Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019, constitutes an emerging human pathogen of zoonotic origin. A critical role in protecting the host against invading pathogens is carried out by interferon-stimulated genes (ISGs), the primary effectors of the type I interferon (IFN) response. All coronaviruses studied thus far have to first overcome the inhibitory effects of the IFN/ISG system before establishing efficient viral replication. However, whether SARS-CoV-2 evades IFN antiviral immunity by manipulating ISG activation remains to be elucidated. Here, we show that the SARS-CoV-2 main protease (Mpro) significantly suppresses the expression and transcription of downstream ISGs driven by IFN-stimulated response elements in a dose-dependent manner, and similar negative regulations were observed in two mammalian epithelial cell lines (simian Vero E6 and human A549). Our analysis shows that to inhibit the ISG production, Mpro cleaves histone deacetylases (HDACs) rather than directly targeting IFN signal transducers. Interestingly, Mpro also abolishes the activity of ISG effector mRNA-decapping enzyme 1a (DCP1A) by cleaving it at residue Q343. In addition, Mpro from different genera of coronaviruses has the protease activity to cleave both HDAC2 and DCP1A, even though the alphacoronaviruse Mpro exhibits weaker catalytic activity in cleaving HDAC2. In conclusion, our findings clearly demonstrate that SARS-CoV-2 Mpro constitutes a critical anti-immune effector that modulates the IFN/ISG system at multiple levels, thus providing a novel molecular explanation for viral immune evasion and allowing for new therapeutic approaches against coronavirus disease 2019 infection.
Keywords: SARS-CoV-2, main protease, interferon-stimulated gene, histone deacetylases, mRNA-decapping enzyme 1a, cleavage
Abbreviations: COVID-19, coronavirus disease 2019; Cp, cleavage product; CP, cleaved peptide; DCP1A, mRNA-decapping enzyme 1a; GBP1, guanylate binding protein 1; HDAC, histone deacetylase; IFIT, interferon-induced protein with tetratricopeptide repeats; IFN, interferon; ISG, interferon-stimulated gene; IRF9, interferon regulatory factor 9; ISGF3, interferon-stimulated gene factor 3; ISRE, interferon-stimulated response element; MIX, mix paired-like homeobox; Mpro, main protease; OAS, 2′,5′-oligoadenylate synthetase; PDCoV, porcine deltacoronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; STAT, signal transducer and activator of transcription; Usp18, ubiquitin specific peptidase 18
The ongoing pandemic of coronavirus disease 2019 (COVID-19) has caused a global health crisis. The agent of this pandemic is the ssRNA virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (1, 2). Since the beginning of COVID-19, a large number of studies on the characteristics and clinical manifestations of the SARS-CoV-2 virus have been conducted. Studies thus far have established that SARS-CoV-2 is an enveloped single-strand positive RNA virus that belongs to the betacoronavirus genus and shares 79% genetic similarity with SARS (3, 4). Similar to other coronaviruses, SARS-CoV-2 infection results in a range of disease severity. Most people infected with SARS-CoV-2 are asymptomatic or only show mild or moderate symptoms, with approximately 15 to 20% of those infected developing severe pneumonia, and approximately 5% developing fatal acute respiratory distress syndrome (5). Some recent studies have shown that severe COVID-19 patients are accompanied by a dysregulated immune response, such as uncontrolled proinflammatory responses and impaired interferon (IFN) responses (6, 7, 8, 9, 10). To date, the development of COVID-19 therapies has been slow due to a lack of detailed studies of SARS-CoV-2, including its pathogenesis and immune evasion. In-depth study of the biological functions of viral virulence factors and the underlying implications of host–pathogen interaction patterns in patients with severe SARS-CoV-2 infection is therefore pivotal for effective treatment of COVID-19 pneumonia and reduction of mortality.
Type I IFNs (such as IFNα/β) constitute highly pleiotropic cytokines that invoke a potent antiviral state in infected and surrounding cells, primarily through the induction of a large set of cellular antiviral proteins, collectively known as IFN-stimulated gene (ISG) proteins (11, 12). Approximately 10% of the genes in the human genome have the potential to be regulated by IFNs (13). Through directly or indirectly interact with IFNs, ISGs achieve one or more cellular outcomes, including antiviral defense (14, 15, 16), antiproliferative activities (17, 18), and stimulation of adaptive immunity (19, 20, 21). Upon binding to the cellular surface IFN receptor, secreted IFNs activate the receptor-associated Janus kinase family (JAK1, and TYK2), which promote recruitment and phosphorylation of signal transduction and transcriptional activation proteins (STAT1, and STAT2). Phosphorylated STAT then forms dimers before interacting with IFN regulatory factor 9 (IRF9), together forming the heterotrimeric complex called IFN-stimulated gene factor 3 (ISGF3). The ISGF3 complex subsequently enters the nucleus and recognizes the IFN-stimulated response element (ISRE) sequences for the expression of ISGs, which act as antivirals (22). Earlier studies suggested that hundreds of ISGs are commonly induced by type I IFNs across different cell backgrounds to target viral gene expression, protein production, and genome amplification (13, 23, 24). For example, ISGs like the IFIT family, including IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) (ISG15), IFIT2 (ISG54), IFIT3, and IFIT5 in humans, bind the translation initiation factor complex to suppress viral protein production (25, 26). Moreover, ISGs such as 2′,5′-oligoadenylate synthetases (OAS proteins) are enzymes that activate cellular ribonuclease L, which efficiently degrades viral genomes (27, 28). However, the signaling leading to the expression of ISGs is globally impaired when histone deacetylases (HDACs) activity is restrained by HDACs inhibitor trichostatin A. Several studies have previously shown that HDAC function does not affect the activation of STAT proteins but is required for ISG transcriptional elongation, and any inhibition of HDAC activity might result in loss of IFN-mediated immune protection (29, 30, 31, 32).
Despite the importance of the IFN/ISG system in defense against viruses, patients severely infected with SARS-CoV-2 receive only low protective immunity from type I IFNs (8, 33). This dysregulated immune response suggests that SARS-CoV-2 has developed mechanisms to interfere with the production of IFNs and ISGs. Interestingly, the inhibition of IFN response mediated by the main protease (also called Mpro, 3CL-protease, Nsp5) has been widely studied in the context of different vertebrate coronaviruses. For instance, the porcine deltacoronavirus (PDCoV) Nsp5 cleaves the nuclear factor-κB essential modulator protein, thus inhibiting IFN synthesis (34); SARS-CoV-2 Mpro prevents the nuclear translocation of phosphorylated IRF3, thus suppressing IFN induction (35). While several studies have investigated the antagonistic functions of Mpro protein in IFN, how exactly SARS-CoV-2 Mpro affects the downstream ISG effectors remains unclear.
Recent studies analyzing the SARS-CoV-2–host proximity interactome have shown that SARS-CoV-2 Mpro interacts with host protein HDAC2, suggesting that Mpro affects the function of HDAC2 (36). However, the precise implication of this interaction for SARS-CoV-2–infected cells remains unknown. Considering the pivotal role of deacetylation in IFN-dependent gene expression, we set out to investigate whether Mpro mediates viral immune evasion by interfering with ISG-related functions of HDACs. In addition, we screened 10 ISGs to test whether Mpro directly targets ISG functions. Here, we report that SARS-CoV-2 Mpro inhibits HDAC-dependent ISG antiviral responses and abolishes the ISG effector mRNA-decapping enzyme 1a (DCP1A) activity. Mpro suppresses IFN response, from ISG induction to ISG activity, thus providing new insights into the pathogenesis and transmission of SARS-CoV-2.
Results
SARS-CoV-2 Mpro antagonizes IFN-stimulated gene production
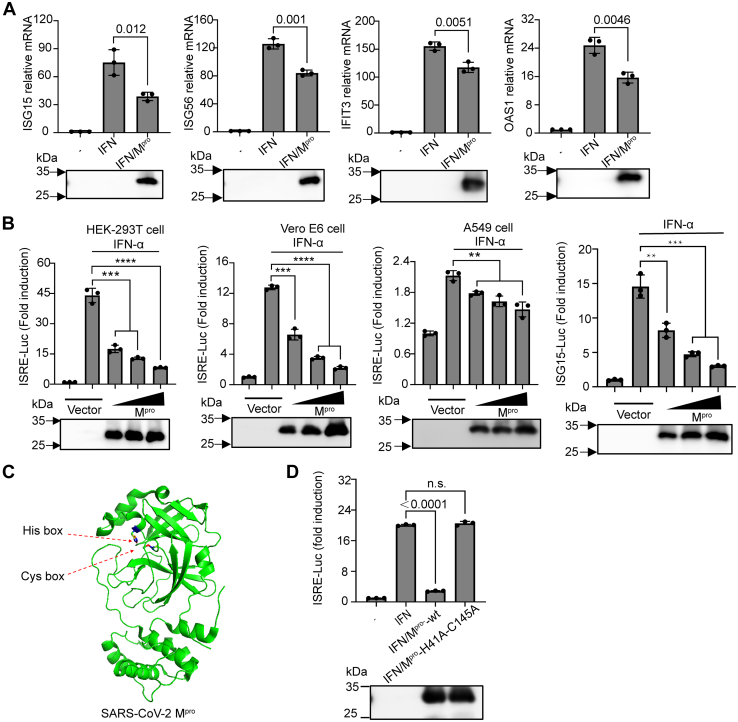
The main protease of coronaviruses, commonly associated with viral polyprotein precursors processing, is essential to antagonize intracellular antiviral defenses (37). To examine how the presence of Mpro affects the expression of IFN-dependent genes, we performed quantitative real-time RT-PCR for four well-characterized ISGs that inhibit SARS-CoV-2 infection, namely ISG15, ISG56, IFIT3, and 2′-5′-oligoadenylate synthetase 1 (OAS1), following overexpression of Mpro in human embryonic kidney cells (HEK-293T) (28, 38, 39).The cells were first treated for 8 h with 0, 250, 500, 1000, or 2000 U/ml of recombinant human IFN-α and then collected for measuring mRNA levels of IFIT3 using quantitative PCR. As shown in Fig. S1A, IFIT3 was significantly induced in an IFN dose-dependent manner and reached a saturation point at approximately 1000 U/ml. Similarly, we treated cells with 1000 U/ml IFN-α for 0, 4, 8, or 12 h to measure IFIT3 transcription. As expected, the expression of IFIT3 increased over time (Fig. S1B). To further investigate the effect of Mpro on the transcription of other ISGs, we selected an effective dose of 1000 U/ml and an intermediate time of 8 h for subsequent analysis. As shown in Figure 1A, gene expression substantially increased in response to IFN stimulation in the control cells. In contrast, IFN stimulation failed to induce the expression of ISGs in cells overexpressing Mpro. Considering the existence of ISREs within the ISG promoter regions, we assessed the ISRE-dependent transcription by dual-luciferase reporter assay in HEK-293T cells, Vero E6 cells, and A549 cells. Consistent with our mRNA results, Mpro strongly suppressed IFN-α-induced ISRE promoter activity in a dose-dependent manner in HEK-293T cells, Vero E6 cells, and A549 cells (Fig. 1B). Next, we examined the expression of ISG15, a gene highly responsive to IFN stimulation (40). As shown in Figure 1B, ISG15-driven gene expression was highly expressed in IFN-stimulated cells. In contrast, overexpression of Mpro prevented IFN-dependent upregulation of ISG15 in a dose-dependent manner. Previous studies showed that histidine 41 and cysteine 145 are catalytic residues which are required for Mpro to achieve effective proteolysis (41, 42) (Fig. 1C). We next investigated whether inactive Mpro impedes the activation of ISRE promoter activity. As shown in Figure 1D, impaired Mpro lost the repression effect on ISRE-driven gene expression due to H41A and C145A double mutations in Mpro, suggesting that the enzymatic activity of Mpro is required for the negative regulation of ISGs.
