Abstract
Osteoarthritis (OA) is a degenerative disease with a series of metabolic changes accompanied by many altered enzymes. Here, we report that the down-regulated dimethylarginine dimethylaminohydrolase-1 (DDAH1) is accompanied by increased asymmetric dimethylarginine (ADMA) in degenerated chondrocytes and in OA samples. Global or chondrocyte-conditional knockout of ADMA hydrolase DDAH1 accelerated OA development in mice. ADMA induces the degeneration and senescence of chondrocytes and reduces the extracellular matrix deposition, thereby accelerating OA progression. ADMA simultaneously binds to SOX9 and its deubiquitinating enzyme USP7, blocking the deubiquitination effects of USP7 on SOX9 and therefore leads to SOX9 degradation. The ADMA level in synovial fluids of patients with OA is increased and has predictive value for OA diagnosis with good sensitivity and specificity. Therefore, activating DDAH1 to reduce ADMA level might be a potential therapeutic strategy for OA treatment.
Asymmetric dimethylarginine (ADMA) accelerates osteoarthritis by interfering with the deubiquitination effects of USP7 on SOX9.
INTRODUCTION
Osteoarthritis (OA), as one of the most prevalent chronic joint diseases, affects up to one-third of the population over the age of 45 (1). It is mainly caused by aging and mechanical injury. OA is characterized by cartilage destruction, synovial inflammation, subchondral bone remodeling, and osteophyte formation. The glycosaminoglycan-rich extracellular matrix (ECM), the main component of cartilage, is produced by chondrocytes. In OA chondrocytes, catabolic regulators release several inflammatory factors, including interleukin-1β (IL-1β), IL-6, and tumor necrosis factor–α (TNF-α), and subsequently result in the activation of matrix metalloproteinases (MMPs) and A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS), eventually leading to ECM degeneration (2). Disrupted cartilage exacerbates the imbalanced mechanical stress in the joints, which traps OA pathogenesis into a vicious cycle.
During OA pathogenesis, numerous enzymes in chondrocytes change, resulting in a series of metabolic changes, which makes it possible for certain metabolites to be OA diagnostic markers. In turn, activation or suppression of certain metabolic pathways may exacerbate or prevent OA progression. Corciulo et al. (3) demonstrated that adenosine, a metabolite of adenosine triphosphate, which decreased under the effect of IL-1β, could protect against OA development in rats. A previous study indicated that osteoarthritic chondrocytes have increased cholesterol metabolism. Meanwhile, cholesterol itself or the critical cholesterol hydroxylases CH25H and CYP7B1 could mediate OA progression via RAR Related Orphan Receptor α (RORα) (4, 5).
Asymmetric dimethylarginine (NG, NG-dimethyl-l-arginine, ADMA) is a naturally occurring analog of l-arginine. It is released into the cytoplasm during posttranslational methylation of arginine residues by a group of methyltransferases called protein arginine methyltransferases and proteolysis (6). The kidneys are responsible for clearance of free ADMA in the circulatory system. In addition to renal excretion, ADMA can also be hydrolyzed by the dimethylarginine dimethylaminohydrolase (DDAH) family (7, 8) and alanine-glyoxylate aminotransferase 2 (AGXT2) (9). As a competitive endogenous nitric oxide synthase (NOS) inhibitor (10), ADMA is able to decrease NOS activity, therefore leading to a reduction of nitric oxide (NO). Research in this area mainly focuses on cardiovascular disease. ADMA is reported as a major and independent risk factor for hypertension (11) and congestive heart failure (12), particularly in patients with renal failure. Furthermore, several studies have revealed that ADMA is associated with insulin resistance, diabetes (13, 14), and cirrhosis (8). Except for NOS activity inhibition, ADMA is able to induce multiple sclerosis via facilitating T helper cell 1(TH1)– and TH17-mediated immune response (15). Our previous study indicated that, in the musculoskeletal system, elevated ADMA suppressed bone formation (16). Patients with OA generally exhibit higher ADMA concentrations in synovial fluids than in their plasma (17). Our data in metabonomics revealed a notable ADMA elevation in the degenerated chondrocytes with the most predominant statistical significance. Therefore, our aim was to analyze the role of ADMA in OA and how it affects OA progression. In addition, we also intend to analyze whether ADMA in synovial fluids from patients with OA could serve as a potential diagnostic marker for OA.
RESULTS
Increased ADMA in OA chondrocytes triggers cartilage degeneration
Metabonomics of chondrocytes from six patients with OA and six relatively healthy patients (knee joint fracture with no history of OA) demonstrated a series of altered metabolites (Fig. 1A). Partial least squares discrimination analysis (PLS-DA) and orthogonal PLS-DA showed a good difference between the groups (fig. S1A), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed substantial enriched pathways (fig. S1B). Of the top 25 increased metabolites, ADMA had the highest statistical significance. Liquid chromatography–mass spectrometry (LC-MS) confirmed the elevated ADMA level in OA chondrocytes, with no substantial change in its isomer symmetric dimethylarginine (SDMA) (Fig. 1B). To investigate whether ADMA induces chondrocyte degeneration, ADMA was added to chondrocyte culture medium. Micromass culture and three-dimensional (3D) agarose culture combined with senescence-associated (SA)-β-galactosidase staining demonstrated that ADMA reduced the ECM deposition and induced senescence in chondrocytes (fig. S1, C to E). We also found down-regulated anabolic factors (COL2A1 and aggrecan), up-regulated catabolic factor MMP13, and senescence marker P21 in ADMA-treated mouse chondrocytes in a dose-dependent manner under short-pulse stimulation or long-duration stimulation with a much lower concentration (fig. S1F). To investigate whether ADMA could induce OA in vivo, we directly injected ADMA into the knee joints of mice 1 week after destabilization of the medial meniscus (DMM) surgery. ADMA significantly accelerated cartilage degeneration and facilitated osteophyte formation with DMM surgery (fig. S1G). Moreover, immunohistochemical staining confirmed decreased expression of type II collagen and aggrecan in cartilage exposed to ADMA (fig. S1H). Hot plate analysis, rotarod assay, and open field assay revealed that ADMA-injected mice had a considerably lower pain threshold and moving ability (fig. S1I). Together, these data reveal that increased ADMA in degenerated chondrocytes triggers cartilage degeneration and senescence.
Fig. 1. Asymmetric dimethylarginine (ADMA) up-regulated by DDAH1 accelerates OA.
(A) Metabolomics between chondrocytes from patients with OA and controls. Cells harvested from cartilages were cultured for 3 days in identical conditions. (B) The ADMA level and its isomer SDMA in chondrocytes from patients with OA and controls evaluated by liquid chromatography–mass spectrometry (LC-MS) analysis and bicinchoninic acid (BCA) protein quantification assay. (C and D) The ADMA level of PD 404182–treated or DDAH1−/− mouse chondrocytes evaluated by LC-MS analysis and BCA protein quantification assay. (E) Micromass culture and quantification of DDAH1−/− or DDAH1+/+ primary chondrocytes. n = 6 per group. Scale bar, 3 mm. (F) 3D agarose culture of DDAH1−/− or DDAH1+/+ primary chondrocytes (alcian blue staining) showing the thickness of the pericellular matrix. n = 6 per group. Scale bar, 20 μm. (G) The safranin O/fast green staining and alcian blue staining of knee joints of DDAH1 global knockout mice that underwent destabilization of the medial meniscus (DMM) surgery or sham. n = 12 per group. Scale bar, 100 μm. (H) Immunoblot detection of type II collagen, aggrecan, and DDAH1 in cartilage of mice indicated above. n = 8 per group. Scale bar, 100 μm; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, (B) to (H), means ± SD, unpaired two-tailed t test; NS, not significant.
Down-regulated DDAH1 is responsible for ADMA elevation in OA
ADMA is released from proteolysis of arginine with methylation residues and subsequently hydrolyzed by the DDAH family or AGXT2. Reverse transcription polymerase chain reaction (RT-PCR) demonstrated that DDAH1 was more abundantly expressed than DDAH2, while AGXT2 was hardly expressed in chondrocytes (fig. S2, A and B). Previous data in GSE113852 confirmed that the RNA expression of DDAH1 was lower in OA tissues, while DDAH2 was higher compared with non-OA tissues (fig. S2C) (18). We next collected cartilage from both sides of the tibial plateau of patients with OA who underwent total knee arthroplasty and found that DDAH1 was significantly down-regulated in damaged cartilage compared with its paired undamaged regions. In contrast, DDAH2 was up-regulated in the damaged region of cartilage, which is in accordance with a previous study (fig. S2, D to F) (19). These results were further confirmed by immunohistochemistry (fig. S2G). Three different OA animal models were subsequently used. First, the protein expression of DDAH1, instead of DDAH2, was decreased in the cartilage of mice subjected to DMM surgery (fig. S2H). Second, in aged mice, DDAH1 was significantly down-regulated in an age-dependent manner, while DDAH2 was unchanged (fig. S2I). Third, STR/Ort mice that spontaneously develop OA, and their CBA/CaCrl controls were harvested at 6 months of age. Middle to severe cartilage lesions were observed in 6-month-old STR/Ort mice. DDAH1 was sharply decreased in STR/Ort mice, with DDAH2 not significantly changed (fig. S2J). A DDAH1 inhibitor, PD 404182, was applied to explore the role of DDAH1 in OA. The ADMA level sharply increased in PD 404182–treated chondrocytes (Fig. 1C). In addition, ADMA level was increased in DDAH1−/− primary chondrocytes (Fig. 1D). These data indicate that DDAH1 is suppressed in OA progression, which increases ADMA levels in chondrocytes. DDAH1+/+ and DDAH1−/− primary chondrocytes were separately exposed to PD 404182, and OA-related proteins were determined. COL2A1 and aggrecan protein levels were down-regulated, while MMP13 was up-regulated in DDAH1+/+ chondrocytes, with no significant change observed in DDAH1−/− chondrocytes, confirming the specificity and efficacy of PD 404182 on DDAH1 (fig. S3A). PD 404182 was injected into the knee joints of mice after DMM surgery and accelerated cartilage degeneration (fig. S3B) and down-regulated the protein expression of type II collagen and aggrecan (fig. S3C).
DDAH1 global mice were gifted by Y. Chen, Minnesota University (fig. S4, A and B), and Western blotting and immunohistochemistry confirmed the DDAH1 deletion (Fig. 1H and fig. S3A). Micromass culture and 3D agarose culture demonstrated low ECM deposition in DDAH1−/− primary chondrocytes (Fig. 1, E and F). Although no significant OA-related phenotypes were observed in 20-week-old DDAH1−/− mice, DDAH1 deletion markedly accelerated cartilage degeneration in DMM-induced OA mice (Fig. 1G). Immunohistochemistry showed decreased type II collagen and aggrecan in the cartilages of DDAH1−/− mice (Fig. 1H). In addition, compared with that of their controls, both 12- and 18-month-old DDAH1−/− mice exhibited substantially more severe cartilage damage (fig. S4C). ADMA at a considerably lower concentration reduced COL2A1 and aggrecan and activated MMP13 in mouse chondrocytes without DDAH1 (fig. S4D). Therefore, it was determined that exogenous ADMA could be rapidly hydrolyzed under the effect of DDAH1. This might partly explain why exogenous ADMA at a concentration far above the physiological concentration is required to induce chondrocyte degeneration.
To further exclude the effect of DDAH2 in OA, DDAH2 global knockout mice were also generated (fig. S4, E and F). No significant difference in extracellular deposition was observed in DDAH2−/− primary chondrocytes compared to that in the control (fig. S4, G and H). Furthermore, the knee joint cartilage of DDAH2−/− did not differ from that of the control with or without DMM surgery (fig. S4I). In conclusion, DDAH1, but not DDAH2, protects against OA progression.
