Abstract
Interactions between chromosomes and LINC (linker of nucleoskeleton and cytoskeleton) complexes in the nuclear envelope (NE) promote homolog pairing and synapsis during meiosis. By tethering chromosomes to cytoskeletal motors, these connections lead to processive chromosome movements along the NE. This activity is usually mediated by telomeres, but in the nematode Caenorhabditis elegans, special chromosome regions called “pairing centers” (PCs) have acquired this meiotic function. Here, we identify a previously uncharacterized meiosis-specific NE protein, MJL-1 (MAJIN-Like-1), that is essential for interactions between PCs and LINC complexes in C. elegans. Mutations in MJL-1 eliminate active chromosome movements during meiosis, resulting in nonhomologous synapsis and impaired homolog pairing. Fission yeast and mice also require NE proteins to connect chromosomes to LINC complexes. Extensive similarities in the molecular architecture of meiotic chromosome-NE attachments across eukaryotes suggest a common origin and/or functions of this architecture during meiosis.
A previously-uncharacterized nuclear envelope protein is required for meiotic chromosome pairing and synapsis in C. elegans.
INTRODUCTION
Sexual reproduction relies on meiosis, the specialized cell division program that produces haploid gametes. During meiosis, homologous chromosomes must pair, synapse, and undergo crossover recombination to segregate accurately. Upon meiotic entry, each replicated chromosome is assembled into an array of loops anchored to a linear structure known as the chromosome axis (1, 2). Pairing of homologs is gradually stabilized by assembly of a protein matrix, the synaptonemal complex (SC), between axes (1–4). SCs promote and regulate crossover recombination, which results in chiasmata, physical linkages between homologous chromosomes that persist until segregation and mediate bipolar alignment on the spindle (3, 5, 6).
Chromosome pairing, synapsis, and recombination are promoted by nuclear envelope (NE)–associated chromosome dynamics during meiosis (7–12). At meiotic entry, chromosomes become tethered to LINC (linker of nucleoskeleton and cytoskeleton) complexes composed of Sad1/UNC-84 homology (SUN) and Klarsicht/ANC-1/Syne-1 homology (KASH) domain proteins that span the two membranes of the NE (13, 14). Cytoskeletal motors interact with LINC complexes on the cytoplasmic face of the NE, resulting in dramatic chromosome movements during early meiotic prophase (15–17). This often leads to clustering of chromosome ends near cytoplasmic microtubule-organizing centers (MTOCs) to form a chromosome configuration known as the “meiotic bouquet” (15, 18, 19). However, in meiocytes that lack focal MTOCs, such as those in the nematode Caenorhabditis elegans, clustering of the NE attachment sites is absent or less prominent (20, 21).
In C. elegans, specialized regions on each chromosome known as “pairing centers” (PCs) promote homolog pairing and initiate synapsis (20, 22–25). Each PC recruits one of four meiosis-specific zinc finger proteins, Zinc finger In Meiosis (ZIM-1), ZIM-2, ZIM-3, or High Incidence of Males (HIM-8), through DNA binding sites present in clusters throughout the PC regions (26–28). During early meiotic prophase, PCs associate with LINC complexes composed of SUN-1 and ZYG-12 (29, 30). ZYG-12 interacts with cytoplasmic dynein and perhaps other microtubule motors to drive processive movement of chromosomes that promotes pairing and synapsis (21, 30–32).
In fission yeast, inner NE proteins Bqt3 and Bqt4 are required for tethering of telomeres to LINC complexes (33, 34). In mouse spermatocytes, the small NE protein MAJIN (membrane-anchored junction protein) connects the meiosis-specific shelterin-binding proteins Telomere Repeat Binding Bouquet Formation Protein 1 (TERB1) and TERB2 to LINC complexes (35, 36). Bqt4 and MAJIN share a similar structure, with a single transmembrane domain near their C termini (35, 37). Homologs of mouse MAJIN have been identified in many metazoans but not in nematodes (38).
Here, we report the identification and characterization of a previously uncharacterized meiosis-specific NE protein that is essential for interactions between PCs and LINC complexes in C. elegans. On the basis of the functions we have characterized, we named it MJL-1 (MAJIN-Like-1).
