Background:
Circulating tumor DNA (ctDNA) positivity has been shown to suggest the presence of minimally residual tumor cells in numerous investigations. We aimed to assess the prognostic value of ctDNA positivity for recurrence-free survival in patients with early-stage colorectal cancer after radical surgery and following adjuvant chemotherapy.
Methods:
We systematically reviewed studies published in English until August 15, 2022, concerning ctDNA and tumor-node-metastasis I to III colorectal cancer after surgery, and quantified the correlation between ctDNA positivity and early-stage (tumor-node-metastasis stage I–III) colorectal cancer using meta-analysis methods.
Results:
In total, the meta-analysis comprised 1713 patients from 6 studies. Patients with ctDNA-positive colorectal cancer after surgery had a significantly higher risk of recurrence than patients with ctDNA-negative colorectal cancer (hazard ratio 4.64, 95% confidence interval 2.17–9.92, z = 3.96; P < .001). After adjuvant chemotherapy, patients who were ctDNA-positive had a significantly higher risk of recurrence than those who were ctDNA-negative (hazard ratio 7.27, 95% confidence interval 4.50–11.75, z = 8.1; P < .001).
Conclusions:
CtDNA positivity may potentially be a predictor for early-stage colorectal tumor recurrence following surgery and adjuvant chemotherapy.
Keywords: adjuvant chemotherapy, colorectal cancer, ctDNA, hazard ratio, surgery
1. Introduction
The incidence of colorectal cancer is increasing annually worldwide, and the mortality rate is among the highest of all cancers. Most early-stage (tumor-node-metastasis [TNM] stage I–III) colorectal cancers are curable with surgery; however, approximately 20% of patients with colorectal cancer experience recurrence in the postoperative period.[1–3] Currently, assessments of microsatellite instability (MSI) status; TNM stage; BRAF, KRAS, and NRAS mutation status; lymphatic vascular infiltration; tumor budding; and tumor differentiation are used to predict the risk of tumor recurrence.[4] Adjuvant chemotherapy, initiated to reduce the risk of recurrence, is mainly based on postoperative clinicopathology and a patient’s physical tolerance.[5,6] Regular postoperative clinical follow-up involves monitoring of tumor markers such as MSI, carcinoembryonic antigen, and CA19-9, with chest computed tomography (CT), abdominal enhanced CT, and colonoscopy used for detection of tumor recurrence.[6] Biomarkers such as MSI are only effective in a small percentage of patients with colon cancer and high MSI (MSI-H).[7] In addition, overuse of chemotherapy leads to unacceptable drug toxicity, and insufficient chemotherapy leads to recurrence.[5,8] These are practical challenges, and finding more accurate biomarkers for monitoring and early detection of recurrence would be helpful.
Circulating tumor DNA (ctNDA) is released from tumor cells and can be detected in body fluids through various technologies. CtDNA strands contain tumor-specific alterations in tumor suppressor genes or oncogenes, along with MSI and DNA hypermethylation.[9,10] An increasing number of studies have shown that patients who are ctDNA-positive postoperatively have up to 80% higher risk of recurrence and lower rates of disease-free survival.[11–13] CtDNA testing is considered noninvasive and ctDNA is a promising biomarker.[14] Moreover, the recurrence rate in patients with negative ctDNA after the first surgery is approximately 20%, although ctDNA may become positive in subsequent long-term monitoring.[15] However, it remains controversial whether ctDNA can be used in scheduled clinical monitoring and in guiding clinical treatment.
In this meta-analysis, we included recent studies on ctDNA and early-stage colorectal cancer to evaluate the correlation between ctDNA and recurrence-free survival (RFS) in patients who had undergone surgery for colorectal cancer.
2. Methods
The Preferred Reporting Items for Systematic Reviews and Meta-Analysis Protocols declaration has been followed. The International Prospective Register of Systematic Reviews registration number for this study is CRD42023389305.