Cartilage-specific DDAH1 is involved in OA progression
DDAH1fl/fl mice were established to further investigate the role of DDAH1 and resemble a high-ADMA state in chondrocytes (fig. S5A). The primary chondrocytes of DDAH1fl/fl were cultured and infected with Cre adenovirus or vector. RT-PCR indicated decreased anabolic factors (COL2A1 and ACAN) and increased catabolic factors (MMP3 and MMP13; Fig. 2A). In addition, the protein expression of aggrecan and COL2A1 was significantly down-regulated following DDAH1 deletion (Fig. 2B). Our previous study demonstrated that DDAH1 in osteoblasts inhibited bone formation via hydrolyzing ADMA (16). To rule out the effects of DDAH1 in bone, we generated DDAH1 conditional knockout mice with conditional deletion of the DDAH1 gene in chondrocytes using Col2-CreERT2 and ACAN-CreERT2 mice, whose genotypes were determined by PCR (fig. S5B). Similar to DDAH1 global knockout mice, DDAH1fl/fl, Col2-CreERT2 and DDAH1fl/fl, ACAN-CreERT2 mice did not exhibit OA phenotypes at 20 weeks of age. However, deletion of DDAH1 in cartilage could cause more severe cartilage damage after DMM surgery compared with that in controls (Fig. 2C and fig. S5C). Moreover, the protein expressions of type II collagen and aggrecan in the cartilages of DDAH1fl/fl, Col2-CreERT2 and DDAH1fl/fl, ACAN-CreERT2 mice were predominantly decreased (Fig. 2D and fig. S5D). Meanwhile, more osteophytes were observed in DDAH1fl/fl, Col2-CreERT2 and DDAH1fl/fl, ACAN-CreERT2 mice (Fig. 2E and fig. S5E). A previous study reported that subchondral bone plate sclerosis can occur in late-stage OA owing to long-term subchondral bone remodeling (20). Therefore, we also explored the subchondral bone density and found that both DDAH1fl/fl, Col2-CreERT2 and DDAH1fl/fl, ACAN-CreERT2 mice exhibited higher subchondral bone volume (Fig. 2F and fig. S5F). Hot plate, rotarod, and open field assays indicated that DDAH1fl/fl, Col2-CreERT2 and DDAH1fl/fl, ACAN-CreERT2 mice were more sensitive to pain and had moving disability to certain degrees (Fig. 2G and fig. S5G). These data suggest that cartilage-specific DDAH1 knockout is critical for OA progression. Vice versa, DDAH1 overexpression exerts therapeutic effects on OA. We articularly injected DDAH1 adeno-associated virus (AAV) 1 week after DMM surgery. Cartilage degeneration was partly reversed 8 weeks after DMM surgery (fig. S6A). The critical anabolic factors type II collagen and aggrecan in knee cartilages were significantly up-regulated after DDAH1 overexpression in the impaired cartilage (fig. S6B). The hot plate, rotarod, and 6-min distance in the open field assays also indicated a predominant elevation in pain threshold and moving ability (fig. S6C). LC-MS confirmed the decreased ADMA level in chondrocytes with DDAH1 overexpression (fig. S6D). These data illustrate that DDAH1 could partly reverse DMM-induced OA in mice.
Fig. 2. Cartilage-specific knockout of DDAH1 accelerates OA progression.
(A) The mRNA expression level of COL2A1, ACAN, MMP3, MMP13, ADAMTS4, and ADAMTS5 in DDAH1fl/fl mouse chondrocytes infected with Cre or vector adenovirus. (B) Immunoblot detection of aggrecan, COL2A1, and DDAH1 in DDAH1fl/fl mouse chondrocytes. Cre or vector adenovirus was added into chondrocytes 48 hours before harvesting. (C) The safranin O/fast green staining and alcian blue staining of knee joints of DDAH1fl/fl and DDAH1fl/fl, Col2-CreERT2 mice that underwent DMM surgery or sham. DDAH1fl/fl (sham), n = 15, DDAH1fl/fl; Col2-CreERT2 (sham), n = 12; DDAH1fl/fl (DMM), n = 15; DDAH1fl/fl, Col2-CreERT2 (DMM), n = 14. Scale bar, 100 μm. (D) Immunoblot detection of type II collagen, aggrecan, and DDAH1 in cartilage of DDAH1fl/fl and DDAH1fl/fl, Col2-CreERT2 mice that underwent DMM surgery or sham, n = 8 per group. Scale bar, 100 μm. (E) The micro–computerized tomography images of knee joints from DDAH1fl/fl and DDAH1fl/fl, Col2-CreERT2 mice. Yellow arrows indicate the osteophytes. Scale bar, 1 mm. (F) The Bone volume/Tissue volume (BV/TV), Trabecular Thickness (TB.Th), and Trabecular Separation (TB.Sp) of subchondral bone in the indicated groups above were calculated. DDAH1fl/fl, n = 4; DDAH1fl/fl, Col2-CreERT2, n = 5. (G) The hot plate, rotarod, and open field assays of the mice indicated above. Data are representative of three independent experiments (B). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, means ± SD, unpaired two-tailed t test.
ADMA activates the ubiquitin-proteosome pathway of SOX9
As an endogenous NOS inhibitor, ADMA can influence the pathophysiological process via NO, particularly in cardiovascular diseases (21–24). We first explored whether ADMA induced cartilage degeneration through the NOS/NO pathway. DDAH1 knockout or ADMA stimulation did not alter the protein expression of NOS1 and NOS3 or activate the phosphorylated forms of these proteins in chondrocytes or in cartilage, while NOS2 was not expressed (fig. S7, A to C). Unexpectedly, NO did not show an obvious change after the deletion of DDAH1 (fig. S7, D and E) possibly because of its low concentration under physiological conditions in chondrocytes. ADMA is elevated by the effect of TNF-α in vein endothelial cells (25) and fibroblast-like synoviocytes (26). Therefore, TNF-α was applied, and an increased secretion of ADMA in chondrocytes was observed, which could be partly reversed by DDAH1 overexpression (fig. S7F). However, NO was not significantly altered in chondrocytes exposed to TNF-α or overexpressed with DDAH1 (fig. S7G). Therefore, ADMA induces chondrocyte degeneration independent of the NOS/NO pathway.
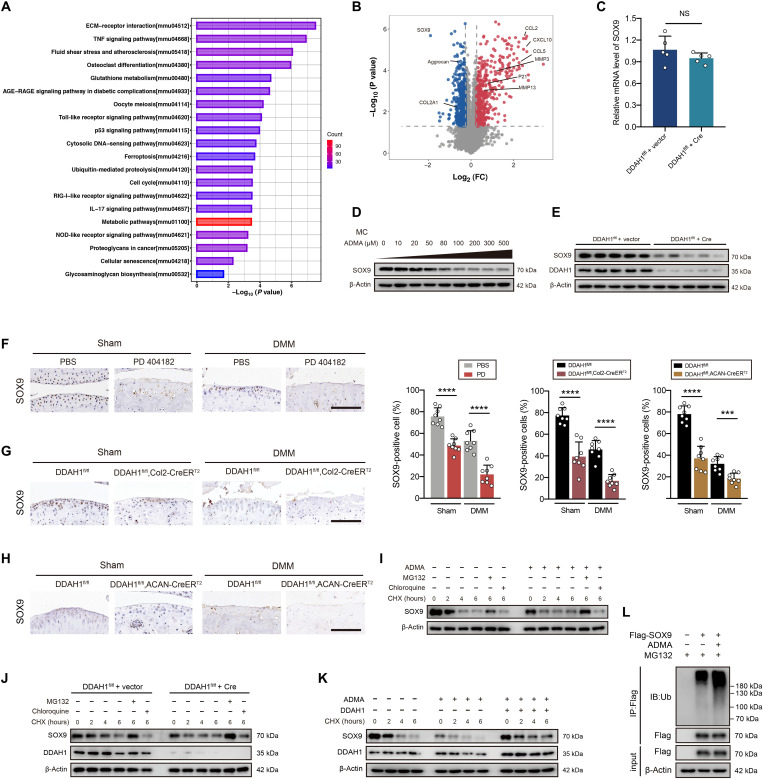
To better understand its mechanism, proteomics of ADMA-treated cells was investigated (fig. S7H). Subcellular localization, Gene Ontology, and Cluster of Orthologous Groups analyses of differentially expressed proteins were performed (fig. S7, I to K). KEGG analysis revealed several enriched OA-related pathways, such as ECM-receptor interaction, TNF signaling pathway, cellular senescence, and glycosaminoglycan biosynthesis (Fig. 3A). Volcano plots demonstrated up-regulated catabolic factors (MMP3 and MMP13), senescence marker (P21), and inflammatory chemokines (CCL2, CCL5, and CXCL10), and down-regulated anabolic factors (aggrecan, COL2A1, and SOX9; Fig. 3B). A previous study demonstrated that SOX9 deficiency resulted in proteoglycan loss in healthy cartilage and led to much severe cartilage erosion after posttraumatic OA using SOX9fl/fl, ACAN-CreERT2 mice (27). Considering that SOX9 is one of the most significantly changed proteins in proteomics and one of the most critical transcription factors in cartilage, we next focused on whether SOX9 is the core protein influenced by ADMA. RT-PCR showed that the deletion of DDAH1 did not significantly change SOX9 RNA levels (Fig. 3C). However, SOX9 protein expression was predominantly down-regulated in chondrocytes exposed to ADMA or DDAH1 deletion (Fig. 3, D and E). Moreover, immunohistochemistry suggested a sharp reduction in SOX9 expression in the cartilage of mice articular-injected with PD 404182 (Fig. 3F) or from DDAH1fl/fl, Col2-CreERT2 and DDAH1fl/fl, ACAN-CreERT2 mice (Fig. 3, G and H). Moreover, SOX9 half-life was shorter in ADMA-exposed cells than in control cells, demonstrating that ADMA results in the instability of SOX9. MG132 (a proteasome inhibitor) treatment blocked the decay of SOX9, while chloroquine (a lysosome inhibitor) did not, indicating that SOX9 degradation is dependent on the proteasome but not the lysosome pathway (Fig. 3, I and J). In addition, ADMA-induced SOX9 decay was partly reversed in chondrocytes overexpressing DDAH1 (Fig. 3K). Notably, ADMA increased the levels of ubiquitinated SOX9 (Fig. 3L). Together, ADMA triggers SOX9 degradation via the ubiquitin-proteasome pathway.
Fig. 3. ADMA induces SOX9 degradation through the ubiquitin-proteasome pathway.
(A and B) KEGG analysis and volcano plots (fold change >1.2 or <0.83, P < 0.05) of the differentially expressed proteins between chondrocytes treated with ADMA or control. (C) The relative mRNA expression of SOX9 in DDAH1fl/fl chondrocytes infected with Cre adenovirus or vector. (D and E) Immunoblot detection of SOX9 or DDAH1 in mouse chondrocytes exposed to ADMA or DDAH1-knockout chondrocytes. (F) Immunoblot detection of SOX9 in cartilage of mice that underwent DMM surgery accompanied by PD 404182 articular injection weekly. n = 8 per group. Scale bar, 100 μm. (G and H) Immunoblot of SOX9 in cartilage of DDAH1fl/fl and DDAH1fl/fl, Col2-CreERT2 or DDAH1fl/fl, ACAN-CreERT2 mice that underwent DMM surgery or sham. n = 8 per group. Scale bar, 100 μm. (I and J) Immunoblot of SOX9 or DDAH1 in mouse chondrocytes exposed to ADMA or with DDAH1 knockout. Where indicated, cells were pretreated with either MG132 or chloroquine 30 min before cycloheximide (CHX) treatment at indicated times. (K) Immunoblot of SOX9 and DDAH1 in mouse chondrocytes. Cells were treated with ADMA, DDAH1 lentivirus (or vector), and CHX at the indicated times. (L) Immunoblot detection of ubiquitinated species of Flag in 293T cells via Flag immunoprecipitation. Cells were transfected with Flag-SOX9 and treated with ADMA for 24 hours and MG132 for 6 hours before harvesting. Data are representative of three independent experiments (D, E, and I to L). ***P < 0.001, ****P < 0.0001, means ± SD, unpaired two-tailed t test.