Figure 1.
SARS-CoV-2 Mproinhibits interferon-stimulated gene production.A, HEK-293T cells were transfected with pCDNA3.1 empty vector or the pCDNA3.1-Mpro-HA plasmid. Twenty-four hours posttransfection, cells were stimulated with 1000 U/ml IFNα for 8 h as indicated. Isolated RNA was analyzed by RT-PCR for the ISG15, ISG56, IFIT3, or OAS1 relative mRNA level and normalized to GAPDH mRNA transcription. B, HEK-239T cells, Vero E6 cells, or A549 cells were transfected with various concentrations of pCDNA3.1-Mpro-HA plasmid, along with pRL-TK plasmid (10 ng) and pISRE-Luc (50 ng) or pISG15-Luc. Twenty-four hours posttransfection, cells were stimulated with or without IFNα (1000 U/ml) for 12 h, followed by a dual-luciferase assay. C, schematic representation of SARS-CoV-2 Mpro (PDB code: 7JYC) with the conserved catalytic residues His41 and Cys145. D, HEK-293T cells were transfected with pISRE-Luc plasmid (50 ng), pRL-TK plasmid (10 ng), along with SARS-CoV-2 Mpro expression constructs (200 ng) or its inactive double mutants H41A and C145A. Twenty-four hours posttransfection, a dual-luciferase assay was performed after treatment IFNα (1000 U/ml) for another 8 h. Western blotting for the expression levels of Mpro were shown below the graph. All presented results represent the means and standard deviations of data from three independent experiments. Statistical significance was calculated using unpaired, two-tailed Student’s t test. Data significance is shown as indicated. ns, not significant; ∗P < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. ∗∗∗∗p < 0.0001. ISG, interferon-stimulated gene; Mpro, main protease; OAS, 2′,5′-oligoadenylate synthetase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
To further assess the levels of ISG transcription after IFN stimulation with or without Mpro, we performed RNA sequencing (RNA-seq) on HEK-293T cells from IFN-vector (n = 3) and IFN-Mpro (n = 3). We found that following IFN stimulation, Mpro significantly changed the transcription profiles of HEK-293T cells, in which 40 genes were upregulated, and 408 genes were downregulated (Fig. S1C). Recent studies assembled a list of 399 ISGs, which have been characterized to possess broad-acting antiviral activities (38, 43, 44). Our gene enrichment analysis of differentially expressed transcripts identified at least 20 ISGs significantly downregulated by Mpro in response to IFN according to the datasets (Fig. S1D). The effects of Mpro overexpression on the transcriptome in IFN-stimulated cells suggested that viral Mpro affects host responses not exclusively by impairing the expression of ISGs. Together, these results strongly indicated that the SARS-CoV-2 Mpro is able to prevent the upregulation of ISGs.
Mpro does not directly target transducers of the IFN-signal pathway
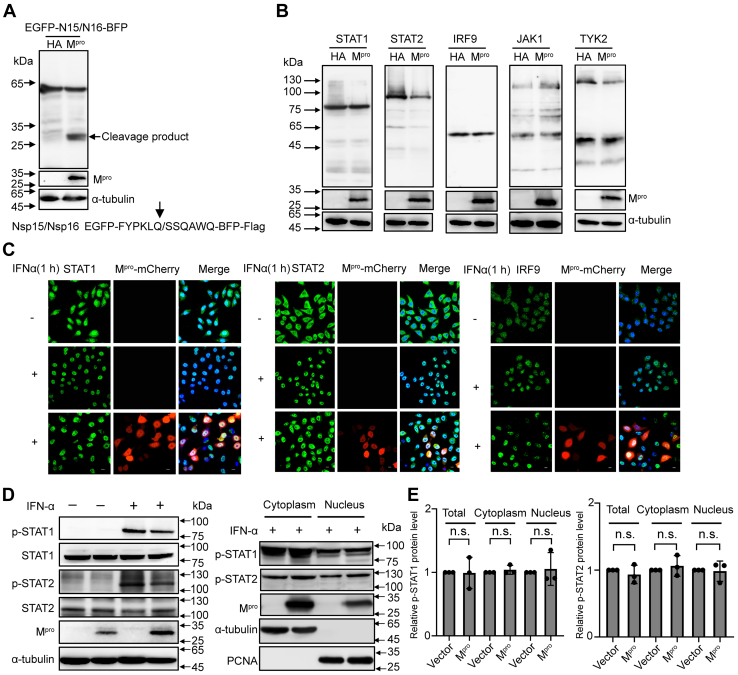
Many coronavirus-encoded factors allow escape from IFN-mediated immune defense by targeting IFN-signal pathway transducers. For instance, PDCoV Mpro antagonizes type I IFN signaling by cleaving STAT2 (45). To investigate whether SARS-CoV-2 Mpro interacts with transcription factors in the signaling pathway, we first tested the activity of Mpro. The known substrate sequence between Nsp15 and Nsp16 in SARS-CoV-2 was inserted into the EGFP and BFP sequences (46). As shown in Figure 2A, the fusion protein of EGFP-N15/N16-BFP-Flag was cleaved into two new products under the overexpression of Mpro, indicating that Mpro possesses protease activity. We next co-expressed Mpro with either STAT1, STAT2, IRF9, JAK1, or TYK2 in HEK-293T cells. However, the five transducers were not cleaved by Mpro, which showed that SARS-CoV-2 Mpro does not target transcription factors as is established for the Mpro of PDCoV (Fig. 2B). In addition to its interaction with transducers, it was previously shown that the SARS-CoV-2 ORF6 blocks the nuclear translocation of STAT1 and STAT2, thus interrupting the host IFN defense (47). To assess whether Mpro interferes with the location of transducers, we assessed the effects of SARS-CoV-2 Mpro expression on the subcellular localization of STAT1, STAT2, and IRF9 using an indirect immunofluorescence assay. We first tested the activity of Mpro-mCherry by Western blotting. As shown in Fig. S2A, the fusion protein of EGFP-N15/N16-BFP-Flag was cleaved by Mpro-mCherry, demonstrating that Mpro-mCherry was catalytically active. However, nuclear translocation of STAT1, STAT2, and IRF9 differed insignificantly following IFN stimulation during Mpro expression (Fig. 2C). As ISGF3 phosphorylation is highly correlated with activation of IFN signaling (48), SARS-CoV-2–encoded Nsp6, Nsp13, ORF7b, and N proteins enable the virus to antagonize the IFN responses by suppressing STAT1 or STAT2 phosphorylation (49, 50), and we next assessed the phosphorylation levels of these two signal transducers (STAT1 and STAT2) by immunoblotting using phospho-specific antibodies. As shown in Figure 2, D and E, Mpro failed to affect IFN-dependent activation of STAT1 and STAT2, both in the nuclear fraction as well as in the cytoplasm. Together, these results indicated that overexpression of Mpro had little effect on IFN-α-responsive signaling from the cytoplasm to the nucleus, suggesting that Mpro suppresses ISGs without targeting ISG3 or upstream components of this pathway.
Figure 2.
Mproshows no interaction with IFN signaling pathway transducers.A, HEK-293T cells were transfected with a C-terminal flag-tagged EGFP-N15/N16-BFP plasmid along with SARS-CoV-2 Mpro or empty vector. Cells were lysed at 24 h after transfection and analyzed by Western blotting. B, HEK-293T cells were cotransfected with a C-terminal flag-tagged STAT1, STAT2, IRF9, JAK1, or TYK2 expression plasmid combined either with SARS-CoV-2 Mpro or empty vector. Whole-cell extracts were lysed 30 h posttransfection and analyzed by Western blotting. C, HeLa cells were transfected with pCDNA3.1-Mpro-mCherry visual construct, and 24 h posttransfection, cells were treated with or without IFNα (1000 U/ml) for 1 h and possessed for indirect immunofluorescence to detect the STAT1, STAT2, and IRF9. Scale bar, 10 μm. D, HEK-293T cells were transfected with SARS-CoV-2 Mpro or empty vector. After 24 h of expressing, cells were cultured with or without IFNα (1000 U/ml) for 1 h. Whole-cell extracts or the nuclear and cytoplasmic fractions were analyzed by Western blotting with specific antibodies for the detection of tyrosine phospho-STAT1 and STAT2 or total STAT1 and STAT2. E, density analysis represents the relative protein levels of phospho-STAT1 or phospho-STAT2 that was normalized to the protein levels of α-tubulin or PCNA. The value of control group was set to 1. The presented results represent the means and standard deviations of data from three independent experiments. Statistical significance was calculated using unpaired, two-tailed Student’s t test. Protein band intensities were quantitated by Image Lab software. IFN, interferon; IRF9, interferon regulatory factor 9; Mpro, main protease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; STAT, signal transducer and activator of transcription.
