Abstract
FIMM database (http://sdmc.krdl.org.sg:8080/fimm) contains data relevant to functional molecular immunology, focusing on cellular immunology. It contains fully referenced data on protein antigens, major histocompatibility complex (MHC) molecules, MHC-associated peptides and relevant disease associations. FIMM has a set of search tools for extraction of information and results are presented as lists or as reports.
INTRODUCTION
T cells of the immune system are involved in cell-mediated immunity, regulation of immune responses and regulation of antibody production (humoral immunity). T cells recognise antigenic structures using T-cell receptors (TcR) to discriminate self versus non-self. Peptides generated by processing of protein antigens bind major histocompatibility complex (MHC) molecules, and are presented on the cell surface for recognition by TcR. MHC-associated peptides which induce T cell responses are termed T-cell epitopes.
The diversity of immune receptors arises by several mechanisms including phenotypic variation (MHC), allelic variation of genes (MHC, TcR), combining of chains in heterodimeric molecules (some MHC molecules, TcR), combinatorial joining of gene segments (TcR) or insertion of nucleotides at gene segment junctions (TcR). The MHC gene family is highly polymorphic—more than 1100 allelic variants of human leukocyte antigens (HLA, human MHC) have been characterised (1). The diversity of immune system receptors allows an immune system to initiate and regulate appropriate responses. Hundreds of disease-specific antigens have been identified and reported. Thousands of peptides have been reported to bind various MHC molecules or stimulate immune responses (2,3). Sets of peptides that are presented by different MHC molecules may overlap to various degrees, or may be exclusive. A number of associations between HLA genes and susceptibility (or protection) to diseases have been described (4). This complexity has created a need for a database that integrates data on functional aspects of molecular immunology.
FIMM contains fully referenced data on protein antigens, MHC molecules, MHC-associated peptides and relevant disease associations. A set of search and querying tools allow users to perform specific queries and combine different views of data. Extracted information is in the form of reports or lists containing hyperlinks to other sources that provide more detailed or specialised information. The reports and lists are designed to facilitate data interpretation and help design related experiments. Data in FIMM originate from various sources including literature, public databases and HLA workshop reports. FIMM is designed to assist both basic and applied research in molecular immunology.
The new entries, since the version 1.0 (5), include 13 diseases, 33 disease associations, 358 HLA-binding peptides, 108 antigens, almost 400 HLA entries, and more than 500 references. New features of the FIMM (version 1.2) are the visualisation of three-dimensional (3D) structures of HLA molecules and integration of searchable database of 38 235 related antigens from the non-redundant SWISS-PROT/TrEMBL database. Each related antigen contains one or more MHC-associated peptides that have FIMM entries. The current FIMM (version 1.2) contains data on 571 protein antigens, 1591 peptides, 1390 HLA sequences, 65 diseases, 52 disease associations and 2815 references. In addition, FIMM version 1.2 contains nearly 60 PDB (6) structures of HLA molecules and 500 entries of 3D structures of empty (without peptide) HLA-A, -B and -C molecules, derived by homology modelling.
DESCRIPTION
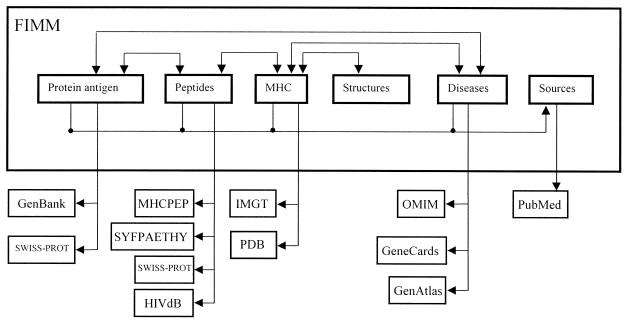
FIMM provides (i) a unique compilation of information relevant to molecular immunology, (ii) a means for extraction of this information, including the analysis of query antigens, and (iii) hyperlink access to related information available in the external sources (Fig. 1). The dimensional data model (7) of FIMM is given in Table 1. The current FIMM data model has six dimensions (or views): protein antigens, peptides, MHC, structures, diseases and publication sources (Fig. 1). FIMM can be queried for specific information within a particular view. A set of generic tools allows keyword searches and sequence comparison analysis. An online documentation provides help for use and the description of the database.
Figure 1.
Data views in FIMM, internal links and links to the external sources.