Interaction between SOX9 and USP7 is blocked by ADMA
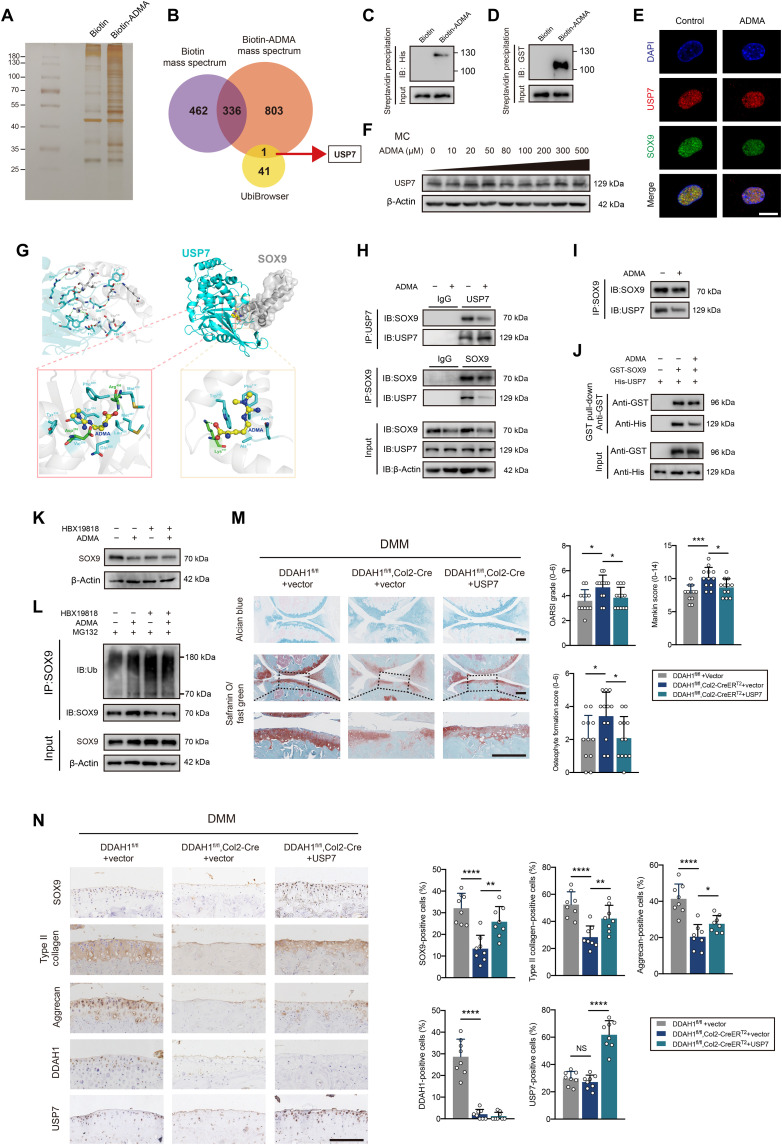
Having shown that ADMA accelerates SOX9 degradation via the ubiquitin-proteasome pathway, we next aimed to understand how ADMA influences SOX9 level. We synthesized biotin-ADMA and incubated it with chondrocyte lysate. Silver staining, together with MS, demonstrated that biotin-ADMA could bind to a series of proteins. We overlapped the proteins specifically interacting with biotin-ADMA and SOX9-related E3 ubiquitin ligases or deubiquitinating enzymes (DUBs) predicted by the UbiBrowser database, finding a possible involvement of DUB USP7 (Fig. 4, A and B). A previous study reported that USP7 knockdown in ATDC5 cells reduces SOX9 expression (28). HBX19818 (a specific USP7 inhibitor) decreased SOX9 protein level in chondrocytes (fig. S8A). Immunoprecipitation showed that USP7 was capable of reducing ubiquitinated SOX9 (fig. S8B). The interaction between USP7 and ADMA was verified by Western blotting (fig. S8C). Unexpectedly, ADMA interacted with SOX9 in the mass spectrum data, which was further confirmed by Western blotting (fig. S8D). To explore whether ADMA directly binds to these two proteins, we applied glutathione S-transferase (GST)–SOX9 recombinant protein and His-USP7 recombinant protein and separately incubated them with biotin-ADMA, finding a direct interaction between SOX9 and ADMA (or USP7 and ADMA; Fig. 4, C and D). Confocal microscopy showed colocalization of USP7 and SOX9. The fluorescence signal of SOX9 weakened after cells were treated with ADMA, while USP7 did not change (Fig. 4E), which was verified by Western blotting (Fig. 4F).
Fig. 4. ADMA blocks the interaction between SOX9 and USP7.
(A) Silver staining of proteins interacting with biotin-ADMA. (B) Venn diagram showing the overlaps between biotin-ADMA specifically conjugated proteins and SOX9-related E3 ubiquitin or de-ubiquitinating enzymes predicted by UbiBrowser. (C and D) Immunoblot of histone and glutathione S-transferase (GST). Biotin-ADMA or biotin was incubated with His-USP7 or GST-SOX9 recombinant protein and precipitated by streptavidin magnetic beads. (E) Confocal microscopy demonstrating the colocalization of USP7, SOX9, and 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 10 μm. (F) Immunoblot of USP7 in chondrocytes exposed to ADMA. (G) Molecular docking predicting the interaction between ADMA, USP7, and SOX9. (H and I) Co-immunoprecipitation of SOX9 and USP7 in mouse chondrocytes exposed to ADMA in normal state or after adjusting the SOX9 protein. (J) GST pull-down showing the interaction between GST-SOX9 and His-USP7 recombinant protein under ADMA or not. (K) Immunoblot of SOX9 in chondrocytes treated with ADMA and HBX19818. (L) Immunoblot of ubiquitinated SOX9 in chondrocytes treated with ADMA, HBX19818, and MG132. (M) The safranin O/fast green and alcian blue staining of joints from DDAH1fl/fl and DDAH1fl/fl, Col2-CreERT2 mice that underwent DMM surgery accompanied by USP7 AAV or vector injection. n = 12 per group. Scale bar, 100 μm. (N) Immunohistochemistry of SOX9, type II collagen, aggrecan, DDAH1, and USP7 of the indicated group. n = 8 per group. Scale bar, 100 μm. Data are representative of three independent experiments (A, C to F, and H to L). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, means ± SD, unpaired two-tailed t test.
Molecular docking was applied to predict the interaction model among ADMA, USP7, and SOX9 (Fig. 4G). USP7 and SOX9 share 12 hydrogen bonds, which results in a strong interaction between these two proteins. On one hand, ADMA binds to the cavity of USP7 consisting of Tyr224, Asp295, Val296, Gln405, Leu406, Met407, Arg408, Phe409, Met410, and Tyr514 sterically, with nitrogen and oxygen atoms in ADMA forming two hydrogen bonds with Asp295 and Arg408 of USP7. Meanwhile, ADMA also forms a strong hydrophobic interaction with Tyr224, Val296, Gln405, Leu406, Met407, Phe409, Met410, and Tyr514 around the cavity of USP7. On the other hand, ADMA perfectly docks into the helix angle cavity of SOX9 formed by Asn110, Ala111, Phe112, Trp143, and Lys151 sterically, with the oxygen atom of ADMA forming a hydrogen bond with Lys151 of SOX9. Meanwhile, a strong hydrophobic interaction between ADMA and Asn110, Ala111, Phe112, and Trp143 around the cavity of SOX9 also existed. Therefore, ADMA is able to bind to the interaction region between SOX9 and USP7, predominantly blocking their binding. Co-immunoprecipitation confirmed the interaction between SOX9 and USP7, which could be weakened by ADMA (Fig. 4H). Considering the unbalanced SOX9 protein levels in immunoblot ladders due to ADMA stimulation, we carefully adjusted the SOX9 protein level to guarantee the equivalent SOX9 initial level and still found a reduced interaction between USP7 and SOX9 in ADMA-treated chondrocytes (Fig. 4I). The GST pull-down assay verified the direct binding between USP7 and SOX9, which could be partly blocked by ADMA (Fig. 4J).
To further test our hypothesis, HBX19818 was used before the cells were treated with ADMA. After inhibiting USP7, ADMA failed to influence SOX9 expression and ubiquitin sticking to SOX9 (Fig. 4, K and L). Moreover, USP7 AAV was articular-injected into the knee joint of DDAH1fl/fl, Col2-CreERT2 mice 1 week after DMM surgery. Notably, cartilage damage was significantly repaired 8 weeks after DMM surgery with increased expression of SOX9, type II collagen, and aggrecan, as determined by immunohistochemistry (Fig. 4, M and N). Furthermore, we tested the protein expression of SOX9 and USP7 in paired damaged and undamaged cartilages from the tibial plateau of patients with OA who underwent total knee arthroplasty. No pronounced correlation of protein expression between SOX9 and USP7 in cartilage was found (fig. S8, E and F), which is partly in accordance with our finding that USP7 enzyme activity, rather than its expression, is involved in OA progression. Together, these data support the idea that ADMA blocks the deubiquitination effects of USP7 on SOX9.
ADMA in synovial fluids is a potential OA diagnosis marker
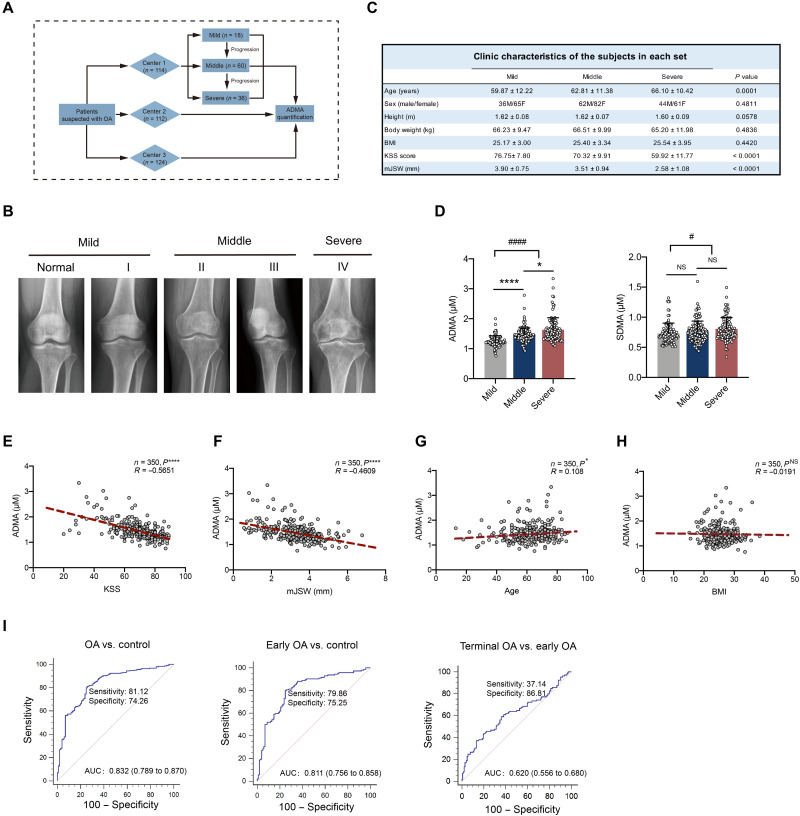
To investigate whether ADMA is increased in human synovial fluids, a retrospective study was performed (Fig. 5A). According to the inclusion criteria, 350 patients with OA (n = 114 in center 1, n = 112 in center 2, and n = 124 in center 3) were included and divided into three stages (mild, middle, and severe) according to Kellgren-Lawrence (KL) grades (Fig. 5B). No significant differences in sex, height, body weight, and body mass index (BMI) were identified among the three stages, although age slightly differed among patients, considering that OA is an age-related degenerative disease (Fig. 5C). As expected, ADMA was predominantly increased in the synovial fluids of patients with OA compared with that of relatively healthy patients, while SDMA did not demonstrate clinical significance. ADMA levels in synovial fluids had a substantially higher elevation in patients with severe OA than in those with middle-stage OA (Fig. 5D), suggesting that ADMA may have a positive correlation with OA progression. According to Pearson correlation analysis, ADMA in synovial fluids was negatively correlated with the Knee Society Score (clinical), a clinical evaluation score for knee OA (Fig. 5E), and minimal medial joint space width, a critical index measuring joint space narrowing and evaluating OA on x-ray (Fig. 5F) (29–31). Moreover, ADMA had a slight positive correlation with age and was not associated with BMI (Fig. 5, G and H). ADMA is closely related to OA both clinically and on radiographs; therefore, diagnostic evaluation was further conducted. The receiver operating characteristic (ROC) curve demonstrated that ADMA in synovial fluids could distinguish patients with OA from relatively healthy patients and be advantageous in early OA diagnosis with good sensitivity and specificity; however, it was ineffective in discriminating terminal OA from early OA (Fig. 5I). These data suggest that ADMA in synovial fluids could be a potential marker for OA diagnosis.