2.1. Search strategy
We searched a range of computerized databases, including Medline and Cochrane Central, for articles published in English up to August 15, 2022. We used a subject and text word strategy, using (colorectal OR colon OR colonic) AND (cancer OR neoplasms) AND (circulating tumor DNA OR ctDNA) as the primary search terms. The search strategy was tailored to each database to ensure a comprehensive search.
2.2. Selection of studies and data extraction
The inclusion criteria were as follows: patients aged ≥18 years diagnosed with nonmetastatic and operable early-stage colorectal cancer, who were involved in studies where ctDNA mutation was assessed using plasma or serum; endpoints included RFS according to ctDNA results after surgery; and sufficient data were presented for determining or calculating the hazard ratio (HR) and 95% confidence interval (CI). Only randomized controlled trials, prospective cohort studies, and retrospective studies were included. We excluded duplicated articles, animal experiments, case reports, review articles or meta-analyses, articles only concerning rectal cancer, and studies with insufficient data.
Two independent reviewers (Fan and Zhang) screened the titles and abstracts identified using an electronic search to identify relevant studies. Relevant studies were further examined to determine whether they met the inclusion criteria. Potentially relevant studies were obtained, and the full-text articles were screened for inclusion by 2 independent reviewers (Fan and Zhang). We resolved disagreements through discussion. The included studies were summarized in data extraction forms, and the authors were contacted when relevant data were missing.
The name of the first author, year of publication, type of study, number of patients, median age, sex, tumor stage, sample origin, status of ctDNA, postoperative ctDNA measuring time, detection of genes, follow-up duration, HR, and 95% CI for RFS according to ctDNA status after surgery and after adjuvant chemotherapy were extracted.
2.3. Quality assessment and data analysis
The quality of all included studies was evaluated using the Newcastle–Ottawa Quality Assessment Scale (NOS).[16] The NOS contains 8 items categorized into 3 dimensions, including selection, comparability, and study type-outcome (cohort studies) or exposure (case-control studies). A star system is used for a semiquantitative assessment of study quality. The NOS score ranges from 0 to 9 stars.
Data were analyzed using Review Manager software, version 5.3 (The Nordic Cochrane Center, Copenhagen, Denmark). We performed the meta-analysis using the genetic inverse-variance method. When heterogeneity between the studies was not significant, we calculated the combined HR using a fixed-effect model; otherwise, a random-effect model was chosen. P > .1 and I2 test < 50% indicated the absence of heterogeneity between studies. An HR value of >1.0 would mean ctDNA positivity was a survival disadvantage in the postoperative arm.
3. Results
3.1. Study characteristics and quality
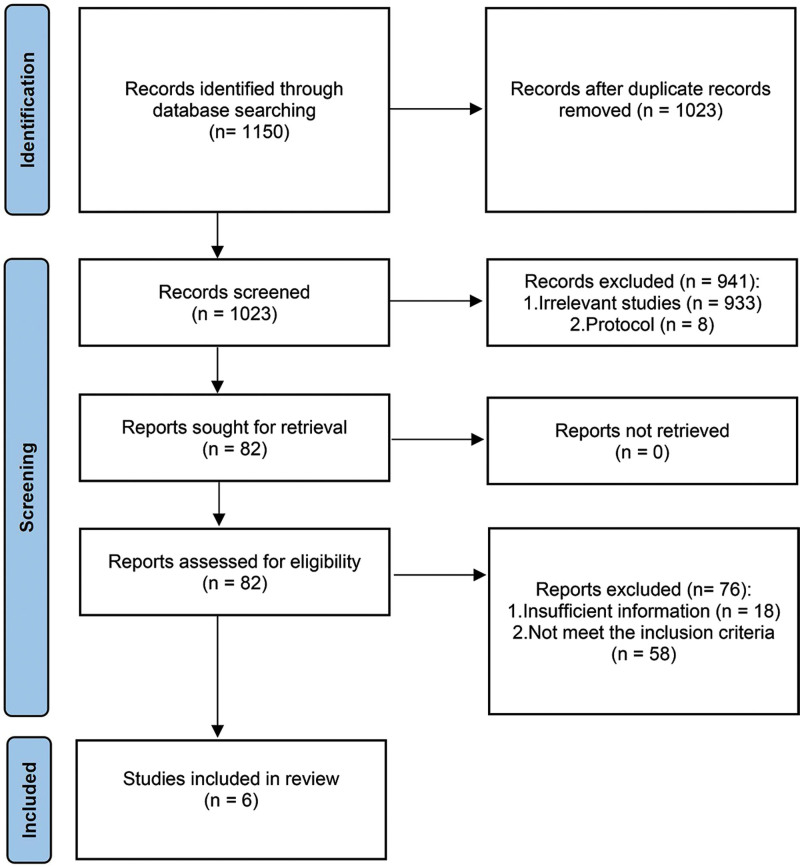