Mpro cleaves HDACs to restrict IFN-stimulated genes expression
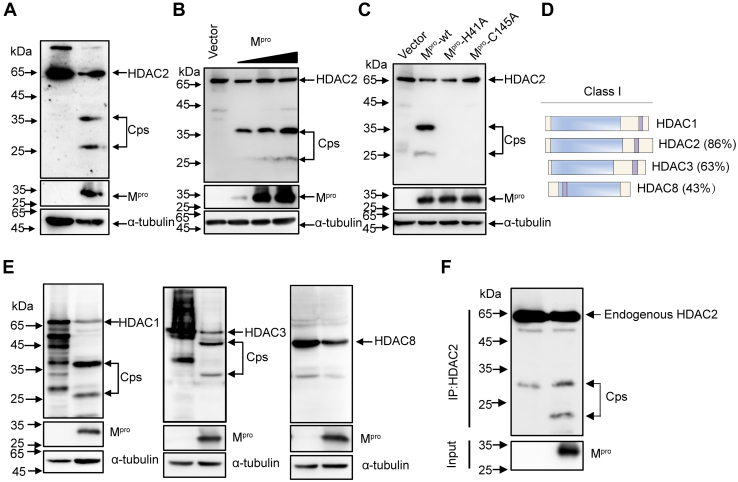
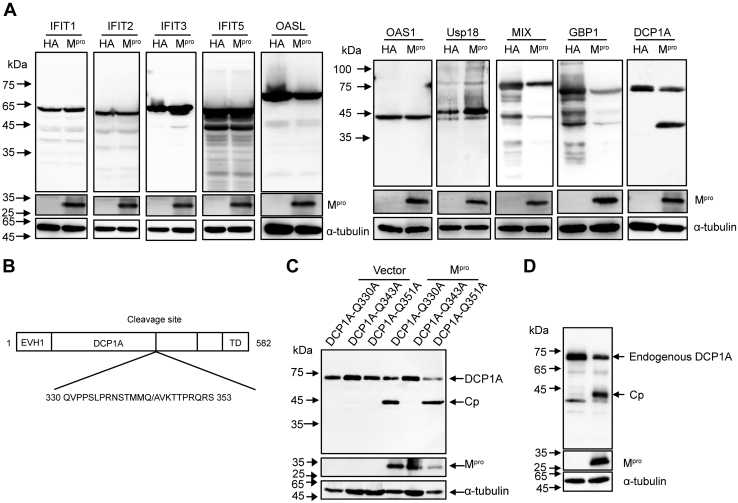
To determine the mechanisms responsible for the low expression levels of ISGs following Mpro overexpression, we evaluated the possible involvement of HDACs in regulation of ISGs. Considering the physical interactions between SARS-CoV-2 Mpro and HDAC2 and the proteolytic function of Mpro (36, 42), we hypothesized that HDAC2 constitutes a substrate of Mpro action. To test this hypothesis, we co-expressed HDAC2 with Mpro in HEK-293T cells, followed by Western blotting with an anti-flag antibody. As shown in Figure 3A, samples co-transfected with pCDNA3.1-HDAC2-Flag and pCDNA3.1-Mpro-HA exhibited two faster-migrating protein bands, representing the possible cleavage productions. This result indicated that Mpro cleaves the host protein HDAC2. To assess whether Mpro-mediated cleavage of HDAC2 occurred in a dose-dependent manner, we co-transfected HEK-293T cells with HDAC2 and various concentrations of SARS-CoV-2 Mpro. As shown in Figure 3B, increased Mpro expression resulted in increased cleaving of HDAC2. The luciferase reporter assay showed that both His 41 and Cys 145 are crucial for Mpro to regulate the expression of ISGs (Fig. 1D). We next assessed whether the inactive Mpro mediates cleavage of HDAC2. As shown in Figure 3C, cells co-transfected with either Mpro -H41A or Mpro -C145A showed no HDAC2 cleaving. Catalytically impaired Mpro was unable to cleave HDAC2, demonstrating that the proteolytic activity of Mpro is necessary for HDAC2 cleaving.
Figure 3.
Mproimpairs ISG induction by cleaving HDACs.A, HEK-293T cells were transfected with pCDNA3.1-HDAC2-Flag plasmids together with or without Mpro for 24 h before Western blotting analysis with the anti-flag antibody. B, HEK-293T cells were transfected with HDAC2 expression construct alone or in the presence of increasing concentrations of the Mpro, and after 30 h transfection, cells were lysed for Western blotting analysis. C, HEK-293T cells were transfected with HDAC2 expression plasmid along with Mpro or its mutants Mpro-H41A, Mpro-C145A. Western blotting was performed after 24 h transfection. D, Schematic representation of Class I HDAC with different similarities to the HDAC1. E, HEK-293T cells were transfected with HDAC1, HDAC3, or HDAC8 separately along with either Mpro or empty vector. Western blotting was performed after 24 h transfection. F, HEK-293T cells were transfected with Mpro or empty vector. Cells were then lysed for immunoprecipitation analysis at 32 h after transfection. HDAC, histone deacetylase; ISG, interferon-stimulated gene; Mpro, main protease.
HDAC2 belongs to the class I HDACs (HDAC1, 2, 3, and 8) and exhibits high homology with all other members of this class (51) (Fig. 3D). Considering that Mpro-mediated cleavage is sequence-dependent, we next tested whether Mpro cleaves HDAC1, HDAC3, or HDAC8. As shown in Figure 3E, HDAC1 presented two cleaved bands almost the same size as HDAC2, suggesting that they share close cleavage sites. In contrast, we observed a slower migrating band for HDAC3, suggesting that an entirely different cleavage site is present in this HDAC form. No cleavage band was observed for HDAC8 upon Mpro expression, possibly due to its poor homology with other members. Considering that HDAC1/2 generally display significant activity within the complexes of proteins to regulate gene expression (52, 53, 54), we concluded that the cleaving reactions caused by Mpro resulted in sufficient impairment of the complex functions, thus further impeding ISG activation.
To assess whether Mpro cleaves the endogenously expressed HDACs, we used specific antibodies against endogenous HDAC1, HDAC2, or HDAC3. However, no specific cleavage fragments of endogenous HDAC1, HDAC2, and HDAC3 were observed in Mpro -overexpressing cells (data not shown). To exclude the possibility that the amount of these cutting bands was below the detection limit, we enriched the target protein products using an immunoprecipitation method. As shown in Figure 3F, the cleavage of endogenous HDAC2 was detected following Mpro overexpression. However, we still failed to observe cleavage fragments of HDAC1 and HDAC3 (Fig. S3A). Previous studies made similar observations, where endogenous cleavage bands were absent, but they observed a reduction of substrate protein abundance upon infection (55, 56, 57, 58).
To rule out the possibility that Mpro degrades HDAC2 through either ubiquitin-proteasome system, autophagy, or apoptosis, we evaluated SARS-CoV-2 Mpro-mediated HDAC2 cleavage following treatment with either the proteasome inhibitor MG132, the caspase inhibitor Z-VAD-FMK, or the autophagy inhibitor NH4Cl. We detected the cleavage products regardless of the inhibitor used, suggesting that SARS-CoV-2 Mpro–mediated HDAC2 cleavage is independently of cellular caspases or proteasomes (Fig. S3B).
HDAC2 activity is lost upon Mpro-dependent cleaving at residues Q261 and Q383
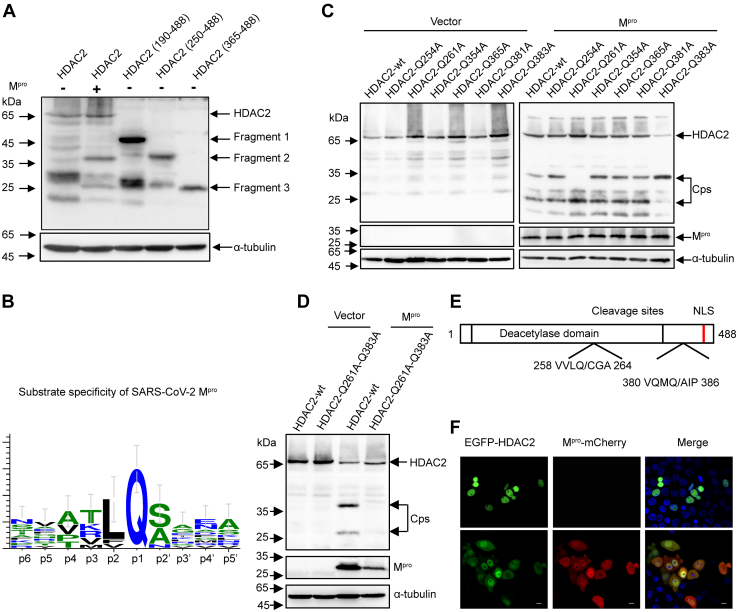
To identify the Mpro cleavage sites within HDAC2, we truncated HDAC2 into three mutant fragments, pHDAC2(190–488)-Flag, pHDAC2(250–488)-Flag, and pHDAC2(365–488)-Flag. As shown in Figure 4A, the cleavage products of C-terminal flag-tagged HDAC2 were approximate 36 kDa and 26 kDa. The bands of pHDAC2(190–488)-Flag, pHDAC2(250–488)-Flag, and pHDAC2(365–488)-Flag corresponded to a molecular mass of 46 kDa, 37 kDa, and 25 kDa, respectively. The migrating protein band of the pHDAC2(250–488)-Flag was slightly slower than that of 36 kDa, and the migrating band of the pHDAC2(365–488)-Flag was almost similar to that of 26 kDa. These results suggested that the recognition sites are located proximal to residues 250 and 365. Previous studies showed that SARS-CoV-2 Mpro prefers to recognize the glutamine (Q) residue at the P1 position for substrate cleavage (42) (Fig. 4B). Based on these findings, we analyzed the glutamines near residues 250 and 365 in HDAC2 and mutated them to alanine (A). As shown in Figure 4C, among the six HDAC2 mutants (Q254A, Q261A, Q354A, Q365A, Q381A, and Q383A), only Q261A and Q383A prevented SARS-CoV-2 Mpro-mediated cleavage. To confirm these putative cleavage sites, we mutated both HDAC2 Q261 and Q383 to alanine and co-expressed these mutants in the absence or presence of Mpro. As shown in Figure 4D, no cleavage products were generated in the presence of the double mutations. Considering that the glutamine residue is conserved at the P1 position and the size of the cleavage products was consistent, we concluded that Q261 and Q383 constitute the Mpro cleavage sites of HDAC2.