Table 1. Data dimensions in FIMM.
| Protein antigens | Peptides | MHC | Structures | Diseases | Sources |
|---|---|---|---|---|---|
| ID | ID | ID | Structure ID | ID | PMID |
| Date | Date | Date | Viewer | Date | Title |
| Name | Sequence | Name | Links | Name | Authors |
| Aliases | MHC | Aliases | MHC | Aliases | Journal |
| Features | T-cell epitope | Sequence | Etiology | Links | |
| Sequence | Naturally processed | Disease assoc. | Disease assoc. | PubMed | |
| Links | Peptide binding | Binding pockets | Links | ||
| Peptides | Links | CD8 binding sites | Protein antigens | ||
| Diseases | Protein antigens | TCR binding sites | MHC | ||
| Sources | MHC | Alignments | Sources | ||
| SWISS-PROT | Sources | Sources | OMIM | ||
| GenBank | MHCPEP | Links | GeneCards | ||
| SYFPAETHY | Peptides | GenAtlas | |||
| HIVdb | Diseases | ||||
| SWISS-PROT | IMGTdb | ||||
| PDB | |||||
| Homology str. |
Peptides
Peptides reported as naturally processed and presented by MHC molecules or as T cell epitopes are factual entries for the dimension ‘Peptides’. Besides common fields, each entry contains information on peptide/MHC association and whether the peptide is naturally processed or a reported T cell epitope. A peptide entry contains the information on peptide binding affinity when available, using the notation from the MHCPEP database (high, moderate or low binding affinity). Major sources of peptide data are MHCPEP database (3), SYFPEITHI database (2), HIV molecular immunology database (8; http://hiv-web.lanl.gov/immunology), and published reports.
Common fields
Each factual entry in the four data dimensions ‘Protein antigens’, ‘Peptides’, ‘MHC’ and ‘Diseases’ has a FIMM unique identifier (ID) and the date of entry (Date). The ‘Sources’ use PubMed identifiers or PMIDs, and ‘Structures’ use HLA allele names as identifiers. Factual entries for ‘Protein antigens’, ‘MHC’ and ‘Diseases’ also have the name of the entry (Name) and the list of alternative names (Aliases). Amino acid sequence information (Sequence) is available for ‘Protein antigens’, ‘Peptides’ and ‘MHC’.
Protein antigens
Proteins that are reported either as sources of T cell epitopes or of naturally processed peptides, are factual entries for the dimension ‘Protein antigens’. For each entry, the most relevant sequence is provided along with links to related entries from the SWISS-PROT (9) and GenBank (10) databases. Sequence features contain information extracted from the descriptions in the SWISS-PROT or GenBank database entries.
MHC view
Factual entries of MHC contain the common fields (as described earlier in the text) and the publication sources. In addition, the ‘MHC’ dimension contains information on MHC binding pocket composition (11), co-receptor (CD8) binding sites (12) and TcR binding sites (13). Binding pocket analysis allows the selection and the inspection of binding pockets for a subset of MHC alleles for a specific phenotype. The major source of MHC sequence information for FIMM is the IMGT/HLA database (14).
HLA structures
FIMM contains 3D structures of HLA molecules. The 3D structures include entries extracted from the PDB database (6). FIMM also contains homology models of the majority of HLA-A, -B and -C structures. These predicted structure entries were generated based on a representative set of published class I MHC crystal structures available in the PDB. Protein models were built using the program modeller (15) and briefly energy minimised in a solvent bath using the molecular mechanics program AMBER (16). The HLA Structure view allows users to display and download available 3D structures using the Chime viewer (www.mdlchime.com/chime). Users can display molecular models in various representations, such as line model, ball-and-stick model or ribbon model.
Diseases
A factual entry from the dimension ‘Diseases’ contains the following information: common fields, etiology (e.g. virus, cancer, etc.) and MHC disease association. External links include OMIM (17), GeneCards (18) and GenAtlas (19) databases.
Sources
The ‘Sources’ dimension contains publication references, namely the author names, title, journal and the links to PubMed (www.ncbi.nlm.nih.gov/PubMed/) records. Users can search references by keywords, author names or title words.
Search tools
FIMM integrates several tools for data searching. Searches using keywords (both full and partial) are available in each dimension except structure. FIMM allows the creation of lists of related entries. The resulting lists contain summaries of factual entries (e.g. diseases, protein antigens, MHC, peptides and reference sources) generated according to various grouping criteria. Grouping criteria are usually defined by user-provided keywords. Alternatively, specific grouping criteria are also predefined in FIMM menus (e.g. list HLA alleles by loci, diseases by etiology, peptides by MHC molecules, etc.).
Users can compare their query sequence to FIMM entries using BLAST2 (20) searches. A query sequence can be compared with either protein antigens or MHC sequences in FIMM. The HLA alignment tool uses CLUSTALW (21) for sequence alignment and MView (22) for alignment coloring. The peptide search tool uses a local string-matching search program or a Smith–Waterman algorithm customised for finding short sequences. These algorithms enable users to search FIMM database for antigens that contain a subsequence identical (MapSequence) or similar (SW) to a query peptide. The binding pocket tool displays the amino acids that form binding pockets in the groove of known HLA molecules. It also enables the alignment of a query MHC sequence to the known HLA sequence and identification of pocket-forming positions in the query sequence.
DATA ANNOTATION
FIMM has been compiled mainly from published reports, journal articles and public databases. Cross-checking of data for both accuracy and redundancy is performed routinely. The inclusion of HLA sequences is based on publications by the HLA nomenclature committee (1). The criteria for the inclusion of a peptide into FIMM are that it has a complete sequence and has been previously published. Disease associations have been compiled from selected publications, such as HLA workshop proceedings (23) or journal articles.