The search strategy identified 1150 potentially relevant articles, and 1068 articles were excluded according to their titles and abstracts. A total of 82 full-text articles were retrieved, of which 76 were excluded. The remaining 6 studies met the inclusion criteria, and a diagram of the search results is shown in Figure 1. The main characteristics of each study are shown in Table 1. The 6 studies[12,13,17–20] were published between 2016 and 2022. Of these, 2 were from Australia,[13,20] and 1 study each was from Spain,[19] Denmark,[12] France,[18] and China.[17] Among the 6 retrieved studies, 5[12,13,17,19,20] were prospective cohort studies, and 1[18] was a randomized controlled trial from France. A total of 1713 patients were included in this meta-analysis. In all 6 studies, researchers analyzed ctDNA in patients’ blood samples after surgery and after adjuvant chemotherapy. All the included studies reported the HRs for the correlation between RFS and positive ctDNA. The quality of the included studies was assessed using the NOS. The NOS assessment results are shown in Table 2. All the studies received at least 7 stars, which we regarded as indicative of high quality.
Figure 1.
PRISMA flow diagram for study selection. PRISMA = Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
Table 1.
The main characteristics of studies.
| Median age, yr (range) | Sex male/female | Tumor stage | Sample origin | ctDNA (+) | ctDNA (−) | ctDNA (+) recurrence number | ctDNA (−) recurrence number | ctDNA measuring time | Gene mutation detected | Median follow time (mo) | Author, yr | Type of study | Detction platform | Numbers of patients |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 65 (23–87) | 131/99 | II | Plasma | 20 | 210 | 14 | 20 | 4–10 wk | SMAD4, TP53, AKT1, APC, BRAF, CTNNB1, ERBB3, FBXW7, HRAS, KRAS, NRAS, PIK3CA, PPP2R1A, RNF43, POLE | 27 | Tie, 2016 | Prospective cohort | SafeSeq-S | 230 |
| 71 (48–93) | 61/33 | I, II, III | Plasma | 14 | 55 | 8 | 10 | 6–8 wk | ACVR2A, AKT1, AMER1, APC, ARID1A, RAF, CTNNB1, EGFR, ERBB3 (HER3), ERBB4 (HER4), FAT4, FLNA, FBXW7, HRAS, KMT2C, KRAS, MEK1, NRAS, PIK3CA, POLE, PP2R1A, PTEN, RNF43, SMAD2, AD4, SOX9, TCF7L2, TGFBR2, and TP53 | 24.7 | Tarazona, 2019 | Prospective cohort | QIA-Seq | 94 |
| 64 (26–82) | 49/47 | III | Plasma | 20 | 76 | 11 | 13 | 42 d | APC, KRAS, TP53, SMAD4, RNF, BRAF, PIK3CA | 28.9 | Tie, 2019 | Prospective cohort | SafeSeq-S | 96 |
| 69 (43–91) | 73/52 | I, II, III | Plasma | 10 | 84 | 7 | 10 | 30 d | TP53, APC, KRAS, BRAF, PIK3CA, FBXW7, SMAD4, TCF7L2, SDK1, HMCN1, RNF43, DMD, ARID1A, FAT2, ABCA12, ANK2, SO X9, YH11, BMPR2, ATM, SPAG17, TPTE, NTNAP2, RNF17, WBSCR17, ITPR2, WDFY3 | 12.5 | Reinert, 2019 | Multicenter, prospective cohort | Hi-Seq | 125 |
| 64 (54–70) | 576/441 | III | Plasma | 140 | 877 | 44 | 151 | 35–50 d | WIF1 and NPY | 79 | Taieb, 2021 | Multicenter, randomized controlled trial | Methylation ddPCR and NGS | 1017 |
| 6 1(25–86) | 90/61 | III | Plasma | 24 | 127 | 13 | 26 | 30 d | 197 cancer-related genes | 33.5 | Li, 2022 | Prospective, observational cohort study | Targeted sequencing panel | 151 |
ctDNA = circulating tumor DNA, ddPCR = droplet-based digital polymerase chain reaction.