Figure 4.
Mprocleavage HDAC2 occurs at residues Q261 and Q383.A, HEK-293T cells were transfected with pCDNA3.1-HDAC2-Flag plasmid or its truncated constructs pHDAC2 (190–488)-Flag, pHDAC2 (250–488)-Flag, pHDAC2 (365–488)-Flag. After 30 h of transfection, the cell lysates were analyzed by Western blotting with anti-flag antibody. B, schematic representation of substrate specificity of SARS-CoV-2 Mpro. C, HEK-293T cells were respectively transfected with HDAC2, HDAC2-Q254A, HDAC2-Q261A, HDAC2-Q354A, HDAC2-Q365A, HDAC2-Q381A, and HDAC2-Q383A together with Mpro or empty vector. Western blotting was performed 24 h posttransfection. D, HEK-293T cells were transfected with double mutants Q261A and Q383A of HDAC2 together with Mpro or empty vector. Western blotting was performed 24 h posttransfection. E, schematic representation of HDAC2 with the position of two cleavage sites. F, HEK-293T cells were transfected with a plasmid expressing EGFP-HDAC2 together with Mpro-mCherry or empty vector. After 24 h expressing, images of transfected cells were recorded with GFP and mCherry excitation and emission spectra. Scale bar, 10 μm. HDAC, histone deacetylase; Mpro, main protease; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Class I HDACs usually perform their catalytic functions inside the nucleus. We therefore assessed whether cleavage affects the subcellular location of HDACs (59). We used HDAC2 as a template to test the localization of cleavage products (Fig. 4E) and ectopically expressed EGFP-HDAC2 in the presence of Mpro-mCherry. As shown in Figure 4F, EGFP-HDAC2 alone localized exclusively in the nucleus; however, we found that partial EGFP-HDAC2 reposited into the cytoplasm during the expression of Mpro-mCherry. To confirm the subcellular distribution of these cleavage products, we fused the cleavage fragments with EGFP, including pEGFP-HDAC2(1–261), pEGFP-HDAC2(262–488), pEGFP-HDAC2(262–383), pEGFP-HDAC2(1–383), and pEGFP-HDAC2(384–488) based on the identified cleaved residues. We found that both EGFP-HDAC2(1–261) and EGFP-HDAC2(1–383) were primarily located within the cytoplasmic regions, while EGFP-HDAC2(262–488) and EGFP-HDAC2(384–488) remained nuclear. Only the EGFP-HDAC2(262–383) was evenly distributed throughout both the nucleus and the cytoplasm (Fig. S4A). Together, these results strongly suggested that Mpro not only cleaves HDAC2 but also prevents nuclear translocation of HDAC2.
Mpro cleaves the IFN-stimulated gene DCP1A at residue Q343
A recent study reported that SARS-CoV-2 papain-like protease attenuates type I IFN responses and regulates NF-κB pathway by cleaving the ubiquitin-like IFN-stimulated gene 15 protein (IFIT1) (60). An earlier study showed that MERS-CoV NS4b interferes with virus-induced expression of type I and type III IFNs. In addition, it cleaves OAS via its phosphodiesterase activity (61). To assess whether the Mpro of SARS-CoV-2 targets ISG functions, we chose 10 ISGs that play important antiviral roles, namely IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OASL, DCP1A, mix paired-like homeobox, guanylate binding protein 1, and ubiquitin-specific peptidase 18 (13, 38), to co-express with Mpro. As shown in Figure 5A, Mpro selectively cleaved the N-terminal flag-tagged DCP1A. One previous study reported that porcine deltacoronavirus (PDCoV) Mpro cleaves DCP1A at residue Q343 resulting in decreased antiviral activity (62) (Fig. 5B). Similarly, we observed a migrating band close to that caused by PDCoV Mpro. To investigate whether SARS-CoV-2 Mpro possesses the identical cleavage site as PDCoV Mpro, we mutated either Q330, Q343, or Q351 to Ala in DCP1A. We found that only Q343A disrupted the cleavage of SARS-CoV-2 Mpro (Fig. 5C). To assess whether Mpro cleaves endogenous DCP1A, we used a specific antibody raised against endogenously expressed DCP1A. Remarkably, a similar cleavage product of DCP1A was detected following the expression of Mpro, revealing a biological effect of this cleavage in virus infection (Fig. 5D). On the basis of earlier findings and our own results, we concluded that the residue Q343 of DCP1A constitutes the cleavage site used by SARS-CoV-2 Mpro.
Figure 5.
Mprocleaves ISG DCP1A at residue Q343.A, HEK-293T cells were transfected with N-terminal flag-tagged IFIT1, IFIT2, IFIT3, IFIT5, OSAL, OAS1, Usp18, MIX, GBP1, or DCP1A plasmids along with SARS-CoV-2 Mpro or empty vector, after 30 h of transfection, followed by Western blotting analysis. B, schematic representation of DCP1A with the position of the cleavage site. C, HEK-293T cells were separately transfected with pDCP1A-Q330A, pDCP1A-Q343A, or pDCP1A-Q351A plasmid together with SARS-CoV-2 Mpro or empty vector, protein extracts were analyzed by Western blotting with anti-flag antibody 24 h posttransfection. D, HEK-293T cells were transfected with Mpro or empty vector. Cells were lysed at 24 h after transfection and analyzed by Western blotting with anti-DCP1A antibody. DCP1A, mRNA-decapping enzyme 1a; GBP1, guanylate binding protein 1; HDAC, histone deacetylase; IFIT, interferon-induced protein with tetratricopeptide repeats; ISG, interferon-stimulated gene; MIX, mix paired-like homeobox; Mpro, main protease; OAS, 2′,5′-oligoadenylate synthetase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Usp18, ubiquitin specific peptidase 18.
Mpro proteolytic efficiency varies among different coronaviruses
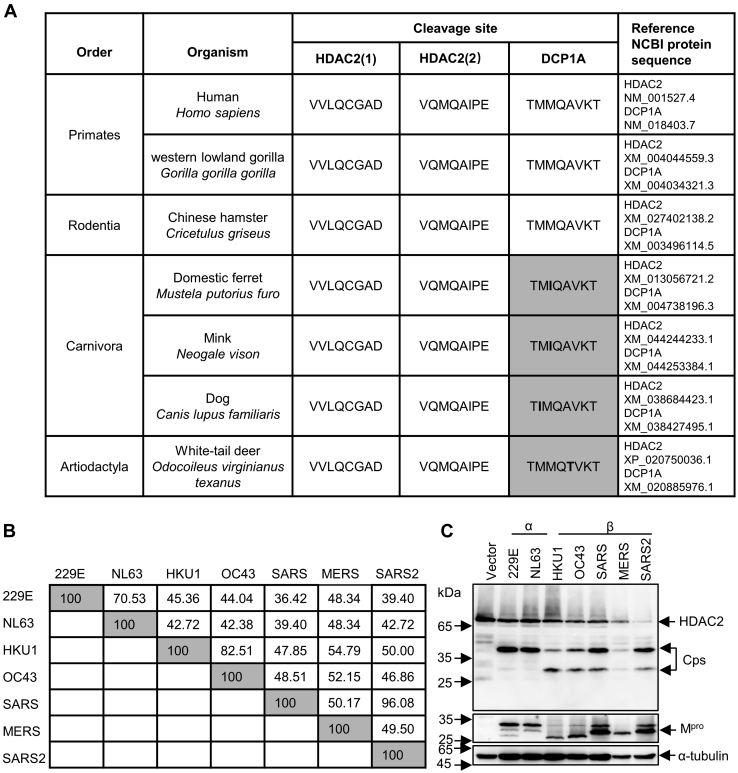
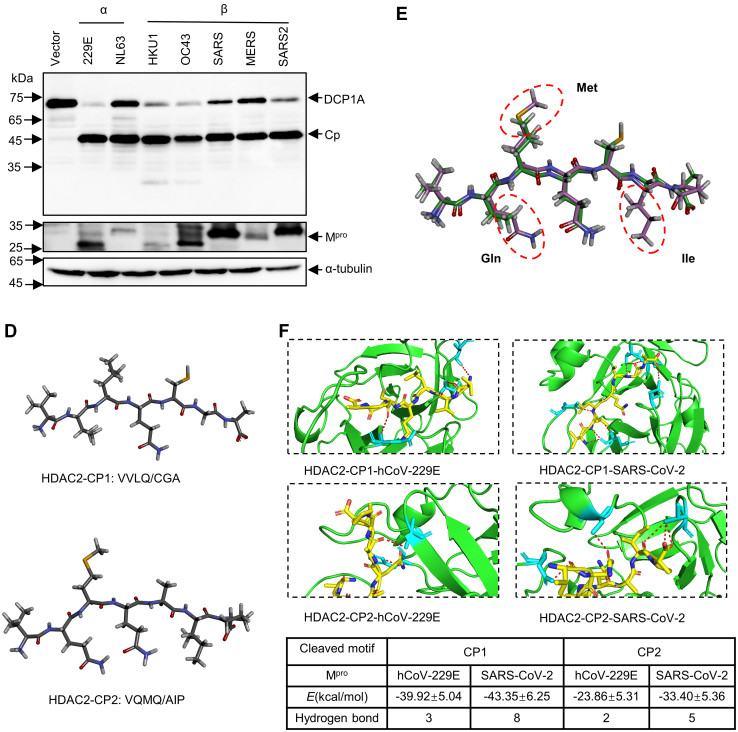
To assess whether these cleavage events mediated by Mpro are ubiquitous in different animal hosts and coronaviruses, we first compared the protein sequences of HDAC2 and DCP1A in different species, especially the sequence proximal to the cleaved motif. As shown in Figure 6A, HDAC2 cleavage sites were highly conserved across different orders. In contrast, we identified a number of small differences near the cleavage site in DCP1A, where Met at P2 or P3 changed into Ile, or Ala was replaced by Thr at P2’. Considering that all mutations were small neutral amino acids, we speculated that these substitutions in DCP1A would not affect the formation of active enzyme-substrate complexes. To investigate whether Mpro from other related human pathogenic coronaviruses cleaves HDAC2 and DCP1A, we analyzed the diversity of Mpro sequences from seven different evolutionary clades members within the coronavirus family: hCoV-229E, hCoV-NL63, hCoV-HKU1, hCoV-OC43, SARS, MERS, SARS-CoV-2. As shown in Figure 6B, the Mpro protein of different coronavirus proteins is relatively conserved, although it exhibits up to 64% amino acid difference across the different Mpro forms. To gain insights into the cleaving ability of different coronavirus Mpro, we co-expressed seven coronaviruses Mpro with either HDAC2 or DCP1A. Interestingly, both HDAC2 and DCP1A were cleaved by seven coronaviruses Mpro. However, the two alphacoronaviruses Mpro only cleaved HDAC2 at residue Q261, while the five betacoronaviruses Mpro cleaved both sites (Fig. 6C).