Fig. 5. ADMA in synovial fluids is predictive for OA diagnosis.
(A) Schematic diagram demonstrating multicenter retrospective analysis for ADMA in synovial fluids of patients with OA. (B) Representative knee radiographs of patients in different OA stages. Patients were divided into five stages according to the tibiofemoral Kellgren-Lawrence (KL) score. Mild OA includes normal and stage I, and middle OA includes stages II and III with stage IV defined as severe OA. (C) Clinical data of patients including age, sex, height, body weight, BMI, Knee Society Score (clinical, KSS), and minimal medial joint space width (mJSW). (D) The level of ADMA and its isomer SDMA in synovial fluids of patients with mild, middle, and severe OA [*P < 0.05, ****P < 0.0001, nonparametric test (Kruskal-Wallis test) followed by Dunn’s multiple comparisons test; #P < 0.05, ####P < 0.0001, unpaired two-tailed t test for comparison between mild and middle + severe]. (E to H) Pearson correlation analysis between ADMA level in synovial fluids and KSS, mJSW of patients with OA, age, and BMI (*P < 0.05, ****P < 0.0001, Pearson analysis). (I) Receiver operating characteristic (ROC) curve for ADMA discriminating control and OA, control and early OA, early OA and terminal OA. Control, early OA, and terminal OA represent patients with mild OA, middle OA, and severe OA, respectively, according to the KL grades. AUC, area under the ROC curve.
DISCUSSION
OA is a degenerated and metabolic disease with a series of metabolic changes during its pathogenesis, such as tricarboxylic acid cycle (32), lipid metabolism (33), and amino acid metabolic alterations (34). In our data from metabonomics, ADMA is increased in degenerated OA chondrocytes. ADMA is mainly involved in cardiovascular diseases, including ventricular hypertrophy (35) and atherosclerosis (21), through its inhibitory effects on NOS. Our data demonstrated that ADMA could accelerate the senescence of chondrocytes and promote cartilage degeneration independent of the NOS/NO pathway. NO is harmful to chondrocytes that can suppress proteoglycan and type II collagen synthesis, activate metalloproteinase, and promote chondrocyte apoptosis (36). However, the basic level of NO without inflammatory stimulation is considerably low. Therefore, the anti-NOS effects of ADMA under physiological conditions may be inconspicuous. Instead, ADMA directly and simultaneously interacts with SOX9, a master transcription factor that participates in chondrogenesis and cartilage anabolism (37), and its DUB USP7. As predicted by molecular docking, the binding sites between ADMA and SOX9 (or ADMA and USP7) are within the interacting region of SOX9 and USP7. Therefore, it is likely that ADMA could stick to and occupy the binding sites of SOX9 and USP7 and subsequently block the deubiquitination process exerted by USP7, resulting in a sharp reduction of SOX9 through the ubiquitin-proteasome pathway. However, it is possible that the inhibitory effect of ADMA is not specific since other potential ADMA-binding proteins, such as DHX9, FN1, and FUS, are also the substrates of USP7. More data are needed to analyze this point.
Free ADMA mainly has two catabolic pathways, which are separately catalyzed by DDAH family and AGXT2 (9). Compared with DDAH1 and DDAH2, AGXT2, has a very low abundance in cartilage tissues. DDAH1 has a relatively higher expression than DDAH2 in cartilage and is down-regulated in human OA cartilage or several murine OA models. DDAH2 was slightly up-regulated possibly because of its compensation for DDAH1 under high local ADMA conditions. Three in vivo interventions (DDAH1 inhibitor PD 404182, DDAH1 global mice, and DDAH1 cartilage-specific conditional knockout mice) were conducted. These mice exhibited similar OA phenotypes to that of mice articular-injected with ADMA in cartilage, particularly after DMM surgery. In comparison, no pronounced changes were found in the knee cartilage of DDAH2 global knockout mice.
In recent decades, OA diagnosis has primarily relied on clinical symptoms and radiology, including x-ray, computed tomography (CT), and magnetic resonance imaging. However, in clinical practice, not all the patients with OA simultaneously demonstrate clinical symptoms and radiological images to the same degree. Certain patients with severe pain or movement limitation may have few detectable lesions on radiographs, and vice versa (38). A survey of large-scale middle-aged community residents indicated that early patients with knee pain for more than 3 months could have no radiological OA images for no longer than 12 years (39). Therefore, such diagnostic criteria might not be able to provide evidence for early OA diagnosis or be sufficient to predict high-risk OA individuals. Local metabolism in cartilage is dynamic and closely associated with OA pathological state, which enables certain metabolites to be potential biomarkers (40). To the best of our knowledge, few studies on biomarkers to evaluate OA at early stages or track its progression have been reported. The plasma lysophosphatidylcholine-to-phosphatidylcholine ratio (41), arginine depletion (42), urine glycolate, hippurate, and trigonelline (43) have been previously considered as possible biomarkers for predicting advanced knee OA. However, since plasma or urea collects metabolites from organs and tissues in the whole body, it is likely that these plasma or urine biomarkers are nonspecific. Our data demonstrate a prominent and progressive increase in ADMA in OA chondrocytes and synovial fluids of patients with OA in three clinical centers among 350 patients. On this basis, ADMA, not its isomer SDMA, is able to distinguish patients with OA from relatively heathy ones and has advantages in early OA diagnosis with good sensitivity and specificity. Although more samples are required, ADMA in synovial fluids may still be a potential biomarker for OA diagnosis. A previous study revealed elevated ADMA levels in the blood sera of patients with severe OA (44). However, considering that serum ADMA also increased in patients with cardiovascular (45) or chronic kidney disease (46), its sensitivity and specificity for OA diagnosis require further investigation.
In conclusion, our study identifies a previously unidentified increased metabolite ADMA, accompanied by down-regulated DDAH1 in degenerated chondrocytes. Regulated by DDAH1, ADMA promotes degeneration and senescence of chondrocytes and cartilage, thus accelerating OA progression independent of the NOS/NO pathway. Instead, ADMA directly binds to SOX9 and USP7 and blocks their interaction region, facilitating SOX9 degradation through the ubiquitin-proteasome pathway. Therefore, targeting DDAH1 might provide a new direction for OA treatment.
METHODS
Study design
In this study, we investigated the role of ADMA in OA progression. This study consists of four major steps. First, we identified a higher ADMA levels and lower DDAH1 expression in OA chondrocytes and cartilages than in normal ones. Second, we revealed that increased ADMA accompanied by down-regulated DDAH1 could induce chondrocyte matrix degradation and accelerate cartilage damage both in vivo and in vitro. Third, we revealed the target of ADMA and explored the underlying mechanism for how ADMA regulates its downstream. Fourth, we investigated the role of ADMA in synovial fluids and its predictive value for early OA diagnosis.
Human subjects
Human cartilage tissues were collected following the protocols approved by the Ethics Committee of Sir Run Run Shaw Hospital, and the experiments were performed in accordance with the approved guidelines. Written informed consent was obtained from all the patients. Individual numbers of patient samples are indicated in the figure legends. Since the medication state of the patient is considered to be an important exclusion criterion for the studies, cartilage samples were strictly chosen from patients with OA with no history of pharmacological OA treatment, including steroid or nonsteroidal anti-inflammatory drug therapy within 3 years of total knee replacement surgery, and with no history of rheumatic arthritis, infectious arthritis, or severe underlying diseases. Damaged cartilage tissues were acquired from the medial part of the tibial plateau from patients with end-stage symptomatic knee OA at the time of total knee replacement surgery, whereas their corresponding undamaged cartilage tissues were obtained from the relatively healthy lateral part. All cartilage tissues used were representatively harvested and confirmed by the Osteoarthritis Research Society International (OARSI) grade (47) and Mankin score (48).
The use of human synovial fluid specimen was approved in accordance with the Ethics Committee of Sir Run Run Shaw Hospital, China Coast Guard Hospital of the People’s Armed Police Force, and Taizhou Hospital of Zhejiang Province, and was performed according to standard guidelines. Written informed consent was obtained from all the patients. Individual numbers of patient samples are indicated in the figure legends. OA was diagnosed by experienced orthopedists and rheumatologists on the basis of clinical data pertinent to the classification criteria of the American College of Rheumatology (49). The medical records of each patient were reviewed for age, sex, height, body weight, radiological findings, and laboratory data. Synovial fluids were previously obtained from patients with end-stage symptomatic knee OA at the time of total knee replacement surgery or from patients undergoing arthrocentesis. All samples were previously centrifuged at 12,000 rpm at 4°C for 5 min, and the supernatants were collected and stored at −80°C. Patients were divided into three stages according to the KL grades (50). Mild, consisting of KL grades 0 and 1, represents patients with very slight OA or normal. Middle, consisting of KL grades 2 and 3, represents patients with moderate OA. Severe, consisting of KL grade 4, represents patients with end-stage OA. LC-MS was used to determine the concentrations of ADMA and SDMA in the synovial fluid. ADMA (D4268, Sigma-Aldrich, St. Louis, MO, USA) and SDMA (D0390, Sigma-Aldrich) standards were used as controls. The linear results of ADMA and SDMA are good and reliable in the range of 0.1 to 1000 ng/ml. The quality control (QC) results of the synovial and standard samples were verified.
Animal studies
The animal handling and experimental procedures were performed with approval from the Institutional Animal Care and Use Committee. DDAH1 global knockout mice (DDAH1−/−) mice were gifted by Y. Chen, Minnesota University (51). Mice with DDAH1 conditionally knocked out in chondrocytes were generated by crossing DDAH1fl/fl mice (a gift given by Y. Chen, Minnesota University) with Col2-CreERT2 mice (the Jackson Laboratory, USA) or ACAN-CreERT2 (gifted by X. Wu, Zhejiang University School of Medicine). DDAH2 global knockout mice (DDAH2−/−) were purchased from Cyagen Biology Technology. The genotypes were confirmed using mouse direct PCR kit (Bimake, China). Wild-type (WT) C57BL/6 (B6) mice were purchased from SLAC Laboratory Animal Company (Shanghai, China). The use of animals was conducted according to the applicable ethical regulations and procedures approved by the Sir Run Run Shaw Hospital Committee for Animal Resources.