RESULTS
MJL-1 is a previously uncharacterized, meiosis-specific NE protein
In C. elegans, defects in meiosis result in a High incidence of males (Him) phenotype due to nondisjunction of the X chromosome (39); most meiotic mutants also produce many inviable embryos due to autosomal aneuploidy. We used an established “Green eggs and Him” screen, which exploits an xol-1p::gfp reporter that is expressed in XO (male) embryos, to identify mutant hermaphrodites with elevated meiotic nondisjunction (40). Molecular lesions in the mutants were identified by outcrossing to the CB4856 Hawaiian strain, reisolating homozygous mutants, whole-genome sequencing of their progeny, and computational analysis to identify likely causal mutations (41–43). Most of the mutations that we identified (50 of 52) were in genes previously shown to be important for meiosis, indicating that such Him screens are nearing saturation. Two mutations were found in previously uncharacterized genes. One of these resulted in a premature stop codon in C17E4.4, which encodes a small protein with a single predicted transmembrane domain. On the basis of previous transcriptome data, this gene is specifically expressed in germline and arcade cells (44–46).
Using CRISPR-Cas9–based genome editing, we inserted a hemagglutinin (HA) epitope tag at the N terminus of the C17E4.4 protein. Hermaphrodites homozygous for this insertion produced a normal number of embryos and a slightly elevated frequency of male self-progeny (~1%, compared to 0.2% in wild-type broods). Immunofluorescence of the HA-tagged protein showed NE-specific localization throughout the meiotic region of the germ line. The protein was undetectable in proliferating germline stem cells but was clearly observed at the NE upon meiotic entry and concentrated to form NE “patches” in transition zone (leptotene-zygotene) nuclei. Following synapsis, the protein was redistributed throughout the NE and persisted until late pachytene (Fig. 1, A and B).
Fig. 1. MJL-1 is a meiosis-specific NE protein that associates with PCs and LINC complexes.
(A) Composite maximum-intensity projection images of whole gonads from ha::mjl-1 hermaphrodites, stained with 4′,6-diamidino-2-phenylindole (DAPI) (top) and anti-HA antibodies (bottom). Scale bar, 20 μm. (B) Examples of nuclei at different stages of meiotic prophase. Scale bar, 5 μm. (C) HA-tagged MJL-1 colocalizes with SUN-1 (top) and phosphorylated PC proteins HIM-8 and ZIM-1, ZIM-2, and ZIM-3 (bottom) in early meiotic nuclei. PC proteins were detected using a phospho-specific antibody that recognizes these proteins when phosphorylated by CHK-2 (Checkpoint kinase 2 homolog) (54). Scale bars, 5 μm.
During pairing and synapsis, the zinc finger proteins HIM-8, ZIM-1, ZIM-2, and ZIM-3 (which we will refer to as “PC proteins”) bind to PCs and interact with the LINC complex proteins SUN-1 and ZYG-12, which concentrate within the NE to form multiple patches. Immunofluorescence revealed that HA-tagged C17E4.4 colocalized with SUN-1 and all four PC proteins during this transient stage (Fig. 1C).
On the basis of structural and functional similarities between C17E4.4 and mouse MAJIN (35), we named the gene mjl-1. Although MJL-1 shares no discernible sequence homology with MAJIN, both are small, single-pass transmembrane proteins with similar meiosis-specific functions (below) (Fig. 2A). MJL-1 is only weakly conserved within the genus Caenorhabditis, and we have not yet identified homologs in other nematode genera, including those that express homologs of the PC proteins (fig. S1) (47).
Fig. 2. Loss of MJL-1 disrupts PC function.
(A) Diagram of the mjl-1 gene, indicating the mutations described in this work (top). Primary structure of MJL-1 in C. elegans and MAJIN in M. musculus (bottom) (TM, transmembrane; DB, DNA binding domain). (B) Number of DAPI-staining bodies in oocyte nuclei at diakinesis in wild-type and mjl-1 mutant hermaphrodites. (C) Loss of MJL-1 disrupts the colocalization of PC proteins and LINC complex. Transition zone nuclei were stained with antibodies against HIM-8 (top) or ZIM-2 (bottom) (green), which mark X chromosome or chromosome V PCs, respectively, and SUN-1 (magenta). Scale bars, 5 μm. (D) Temporal projection of 75-s time course displacement track of GFP::HIM-8 in representative transition zone nuclei in wild type and mjl-1(ie142). Scale bar, 5 μm. (E) Average speed of GFP::HIM-8 foci in transition zone nuclei in wild-type (WT), mjl-1(ie142) (****P < 0.0001), and sun-1(jf18) (****P < 0.0001) hermaphrodites. Each point represents a single nucleus. P values were computed using Student’s t test with Bonferroni post hoc correction.
MJL-1 is required for association of PCs with LINC complexes
The mutation identified in our screen, mjl-1(ie142), is likely a null allele because it results in a stop codon before the transmembrane domain. We also obtained a deletion allele, mjl-1(tm1651), from the Japanese National BioResource Project (Fig. 2A). Hermaphrodites homozygous for either mutant allele produced very few viable self-progeny (<1%), and among these, many were males (29 and 33%, statistically indistinguishable), indicative of extensive meiotic nondisjunction. At diakinesis, most oocytes in mjl-1(ie142) and mjl-1(tm1651) hermaphrodites displayed 10 to 12 4′,6-diamidino-2-phenylindole (DAPI)–staining bodies (Fig. 2B), indicating that chromosomes failed to undergo crossing-over.