Table 2.
Results of the Newcastle–Ottawa Scale (NOS) quality assessment.
| Authors | Yr | Selection | Comparability | Outcome | Total |
|---|---|---|---|---|---|
| Tie | 2016 | **** | – | *** | ******* |
| Tarazona | 2019 | **** | – | *** | ******* |
| Tie | 2019 | **** | * | *** | ******** |
| Reinert | 2019 | **** | – | *** | ******* |
| Taieb | 2021 | **** | ** | *** | ********* |
| Li | 2022 | **** | – | *** | ******* |
3.2. RFS analysis according to ctDNA status
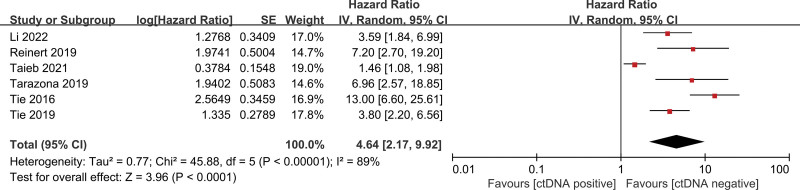
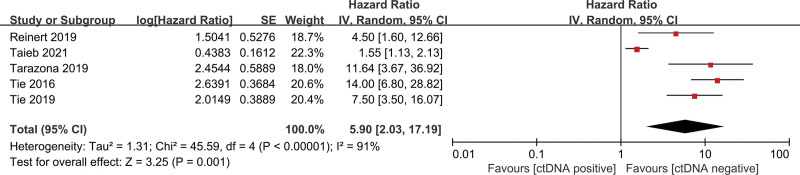
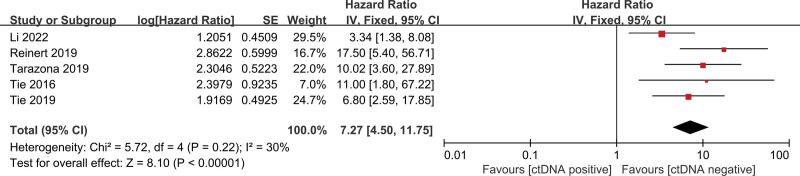
The pooled univariate HR for RFS in patients with postoperative ctDNA-positive colorectal cancer compared with ctDNA-negative patients was 4.64 (95% CI 2.17–9.92, z = 3.96, P < .001) (Fig. 2), indicating an increased risk of recurrence in ctDNA-positive patients. Heterogeneity existed in all studies (I2 = 89%, P < .001) (Fig. 2). We, therefore, performed further analyses using a random-effect model. Similarly, the pooled multivariate HR for RFS in patients with postoperative ctDNA-positive colorectal cancer compared with ctDNA-negative patients was 5.90 (95% CI 2.03–17.19, z = 3.25, P < .001) (Fig. 3), indicating an increased risk of recurrence in ctDNA-positive patients. There was significant heterogeneity in all studies. After adjuvant chemotherapy, the pooled HR for RFS in ctDNA-positive patients compared with ctDNA-negative patients was 7.27 (95% CI 4.50–11.75, z = 8.10, P < .001) (Fig. 4), indicating an increased risk of recurrence in ctDNA-positive patients, with no significant heterogeneity.
Figure 2.
Forest plots for univariate RFS in patients with early-stage colorectal cancer after surgery. RFS = recurrence-free survival.
Figure 3.