For the DMM model, the medial meniscotibial ligament was transected in the articular joint of 12-week-old male C57BL/6 mice according to a previous study (52). Control mice were sham-operated. The mice were euthanized 8 weeks after surgery. For the spontaneous OA model, spontaneous OA STR/Ort mice and their control CBA/CaCrl mice (53, 54) were donated by the Research Center of Regenerative Medicine, Guangxi Medical University. Mice were euthanized at 6 months of age. For the senile OA model, C57BL/6 mice were fed normally and euthanized at 12, 18, or 24 months of age, with 4-month-old mice serving as controls. All the articular joints of mice in the corresponding OA models were confirmed by the OARSI grade (47) and Mankin score (48).
For animal praxeology assays, this study used the hot plate, rotarod, and 6-min distance in an open field assay. The hot plate and rotarod assays have been previously described (55). For the 6-min distance, an open field system (ViewPoint, Civrieux, France) was applied. The mice were acclimated to the test room for 30 min before starting the experiment. Subsequently, we placed the mice in the center of the square chambers and allowed the mice to explore the chamber freely for 6 min. The distance was calculated automatically by the system.
AAV was purchased from Hanbio Tech (Shanghai, China). Virus [5 × 109 plaque-forming units (PFUs) in a total volume of 10 μl] was injected into the knee joints of mice using a 10-μl microsyringe with a 34G needle (Hamilton Company, Reno, NV, USA) at the indicated time or 1 week after DMM surgery. PD 404182 (150 μM in a total volume of 10 μl) was injected into the knee joints of mice using a 10-μl microsyringe with a 34G needle once a week. Injection started 1 week after DMM surgery.
Cell and cell lines
For mouse articular chondrocytes, primary cells were carefully isolated from the femoral condyles and tibial plateaus of 5-day-old WT, DDAH1−/−, DDAH2−/−, DDAH1fl/fl, or their indicated control mice by 0.2% collagenase digestion (56). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Amarillo, TX, USA) supplemented with 10% fetal bovine serum (FBS, catalog no. 10100147C, Gibco) and 1% penicillin-streptomycin (catalog no. 60162, Yeasen, Shanghai, China). Passage 1 cells were used for the analyses. For human articular chondrocytes, the cells were isolated from tibial plateaus and femoral condyles of human cartilage, digested by collagenase overnight, and then cultured in DMEM with 10% FBS and 1% penicillin-streptomycin. Passage 0 cells were used for metabolomics analysis. C28/I2 normal chondrocyte cells were purchased from Sigma-Aldrich and cultured in DMEM/F-12 supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin. 293T cells were purchased from the American Type Culture Collection and cultured in DMEM supplemented with 10% FBS (Gibco) and 1% penicillin G and streptomycin. All cells were cultured at 37°C in a humidified atmosphere consisting of 5% CO2 and 95% air.
For lentivirus infection, chondrocytes in a six-well plate were cultured and infected with DDAH1 lentivirus (Tsingke Biotechnology Co. Ltd., Beijing, China) at a density around 50% in 1 ml of culture medium for 12 hours. After that, cells were added with another 1 ml of culture medium and cultured for another 36 hours. Polybrene (Tsingke Biotechnology Co. Ltd., Beijing, China) at a final concentration of 10 μg/ml was added into the cell culture medium. Virus was subsequently replaced by fresh culture medium.
For adenovirus infection, chondrocytes in a six-well plate were cultured and infected with Cre adenovirus (5 × 107 PFUs) at a density around 50% in 1 ml of culture medium for 4 hours. After that, cells were added with another 1 ml of culture medium and cultured for another 4 to 6 hours. Virus was subsequently replaced by fresh culture medium.
Untargeted metabolomics
Untargeted metabolomics were performed by Luming Biotech (Shanghai, China). Briefly, for cell sample preparation, 2-chloro-l-phenylalanine (0.3 mg/ml) dissolved in methanol was used as an internal standard. Methanol/water (4:1) was added to each sample, and trichloromethane was added to each aliquot. An ultrasonic homogenizer was used to break up the cells for 6 min at 500 W. Mixtures of each sample were transferred to 1.5-ml Eppendorf tubes and then extracted by ultrasonication for 20 min in an ice water bath. The extract was centrifuged at 13,000 rpm at 4°C for 10 min. The supernatant (1 ml) was dried in a freeze-concentration centrifugal dryer. A mixture of methanol and water (1:4, v/v) was added to each sample. Samples were vortexed for 30 s, placed at 4°C for 2 min, and centrifuged at 13,000 rpm at 4°C for 15 min. The supernatants from each tube were collected using crystal syringes, filtered through 0.22-μm microfilters, and transferred to LC vials. The vials were stored at −80°C until LC-MS analysis. QC samples were prepared by mixing aliquots of all samples to form a pooled sample.
For sample detection, a Dionex Ultimate 3000 RS ultrahigh-performance liquid chromatography system fitted with a Q-Exactive quadrupole-Orbitrap mass spectrometer equipped with a heated electrospray ionization (ESI) source (Thermo Fisher Scientific, Waltham, MA, USA) was used to analyze the metabolic profiles in both ESI positive and negative ion modes. An ACQUITY UPLC BEH C18 column (1.7 μm, 2.1 mm by 100 mm) was used in both positive and negative modes. The binary gradient elution system consisted of (A) water (containing 0.1% formic acid, v/v) and (B) acetonitrile (ACN) (containing 0.1% formic acid, v/v), and separation was achieved using the following gradient: 5 to 20% B over 0 to 2 min, 20 to 60% B over 2 to 4 min, and 60 to 100% B over 4 to 11 min; the composition was held at 100% B for 2 min, then 13 to 13.5 min, 100 to 5% B, and 13.5 to 14.5 min holding at 5% B. The flow rate was 0.4 ml/min, and the column temperature was 45°C. All samples were stored at 4°C during the analysis. The injection volume was 5 μl. The mass range was from m/z (mass/charge ratio) 66.7 to 1000.5. The resolution was set at 70,000 for the full MS scans and 35,000 for the high energy collision dissociation (HCD) tandem MS (MS/MS) scans. The collision energies were set at 10, 20, and 40 eV. The mass spectrometer operated as follows: spray voltage, 3000 V (+) and 2500 V (−); sheath gas flow rate, 45 arbitrary units; auxiliary gas flow rate, 15 arbitrary units; and capillary temperature, 350°C. The QCs were injected every 10 samples throughout the analytical run to provide a set of data from which repeatability could be assessed.
ADMA detection for cells
The ADMA levels of chondrocytes were measured using LC-MS analysis and a bicinchoninic acid (BCA) protein quantification kit (Fudebio, Hangzhou, China). Cells were harvested, washed, and resuspended in an appropriate volume of phosphate-buffered saline (PBS). Quick freeze-thaw cycles combined with sonication were performed on the sample to release cell metabolites and proteins. Samples were then centrifuged, and the supernatants were collected. Protein concentration was measured using a BCA protein quantification kit (Fudebio, Hangzhou, China) as an internal reference for normalization. ADMA levels were measured using LC-MS analysis. The results are shown as ADMA levels/total protein.
ADMA in the culture medium was evaluated using ADMA direct (mouse/rat) enzyme-linked immunosorbent assay (ELISA kit, Enzo Life Sciences, Farmingdale, NY, USA) following the manufacturer’s instructions. Briefly, samples, standards, and controls were mixed with the reaction buffer and derivatization reagent in vials. After a 45-min incubation at room temperature, a dilution buffer was added to the vials. Subsequently, samples, standards, and controls were added to the wells of the microtiter plate. The ADMA antibody was then added to the wells and incubated at 4°C overnight. Each well was washed with wash buffer five times the next day by firmly tapping the plate on absorbent paper. Conjugate was added to each well, and the plate was incubated for 1 hour at room temperature. The plate was washed five times with a wash buffer. Last, the substrate was added to each well of the plate. After incubation for 10 min, a stop solution was applied. Each well was determined using an ELISA reader at 450 against 620 nm as a reference.
Histology and immunohistochemistry
The human cartilage or mouse articular joint specimens were fixed in 4% paraformaldehyde, dehydrated, decalcified in 0.5 M EDTA, embedded in paraffin, and sectioned at 3 μm. The sections were deparaffinized in xylene and hydrated with graded ethanol. For safranin O/fast green staining, the sections were first stained with 1% fast green (Sigma-Aldrich) for 3 to 5 min, rinsed with 1% acetic acid for 10 s, and then stained with 1% safranin O (Sigma-Aldrich) for 3 to 5 min. For alcian blue staining, the sections were stained with 1% alcian blue (Solarbio) for 30 min and then stained with nuclear fast red (Solarbio) for 10 min. Cartilage destruction was evaluated by OARSI grade (47), Mankin score (48), and osteophyte formation score (57) consisting of two domains: size and maturity. For immunohistochemistry, the sections were deparaffinized in xylene and hydrated with graded ethanol. Next, the sections were incubated with 3% H2O2 and 5% bovine serum albumin (BSA) at room temperature and with the indicated antibodies overnight. Subsequently, a secondary antibody conjugated with horseradish peroxidase (HRP) was added to the sections, followed by 3,3'-diaminobenzidine (DAB) staining (Sigma-Aldrich). The sections were finally stained with hematoxylin (Beyotime), washed and scanned using a KFBIO scan and analysis system (KFBIO, Zhejiang, China).
Micro-CT analysis
The knee joints of the mice were harvested with soft tissues dissected and fixed in 4% paraformaldehyde. CT scanning was performed using a high-resolution μCT (SHIMADZU, Kyoto, Japan). A 50-kVp voltage, 200-μA current, and 5.7-μm resolution per pixel were set for the scanner. 3D reconstructions of the CT scanning, including transverse, coronal, and sagittal sections, and subchondral bone parameters were analyzed using vgstudiomax 34 software according to the standard protocol under the same conditions. Each tissue was labeled with a random number for the blind assessment.
Micromass culture and chondrocyte 3D agarose culture
Micromass culture was used to evaluate ECM deposition, according to previous studies (58). Approximately 150,000 primary mouse chondrocytes were seeded in the center of 12-well plates and cultured for 7 days. Next, cells were fixed with 4% paraformaldehyde and stained with alcian blue 8 GS (Solarbio, China) at pH 0.2. ECM deposition was quantified using ImageJ software.
Chondrocyte 3D agarose culture was conducted as previously described (59). Briefly, we mixed 4 volumes of 2.5% agarose with 1 volume 5× culture medium, containing 10% FBS, 5% Hepes, and 5% penicillin-streptomycin to obtain a concentration of 2% agarose and 1× culture medium. The cell suspension was added to the medium/agarose solution to a final concentration of 2 × 106 cells/ml. Then, 700 μl of the mixture was added to a 24-well plate and incubated for approximately 15 min until gelling. The culture medium was subsequently added to the 24-well plate and replaced every day. After 14 days of culture, the hydrogel was fixed with 4% paraformaldehyde, embedded in paraffin, sectioned at 7 μm, and stained with alcian blue.
Proteolysis investigation and Western blotting
For proteolysis investigation, chondrocytes were pretreated with either MG132 (1 μM), a proteasome inhibitor, or chloroquine (10 μM), a lysosome inhibitor, for 30 min. Cells were then treated with cycloheximide (CHX, 1 μM) in a time-dependent manner (0, 2, 4, and 6 hours) and subsequently collected for protein extraction.
For total protein extraction, cells were lysed in strong radioimmunoprecipitation assay (RIPA) lysis buffer (Fudebio, Hangzhou, China) supplemented with protease inhibitor cocktails (Fudebio). Protein concentrations were quantified using a BCA protein assay kit (Fudebio). Equivalent protein amounts were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) under constant 120 V for approximately 50 min and subsequently transferred onto 0.22-μm polyvinylidene difluoride membranes (Merck KGaA) under constant 260 mA for 90 to 120 min. The membranes were then incubated in 5% BSA for 1 hour. After overnight incubation with a high-affinity antibody, the membranes were washed with Tris-HCl solution+Tween (TBST) and incubated with an HRP-conjugated secondary antibody (FDM007 and FDR007, Fudebio). After washing, the signals were detected using an enhanced chemiluminescence kit (FD8030, Fudebio). The antibodies were diluted in primary antibody dilution buffer (Fudebio).