Figure 6.
Mprocleavage efficiency differs in different coronaviruses.A, analysis of protein sequences of HDAC2 and DCP1A in cleaved sites across species. B, amino acid percent identity matrix of Mpro proteins in different coronaviruses. C, HEK-293T cells were individually transfected with empty vector, 229E-Mpro, NL63-Mpro, HKU1-Mpro, OC43-Mpro, SARS-Mpro, MERS-Mpro, or SARS2-Mpro plasmids along with HDAC2 or DCP1A. Western blotting was performed 30 h posttransfection. D, generation of peptide conformations based on sequence of two cleaved motifs in HDAC2. CP1 represents the first cleaved peptide, and CP2 means the second cleaved peptide. E, comparison of predicted two heptapeptides structures using superimpose in discovery studio. F, binding energy and hydrogen bonds formed between Mpro and cleaved motifs. The hydrogen bonds (Red) formed between the two cleaved peptides and Mpro are presented as dash lines labeled with donor residual (yellow) and corresponding acceptor residual (cyan) in Mpro. CP, cleaved peptide; DCP1A, mRNA-decapping enzyme 1a; HDAC, histone deacetylase; Mpro, main protease.
To clarify why the cleavage efficiency of Mpro differs in alphacoronavirus and betacoronavirus, we simulated the interactions between Mpro and cleaved motifs in HDAC2 by molecular docking. We chose the hCoV-229E Mpro (alphacoronavirus) and SARS-CoV-2 Mpro (betacoronavirus) for subsequently large-scale conformational modeling. We first prepared the structure of these two cleaved heptapeptides on Yinfo Cloud Computing Platform (http://www.yinfotek.com/) (Fig. 6D). To compare the structure difference between these two motifs, we performed molecular superpositions. As shown in Figure 6E, the backbones and several functional groups of these two short heptapeptides were ideally superimposed, except that the residues of Gln, Met, and Ile in the second cleaved peptide (CP2) showed greater steric hindrance, which indicated that Mpro might have more difficulty accessing the active site of CP2 when they interact. We next molecularly docked these two short peptides with either hCoV-229E Mpro or SARS-CoV-2 Mpro by using the CABS-dock server (http://biocomp.chem.uw.edu.pl/CABSdock/?from=groupmessage). In each case, the calculations yielded 10 results for the complexes. We chose the most probable model one, which was ranked and numbered according to their occurrence in docking trajectory, for subsequent analysis (Fig. S5A). We next performed molecular dynamics simulations of these four selected complexes and calculated their binding free energy on the Yinfo Cloud Computing Platform. As shown in Figure 6F, the binding of hCoV-229E Mpro and CP2 required more energy in comparison, suggesting that hCoV-229E Mpro failed to bind CP2. Moreover, the binding energy of the two receptor proteins for the first cleaved motif was generally lower than those of the second, suggesting that the more canonical sequence of CP1 (VLQ/CG) is easier to be targeted. The computational modeling of protein–peptide interactions showed that 229E-Mpro only formed two hydrogen bonds with the second cleavage motif. However, SARS-CoV-2 Mpro and CP2 were stabilized by five hydrogen bonds (Fig. 6F). Collectively, these detailed molecular interactions between the two substrates and Mpro explain why the Mpro of alphacoronavirus only cleaves HDAC2 at residue Q261, which aid in the design of targeted drugs against SARS-CoV-2 Mpro.
To assess whether cleavage of HDAC2 is CoVs-restricted, we tested three different types of viral proteases, namely PR (HIV), NS4AB (HCV), and NS2B-NS3 (ZIKV). We co-expressed these proteases with HDAC2 and assessed whether these proteases cleave HDAC2. We found that none of these proteases cut HDAC2 (Fig. S5B). Together, these findings clearly demonstrated that the cleavage mediated by Mpro is highly conserved among coronaviruses, even though the efficiencies in the cleaving of HDAC2 differed significantly.
Discussion
Here, we found that intranuclear HDACs present a possible viral target during SARS-CoV-2 infection. We show that following host infection, SARS-CoV-2 deploys Mpro to cleave HDAC2 at residues Q261 and Q383, which sequesters inactive HDAC fragments outside the nucleus. Similar cleavage events by Mpro occurred in its cognate members HDAC1 and HDAC3, severely compromising the functions of HDACs. In addition, Mpro selectively cleaves ISG direct effector DCP1A at residue Q343. These results reveal unknown intracellular antagonism mechanisms driven by virus–host interactions, which provide new evidence explaining why patients with severe COVID-19 infections often exhibit limited ISG response.
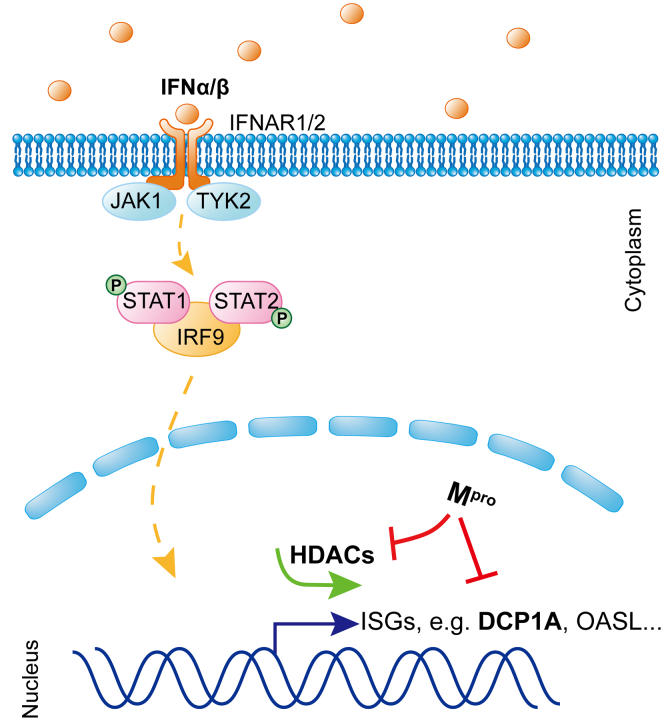
The IFN/ISG system is an early and highly efficient antiviral defense. Our results suggest that SARS-CoV-2 Mpro is primarily targeting transcriptional regulatory protein HDACs positioned further downstream, rather than the upstream common targets during an IFN response to prevent the upregulation of ISGs (Figure 1, Figure 2, Figure 3). RNA-seq data indicated that overexpression of Mpro upon IFN stimulation significantly downregulated transcription of multiple ISGs, including IRF9, MX2, REC8 meiotic recombination protein, OASL, and mitogen-activated protein kinase kinase kinase 14 (Fig. S1D). In addition to being the direct antiviral effectors of the IFN response, many ISGs themselves function as either signaling transducers or regulatory proteins of innate and adaptive immunity. For instance, IRF9 acts as a signaling transducer, which in turn leads to further regulation and development of immune responses (63). Ectopically expression of transcription factor REC8 meiotic recombination protein significantly increases ISG54 mRNA level and reduces the entry of the SARS-CoV-2-pseudotyped VSV-spike (38). Mitogen-activated protein kinase kinase kinase 14 is involved in a broad range of NF-κB activation pathways, which associate with the inflammation process (64, 65). Therefore, our RNA transcriptome data suggest that SARS-CoV-2 inhibition of the IFN signaling not only directly interferes with the antiviral actions of numerous ISGs but also indirectly influences the response of various innate and adaptive immune cells.
HDACs exert pleiotropic effects in regulating host immunity (66, 67). Interestingly, all three members in class I HDACs were cleaved by Mpro at two residues, indicating the broad spectrum of this enzyme (Fig. 3). Multiple-sequence alignment of HDACs showed that HDAC1/2 might contain similar cleavage sites at residues Q261 and Q383. However, in addition to residue Q261, Mpro may also cleave HDAC3 at residue Q113 as predicted in NetCorona 1.0 (Fig. S6A). It is well established that structural integrity is essential for HDAC1 to function in the complex, especially the N-terminal HDAC association domain (residues 1–53) (68). Our results suggest that the cleavage of Q261 is considered to break the integrity and activity of highly conserved deacetylase domain (residues 9–322). The second cleavage site, Q383, is located between the catalytic domain and the nuclear location sequence (69), and its cleavage releases the C-terminal nuclear location sequence of HDAC2, explaining the retention of the inactive HDAC fragments in the cytoplasm (Fig. 4F). A recent study claimed that SARS-CoV-2 Mpro interacts with HDAC2 but Mpro inhibits the expression of IFNβ gene and IFN-signaling pathway in an HDAC2-independent manner (70). However, cleavage of HDAC1/2 by Mpro observed in our experiments negatively affects IFN-α-stimulated ISG production.
