), 397–407. 10.1016/s0248-4900(00)01099-6. [DOI] [PubMed] [Google Scholar]
- Landon F, Lemonnier M, Benarous R, Huc C, Fiszman M, Gros F, Portier MM, 1989. Multiple mRNAs encode peripherin, a neuronal intermediate filament protein. EMBO J. 8 (6), 1719–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Bossers K, Janky R, Salta E, Frigerio CS, Barbash S, Rothman R, Sierksma AS, Thathiah A, Greenberg D, et al. , 2013. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 5 (10), 1613–1634. 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavedan C, Buchholtz S, Nussbaum RL, Albin RL, Polymeropoulos MH, 2002. A mutation in the human neurofilament M gene in Parkinson’s disease that suggests a role for the cytoskeleton in neuronal degeneration. Neurosci. Lett. 322 (1), 57–61. 10.1016/s0304-3940(01)02513-7. [DOI] [PubMed] [Google Scholar]
- Lecomte MJ, Basseville M, Landon F, Karpov V, Fauquet M, 1998. Transcriptional activation of the mouse peripherin gene by leukemia inhibitory factor: involvement of STAT proteins. J. Neurochem 70 (3), 971–982. [PubMed] [Google Scholar]
- Lee SK, Hollenbeck PJ, 2003. Organization and translation of mRNA in sympathetic axons. J. Cell Sci. 116 (Pt 21), 4467–4478. 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- Leonard DG, Gorham JD, Cole P, Greene LA, Ziff EB, 1988. A nerve growth factor-regulated messenger RNA encodes a new intermediate filament protein. J. Cell Biol. 106 (1), 181–193. 10.1083/jcb.106.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CL, He CZ, Kaufmann P, Chin SS, Naini A, Liem RK, Mitsumoto H, Hays AP, 2004. A pathogenic peripherin gene mutation in a patient with amyotrophic lateral sclerosis. Brain Pathol. 14 (3), 290–296. 10.1111/j.1750-3639.2004.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhai J, Schlaepfer WW, 2005. RNA-binding protein is involved in aggregation of light neurofilament protein and is implicated in the pathogenesis of motor neuron degeneration. Hum. Mol. Genet. 14 (23), 3643–3659. 10.1093/hmg/ddi392. [DOI] [PubMed] [Google Scholar]
- Lin H, Zhai J, Canete-Soler R, Schlaepfer WW, 2004. 3’ untranslated region in a light neurofilament (NF-L) mRNA triggers aggregation of NF-L and mutant superoxide dismutase 1 proteins in neuronal cells. J. Neurosci. 24 (11), 2716–2726. 10.1523/JNEUROSCI.5689-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhai J, Nie Z, Wu J, Meinkoth JL, Schlaepfer WW, Canete-Soler R, 2003. Neurofilament RNA causes neurodegeneration with accumulation of ubiquitinated aggregates in cultured motor neurons. J. Neuropathol. Exp. Neurol. 62 (9), 936–950. 10.1093/jnen/62.9.936. [DOI] [PubMed] [Google Scholar]
- Lindenbaum MH, Carbonetto S, Grosveld F, Flavell D, Mushynski WE, 1988. Transcriptional and post-transcriptional effects of nerve growth factor on expression of the three neurofilament subunits in PC-12 cells. J. Biol. Chem. 263 (12), 5662–5667. [PubMed] [Google Scholar]
- Liu Y, Szaro BG, 2011. hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development 138 (14), 3079–3090. 10.1242/dev.066993. [DOI] [PubMed] [Google Scholar]
- Liu Y, Gervasi C, Szaro BG, 2008. A crucial role for hnRNP K in axon. Dev. Xenopus Laevis. Dev. 135 (18), 3125–3135. 10.1242/dev.022236. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yu H, Deaton SK, Szaro BG, 2012. Heterogeneous nuclear ribonucleoprotein K, an RNA-binding protein, is required for optic axon regeneration in Xenopus laevis. J. Neurosci. 32 (10), 3563–3574. 10.1523/JNEUROSCI.5197-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffreda A, Rigamonti A, Barabino SM, Lenzken SC, 2015. RNA-binding proteins in the regulation of miRNA activity: a focus on neuronal functions. Biomolecules 5 (4), 2363–2387. 