The following antibodies were used in this study: anti–β-actin (M1210-2, HUABIO), anti-P21 antibody (ab188224, Abcam, Cambridge, UK), MMP13 antibody (ab39032, Abcam), anti-COL2A1 antibody (MAB8887, Millipore, Burlington, MA, USA), anti-aggrecan antibody (C8035, Millipore, Burlington, MA, USA), anti-SOX9 (ab185966, Abcam; Western blotting and immunofluorescence), anti-SOX9 [ab185230, Abcam; immunoprecipitation (IP)], anti-DDAH1 (37368, SAB), anti-DDAH2 (ab184166, Abcam), anti-NOS1 (R1510-28, HUABIO), anti–p-NOS1S1417 (ab5583, Abcam), anti-NOS2 (ER1706-89, HUABIO), anti-NOS3 (ab199956, Abcam), anti–p-NOS3S1177 (ab215717, Abcam), anti-ubiquitin (ab134953, Abcam), anti-USP7 (66514-1-Ig, Proteintech, Rosemont, IL, USA; Western blotting), anti-USP7(sc-137008, Santa Cruz Biotechnology, Dallas, TX, USA; immunofluorescence), anti-GST (ab111947, Abcam), and anti-His (12698, CST).
RNA extraction and quantitative real-time PCR analysis
RNAEX reagent (Accurate Biotechnology, Hunan, China) and the SteadyPure Universal RNA Extraction Kit (Accurate Biotechnology, Hunan, China) were used to extract total RNA from cells according to the manufacturer’s instructions. For mRNA analysis, Evo M-MLV RT Premix for quantitative PCR (qPCR) (Accurate Biotechnology, Hunan, China) and Hieff qPCR SYBR Green Master Mix (catalog no. 11201, Yeasen, Shanghai, China) were used according to the manufacturer’s instructions, and the reactions were then measured using a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer’s instructions. The reactions were normalized to the mRNA housekeeping gene β-actin. Primers were obtained from Tsingke (Beijing, China).
Cell counting kit-8 assay
Chondrocytes were seeded in a 96-well plate and subsequently treated with ADMA in a dose-dependent manner after attachment. After 24 or 48 hours, the supernatant was replaced with fresh medium with 10% cell counting kit-8 solution (Yeasen, Shanghai, China), and the plate was incubated for 1 to 4 hours. The optical density at 450 nm of each well was detected using a Multiskan FC System (Thermo Fisher Scientific).
Flow cytometry assay
Cytometry was performed as previously described (55). Briefly, the cells were collected using trypsin without EDTA (Gibco). After washing twice, the cells were stained with annexin V–fluorescein isothiocyanate (FITC)/propidium iodide (PI) using annexin V-FITC/PI apoptosis detection kit (catalog no. A211-02, Vazyme Biotech Co. Ltd). Last, the specimens were analyzed using a BD LSRFortessa flow cytometer (BD Biosciences).
NO assay
NO in the cell supernatant was detected using an NO detection assay (Beyotime, Jiangsu, China) according to the manufacturer’s instructions. Briefly, standards were diluted to concentrations of 0, 1, 2, 5, 10, 20, 40, 60, and 100 μM. Samples were collected and added to the wells of 96-well plates. Griess Regent I and Griess Regent II were added separately to each well. The samples and standards were detected at 540 nm using a Multiskan FC System (Thermo Fisher Scientific).
Silver staining and nano–LC-MS/MS
Cell lysates were incubated with biotin-ADMA or biotin at 4°C overnight. The following day, Pierce streptavidin magnetic beads (Thermo Fisher Scientific) were added to the mixture and incubated for 1.5 hours at room temperature. After washing five times with 1% Triton X-100 in PBS, the samples were denatured at 95°C in 1× loading buffer for 5 min. The differentially expressed proteins were determined using SDS-PAGE gel electrophoresis Fast Silver Stain kit (Beyotime) according to the manufacturer’s instructions. The protein bands were subsequently cut into pieces. After destaining with 50 mM NaS2O3/15 mM K3Fe(CN) (1:1), a volume of 10 mM dithiothreitol (DTT) in NH4HCO3 sufficient to cover the gel pieces was added, and proteins were reduced for 30 min at 56°C. The DTT solution was replaced with approximately the same volume of 55 mM iodoacetamide in 100 mM NH4HCO3. After 45 min of incubation at ambient temperature in the dark with occasional vortexing, the gel pieces were washed with 100 mM NH4HCO3, ACN, and 100 mM NH4HCO3, and shrunk again by adding the same volume of ACN. The liquid phase was removed, and the gel pieces were completely dried in a vacuum centrifuge. The gel pieces were covered with trypsin at 4°C (12.5 ng/μl of trypsin in 50 mM ammonium bicarbonate) and placed in an air-dry incubator (37°C) for 16 to 18 hours. Next, they were spun for 1 min; the supernatant was collected in a PCR tube and extracted with buffer (5% formic acid and 50% AcN). The supernatant was then collected in a PCR tube and dried in a vacuum centrifuge.
The peptide solution was then transferred to a ZIPTIP C18 (Millipore) for desalting and clean-up of the sample. Peptide samples were resuspended in water with 0.1% formic acid (v/v) and analyzed by nano–LC-MS/MS. LC separations were conducted on an Easy nano LC system (Thermo Fisher Scientific) using a 70-min LC gradient. The chromatography solvents used were water (A) and ACN (B), both with 0.1% formic acid. Peptide samples were concentrated and washed on a reversed-phase trap column (75 μm by 2 cm, 5 μm, 100 Å, C-18; Thermo Fisher Scientific) with 0.1% formic acid and then eluted from the analytical column (75 μm by 15 cm, 3 μm, 100 Å, C-18; Thermo Fisher Scientific) with the following gradient: 5 to 40% B (109 min). At 111 min, the gradient was increased to 90% B and maintained for 9 min. Eluting peptides were directly analyzed via MS/MS on a Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific). The MS1 full scan was set at a resolution of 60,000 at m/z 200, followed by “top 20” MS2 scans generated by HCD fragmentation at a resolution of 15,000 at m/z 200. The normalized collision energy (NCE) was set at NCE 28%, and the dynamic exclusion time was 45 s. All raw Xcalibur files acquired from MS runs were analyzed using the default settings of Proteome Discoverer 2.4 (Thermo Fisher Scientific) with minor modifications. The enzyme specification used during the search was trypsin. Carbamidomethylation of cysteine was selected as a fixed modification, whereas oxidation of methionine and N-terminal acetylation were selected as variable modifications. Mass tolerances for precursor and fragment ions were set at 10 parts per million (ppm) and 0.5 Da, respectively. The SEQUEST search algorithm was used to search MS/MS spectra against a composite database comprising all the UniProt Sus scrofa (Pig) database (10/2016, 34361 entries). The minimum cutoff for peptide length was set at seven amino acids, and the maximum permissible missed cleavage was set at two. Maximal false discovery rate (FDR) for peptide spectral match, proteins, and site was set to 0.01.
Proteomics
Proteomics was performed by Biotree Tech (Shanghai, China). Briefly, proteins were extracted using RIPA lysis buffer with sonication at 4°C and quantified using the BCA assay (Fudebio) following the manufacturer’s instructions. Samples were diluted to 1 mg/ml, mixed with ice acetone, and incubated overnight at −20°C. The samples were then centrifuged at 13,000 rpm at 4°C for 10 min and subsequently washed with 80% acetone. The proteins were redissolved, and DTT and indole-3-acetic acid were added, followed by protein digestion using trypsin. Equal amounts of proteins were labeled with Tandem Mass Tags (TMT) according to the protocol of the TMT label kit (Thermo Fisher Scientific). SDS was then deposited and removed using 2% trifluoroacetic acid. Peptide desalting was subsequently performed using C18 mini tubes, and the eluents were collected and dried at 4°C in a vacuum.
For nano–LC-MS/MS analysis, total peptides were separated and analyzed with a nano-UPLC (EASY-nLC1200) coupled to a Q Exactive HFX Orbitrap instrument (Thermo Fisher Scientific) with a nano–electrospray ion source. Separation was performed using a reversed-phase column (100 μm inside diameter ×15 cm, Reprosil-Pur 120 C18-AQ, 1.9 μm; Dr. Maisch). The mobile phases were H2O with 0.1% formic acid (FA), 2% ACN (phase A) and 80% ACN, 0.1% FA (phase B). Sample separation was executed with a 90-min gradient at a 300 nl/min flow rate. Gradient B was 2 to 5% for 2 min, 5 to 22% for 68 min, 22 to 45% for 16 min, 45 to 95% for 2 min, and 95% for 2 min. Data-dependent acquisition was performed in profile and positive mode with an Orbitrap analyzer at a resolution of 120,000 (at 200 m/z) and an m/z range of 350 to 1600 for MS1; for MS2, the resolution was set to 45k with a fixed first mass of 110 m/z. The automatic gain control target for MS1 was set to 3 × 106 with max injection time (IT) 30 ms, and 1 × 105 for MS2 with max IT 96 ms. The top 20 most intense ions were fragmented by HCD with an NCE of 32% and an isolation window of 0.7 m/z. The dynamic exclusion time window was 45 s; single-charged peaks and peaks with charge exceeding six were excluded from the data-dependent acquisition procedure.
For the proteome discoverer database search, Vendor’s raw MS files were processed using Proteome Discoverer software (version 2.4.0.305) and the built-in Sequest HT search engine. MS spectra lists were searched against their species-level UniProt FASTA databases, with carbamidomethyl (C), TMT 6 plex (K), and TMT 6 plex (N-term) as fixed modifications and oxidation (M) and acetyl (protein N-term) as variable modifications. Trypsin was used as the protease. A maximum of two missed cleavages was allowed. The FDR was set to 0.01 for both peptide-spectrum match (PSM) and peptide levels. Peptide identification was performed with an initial precursor mass deviation of up to 10 ppm and a fragment mass deviation of 0.02 Da. Unique peptides and razor peptides were used for protein quantification and total peptide amount for normalization. All other parameters were reserved as defaults.
Immunofluorescence
Cells cultured in confocal dishes were fixed in 4% paraformaldehyde for 20 min and then permeabilized in 0.5% Triton X-100 in PBS for 30 min. After washing with PBS, the cells were blocked with 5% BSA for 60 min. Primary antibodies against USP7 and SOX9 and secondary antibodies [Alexa Fluor 594 goat anti-mouse immunoglobulin G (IgG) and Alexa Fluor 488 goat anti-rabbit IgG, Fudebio] were diluted in 5% BSA. Cells were first incubated with primary antibodies at 4°C overnight and subsequently incubated with secondary antibodies for 1 hour at room temperature. After washing three times in PBS, the cells were incubated in 4′,6-diamidino-2-phenylindole (Beyotime). Representative images were taken using a Nikon A1 Ti confocal microscope (Tokyo, Japan).
Co-immunoprecipitation assay
Cells were lysed in cold Pierce IP lysis buffer (87787, Thermo Fisher Scientific), sonicated in an ultrasonic processor, and subsequently centrifuged at 12,000 rpm 4°C for 5 min to collect the supernatant (input). An antibody specific for the target protein was added to the lysis buffer and incubated overnight at 4°C. On the second day, Pierce protein A/G magnetic beads were washed three times with PBS and incubated with cell lysis buffer containing the indicated antibody for 1.5 hours at room temperature. The magnetic beads were washed three times with PBS containing 1% Triton X-100 and then boiled at 95°C in 1× loading buffer for 5 min. The indicated proteins were detected by Western blotting using specific antibodies.