Forest plots for multivariate RFS in patients with early-stage colorectal cancer after surgery. RFS = recurrence-free survival.
Figure 4.
Forest plots for RFS in patients with early-stage colorectal cancer after adjuvant chemotherapy. RFS = recurrence-free survival.
4. Discussion
Currently, TNM stage, microsatellite status, tumor grade, and lymphovascular infiltration, are mainly used to assess the extent and prognosis of colorectal cancers.[4] Nevertheless, despite advancements in the treatment of colorectal cancers, survival rates remain highly variable for different patients, even within the same TNM stage.[4] Tumor heterogeneity may be a crucial factor, which has been investigated on different levels including genomics, transcriptomics, histopathologic features, and characterization of the inflammatory infiltrate.[4,21] Therefore, there is a need to improve the capability of identifying people at high risk of relapse. In recent years, studies have been conducted involving ctDNA to detect minimum residual disease, and some researchers consider that this approach offers a potential alternative to treatment strategies based on postoperative pathological typing.[13,19,20,22] However, some researchers consider that ctDNA test results are affected by the specimen, test genes, and testing platforms, with false positives and false negatives having been reported.[15,23] Therefore, it is controversial to conclude that patients with positive ctDNA are at high risk of recurrence. Based on this situation, we aimed to determine the relationship between ctDNA and nonmetastatic operable colorectal cancer in this study, and the results suggest that patients with positive ctDNA after surgery have significantly lower RFS rates. To our knowledge, this meta-analysis provides updated evidence on ctDNA testing for the prognosis of early-stage colorectal cancer after surgery.
Early-stage colorectal cancer has a higher survival rate, and surgery can cure most patients. However, some patients remain at risk of recurrence. Postoperative adjuvant chemotherapy is currently recommended for high-risk patients with stage II and III tumors to reduce the risk of recurrence.[6,24] For such patients, the main goal is to find a better biomarker for detecting recurrence earlier to improve disease-free survival. This biomarker would be important if it could avoid or reduce the incidence of underutilization or overutilization of chemotherapy and the side effects of chemotherapy. CtDNA is noninvasive and can detect residual tumors earlier than CT scans and carcinoembryonic antigens.[13] Although the sensitivity and cost of testing for ctDNA need to be improved, this approach remains promising.[22]
In this meta-analysis, the results of univariate and multivariate analyses were consistent. Patients with detectable ctDNA after colorectal cancer surgery had significantly lower RFS rates than patients with undetectable ctDNA (HR 4.64, 95% CI 2.17–9.92, z = 3.96, P < .001). This result was slightly lower than that reported in Emre Yekeduz’s study,[25] in which no large samples from randomized controlled trials were included. In pooled multivariate analyses, significant heterogeneity was found, which may be primarily related to Taieb et al’s study.[18] The I2 value would decrease from 91% to 16% if Taieb et al’s findings were excluded. The reason for this heterogeneity may be that, when compared with the other studies, Taieb et al’s study involved the largest number of patients and was a randomized controlled trial. Furthermore, the ctDNA detection method in that trial adopted multiplex droplet-based digital polymerase chain reaction (PCR) to test WIF1 and NPY only. Moreover, the blood samples were not originally intended for ctDNA testing, which may have led to a slightly lower detection rate. In their study,[18] ctDNA was detected in 95 patients by next-generation sequencing, which was not significantly different from droplet-based digital PCR. Once their results were eliminated, the pooled HR was 9.03 (95% CI 5.59–14.58, z = 9.00, P < .00001), indicating that the statistical results were stable and continued to show statistical significance.
This meta-analysis showed that ctDNA is also a good predictor of recurrence after postoperative adjuvant chemotherapy. Patients with positive ctDNA after chemotherapy had a 7-fold higher risk of recurrence than patients with negative ctDNA. Monitoring ctDNA changes after adjuvant chemotherapy can help predict disease recurrence and aid in evaluating the effect of chemotherapy.[15,25,26] There was no heterogeneity among the included articles. Taieb et al[18] compared differences between different chemotherapy cycles (3 months vs 6 months), so it was not included in the post-chemotherapy subgroup analysis. In their study,[18] ctDNA was found to be a prognostic factor for DFS in patients treated for 3 months but not for 6 months. It would be interesting to know whether the change from ctDNA positivity to negativity could mean that chemotherapy might be stopped. Further studies with large samples are needed in this regard.