10.3390/biom5042363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. , 2017. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron 95 (4), 808–816. 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Scott WK, Nance MA, Watts RL, Hubble JP, Koller WC, Lyons K, Pahwa R, Stern MB, Colcher A, et al. , 2001. Association of single-nucleotide polymorphisms of the tau gene with late-onset Parkinson disease. JAMA 286 (18), 2245–2250. 10.1001/jama.286.18.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziuk BF, Apicco DJ, Cruz AL, Jiang L, Ash PEA, da Rocha EL, Zhang C, Yu WH, Leszyk J, Abisambra JF, et al. , 2018. RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol. Commun. 6 (1), 71. 10.1186/s40478-018-0574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw TS, Mickle JP, Shaw G, Streit WJ, 2002. Axonally transported peripheral signals regulate alpha-internexin expression in regenerating motoneurons. J. Neurosci. 22 (12), 4955–4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier S, Bell M, Lyons DN, Rodriguez-Rivera J, Ingram A, Fontaine SN, Mechas E, Chen J, Wolozin B, LeVine H 3rd, et al. , 2016. Pathological tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J. Neurosci. 36 (3), 1001–1007. 10.1523/JNEUROSCI.3029-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneses A, Koga S, O’Leary J, Dickson DW, Bu G, Zhao N, 2021. TDP-43 pathology in Alzheimer’s disease. Mol. Neurodegener. 16 (1), 84. 10.1186/s13024-021-00503-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Grierson AJ, Cookson MR, Heath PR, Tomkins J, Figlewicz DA, Ince PG, Shaw PJ, 2002. Selective loss of neurofilament expression in Cu/Zn superoxide dismutase (SOD1) linked amyotrophic lateral sclerosis. J. Neurochem 82 (5), 1118–1128. 10.1046/j.1471-4159.2002.01045.x. [DOI] [PubMed] [Google Scholar]
- Mercken M, Fischer I, Kosik KS, Nixon RA, 1995. Three distinct axonal transport rates for tau, tubulin, and other microtubule-associated proteins: evidence for dynamic interactions of tau with microtubules in vivo. J. Neurosci. 15 (12), 8259–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersiyanova IV, Perepelov AV, Polyakov AV, Sitnikov VF, Dadali EL, Oparin RB, Petrin AN, Evgrafov OV, 2000. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am. J. Hum. Genet 67 (1), 37–46. 10.1086/302962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y, Hoshi M, Nishida E, Minami Y, Sakai H, 1986. Binding of microtubule-associated protein 2 and tau to the intermediate filament reassembled from neurofilament 70-kDa subunit protein. Its regulation by calmodulin. J. Biol. Chem. 261 (28), 13026–13030. [PubMed] [Google Scholar]
- Morita T, Sobue K, 2009. Specification of neuronal polarity regulated by local translation of CRMP2 and Tau via the mTOR-p70S6K pathway. J. Biol. Chem. 284 (40), 27734–27745. 10.1074/jbc.M109.008177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschner K, Sundermann F, Meyer H, da Graca AP, Appel N, Paululat A, Bakota L, Brandt R, 2014. RNA protein granules modulate tau isoform expression and induce neuronal sprouting. J. Biol. Chem. 289 (24), 16814–16825. 10.1074/jbc.M113.541425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz PF, Oblinger MM, 1994. Mechanisms regulating the stability of cytoskeletal mRNAs in regerating rat sensory neurons. Soc. Neurosci. Abstr. 24, 1294. [Google Scholar]
- Moskowitz PF, Oblinger MM, 1995. Transcriptional and post-transcriptional mechanisms regulating neurofilament and tubulin gene expression during normal development of the rat brain. Brain Res. Mol. Brain Res. 30 (2), 211–222. 10.1016/0169-328x(95)00006-e. [DOI] [PubMed] [Google Scholar]