GST pull-down
Recombinant human His-USP7 protein was purchased from Novus (E-519, Littleton, CO, USA), and recombinant human GST-SOX9 protein was purchased from Abnova (H00006662-P01, Taipei, Taiwan). The proteins were mixed and incubated in binding buffer at 4°C for 4 hours. Subsequently, anti-GST magnetic beads (P2138, Beyotime) were added to the buffer and incubated for another 2 hours. After washing five times, the beads were boiled and denatured at 95°C in 1× loading buffer for 5 min. The protein bands were detected by Western blotting using anti-GST (ab111947, Abcam) and anti-His (12698, CST) antibodies.
Molecular docking
The crystal structures of SOX9 and USP7 were obtained from the Protein Data Bank (https://rcsb.org). The SOX9 and USP7 docking was predicted by ZDOCK 3.0.2, according to a previous study (60). The chemical structure of ADMA was obtained from PubChem and processed by MOPAC and AutoDock Tools 1.5.6. The docking between ADMA and the interaction region of proteins was processed using AutoDock 4.2.6. Docking was energy-optimized using the Amber force including the steepest descent and conjugate gradient methods.
Statistics
Statistical analyses were performed using GraphPad Prism 8 or MedCalc V 19.0.4. Data are presented as the means ± SD. Statistical comparisons of two groups were performed using the Shapiro-Wilk test for normality, Levene’s test for homogeneity of variance, and paired or unpaired two-tailed t test. Multiple comparisons were made using the Shapiro-Wilk test, Levene’s test, and one-way analysis of variance (ANOVA) with Dunnett test. Data based on ADMA levels in synovial fluids were analyzed using Kolmogorov-Smirnov test and nonparametric test (Kruskal-Wallis test) with Dunn’s multiple comparisons test. Data based on ADMA clinical correlation were made using Pearson correlation analysis. The predicted probability of being diagnosed with OA was used as a surrogate marker to construct an ROC curve. Area under the ROC curve (95% confidence intervals) was used as an accuracy index for evaluating the diagnostic performance of ADMA. Statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001) was indicated in figures.
Acknowledgments
Funding: This work was supported by the China Postdoctoral Science Foundation (2022 M720125), the National Key R&D Program of China (2020YFC1107104), the Medical Healthy Scientific Technology project of Zhejiang Province (WKJ-ZJ-1906 and 2022502358), the National Natural Science Foundation of China (82102591 and 92068102), and the Natural Science Foundation of Zhejiang Province (LQ22H060001, Z20H060003, and LR22H060001).
Author contributions: S.F., A.Q., and Z.X. designed the experiments; Y.W., S.S., and Jiaxin Chen carried out most of the experiments; W.N., H. Zhou, W.X., Junxin Chen, Z.M., S.W., K.W., and H.W. helped to conduct animal experiments; Y.W., Q.W., Y.M., H.Zhang, X.S., Z.H.., P.S., and Z.J. helped to collect the materials; S.F., A.Q., Z.X., Y.W., S.S., and Jiaxin Chen supervised the experiments and analyzed results; Y.W., S.F., A.Q., and Z.X. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.T. Neogi, The epidemiology and impact of pain in osteoarthritis. Osteoarthr. Cartil. 21, 1145–1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. Kapoor, J. Martel-Pelletier, D. Lajeunesse, J. P. Pelletier, H. Fahmi, Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol. 7, 33–42 (2011). [DOI] [PubMed] [Google Scholar]
- 3.C. Corciulo, M. Lendhey, T. Wilder, H. Schoen, A. S. Cornelissen, G. Chang, O. D. Kennedy, B. N. Cronstein, Endogenous adenosine maintains cartilage homeostasis and exogenous adenosine inhibits osteoarthritis progression. Nat. Commun. 8, 15019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. Farnaghi, I. Prasadam, G. Cai, T. Friis, Z. Du, R. Crawford, X. Mao, Y. Xiao, Protective effects of mitochondria-targeted antioxidants and statins on cholesterol-induced osteoarthritis. FASEB J. 31, 356–367 (2017). [DOI] [PubMed] [Google Scholar]
- 5.W. S. Choi, G. Lee, W. H. Song, J. T. Koh, J. Yang, J. S. Kwak, H. E. Kim, S. K. Kim, Y. O. Son, H. Nam, I. Jin, Z. Y. Park, J. Kim, I. Y. Park, J. I. Hong, H. A. Kim, C. H. Chun, J. H. Ryu, J. S. Chun, The CH25H-CYP7B1-RORα axis of cholesterol metabolism regulates osteoarthritis. Nature 566, 254–258 (2019). [DOI] [PubMed] [Google Scholar]
- 6.J. Leiper, M. Nandi, The therapeutic potential of targeting endogenous inhibitors of nitric oxide synthesis. Nat. Rev. Drug Discov. 10, 277–291 (2011). [DOI] [PubMed] [Google Scholar]
- 7.T. Ogawa, M. Kimoto, K. Sasaoka, Purification and properties of a new enzyme, NG,NG-dimethylarginine dimethylaminohydrolase, from rat kidney. J. Biol. Chem. 264, 10205–10209 (1989). [PubMed] [Google Scholar]
- 8.R. P. Mookerjee, G. Mehta, V. Balasubramaniyan, Z. Mohamed Fel, N. Davies, V. Sharma, Y. Iwakiri, R. Jalan, Hepatic dimethylarginine-dimethylaminohydrolase1 is reduced in cirrhosis and is a target for therapy in portal hypertension. J. Hepatol. 62, 325–331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.T. Ogawa, M. Kimoto, K. Sasaoka, Dimethylarginine:pyruvate aminotransferase in rats. Purification, properties, and identity with alanine:glyoxylate aminotransferase 2. J. Biol. Chem. 265, 20938–20945 (1990). [PubMed] [Google Scholar]
- 10.R. H. Böger, S. M. Bode-Böger, P. S. Tsao, P. S. Lin, J. R. Chan, J. P. Cooke, An endogenous inhibitor of nitric oxide synthase regulates endothelial adhesiveness for monocytes. J. Am. Coll. Cardiol. 36, 2287–2295 (2000). [DOI] [PubMed] [Google Scholar]
- 11.F. Perticone, A. Sciacqua, R. Maio, M. Perticone, R. Maas, R. H. Boger, G. Tripepi, G. Sesti, C. Zoccali, Asymmetric dimethylarginine, L-arginine, and endothelial dysfunction in essential hypertension. J. Am. Coll. Cardiol. 46, 518–523 (2005). [DOI] [PubMed] [Google Scholar]
- 12.M. Saitoh, T. Osanai, T. Kamada, T. Matsunaga, H. Ishizaka, H. Hanada, K. Okumura, High plasma level of asymmetric dimethylarginine in patients with acutely exacerbated congestive heart failure: Role in reduction of plasma nitric oxide level. Heart Vessels 18, 177–182 (2003). [DOI] [PubMed] [Google Scholar]
- 13.M. C. Stühlinger, F. Abbasi, J. W. Chu, C. Lamendola, T. L. McLaughlin, J. P. Cooke, G. M. Reaven, P. S. Tsao, Relationship between insulin resistance and an endogenous nitric oxide synthase inhibitor. JAMA 287, 1420–1426 (2002). [DOI] [PubMed] [Google Scholar]
- 14.K. Sydow, C. E. Mondon, J. P. Cooke, Insulin resistance: Potential role of the endogenous nitric oxide synthase inhibitor ADMA. Vasc. Med. 10 (Suppl. 1), S35–S43 (2005). [DOI] [PubMed] [Google Scholar]
- 15.I. Singh, J. Kim, N. Saxena, S. Choi, S. M. T. Islam, A. K. Singh, M. Khan, J. Won, Vascular and immunopathological role of asymmetric dimethylarginine (ADMA) in experimental autoimmune encephalomyelitis. Immunology 164, 602–616 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Z. Xie, L. Hou, S. Shen, Y. Wu, J. Wang, Z. Jie, X. Zhao, X. Li, X. Zhang, J. Chen, W. Xu, L. Ning, Q. Ma, S. Wang, H. Wang, P. Yuan, X. Fang, A. Qin, S. Fan, Mechanical force promotes dimethylarginine dimethylaminohydrolase 1-mediated hydrolysis of the metabolite asymmetric dimethylarginine to enhance bone formation. Nat. Commun. 13, 50 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.S. Arlt, F. Schulze, M. Eichenlaub, R. Maas, J. T. Lehmbeck, E. Schwedhelm, H. Jahn, R. H. Böger, Asymmetrical dimethylarginine is increased in plasma and decreased in cerebrospinal fluid of patients with Alzheimer’s disease. Dement. Geriatr. Cogn. Disord. 26, 58–64 (2008). [DOI] [PubMed] [Google Scholar]
- 18.H. Zhang, C. Chen, Y. Cui, Y. Li, Z. Wang, X. Mao, P. Dou, Y. Li, C. Ma, lnc-SAMD14-4 can regulate expression of the COL1A1 and COL1A2 in human chondrocytes. Peer J. 7, e7491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.B. Cillero-Pastor, J. Mateos, C. Fernández-López, N. Oreiro, C. Ruiz-Romero, F. J. Blanco, Dimethylarginine dimethylaminohydrolase 2, a newly identified mitochondrial protein modulating nitric oxide synthesis in normal human chondrocytes. Arthritis Rheum. 64, 204–212 (2012). [DOI] [PubMed] [Google Scholar]
- 20.H. Jia, X. Ma, Y. Wei, W. Tong, R. J. Tower, A. Chandra, L. Wang, Z. Sun, Z. Yang, F. Badar, K. Zhang, W. J. Tseng, I. Kramer, M. Kneissel, Y. Xia, X. S. Liu, J. H. C. Wang, L. Han, M. Enomoto-Iwamoto, L. Qin, Loading-induced reduction in sclerostin as a mechanism of subchondral bone plate sclerosis in mouse knee joints during late-stage osteoarthritis. Arthritis Rheumatol. 70, 230–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. Antoniades, C. Shirodaria, P. Leeson, A. Antonopoulos, N. Warrick, T. Van-Assche, C. Cunnington, D. Tousoulis, R. Pillai, C. Ratnatunga, C. Stefanadis, K. M. Channon, Association of plasma asymmetrical dimethylarginine (ADMA) with elevated vascular superoxide production and endothelial nitric oxide synthase uncoupling: Implications for endothelial function in human atherosclerosis. Eur. Heart J. 30, 1142–1150 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Y. T. Ghebremariam, D. A. Erlanson, J. P. Cooke, A novel and potent inhibitor of dimethylarginine dimethylaminohydrolase: A modulator of cardiovascular nitric oxide. J. Pharmacol. Exp. Ther. 348, 69–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.X. Liu, X. Xu, R. Shang, Y. Chen, Asymmetric dimethylarginine (ADMA) as an important risk factor for the increased cardiovascular diseases and heart failure in chronic kidney disease. Nitric Oxide 78, 113–120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L. Rochette, J. Lorin, M. Zeller, J. C. Guilland, L. Lorgis, Y. Cottin, C. Vergely, Nitric oxide synthase inhibition and oxidative stress in cardiovascular diseases: Possible therapeutic targets? Pharmacol. Ther. 140, 239–257 (2013). [DOI] [PubMed] [Google Scholar]
- 25.J. T. Kielstein, S. M. Bode-Böger, J. C. Frölich, H. Haller, R. H. Böger, Relationship of asymmetric dimethylarginine to dialysis treatment and atherosclerotic disease. Kidney Int. Suppl. 78, S9–S13 (2001). [DOI] [PubMed] [Google Scholar]