The main pathways involved in colorectal cancer development are the adenoma-carcinoma pathway and the serrated pathway, which involve multiple genetic mutations, such as those in MMR genes, APC, and KRAS.[27] Colorectal cancer is heterogeneous, and even patients at the same stage may have a very different prognosis due to different genetic mutations involved.[4,21] While it may appear challenging to identify all cancerous genes, the presence of small residual tumors can be detected by examining the ctDNA of these mutated genes. Although the ctDNA mutation genes detected in the included studies differed, which may affect the positive rate of detection, we consider that the significance of ctDNA positivity in the prognosis of colorectal cancer cannot be denied.
CtDNA detection methods include refining conventional PCR techniques to target specific mutated genes and next-generation gene sequencing that can target hundreds of cancer-related genes.[28] Targeting specific mutated genes has low sensitivity, is not costly, and involves a short detection time, whereas next-generation gene sequencing has high sensitivity, is expensive, and involves a long detection time. Both have their advantages and disadvantages and need to be improved. Jin et al[26] developed a simple methylation-specific quantitative PCR assay for ctDNA analysis, and 14 patients (70%) had detectable ctDNA before recurrence, with a median lead time of 8.0 months earlier than seen with radiologic imaging.[26] However, because that study included patients with stage IV colorectal cancer, it was not included in this meta-analysis. Further studies need to be conducted to reduce the false-positive rate, which would be beneficial for improving the positive ctDNA detection rate and reducing the difficulty of detection.
This study had some limitations. First, only 6 eligible studies were identified, with only 1 high-quality randomized controlled trial included. Our findings need to be verified with more large-sample studies. Second, in the included studies, different gene ctDNA mutations were detected, which may have introduced bias. Finally, heterogeneity may also result from different detection platforms.
5. Conclusions
In patients with early-stage colorectal cancer after surgery, ctDNA is a promising biomarker for predicting recurrence and evaluating the effect of chemotherapy. However, more randomized controlled trials are needed to verify whether ctDNA can be used as an independent factor in determining whether to initiate postoperative chemotherapy.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
Author contributions
Conceptualization: Xiaoyuan Fan.
Data curation: Xiaoyuan Fan.
Formal analysis: Xiaoyuan Fan.
Investigation: Xiaoyuan Fan.
Methodology: Jiakai Zhang.
Project administration: Xiaoyuan Fan, Dewen Lu.
Software: Jiakai Zhang.
Supervision: Dewen Lu.
Validation: Jiakai Zhang, Dewen Lu.
Visualization: Jiakai Zhang.
Writing – original draft: Xiaoyuan Fan.
Writing – review & editing: Xiaoyuan Fan.
Abbreviations:
- CI
- confidence interval
- CT
- computed tomography
- ctDNA
- circulating tumor DNA
- HR
- hazard ratio
- MSI
- microsatellite instability
- NOS
- Newcastle–Ottawa Quality Assessment Scale
- PCR
- polymerase chain reaction
- RFS
- recurrence-free survival
- TNM
- tumor-node-metastasis
Ethical approval was not required as this study was based on existing literature.
The authors have no funding and conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
How to cite this article: Fan X, Zhang J, Lu D. CtDNA’s prognostic value in patients with early-stage colorectal cancer after surgery: A meta-analysis and systematic review. Medicine 2023;102:6(e32939).
Contributor Information
Jiakai Zhang, Email: gary0125@163.com.
Dewen Lu, Email: ludw111111@163.com.