- Moszczynski AJ, Harvey M, Fulcher N, de Oliveira C, McCunn P, Donison N, Bartha R, Schmid S, Strong MJ, Volkening K, 2019. Synergistic toxicity in an in vivo model of neurodegeneration through the co-expression of human TDP-43 (M337V) and tau(T175D) protein. Acta Neuropathol. Commun. 7 (1), 170. 10.1186/s40478-019-0816-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RL, Combs B, Alhadidy MM, Brady ST, Morfini GA, Kanaan NM, 2021. Tau: a signaling hub protein. Front Mol. Neurosci. 14, 647054 10.3389/fnmol.2021.647054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mul K, Schouten MI, van der Looij E, Dooijes D, Hennekam FAM, Notermans NC, Praamstra P, van Gaalen J, Kamsteeg EJ, Verbeek NE, et al. , 2020. A hereditary spastic paraplegia predominant phenotype caused by variants in the NEFL gene. Park. Relat. Disord. 80, 98–101. 10.1016/j.parkreldis.2020.09.016. [DOI] [PubMed] [Google Scholar]
- Murrell JR, Spillantini MG, Zolo P, Guazzelli M, Smith MJ, Hasegawa M, Redi F, Crowther RA, Pietrini P, Ghetti B, et al. , 1999. Tau gene mutation G389R causes a tauopathy with abundant pick body-like inclusions and axonal deposits. J. Neuropathol. Exp. Neurol. 58 (12), 1207–1226. 10.1097/00005072-199912000-00002. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Ikenaka K, Wada K, Tamura T, Furuichi T, Mikoshiba K, 1990. Structure of the 68-kDa neurofilament gene and regulation of its expression. J. Biol. Chem. 265 (32), 19786–19791. [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, et al. , 2014. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46 (9), 989–993. 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Wu J, Zhai J, Lin H, Ge W, Schlaepfer WW, Canete-Soler R, 2002. Untranslated element in neurofilament mRNA has neuropathic effect on motor neurons of transgenic mice. J. Neurosci. 22 (17), 7662–7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oblinger MM, Wong J, Parysek LM, 1989. Axotomy-induced changes in the expression of a type III neuronal intermediate filament gene. J. Neurosci. 9 (11), 3766–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara O, Gahara Y, Miyake T, Teraoka H, Kitamura T, 1993. Neurofilament deficiency in quail caused by nonsense mutation in neurofilament-L gene. J. Cell Biol. 121 (2), 387–395. 10.1083/jcb.121.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco D, Tahirovic S, Rentzsch K, Schwenk BM, Haass C, Edbauer D, 2012. Loss of fused in sarcoma (FUS) promotes pathological Tau splicing. EMBO Rep. 13 (8), 759–764. 10.1038/embor.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachter JS, Liem RK, 1984. The differential appearance of neurofilament triplet polypeptides in the developing rat optic nerve. Dev. Biol. 103 (1), 200–210. 10.1016/0012-1606(84)90021-6. [DOI] [PubMed] [Google Scholar]
- Pachter JS, Liem RK, 1985. alpha-Internexin, a 66-kD intermediate filament-binding protein from mammalian central nervous tissues. J. Cell Biol. 101 (4), 1316–1322. 10.1083/jcb.101.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ, 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet 40 (12), 1413–1415. 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Papanikolopoulou K, Roussou IG, Gouzi JY, Samiotaki M, Panayotou G, Turin L, Skoulakis EMC, 2019. Drosophila tau negatively regulates translation and olfactory long-term memory, but facilitates footshock habituation and cytoskeletal homeostasis. J. Neurosci. 39 (42), 8315–8329. 10.1523/JNEUROSCI.0391-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parysek LM, Goldman RD, 1988. Distribution of a novel 57 kDa intermediate filament (IF) protein in the nervous system. J. Neurosci. 8 (2), 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor P, Pastor E, Carnero C, Vela R, Garcia T, Amer G, Tolosa E, Oliva R, 2001. Familial atypical progressive supranuclear palsy associated with homozigosity for the delN296 mutation in the tau gene. Ann. Neurol. 49 (2), 263–267. . [DOI] [PubMed] [Google Scholar]