DATABASE ACCESS
FIMM is available via the World Wide Web (http://sdmc.krdl.org.sg:8080/fimm). FIMM has been designed to be robust and user-friendly. The user interface uses a set of simple Graphical User Interface forms. The data are stored in a flat (semi-structured), comprehensible and easy to access database files. Methods for searching the databases and displaying selected tables are built with a combination of Java, Perl, HTML, C shell scripts, HTML and C programs. FIMM system is implemented in a UNIX environment.
Authors whose research has been assisted by using FIMM should cite this article as the reference.
FUTURE WORK
Version 1.2 of FIMM contains information on human MHC (HLA) and human diseases. Future work will include relevant information from other organisms including laboratory animals and livestock. Additional data dimensions and facilities planned for future FIMM developments include antigen processing, TcR interactions, cytokines and various prediction tools.
REFERENCES
- 1.Marsh S.G., Bodmer,J.G., Albert,E.D., Bodmer,W.F., Bontrop,R.E., Dupont,B., Erlich,H.A., Hansen,J.A., Mach,B., Mayr,W.R. et al. (2001) Nomenclature for factors of the HLA system, 2000. Hum. Immunol., 62, 419–468. [DOI] [PubMed] [Google Scholar]
- 2.Rammensee H., Bachmann,J., Emmerich,N.P., Bachor,O.A. and Stevanovic,S. (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics, 50, 213–219. [DOI] [PubMed] [Google Scholar]
- 3.Brusic V., Rudy,G. and Harrison,L.C. (1998) MHCPEP, a database of MHC-binding peptides: update 1997. Nucleic Acids Res., 26, 368–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorsby E. (1997) Invited anniversary review: HLA associated diseases. Hum. Immunol., 53, 1–11. [DOI] [PubMed] [Google Scholar]
- 5.Schönbach C., Koh,J.L., Sheng,X., Wong,L. and Brusic,V. (2000) FIMM, a database of functional molecular immunology. Nucleic Acids Res., 28, 222–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. Updated article in this issue: Nucleic Acids Res. (2002), 30, 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballard C., Herreman,D., Schau,D., Bell,R., Kim,E. and Valencic,A. (1998) Data Modeling Techniques for Data Warehousing. IBM International Support Organization, San Jose, CA.
- 8.Korber B.T.M., Moore,J.P., Brander,C., Walker,B.D., Haynes,B.F. and Koup,R. (1998) HIV Molecular Immunology Compendium. Los Alamos National Laboratory: Theoretical Biology and Biophysics, Los Alamos, NM.
- 9.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson D.A., Karsch-Mizrachi,I., Lipman,D.J., Ostell,J., Rapp,B.A. and Wheeler,D.L. (2000) GenBank. Nucleic Acids Res., 28, 15–18. Updated article in this issue: Nucleic Acids Res., 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelvanayagam G. (1997) A roadmap for HLA-DR peptide binding specificities. Hum. Immunol., 58, 61–69. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan J.A., Oettinger,H.F., Sachs,D.H. and Edge,A.S. (1997) Analysis of polymorphism in porcine MHC class I genes: alterations in signals recognized by human cytotoxic lymphocytes. J. Immunol., 159, 2318–2326. [PubMed] [Google Scholar]
- 13.Hashimoto K., Okamura,K., Yamaguchi,H., Ototake,M., Nakanishi,T. and Kurosawa,Y. (1999) Conservation and diversification of MHC class I and its related molecules in vertebrates. Immunol. Rev., 167, 81–100. [DOI] [PubMed] [Google Scholar]
- 14.Robinson J., Waller,M.J., Parham,P., Bodmer,J.G. and Marsh,S.G. (2001) IMGT/HLA Database—a sequence database for the human major histocompatibility complex. Nucleic Acids Res., 29, 210–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sali A. and Blundell,T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol., 234, 779–815. [DOI] [PubMed] [Google Scholar]
- 16.Cornell W.D., Cieplak,P., Bayly,C.I., Gould,I.R., Merz,K.M.,Jr, Ferguson,D.M., Spellmeyer,D.C., Fox,T., Caldwell,J.W. and Kollman,P.A. (1995) A second generation force field for the simulation of proteins and nucleic acids. J. Am. Chem. Soc., 117, 5179–5197. [Google Scholar]
- 17.McKusick V.A. (1998) Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders. Johns Hopkins University Press, Baltimore, MD.
- 18.Rebhan M., Chalifa-Caspi,V., Prilusky,J. and Lancet,D. (1998) GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics, 14, 656–664. [DOI] [PubMed] [Google Scholar]
- 19.Frezal J. (1998) Genatlas database, genes and development defects. C. R. Acad. Sci. III, 321, 805–817. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown N.P., Leroy,C. and Sander,C. (1998) MView: a web-compatible database search or multiple alignment viewer. Bioinformatics, 14, 380–381. [DOI] [PubMed] [Google Scholar]
- 23.Charron D. (ed.) (1997) Proceedings of the Twelfth International Histocompatibility Workshop and Conference. EDK, Paris, France.