DCP1A, as a key member of processing bodies as well as an IFN responsive gene, actively participates in the antivirus process (71). Thus, DCP1A is targeted by several different classes of viruses. In this study, we found that both human-CoVs and porcine-CoV Mpro cleave DCP1A at residue Q343 (Figs. 5 and 6). In contrast, poliovirus 3C protease results in the loss of a fragment of 6 kDa at either the N or the C terminus of DCP1A disrupting cytoplasmic processing bodies (72). Similarly, porcine reproductive and respiratory syndrome virus splices DCP1A at residues E238 attenuating its antiviral activity (73). Viruses target different sites in DCP1A, suggesting that the cleavage of DCP1A is a common strategy utilized by viruses. Additionally, the cleavage site Q343 in DCP1A might be CoVs-restricted. This cleavage site may serve as a reference for designing a CoVs Mpro inhibitor.
The Mpro-mediated cleavage for HDAC2 and DCP1A was also found to exist in cross-species, such as hamsters, mink, and white-tail deer (Fig. 6A). However, the report which found the cleavage of TAB1 and NLRP12 by Mpro described that an Ala to Val mutation presented in the second cleaved motif of mouse NLRP12 disrupts cleavage, suggesting that subtle differences around the substrate cleavage site protect host proteins from being targeted by viruses (74). Whether this species difference constitutes an evolutionary advantage or is just a random event remains under debate. Investigating the different responses among different species during SARS-CoV-2 infection will help to find better animal models to study the pathogenesis and transmission of SARS-CoV-2. And yet, the antagonism response by SARS-CoV-2 Mpro shares multiple similarities with that of other coronaviruses. However, a recent study showed that both SARS-CoV-2 Mpro and SARS-CoV Mpro cleave nuclear factor-κB essential modulator at multiple sites, and that the Ser/Ala polymorphism at position 46 of Mpro results in stronger catalytic activity as well as stronger IFN antagonism in SARS-CoV-2 when compared to SARS-CoV (75). These findings strongly suggest that the structure of Mpro is important for proteolytic activity and substrate recognition. Additionally, we found that betacoronavirus Mpro exhibits a stronger ability to cleave HDAC2 than that of alphacoronaviruses, which might also relate to the manifestations of diseases upon different CoVs infections. Sequence and structural analysis showed that the first cleaved motif (VLQ/CG) in HDAC2 is wrapped in the center which seems hard to access, and the second cleaved site Q383 (QMQ/AI) is exposed on the surface of the C-terminal disordered region (Fig. S6B). Interestingly, the Mpro of betacoronavirus cleaves both two sites, whereas the Mpro of alphacoronavirus is unable to cleave the exposed Q383 in HDAC2 (Fig. 6C). Molecular docking analysis shows that there are two reasons to ensure the Mpro cleavage efficiency: one is the formation of binding energy and hydrogen bond, making it easier to form active enzyme–substrate complexes; the other is that smaller volume of amino acids are distributed near the substrate cleavage sites, which is conducive to reducing the steric hindrance of Mpro-substrate interaction.
Although the interactions between Mpro and class I HDACs and DCP1A are well understood in this study, little is known about the substrate cleavage in a physiological context. In addition to the positive role of HDAC for the induction of ISGs, HDAC activity is also related to various cellular processes, such as gene repression, the cell cycle, and innate and adaptive immune responses (76, 77, 78, 79, 80). Whether the cleavage of HDACs leads to other disorders in the host remains unknown. Investigation of possible impaired immune responsiveness in the biological consequences of substrate cleavage is also warranted.
In conclusion, we showed that SARS-CoV-2 Mpro attenuates immune defense of antiviral effectors encoded by ISGs through cleaving host HDACs and DCP1A. The results are intriguing, given that HDACs were directly cleaved by SARS-CoV-2 Mpro rather than degraded or phosphorylated as previously reported (32, 81). Our findings expand the knowledge about the role of Mpro during CoV infection, as shown in the schematic diagram of the molecular mechanism (Fig. 7). Investigating the precise molecular mechanism of IFN antagonism should further advance our understanding of these important human pathogens, which is necessary for developing viable therapeutic approaches against SARS-CoV-2 infections.
Figure 7.
Mproinhibits the IFN-I signaling at multiple steps. Schematic representation of how Mpro antagonizes ISG production. DCP1A, mRNA-decapping enzyme 1a; HDAC, histone deacetylase; IFN, interferon; ISG, interferon-stimulated gene; IRF9, interferon regulatory factor 9; Mpro, main protease; STAT, signal transducer and activator of transcription.
Experimental procedures
Plasmid construction
Seven coronavirus Mpro fragments were synthesized by Sangon Biotech with codon optimization and inserted into the pCDNA3.1- (+) plasmid with a C-terminal HA tag. The promoter sequence of ISG15 was amplified from HeLa genomic DNA and constructed into pGL3-basic plasmid by using classical molecular cloning methods (82). The plasmids pISRE-Luc and pRL-TK were purchased from Beyotime (China). The coding sequences of HDAC1, HDAC2, HDAC3, HDAC8, STAT1, STAT2, IRF9, JAK1, and TYK2 were all amplified from cDNA of HEK-293T cells and inserted into pCDNA3.1- (+) expression vectors with a C-terminal flag tag. The ISGs containing IFIT1, IFIT2, IFIT3, IFIT5, OAS1, OASL, mix paired-like homeobox, ubiquitin-specific peptidase 18, guanylate binding protein 1, and mRNA-decapping enzyme 1a (DCP1A) were also amplified from cDNA of HEK-293T cells and inserted into pCDNA3.1- (+) expression vectors with a N-terminal flag tag. The truncated plasmids of pCDNA3.1-HDAC2(190–488)-Flag, pCDNA3.1-HDAC2(250–488)-Flag, pCDNA3.1-HDAC2(365–488)-Flag and the plasmids of pCDNA3.1-EGFP-HDAC2, pCDNA3.1-Mpro-mCherry, pEGFP-HDAC2(1–261), pEGFP-HDAC2(262–488), pEGFP-HDAC2(262–383), pEGFP-HDAC2(1–383), pEGFP-HDAC2(384–488), pEGFP-N15/N16-BFP-Flag were constructed using classical restriction enzyme cloning, or Gibson Assembly. Site-directed mutagenesis was performed to generate the seven mutants of HDAC2 with a C-terminal flag tag, including pCDNA3.1-HDAC2-Q254A, -Q261A, Q354A, Q365A, Q381A, Q383A, -Q261A-Q383A and the three mutants of DCP1A with a N-terminal flag tag, containing pCDNA3.1-DCP1A-Q330A, -Q343A, and -Q351A. The NS4AB of HCV and the PR of HIV were synthesized by Sangon Biotech with codon optimization. The fragment of NS2B-NS3 of ZIKV was amplified from the plasmid pACYC177-Natal-RGN (83). The above three proteases were all inserted into pCDNA3.1- (+) expression vectors with a C-terminal HA tag.
Cell culture, drug treatments, and transfection
HEK-293T cells and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (GenStar) and antibiotics. Vero E6 cells and A549 cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Gibco). Transfection of cells was performed using Lipofectamine 3000 (Life Technologies) in Opti-MEM (Life Technologies) according to the manufacturer’s instructions. Cells were treated with IFN-α2a (catalog no. 11101–2; PBL Assay Science) at 1000 units/ml, MG132 (MCE) at 20 μM, Z-VAD-FMK (APE x BIO) at 20 μM, and NH4Cl (sigma-Aldrich) at 10 mM.
Real time quantitative PCR
HEK-293T cells were seeded into 6-well plates and transfected with pCDNA3.1-Mpro-HA or pCDNA3.1-vector. After 24 h transfection, cells were stimulated with or without 1000 U/ml IFNα for another 8 h. Total RNA was isolated with TRIzol Reagent (Invitrogen) and reverse-transcribed into cDNA with a transcript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). Real-time quantitative PCR (RT-qPCR) assay were performed using a SYRB Green-based RT-qPCR kit (Takara). RT-qPCR experiments were performed in triplicates. The relative abundances of the mRNA transcripts were normalized to GAPDH. Primers used for qPCR experiments were as follows: qISG15-F: 5′-CGCAGATCACCCAGAAGATCG-3′; qISG15-R: 5′-TTCGTCGCATTTGTCCACCA-3′; qISG56-F: 5′-AGAAGCAGGCAATCACAGAAAA-3′; qISG56-R: 5′-CTGAAACCGACCATAGTGGAAAT-3′; qIFIT3-F: AAAAGCCCAACAACCCAGAAT; qIFIT3-R: CGTATTGGTTATCAGGACTCAGC; qOAS1-F: 5′- TGTCCAAGGTGGTAAAGGGTG-3′; qOAS1-R: 5′-CCGGCGATTTAACTGATCCTG-3′; qGAPDH-F: 5′- CCCTTCATTGACCTCAACT-3′; qGAPDH-R: 5′-GACGCCAGTGGACTCCA-3′.
Luciferase reporter assays
To assess the activity of the reporter plasmid of ISRE-Luc or ISG15-Luc, a dual-luciferase reporter assay was performed according to the manufacturer’s instructions (Promega). In brief, HEK-293T cells, Vero E6 cells, or A549 cells were seeded into 48-well plate (5 × 10ˆ4) in growth medium and cultured overnight. Cells were then co-transfected with 50 ng of reporter plasmid, 10 ng of pRL-TK, and either various concentrations (50 ng, 100 ng, or 200 ng) of pCDNA3.1-Mpro-HA or pCDNA3.1-vector. After 24 h, cells were stimulated with 1000 U/ml IFNα for another 12 h. The cell culture medium was removed following cytokine stimulation, then cells were lysed with 100 μl passive lysis buffer (Promega). To measure the luciferase activity, 50 μl firefly luciferase reagent was first added to 10 μl cell lysate, followed by 50 μl Renilla luciferase reagent. The luminescence value was measured with a Cytation3 Plate Reader and Imager (BioTek). Firefly luciferase values were normalized to the Renilla luciferase values, and experiments were performed in triplicate.