- Perrot R, Julien JP, 2009. Real-time imaging reveals defects of fast axonal transport induced by disorganization of intermediate filaments. FASEB J. 23 (9), 3213–3225. 10.1096/fj.09-129585. [DOI] [PubMed] [Google Scholar]
- Petrozziello T, Amaral AC, Dujardin S, Farhan SMK, Chan J, Trombetta BA, Kivisakk P, Mills AN, Bordt EA, Kim SE, et al. , 2022. Novel genetic variants in MAPT and alterations in tau phosphorylation in amyotrophic lateral sclerosis postmortem motor cortex and cerebrospinal fluid. Brain Pathol. 32 (2), e13035 10.1111/bpa.13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorkaj P, Muma NA, Zhukareva V, Cochran EJ, Shannon KM, Hurtig H, Koller WC, Bird TD, Trojanowski JQ, Lee VM, et al. , 2002. An R5L tau mutation in a subject with a progressive supranuclear palsy phenotype. Ann. Neurol. 52 (4), 511–516. 10.1002/ana.10340. [DOI] [PubMed] [Google Scholar]
- Portier MM, de Nechaud B, Gros F, 1983b. Peripherin, a new member of the intermediate filament protein family. Dev. Neurosci. 6 (6), 335–344. 10.1159/000112360. [DOI] [PubMed] [Google Scholar]
- Portier MM, Brachet P, Croizat B, Gros F, 1983a. Regulation of peripherin in mouse neuroblastoma and rat PC 12 pheochromocytoma cell lines. Dev. Neurosci. 6 (4–5), 215–226. 10.1159/000112348. [DOI] [PubMed] [Google Scholar]
- Qian W, Iqbal K, Grundke-Iqbal I, Gong CX, Liu F, 2011. Splicing factor SC35 promotes tau expression through stabilization of its mRNA. FEBS Lett. 585 (6), 875–880. 10.1016/j.febslet.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Mohan PS, Kumar A, Yuan A, Montagna L, Campbell J, Veeranna, Espreafico EM, Julien JP, Nixon RA, 2011. The myosin Va head domain binds to the neurofilament-L rod and modulates endoplasmic reticulum (ER) content and distribution within axons. PLoS One 6 (2), e17087. 10.1371/journal.pone.0017087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AP, Abrams AJ, Cottenie E, Horga A, Gonzalez M, Bis DM, Sanchez-Mejias A, Pinto M, Buglo E, Markel K, et al. , 2016. Cryptic amyloidogenic elements in the 3’ utrs of neurofilament genes trigger axonal neuropathy. Am. J. Hum. Genet 98 (4), 597–614. 10.1016/j.ajhg.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J, Doroudchi MM, Nguyen MD, Durham HD, Strong MJ, Shaw G, Julien JP, Mushynski WE, 2003. A neurotoxic peripherin splice variant in a mouse model of ALS. J. Cell Biol. 160 (6), 939–949. 10.1083/jcb.200205027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrabhatla P, Jaffe H, Pant HC, 2011. Direct evidence of phosphorylated neuronal intermediate filament proteins in neurofibrillary tangles (NFTs): phosphoproteomics of Alzheimer’s NFTs. FASEB J. 25 (11), 3896–3905. 10.1096/fj.11-181297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrabhatla P, Grant P, Jaffe H, Strong MJ, Pant HC, 2010. Quantitative phosphoproteomic analysis of neuronal intermediate filament proteins (NF-M/H) in Alzheimer’s disease by iTRAQ. FASEB J. 24 (11), 4396–4407. 10.1096/fj.10-157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, 1993. Messenger RNA degradation in eukaryotes. Cell 74 (3), 413–421. 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Sadot E, Barg J, Rasouly D, Lazarovici P, Ginzburg I, 1995. Short- and long-term mechanisms of tau regulation in PC12 cells. J. Cell Sci. 108 (Pt 8), 2857–2864. [DOI] [PubMed] [Google Scholar]