- 26.X. M. Chen, J. Xia, T. Zhou, Q. Yuan, W. F. Zhang, C. P. Hu, Y. J. Li, J. L. Jiang, Involvement of DDAH/ADMA pathway in the pathogenesis of rheumatoid arthritis in rats. Int. Immunopharmacol. 16, 322–331 (2013). [DOI] [PubMed] [Google Scholar]
- 27.A. Haseeb, R. Kc, M. Angelozzi, C. de Charleroy, D. Rux, R. J. Tower, L. Yao, R. Pellegrino da Silva, M. Pacifici, L. Qin, V. Lefebvre, SOX9 keeps growth plates and articular cartilage healthy by inhibiting chondrocyte dedifferentiation/osteoblastic redifferentiation. Proc. Natl. Acad. Sci. U.S.A. 118, e2019152118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.X. Dong, X. Xu, C. Yang, Y. Luo, Y. Wu, J. Wang, USP7 regulates the proliferation and differentiation of ATDC5 cells through the Sox9-PTHrP-PTH1R axis. Bone 143, 115714 (2021). [DOI] [PubMed] [Google Scholar]
- 29.M. Attur, H. Zhou, J. Samuels, S. Krasnokutsky, M. Yau, J. U. Scher, M. Doherty, A. G. Wilson, J. Bencardino, M. Hochberg, J. M. Jordan, B. Mitchell, V. B. Kraus, S. B. Abramson, Interleukin 1 receptor antagonist (IL1RN) gene variants predict radiographic severity of knee osteoarthritis and risk of incident disease. Ann. Rheum. Dis. 79, 400–407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.M. Attur, A. Statnikov, J. Samuels, Z. Li, A. V. Alekseyenko, J. D. Greenberg, S. Krasnokutsky, L. Rybak, Q. A. Lu, J. Todd, H. Zhou, J. M. Jordan, V. B. Kraus, C. F. Aliferis, S. B. Abramson, Plasma levels of interleukin-1 receptor antagonist (IL1Ra) predict radiographic progression of symptomatic knee osteoarthritis. Osteoarthr. Cartil. 23, 1915–1924 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.M. Attur, S. Krasnokutsky, A. Statnikov, J. Samuels, Z. Li, O. Friese, M. P. Hellio Le Graverand-Gastineau, L. Rybak, V. B. Kraus, J. M. Jordan, C. F. Aliferis, S. B. Abramson, Low-grade inflammation in symptomatic knee osteoarthritis: Prognostic value of inflammatory plasma lipids and peripheral blood leukocyte biomarkers. Arthritis Rheumatol. 67, 2905–2915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.E. Maneiro, M. A. Martín, M. C. de Andres, M. J. López-Armada, J. L. Fernández-Sueiro, P. del Hoyo, F. Galdo, J. Arenas, F. J. Blanco, Mitochondrial respiratory activity is altered in osteoarthritic human articular chondrocytes. Arthritis Rheum. 48, 700–708 (2003). [DOI] [PubMed] [Google Scholar]
- 33.M. Loef, A. Ioan-Facsinay, D. O. Mook-Kanamori, K. Willems van Dijk, R. de Mutsert, M. Kloppenburg, F. R. Rosendaal, The association of plasma fatty acids with hand and knee osteoarthritis: The NEO study. Osteoarthr. Cartil. 28, 223–230 (2020). [DOI] [PubMed] [Google Scholar]
- 34.T. Piepoli, L. Mennuni, S. Zerbi, M. Lanza, L. C. Rovati, G. Caselli, Glutamate signaling in chondrocytes and the potential involvement of NMDA receptors in cell proliferation and inflammatory gene expression. Osteoarthr. Cartil. 17, 1076–1083 (2009). [DOI] [PubMed] [Google Scholar]
- 35.X. Xu, P. Zhang, D. Kwak, J. Fassett, W. Yue, D. Atzler, X. Hu, X. Liu, H. Wang, Z. Lu, H. Guo, E. Schwedhelm, R. H. Böger, P. Chen, Y. Chen, Cardiomyocyte dimethylarginine dimethylaminohydrolase-1 (DDAH1) plays an important role in attenuating ventricular hypertrophy and dysfunction. Basic Res. Cardiol. 112, 55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. B. Abramson, A. R. Amin, R. M. Clancy, M. Attur, The role of nitric oxide in tissue destruction. Best Pract. Res. Clin. Rheumatol. 15, 831–845 (2001). [DOI] [PubMed] [Google Scholar]
- 37.H. Song, K. H. Park, Regulation and function of SOX9 during cartilage development and regeneration. Semin. Cancer Biol. 67, 12–23 (2020). [DOI] [PubMed] [Google Scholar]
- 38.A. Abhishek, M. Doherty, Diagnosis and clinical presentation of osteoarthritis. Rheum. Dis. Clin. North Am. 39, 45–66 (2013). [DOI] [PubMed] [Google Scholar]
- 39.C. A. Thorstensson, M. L. Andersson, H. Jönsson, T. Saxne, I. F. Petersson, Natural course of knee osteoarthritis in middle-aged subjects with knee pain: 12-year follow-up using clinical and radiographic criteria. Ann. Rheum. Dis. 68, 1890–1893 (2009). [DOI] [PubMed] [Google Scholar]
- 40.E. B. de Sousa, G. C. J. Dos Santos, M. E. L. Duarte, V. N. Moura, D. P. Aguiar, Metabolomics as a promising tool for early osteoarthritis diagnosis. Braz. J. Med. Biol. Res. 50, e6485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.W. Zhang, G. Sun, D. Aitken, S. Likhodii, M. Liu, G. Martin, A. Furey, E. Randell, P. Rahman, G. Jones, G. Zhai, Lysophosphatidylcholines to phosphatidylcholines ratio predicts advanced knee osteoarthritis. Rheumatology 55, 1566–1574 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.W. Zhang, G. Sun, S. Likhodii, M. Liu, E. Aref-Eshghi, P. E. Harper, G. Martin, A. Furey, R. Green, E. Randell, P. Rahman, G. Zhai, Metabolomic analysis of human plasma reveals that arginine is depleted in knee osteoarthritis patients. Osteoarthr. Cartil. 24, 827–834 (2016). [DOI] [PubMed] [Google Scholar]
- 43.R. F. Loeser, W. Pathmasiri, S. J. Sumner, S. McRitchie, D. Beavers, P. Saxena, B. J. Nicklas, J. Jordan, A. Guermazi, D. J. Hunter, S. P. Messier, Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: An exploratory study. Osteoarthr. Cartil. 24, 1479–1486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.S. Kim, J. Hwang, J. Kim, J. K. Ahn, H. S. Cha, K. H. Kim, Metabolite profiles of synovial fluid change with the radiographic severity of knee osteoarthritis. Joint Bone Spine 84, 605–610 (2017). [DOI] [PubMed] [Google Scholar]
- 45.R. H. Böger, R. Maas, F. Schulze, E. Schwedhelm, Asymmetric dimethylarginine (ADMA) as a prospective marker of cardiovascular disease and mortality--an update on patient populations with a wide range of cardiovascular risk. Pharmacol. Res. 60, 481–487 (2009). [DOI] [PubMed] [Google Scholar]
- 46.J. Rysz, A. Gluba-Brzózka, B. Franczyk, Z. Jabłonowski, A. Ciałkowska-Rysz, Novel biomarkers in the diagnosis of chronic kidney disease and the prediction of its outcome. Int. J. Mol. Sci. 18, 1702 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.K. P. Pritzker, S. Gay, S. A. Jimenez, K. Ostergaard, J. P. Pelletier, P. A. Revell, D. Salter, W. B. van den Berg, Osteoarthritis cartilage histopathology: Grading and staging. Osteoarthr. Cartil. 14, 13–29 (2006). [DOI] [PubMed] [Google Scholar]
- 48.J. A. van der Sluijs, R. G. Geesink, A. J. van der Linden, S. K. Bulstra, R. Kuyer, J. Drukker, The reliability of the Mankin score for osteoarthritis. J. Orthop. Res. 10, 58–61 (1992). [DOI] [PubMed] [Google Scholar]
- 49.R. Altman, E. Asch, D. Bloch, G. Bole, D. Borenstein, K. Brandt, W. Christy, T. D. Cooke, R. Greenwald, M. Hochberg, D. H. MPH, D. Kaplan, W. Koopman, S. Longley III, H. Mankin, D. J. McShane, T. Medsger Jr., R. Meenan, W. Mikkelsen, R. Moskowitz, W. Murphy, B. Rothschild, M. Segal, L. Sokoloff, F. Wolfe, Development of criteria for the classification and reporting of osteoarthritis: Classification of osteoarthritis of the knee. Arthritis Rheum. 29, 1039–1049 (1986). [DOI] [PubMed] [Google Scholar]
- 50.N. Holzer, D. Salvo, A. C. Marijnissen, K. L. Vincken, A. C. Ahmad, E. Serra, P. Hoffmeyer, R. Stern, A. Lübbeke, M. Assal, Radiographic evaluation of posttraumatic osteoarthritis of the ankle: The Kellgren-Lawrence scale is reliable and correlates with clinical symptoms. Osteoarthr. Cartil. 23, 363–369 (2015). [DOI] [PubMed] [Google Scholar]
- 51.X. Hu, D. Atzler, X. Xu, P. Zhang, H. Guo, Z. Lu, J. Fassett, E. Schwedhelm, R. H. Böger, R. J. Bache, Y. Chen, Dimethylarginine dimethylaminohydrolase-1 is the critical enzyme for degrading the cardiovascular risk factor asymmetrical dimethylarginine. Arterioscler. Thromb. Vasc. Biol. 31, 1540–1546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.S. S. Glasson, T. J. Blanchet, E. A. Morris, The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthr. Cartil. 15, 1061–1069 (2007). [DOI] [PubMed] [Google Scholar]
- 53.K. A. Staines, B. Poulet, D. N. Wentworth, A. A. Pitsillides, The STR/ort mouse model of spontaneous osteoarthritis - an update. Osteoarthr. Cartil. 25, 802–808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. G. Chambers, L. Cox, L. Chong, N. Suri, P. Cover, M. T. Bayliss, R. M. Mason, Matrix metalloproteinases and aggrecanases cleave aggrecan in different zones of normal cartilage but colocalize in the development of osteoarthritic lesions in STR/ort mice. Arthritis Rheum. 44, 1455–1465 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Y. Wu, Z. Hong, W. Xu, J. Chen, Q. Wang, J. Chen, W. Ni, Z. Mei, Z. Xie, Y. Ma, J. Wang, J. Lu, C. Chen, S. Fan, S. Shen, Circular RNA circPDE4D protects against osteoarthritis by binding to miR-103a-3p and regulating FGF18. Mol. Ther. 29, 308–323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. Gosset, F. Berenbaum, S. Thirion, C. Jacques, Primary culture and phenotyping of murine chondrocytes. Nat. Protoc. 3, 1253–1260 (2008). [DOI] [PubMed] [Google Scholar]
- 57.S. S. Glasson, M. G. Chambers, W. B. Van Den Berg, C. B. Little, The OARSI histopathology initiative—Recommendations for histological assessments of osteoarthritis in the mouse. Osteoarthr. Cartil. 18 (Suppl. 3), S17–S23 (2010). [DOI] [PubMed] [Google Scholar]
- 58.S. E. Eldridge, A. Barawi, H. Wang, A. J. Roelofs, M. Kaneva, Z. Guan, H. Lydon, B. L. Thomas, A. S. Thorup, B. F. Fernandez, S. Caxaria, D. Strachan, A. Ali, K. Shanmuganathan, C. Pitzalis, J. R. Whiteford, F. Henson, A. W. McCaskie, C. De Bari, F. Dell’Accio, Agrin induces long-term osteochondral regeneration by supporting repair morphogenesis. Sci. Transl. Med. 12, eaax9086 (2020). [DOI] [PubMed] [Google Scholar]
- 59.C. Bougault, A. Paumier, E. Aubert-Foucher, F. Mallein-Gerin, Investigating conversion of mechanical force into biochemical signaling in three-dimensional chondrocyte cultures. Nat. Protoc. 4, 928–938 (2009). [DOI] [PubMed] [Google Scholar]
- 60.H. Hwang, B. Pierce, J. Mintseris, J. Janin, Z. Weng, Protein-protein docking benchmark version 3.0. Proteins 73, 705–709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S4