Western blot
HEK-293T cells were cultured in 6-well plates and collected with lysis buffer (RIPA Lysis Buffer, Millipore). Equivalent amounts of proteins were determined using an Enhanced BCA Protein Assay Kit (Beyotime) and then separated by SDS-PAGE and transferred using the Trans-Blot Turbo Transfer system (Bio-Rad) or wet transfer system. The following antibodies were used: Mouse (Ms) anti-FLAG (Sigma-Aldrich, F1804), Ms anti-HA (Cell Signaling, 2367), Ms anti-HA (Biolegend, 901501), Ms HRP-conjugated anti-Alpha Tubulin (Proteintech, HRP-66031), Ms HRP-conjugated anti-GAPDH (Yesen, 30203ES50), Rabbit (Rb) anti-STAT1 (Cell Signaling, 14994), Rb anti-STAT2 (Proteintech, 16674-1-AP), Ms anti-p-STAT1 (Santa Cruz, sc-136229), Rb anti-p-STAT2 (Cell Signaling, 4441), and Ms anti-IRF9 (Santa Cruz, sc-365893); Ms anti-DCP1A (Santa Cruz, sc-100706); Rb anti-mCherry (Abcam, ab213511); Rb anti-PCNA (Sino biological, 101239-T46).
Immunoprecipitation
HEK-293T cells were cultured in 10 cm plates and collected with lysis buffer (RIPA Lysis Buffer, Millipore). Cell lysates were first incubated with a specific antibody overnight at 4 °C with rotation. The lysate and antibody complex solution were then incubated with protein A/G magnetic beads (MCE, HY-K0202) for 4 h at 4 °C with rotation. After washing four times with PBS, samples were analyzed by Western blotting. The following antibodies were used for immunoprecipitation analysis: Rb anti-HDAC2 (Cell Signaling, 57156), Ms anti-HDAC2 (Santa Cruz, sc-9959), Rb anti-HDAC1 (Abcam, ab280198), Ms anti-HDAC1 (Cell Signaling, 5356), Rb anti-HDAC3 (Proteintech, 10255-1-AP), and Ms anti-HDAC3 (Proteintech, 67151).
Indirect immunofluorescence
HeLa cells were grown on glass bottom cell culture dish (NEST) and transfected with Mpro-mCherry construct, and after 24 h of expressing, cells were treated with 1000 U/ml IFNα for 1 h, then cells were fixed in 1 ml of 4% formaldehyde for 30 min and permeabilized in 0.5% TritonX-100 for 10 min. After washing with PBS, samples were blocked with 10% goat serum for 1 h and incubated with the anti-STAT1(Cell Signaling, 14994), anti-STAT2 (Proteintech, 16674-1-AP), or anti-IRF9 (Santa Cruz, sc-365893) antibody overnight at 4 °C. Goat anti-Rabbit IgG (Alexa Fluor 488, ab150081) or Goat Anti-Mouse IgG (Alexa Fluor 488, ab150113) was used as the secondary antibody at 1:1000 dilution. DAPI (Beyotime) was used as nuclear counterstain. Images were captured by using Nikon A1R+ Inverted Microscope with a 60 × 1.4 NA oil-immersion objective.
RNA sequencing
Total RNA was extracted using a Trizol reagent kit (Invitrogen) according to the manufacturer’s instructions. RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) and checked by RNase-free agarose gel electrophoresis. After extraction of total RNA, eukaryotic mRNA was enriched by removing rRNA by Ribo-ZeroTM Magnetic Kit (Epicentre). Enriched mRNA was then processed into short fragments using fragmentation buffer before being reverse-transcripted into cDNA using random primers. Second-strand cDNA was synthesized using DNA polymerase I, RNase H, dNTP, and buffer. The cDNA fragments were then purified using a QiaQuick PCR extraction kit (Qiagen), end repaired, poly(A) added, and ligated to Illumina sequencing adapters. The ligation products were size-selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeq2500 by Gene Denovo Biotechnology Co.
Molecular docking analysis
Protein–peptide interactions were modeled using the CABS-dock web server (http://biocomp.chem.uw.edu.pl/CABSdock/). We set the contact information in web with 41: A 4: PEP 5.0 1.0, 145: A 4: PEP 5.0 1.0 for SARS-CoV-2 (PDB code: 7jyc) and 41: A 4: PEP 5.0 1.0, 144: A 4: PEP 5.0 1.0 for hCoV-229E (PDB code: 2zu2). Molecular dynamics simulations and binding energy calculations were performed on Yinfo Cloud Computing Platform (https://cloud.yinfotek.com/) using the AmberTools 20 package. AMBER ff19SB force filed was used for protein according to the manufacturer’s instructions.
Statistical analysis
All data are presented as one replicate from three independent experiments. Statistical analysis between groups was performed by unpaired two-tailed Student’s t test. Data are reported as mean±SD. For all experiments, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, ∗∗∗∗ p < 0.0001 were considered to be statistically significant.
Data availability
All other data are included in this article and its supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare there are no conflicts of interest with the contents of this article.
Acknowledgments
We thank the Genedenovo Biotechnology Co for technical support with the RNA sequencing. Special thanks to YINFO TECHNOLOGY for technical support with molecular dynamics simulation. We thank T. Juelich (UCAS, Beijing) for linguistic assistance during the preparation of this manuscript.
Author contributions
L. S. and X.-E. Z. conceptualization; L. S. and X.-E. Z. methodology; L. S. and X.-E. Z. writing–original draft; L. S. investigation; L. S., X.-E. Z., G. A., D. B. W., M. M. C., M. L., Z. L. L., and J. F. W. writing–review and editing.
Funding and additional information
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB29050100), the National Key Research and Development Program of China (Grant No. 2020YFC0861100), the Program of the Chinese Academy of Sciences (Grant No. 2020YJFK-Z-0150), and the National Natural Science Foundation of China (Grant No. 21890743).
Reviewed by members of the JBC Editorial Board. Edited by Craig Cameron
Contributor Information
Jufang Wang, Email: jufwang@scut.edu.cn.
Xian-En Zhang, Email: zhangxe@ibp.ac.cn.
Supporting information
References
- 1.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pormohammad A., Ghorbani S., Baradaran B., Khatami A., R J.T., Mansournia M.A., et al. Clinical characteristics, laboratory findings, radiographic signs and outcomes of 61,742 patients with confirmed COVID-19 infection: a systematic review and meta-analysis. Microb. Pathog. 2020;147:104390. doi: 10.1016/j.micpath.2020.104390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramlall V., Thangaraj P.M., Meydan C., Foox J., Butler D., Kim J., et al. Immune complement and coagulation dysfunction in adverse outcomes of SARS-CoV-2 infection. Nat. Med. 2020;26:1609–1615. doi: 10.1038/s41591-020-1021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucas C., Wong P., Klein J., Castro T.B., Silva J., Sundaram M., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fensterl V., Sen G.C. Interferons and viral infections. Biofactors. 2009;35:14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 12.McNab F., Mayer-Barber K., Sher A., Wack A., O'Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 14.Liu S.Y., Sanchez D.J., Aliyari R., Lu S., Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane M., Zang T.M., Rihn S.J., Zhang F., Kueck T., Alim M., et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe. 2016;20:392–405. doi: 10.1016/j.chom.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoggins J.W. Interferon-stimulated genes: roles in viral pathogenesis. Curr. Opin. Virol. 2014;6:40–46. doi: 10.1016/j.coviro.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maher S.G., Sheikh F., Scarzello A.J., Romero-Weaver A.L., Baker D.P., Donnelly R.P., et al. IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol. Ther. 2008;7:1109–1115. doi: 10.4161/cbt.7.7.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuno T., Mejido J., Zhao T., Schmeisser H., Morrow A., Zoon K.C. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha. J. Immunother. 2009;32:803–816. doi: 10.1097/CJI.0b013e3181ad4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egli A., Santer D.M., O'Shea D., Tyrrell D.L., Houghton M. The impact of the interferon-lambda family on the innate and adaptive immune response to viral infections. Emerg. Microbes Infect. 2014;3:e51. doi: 10.1038/emi.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kroetz D.N., Allen R.M., Schaller M.A., Cavallaro C., Ito T., Kunkel S.L. Type I interferon induced epigenetic regulation of macrophages suppresses innate and adaptive immunity in acute respiratory viral infection. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tough D.F. Type I interferon as a link between innate and adaptive immunity through dendritic cell stimulation. Leuk. Lymphoma. 2004;45:257–264. doi: 10.1080/1042819031000149368. [DOI] [PubMed] [Google Scholar]
- 22.Au-Yeung N., Mandhana R., Horvath C.M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2 doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Veer M.J., Holko M., Frevel M., Walker E., Der S., Paranjape J.M., et al. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 24.Shaw A.E., Hughes J., Gu Q., Behdenna A., Singer J.B., Dennis T., et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017;15 doi: 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vladimer G.I., Gorna M.W., Superti-Furga G. IFITs: emerging roles as key anti-viral proteins. Front Immunol. 2014;5:94. doi: 10.3389/fimmu.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakrabarti A., Jha B.K., Silverman R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011;31:49–57. doi: 10.1089/jir.2010.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danziger O., Patel R.S., DeGrace E.J., Rosen M.R., Rosenberg B.R. Inducible CRISPR activation screen for interferon-stimulated genes identifies OAS1 as a SARS-CoV-2 restriction factor. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang H.-M., Paulson M., Holko M., Rice C.M., Williams B.R., Marié I., et al. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9578–9583. doi: 10.1073/pnas.0400567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusinzon I., Horvath C.M. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:14742–14747. doi: 10.1073/pnas.2433987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marie I.J., Chang H.M., Levy D.E. HDAC stimulates gene expression through BRD4 availability in response to IFN and in interferonopathies. J. Exp. Med. 2018;215:3194–3212. doi: 10.1084/jem.20180520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y., Stuart J.H., Talbot-Cooper C., Agrawal-Singh S., Huntly B., Smid A.I., et al. Histone deacetylase 4 promotes type I interferon signaling, restricts DNA viruses, and is degraded via vaccinia virus protein C6. Proc. Natl. Acad. Sci. U. S. A. 2019;116:11997–12006. doi: 10.1073/pnas.1816399116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galani I.E., Rovina N., Lampropoulou V., Triantafyllia V., Manioudaki M., Pavlos E., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X., Fang L., Wang D., Yang Y., Chen J., Ye X., et al. Porcine deltacoronavirus nsp5 inhibits interferon-β production through the cleavage of NEMO. Virology. 2017;502:33–38. doi: 10.1016/j.virol.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung S.Y., Siu K.L., Lin H., Yeung M.L., Jin D.Y. SARS-CoV-2 main protease suppresses type I interferon production by preventing nuclear translocation of phosphorylated IRF3. Int. J. Biol. Sci. 2021;17:1547–1554. doi: 10.7150/ijbs.59943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roe M.K., Junod N.A., Young A.R., Beachboard D.C., Stobart C.C. Targeting novel structural and functional features of coronavirus protease nsp5 (3CL(pro), M(pro)) in the age of COVID-19. J. Gen. Virol. 2021;102:001558. doi: 10.1099/jgv.0.001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Sancho L., Lewinski M.K., Pache L., Stoneham C.A., Yin X., Becker M.E., et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell. 2021;81:2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickenhagen A., Sugrue E., Lytras S., Kuchi S., Noerenberg M., Turnbull M.L., et al. A prenylated dsRNA sensor protects against severe COVID-19. Science. 2021;374 doi: 10.1126/science.abj3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tecalco Cruz A.C. Free ISG15 and protein ISGylation emerging in SARS-CoV-2 infection. Curr. Drug Targets. 2022;23:686–691. doi: 10.2174/1389450123666220316094720. [DOI] [PubMed] [Google Scholar]
- 41.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubel P., Urban C., Bergant V., Schneider W.M., Knauer B., Stukalov A., et al. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 2019;20:493–502. doi: 10.1038/s41590-019-0323-3. [DOI] [PubMed] [Google Scholar]
- 45.Zhu X., Wang D., Zhou J., Pan T., Chen J., Yang Y., et al. Porcine deltacoronavirus nsp5 antagonizes type I interferon signaling by cleaving STAT2. J. Virol. 2017;91:e00003–e00017. doi: 10.1128/JVI.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J., Kenward C., Worrall L.J., Vuckovic M., Gentile F., Ton A.T., et al. X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation. Nat. Commun. 2022;13:5196. doi: 10.1038/s41467-022-32854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schindler C., Levy D.E., Decker T. JAK-STAT signaling: from interferons to cytokines. J. Biol. Chem. 2007;282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 49.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.RUIJTER A.J. M.d., GENNIP A.H.v., CARON H.N., KEMP S., KUILENBURG A.B. P.v. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Icardi L., Lievens S., Mori R., Piessevaux J., De Cauwer L., De Bosscher K., et al. Opposed regulation of type I IFN-induced STAT3 and ISGF3 transcriptional activities by histone deacetylases (HDACS) 1 and 2. FASEB J. 2012;26:240–249. doi: 10.1096/fj.11-191122. [DOI] [PubMed] [Google Scholar]
- 53.Icardi L., Mori R., Gesellchen V., Eyckerman S., De Cauwer L., Verhelst J., et al. The Sin3a repressor complex is a master regulator of STAT transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 2012;109:12058–12063. doi: 10.1073/pnas.1206458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelly R.D., Cowley S.M. The physiological roles of histone deacetylase (HDAC) 1 and 2: Complex co-stars with multiple leading parts. Biochem. Soc. Trans. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- 55.Qian S., Fan W., Liu T., Wu M., Zhang H., Cui X., et al. Seneca valley virus suppresses host type I interferon production by targeting adaptor proteins MAVS, TRIF, and TANK for cleavage. J. Virol. 2017;91 doi: 10.1128/JVI.00823-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J., Wang D., Sun Z., Gao L., Zhu X., Guo J., et al. Arterivirus nsp4 antagonizes interferon beta production by proteolytically cleaving NEMO at multiple sites. J. Virol. 2019;93 doi: 10.1128/JVI.00385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D., Fang L., Li K., Zhong H., Fan J., Ouyang C., et al. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 2012;86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., et al. Porcine epidemic diarrhea virus 3C-like protease regulates its interferon antagonism by cleaving NEMO. J. Virol. 2016;90:2090–2101. doi: 10.1128/JVI.02514-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park S.Y., Kim J.S. A short guide to histone deacetylases including recent progress on class II enzymes. Exp. Mol. Med. 2020;52:204–212. doi: 10.1038/s12276-020-0382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thornbrough J.M., Jha B.K., Yount B., Goldstein S.A., Li Y., Elliott R., et al. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio. 2016;7 doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu X., Chen J., Tian L., Zhou Y., Xu S., Long S., et al. Porcine deltacoronavirus nsp5 cleaves DCP1A to decrease its antiviral activity. J. Virol. 2020;94 doi: 10.1128/JVI.02162-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kraus T.A., Lau J.F., Parisien J.P., Horvath C.M. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J. Biol. Chem. 2003;278:13033–13038. doi: 10.1074/jbc.M212972200. [DOI] [PubMed] [Google Scholar]
- 64.Chen D., Xu L.-G., Chen L., Li L., Zhai Z., Shu H.-B. NIK is a component of the EGF/heregulin receptor signaling complexes. Oncogene. 2003;22:4348–4355. doi: 10.1038/sj.onc.1206532. [DOI] [PubMed] [Google Scholar]
- 65.Malinin N.L., Boldin M.P., Kovalenko A.V., Wallach D. MAP3K-related kinase involved in NF-kB induction by TNF, CD95 and IL-1. Nature. 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 66.Shakespear M.R., Halili M.A., Irvine K.M., Fairlie D.P., Sweet M.J. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 67.Hull E.E., Montgomery M.R., Leyva K.J. HDAC inhibitors as epigenetic regulators of the immune system: impacts on cancer therapy and inflammatory diseases. Biomed. Res. Int. 2016;2016:8797206. doi: 10.1155/2016/8797206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taplick J., Kurtev V., Kroboth K., Posch M., Lechner T., Seiser C. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J. Mol. Biol. 2001;308:27–38. doi: 10.1006/jmbi.2001.4569. [DOI] [PubMed] [Google Scholar]
- 69.Micelli C., Rastelli G. Histone deacetylases: structural determinants of inhibitor selectivity. Drug Discov. Today. 2015;20:718–735. doi: 10.1016/j.drudis.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 70.Naik N.G., Lee S.-C., Veronese B.H., Ma Z., Toth Z. Interaction of HDAC2 with SARS-CoV-2 NSP5 and IRF3 is not required for NSP5-mediated inhibition of type I interferon signaling pathway. Microbiol. Spectr. 2022;10:e02322–02322. doi: 10.1128/spectrum.02322-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dougherty J.D., Reineke L.C., Lloyd R.E. mRNA decapping enzyme 1a (Dcp1a)-induced translational arrest through protein kinase R (PKR) activation requires the N-terminal enabled vasodilator-stimulated protein homology 1 (EVH1) domain. J. Biol. Chem. 2014;289:3936–3949. doi: 10.1074/jbc.M113.518191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dougherty J.D., White J.P., Lloyd R.E. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 2011;85:64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao R., Fang L., Bai D., Ke W., Zhou Y., Wang D., et al. Porcine reproductive and respiratory syndrome virus nonstructural protein 4 cleaves porcine DCP1a to attenuate its antiviral activity. J. Immunol. 2018;201:2345–2353. doi: 10.4049/jimmunol.1701773. [DOI] [PubMed] [Google Scholar]
- 74.Moustaqil M., Ollivier E., Chiu H.P., Van Tol S., Rudolffi-Soto P., Stevens C., et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infect. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J., Li Z., Guo J., Xu S., Zhou J., Chen Q., et al. SARS-CoV-2 nsp5 exhibits stronger catalytic activity and interferon antagonism than its SARS-CoV ortholog. J. Virol. 2022;96 doi: 10.1128/jvi.00037-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glaser K.B., Staver M.J., Waring J.F., Stender J., Ulrich R.G., Davidsen S.K. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 77.Wilting R.H., Yanover E., Heideman M.R., Jacobs H., Horner J., van der Torre J., et al. Overlapping functions of Hdac1 and Hdac2 in cell cycle regulation and haematopoiesis. EMBO J. 2010;29:2586–2597. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Telles E., Seto E. Modulation of cell cycle regulators by HDACs. Front. Biosci. 2012;4:831. doi: 10.2741/s303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roger T., Lugrin J., Le Roy D., Goy G., Mombelli M., Koessler T., et al. Histone deacetylase inhibitors impair innate immune responses to toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 80.Bodas M., Mazur S., Min T., Vij N. Inhibition of histone-deacetylase activity rescues inflammatory cystic fibrosis lung disease by modulating innate and adaptive immune responses. Respir. Res. 2018;19:2. doi: 10.1186/s12931-017-0705-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Walters M.S., Kinchington P.R., Banfield B.W., Silverstein S. Hyperphosphorylation of histone deacetylase 2 by alphaherpesvirus US3 kinases. J. Virol. 2010;84:9666–9676. doi: 10.1128/JVI.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reich N., Evans B., Levy D., Fahey D., Knight E., Jr., Darnell J.E., Jr. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li S., Wang D., Ghulam A., Li X., Li M., Li Q., et al. Tracking the replication-competent Zika virus with tetracysteine-tagged capsid protein in living cells. J. Virol. 2022;96 doi: 10.1128/jvi.01846-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All other data are included in this article and its supporting information.