- Santa-Maria I, Alaniz ME, Renwick N, Cela C, Fulga TA, Van Vactor D, Tuschl T, Clark LN, Shelanski ML, McCabe BD, et al. , 2015. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J. Clin. Invest. 125 (2), 681–686. 10.1172/JCI78421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara Y, Kobayashi T, Onodera H, Onoda M, Ohnishi M, Kato S, Kusuda K, Shima H, Nagao M, Abe H, et al. , 1996. Okadaic acid suppresses neural differentiation-dependent expression of the neurofilament-L gene in P19 embryonal carcinoma cells by post-transcriptional modification. J. Biol. Chem. 271 (42), 25950–25957. [PubMed] [Google Scholar]
- Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P, 2007. Cerebellar neurodegeneration in the absence of microRNAs. J. Exp. Med. 204 (7), 1553–1558. 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz ML, Shneidman PS, Bruce J, Schlaepfer WW, 1990. Axonal dependency of the postnatal upregulation in neurofilament expression. J. Neurosci. Res. 27 (2), 193–201. 10.1002/jnr.490270209. [DOI] [PubMed] [Google Scholar]
- Schwartz ML, Shneidman PS, Bruce J, Schlaepfer WW, 1992. Actinomycin prevents the destabilization of neurofilament mRNA in primary sensory neurons. J. Biol. Chem. 267 (34), 24596–24600. [PubMed] [Google Scholar]
- Schwartz ML, Shneidman PS, Bruce J, Schlaepfer WW, 1994. Stabilization of neurofilament transcripts during postnatal development. Brain Res. Mol. Brain Res. 27 (2), 215–220. 10.1016/0169-328x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Schwartz ML, Bruce J, Shneidman PS, Schlaepfer WW, 1995. Deletion of 3’-untranslated region alters the level of mRNA expression of a neurofilament light subunit transgene. J. Biol. Chem. 270 (44), 26364–26369. 10.1074/jbc.270.44.26364. [DOI] [PubMed] [Google Scholar]
- Shaw G, Weber K, 1982. Differential expression of neurofilament triplet proteins in brain development. Nature 298 (5871), 277–279. 10.1038/298277a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS, 2011. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 20 (20), 4016–4024. 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- Song Y, Hu M, Zhang J, Teng ZQ, Chen C, 2019. A novel mechanism of synaptic and cognitive impairments mediated via microRNA-30b in Alzheimer’s disease. EBioMedicine 39, 409–421. 10.1016/j.ebiom.2018.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira JR, Calliari A, Kun A, Benech JC, Sanguinetti C, Chalar C, Sotelo JR, 2000. Neurofilament mRNAs are present and translated in the normal and severed sciatic nerve. J. Neurosci. Res. 62 (1), 65–74. . [DOI] [PubMed] [Google Scholar]
- Stefanizzi I, Canete-Soler R, 2007. Coregulation of light neurofilament mRNA by poly (A)-binding protein and aldolase C: implications for neurodegeneration. Brain Res. 1139, 15–28. 10.1016/j.brainres.2006.12.092. [DOI] [PubMed] [Google Scholar]
- Sterneck E, Kaplan DR, Johnson PF, 1996. Interleukin-6 induces expression of peripherin and cooperates with Trk receptor signaling to promote neuronal differentiation in PC12 cells. J. Neurochem. 67 (4), 1365–1374. 10.1046/j.1471-4159.1996.67041365.x. [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Kedersha N, 2013. Relationship of GW/P-bodies with stress granules. Adv. Exp. Med Biol. 768, 197–211. 10.1007/978-1-4614-5107-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong MJ, Volkening K, Hammond R, Yang W, Strong W, Leystra-Lantz C, Shoesmith C, 2007. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell Neurosci. 35 (2), 320–327. 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Stroynowska-Czerwinska A, Fiszer A, Krzyzosiak WJ, 2014. The panorama of miRNA-mediated mechanisms in mammalian cells. Cell Mol. Life Sci. 71 (12), 2253–2270. 10.1007/s00018-013-1551-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura R, Kanai Y, Hirokawa N, 1991. In situ localization of tau mRNA in developing rat brain. Neuroscience 44 (2), 393–407. 10.1016/0306-4522(91)90064-u. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Lee E, Lawe D, Gizang-Ginsberg E, Ziff EB, 1992. Nerve growth factor-induced derepression of peripherin gene expression is associated with alterations in proteins binding to a negative regulatory element. Mol. Cell Biol. 12 (6), 2501–2513. 10.1128/mcb.12.6.2501-2513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG, 2004. Phylogenetically conserved binding of specific K homology domain proteins to the 3’-untranslated region of the vertebrate middle neurofilament mRNA. J. Biol. Chem. 279 (48), 49680–49688. 10.1074/jbc.M408915200. [DOI] [PubMed] [Google Scholar]