References
- [1].Elferink MA, de Jong KP, Klaase JM, et al. Metachronous metastases from colorectal cancer: a population-based study in North-East Netherlands. Int J Colorectal Dis. 2015;30:205–12. [DOI] [PubMed] [Google Scholar]
- [2].Qaderi SM, Galjart B, Verhoef C, et al. Disease recurrence after colorectal cancer surgery in the modern era: a population-based study. Int J Colorectal Dis. 2021;36:2399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].van Gestel YR, de Hingh IH, van Herk-Sukel MP, et al. Patterns of metachronous metastases after curative treatment of colorectal cancer. Cancer Epidemiol. 2014;38:448–54. [DOI] [PubMed] [Google Scholar]
- [4].Sagaert X, Vanstapel A, Verbeek S. Tumor heterogeneity in colorectal cancer: what do we know so far? Pathobiology. 2018;85:72–84. [DOI] [PubMed] [Google Scholar]
- [5].Benson AB, Venook AP, Al-Hawary MM, et al. NCCN guidelines insights: colon cancer, version 2.2018. J Natl Compr Canc Netw. 2018;16:359–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Benson AB, Venook AP, Al-Hawary MM, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19:329–59. [DOI] [PubMed] [Google Scholar]
- [7].Hou W, Yi C, Zhu H. Predictive biomarkers of colon cancer immunotherapy: present and future. Front Immunol. 2022;13:1032314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Varghese A. Chemotherapy for stage II colon cancer. Clin Colon Rectal Surg. 2015;28:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Heitzer E, Auer M, Ulz P, et al. Circulating tumor cells and DNA as liquid biopsies. Genome Med. 2013;5:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res. 2021;27:5586–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20:71–88. [DOI] [PubMed] [Google Scholar]
- [15].Morris VK, George TJ. Using circulating tumor DNA for colon cancer adjuvant therapy: to be or not to be? Clin Cancer Res. 2022;28:438–40. [DOI] [PubMed] [Google Scholar]
- [16].Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [17].Li Y, Mo S, Zhang L, et al. Postoperative circulating tumor DNA combined with consensus molecular subtypes can better predict outcomes in stage III colon cancers: a prospective cohort study. Eur J Cancer. 2022;169:198–209. [DOI] [PubMed] [Google Scholar]
- [18].Taieb J, Taly V, Henriques J, et al. Prognostic value and relation with adjuvant treatment duration of ctDNA in stage III colon cancer: a post hoc analysis of the PRODIGE-GERCOR IDEA-France trial. Clin Cancer Res. 2021;27:5638–46. [DOI] [PubMed] [Google Scholar]
- [19].Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30:1804–12. [DOI] [PubMed] [Google Scholar]
- [20].Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cuyle PJ, Prenen H. Current and future biomarkers in the treatment of colorectal cancer. Acta Clin Belg. 2017;72:103–15. [DOI] [PubMed] [Google Scholar]
- [22].Peng Y, Mei W, Ma K, et al. Circulating tumor DNA and Minimal Residual Disease (MRD) in solid tumors: current horizons and future perspectives. Front Oncol. 2021;11:763790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Solar Vasconcelos JP, Boutin M, Loree JM. Circulating tumor DNA in early-stage colon cancer: ready for prime time or needing refinement? Ther Adv Med Oncol. 2022;14:17588359221143975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chakrabarti S, Peterson CY, Sriram D, et al. Early stage colon cancer: current treatment standards, evolving paradigms, and future directions. World J Gastrointest Oncol. 2020;12:808–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yekeduz E, Koksoy EB, Akbulut H, et al. ctDNA as a prognostic factor in operable colon cancer patients: a systematic review and meta-analysis. Future Oncol. 2021;17:349–57. [DOI] [PubMed] [Google Scholar]
- [26].Jin S, Zhu D, Shao F, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy. Proc Natl Acad Sci U S A. 2021;118:e2017421118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jebelli A, Baradaran B, Mosafer J, et al. Recent developments in targeting genes and pathways by RNAi-based approaches in colorectal cancer. Med Res Rev. 2021;41:395–434. [DOI] [PubMed] [Google Scholar]
- [28].Pessoa LS, Heringer M, Ferrer VP. ctDNA as a cancer biomarker: a broad overview. Crit Rev Oncol Hematol. 2020;155:103109. [DOI] [PubMed] [Google Scholar]