- Thyagarajan A, Szaro BG, 2008. Dynamic endogenous association of neurofilament mRNAs with K-homology domain ribonucleoproteins in developing cerebral cortex. Brain Res. 1189, 33–42. 10.1016/j.brainres.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, McKenna-Yasek D, Sapp PC, Silani V, Bosco DA, Shaw CE, et al. , 2011. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 156B (3), 285–290. 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- Troy CM, Muma NA, Greene LA, Price DL, Shelanski ML, 1990. Regulation of peripherin and neurofilament expression in regenerating rat motor neurons. Brain Res. 529 (1–2), 232–238. 10.1016/0006-8993(90)90832-v. [DOI] [PubMed] [Google Scholar]
- Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, Van den Broeck M, Mattheijssens M, Peeters K, De Deyn PP, et al. , 2010. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology 366–371. 10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- Villace P, Marion RM, Ortin J, 2004. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 32 (8), 2411–2420. 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintilescu CR, Afreen S, Rubino AE, Ferreira A, 2016. The neurotoxic TAU45–230 fragment accumulates in upper and lower motor neurons in amyotrophic lateral sclerosis subjects. Mol. Med. 22, 477–486. 10.2119/molmed.2016.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkening K, Leystra-Lantz C, Strong MJ, 2010. Human low molecular weight neurofilament (NFL) mRNA interacts with a predicted p190RhoGEF homologue (RGNEF) in humans. Amyotroph. Lateral Scler. 11 (1–2), 97–103. 10.3109/17482960902995584. [DOI] [PubMed] [Google Scholar]
- Wang C, Szaro BG, 2016. Post-transcriptional regulation mediated by specific neurofilament introns in vivo. J. Cell Sci. 129 (7), 1500–1511. 10.1242/jcs.185199. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB, 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456 (7221), 470–476. 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gao QS, Wang Y, Lafyatis R, Stamm S, Andreadis A, 2004. Tau exon 10, whose missplicing causes frontotemporal dementia, is regulated by an intricate interplay of cis elements and trans factors. J. Neurochem 88 (5), 1078–1090. 10.1046/j.1471-4159.2003.02232.x. [DOI] [PubMed] [Google Scholar]
- Wang S, Ji D, Yang Q, Li M, Ma Z, Zhang S, Li H, 2019. NEFLb impairs early nervous system development via regulation of neuron apoptosis in zebrafish. J. Cell Physiol. 234 (7), 11208–11218. 10.1002/jcp.27771. [DOI] [PubMed] [Google Scholar]
- Wang Y, Mandelkow E, 2016. Tau in physiology and pathology. Nat. Rev. Neurosci. 17 (1), 5–21. 10.1038/nrn.2015.1. [DOI] [PubMed] [Google Scholar]
- Weingarten MD, Lockwood AH, Hwo SY, Kirschner MW, 1975. A protein factor essential for microtubule assembly. Proc. Natl. Acad. Sci. USA 72 (5), 1858–1862. 10.1073/pnas.72.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B, Ivanov P, 2019. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 20 (11), 649–666. 10.1038/s41583-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Oblinger MM, 1990. Differential regulation of peripherin and neurofilament gene expression in regenerating rat DRG neurons. J. Neurosci. Res 27 (3), 332–341. 10.1002/jnr.490270312. [DOI] [PubMed] [Google Scholar]
- Wong NK, He BP, Strong MJ, 2000. Characterization of neuronal intermediate filament protein expression in cervical spinal motor neurons in sporadic amyotrophic lateral sclerosis (ALS). J. Neuropathol. Exp. Neurol. 59 (11), 972–982. 10.1093/jnen/59.11.972. [DOI] [PubMed] [Google Scholar]
- Yadav P, Selvaraj BT, Bender FL, Behringer M, Moradi M, Sivadasan R, Dombert B, Blum R, Asan E, Sauer M, et al. , 2016. Neurofilament depletion improves microtubule dynamics via modulation of Stat3/stathmin signaling. Acta Neuropathol. 132 (1), 93–110. 10.1007/s00401-016-1564-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB, 2004. Variation in alternative splicing across human tissues. Genome Biol. 5 (10), R74. 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Nixon RA, 2016. Specialized roles of neurofilament proteins in synapses: relevance to neuropsychiatric disorders. Brain Res Bull. 126 (Pt 3), 334–346. 10.1016/j.brainresbull.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Nixon RA, 2021. Neurofilament proteins as biomarkers to monitor neurological diseases and the efficacy of therapies. Front Neurosci. 15, 689938 10.3389/fnins.2021.689938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Rao MV, Kumar A, Julien JP, Nixon RA, 2003. Neurofilament transport in vivo minimally requires hetero-oligomer formation. J. Neurosci. 23 (28), 9452–9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Sasaki T, Kumar A, Peterhoff CM, Rao MV, Liem RK, Julien JP, Nixon RA, 2012. Peripherin is a subunit of peripheral nerve neurofilaments: implications for differential vulnerability of CNS and peripheral nervous system axons. J. Neurosci. 32 (25), 8501–8508. 10.1523/JNEUROSCI.1081-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Rao MV, Sasaki T, Chen Y, Kumar A, Veeranna, Liem RK, Eyer J, Peterson AC, Julien JP, et al. , 2006. Alpha-internexin is structurally and functionally associated with the neurofilament triplet proteins in the mature CNS. J. Neurosci. 26 (39), 10006–10019. 10.1523/JNEUROSCI.2580-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Sershen H, Veeranna, Basavarajappa BS, Kumar A, Hashim A, Berg M, Lee JH, Sato Y, Rao MV, et al. , 2015. Neurofilament subunits are integral components of synapses and modulate neurotransmission and behavior in vivo. Mol. Psychiatry 20 (8), 986–994. 10.1038/mp.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan A, Veeranna, Sershen H, Basavarajappa BS, Smiley JF, Hashim A, Bleiwas C, Berg M, Guifoyle DN, Subbanna S, et al. , 2018. Neurofilament light interaction with GluN1 modulates neurotransmission and schizophrenia-associated behaviors. Transl. Psychiatry 8 (1), 167. 10.1038/s41398-018-0194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yum SW, Zhang J, Mo K, Li J, Scherer SS, 2009. A novel recessive Nefl mutation causes a severe, early-onset axonal neuropathy. Ann. Neurol. 66 (6), 759–770. 10.1002/ana.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gao F, Wang D, Li C, Fu Y, He W, Zhang J, 2018. Tau pathology in Parkinson’s disease. Front Neurol. 9, 809. 10.3389/fneur.2018.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LT, Ye SH, Yang HX, Zhou YT, Zhao QH, Sun WW, Gao MM, Yi YH, Long YS, 2017. A novel role of fragile X mental retardation protein in pre-mRNA alternative splicing through RNA-binding protein 14. Neuroscience 349, 64–75. 10.1016/j.neuroscience.2017.02.044. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Couillard-Despres S, Julien JP, 1997. Delayed maturation of regenerating myelinated axons in mice lacking neurofilaments. Exp. Neurol. 148 (1), 299–316. 10.1006/exnr.1997.6654. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.