We tested whether MJL-1 is required for colocalization of PCs and LINC complexes. Immunofluorescence revealed that in mjl-1 mutants, SUN-1 did not concentrate with PC proteins or form NE patches in transition zone nuclei (Fig. 2C). However, PC proteins still appeared to associate with the NE, suggesting that they may interact directly with the membrane or another NE protein (fig. S2).
We crossed mjl-1(ie142) mutants to a strain expressing green fluorescent protein (GFP)–tagged HIM-8 (21) to analyze X chromosome movement. The average speed of HIM-8 foci (X PCs) in early meiotic nuclei was greatly reduced in the absence of MJL-1, from 59.8 nm/s in wild-type oocytes to 22.7 nm/s in mjl-1(ie142), similar to our measurements for sun-1(jf18), which results in a missense mutation (G311V) near the SUN domain (18.7 nm/s) (Fig. 2, D and E). This agrees with previous analysis of sun-1(jf18) (31, 32) and supports the conclusion that the residual chromosome movement in mjl-1(ie142) and sun-1(jf18) mutants likely results from diffusion rather than active motility (21, 32, 48). Together, we conclude that the normal interactions between PCs and LINC complexes and the resulting chromosome dynamics are disrupted in mjl-1 mutants.
MJL-1 is required to regulate synapsis
Loss of individual PC proteins delays synapsis of the corresponding chromosomes (26, 27). However, in mjl-1 mutants, we observed extensive SC assembly during early meiotic prophase despite very low levels of homolog pairing. Labeling of specific chromosome loci confirmed that this synapsis occurs between nonhomologous chromosomes (Fig. 3, A and B). Similar promiscuous synapsis was observed in sun-1(jf18) mutants and following auxin-induced degradation of ZYG-12 (Fig. 3, A and B), consistent with other evidence that the interaction between PCs and LINC complexes regulates synapsis so that it occurs only between homologous chromosomes (29, 30). How these interactions prevent nonhomologous synapsis remains an open question (30).
Fig. 3. Deletion of mjl-1 results in promiscuous nonhomologous synapsis.
(A) Extensive nonhomologous synapsis is observed in mjl-1(tm1651) and sun-1(jf18) mutants and, to a lesser extent, following depletion of AID::ZYG-12 by treatment with auxin for 12 hours. Mid-pachytene nuclei are stained with antibodies against HIM-8 (green) and SYP-2 (orange). Scale bars, 5 μm. (B) Quantification of chromosome pairing in wild-type and mutant hermaphrodites using immunofluorescence (HIM-8) (top) and fluorescence in situ hybridization (FISH) (Chr IV) (bottom) (*P < 0.5; ***P < 0.001; ****P < 0.0001). Gonads were divided into four zones (zone 1, premeiotic cells; zones 2 to 4, region spanning early prophase through pachytene, divided into three zones of equal length). P values were calculated by one-way analysis of variance (ANOVA) with pairwise Bonferroni post hoc correction.
MJL-1 depends on SUN-1 for its NE localization
Unexpectedly, we did not detect MJL-1 by immunofluorescence in sun-1(ok1282) null mutants or following auxin-induced degradation of SUN-1 (Fig. 4A). The abundance of MJL-1 detected on Western blots was also strongly reduced following auxin-induced degradation of SUN-1 (fig. S3). These findings indicate that SUN-1 is required for NE localization and/or stabilization of MJL-1. In contrast, neither the sun-1(jf18) missense mutation nor auxin-induced degradation of the KASH domain protein ZYG-12 disrupted the NE localization of MJL-1 (Fig. 4A), indicating that neither chromosome movement nor ZYG-12 is required for MJL-1 localization and, by extension, for association between MJL-1 and SUN-1. Coimmunoprecipitation of SUN-1 with MJL-1 further supports the idea that these proteins directly interact with each other (Fig. 4B).
Fig. 4. SUN-1 is required for NE localization of MJL-1.
(A) Transition zone nuclei in wild-type and mutant hermaphrodites expressing HA::MJL-1, stained with anti-HA antibodies (magenta in merged images). Scale bar, 5 μm. (B) Coimmunoprecipitation (CoIP) of SUN-1 and MJL-1. HA::MJL-1 was immunoprecipitated using an anti-HA antibody from lysates prepared from ha::mjl-1; v5::sun-1 hermaphrodites. Precipitated proteins were resolved by SDS–polyacrylamide gel electrophoresis and detected using anti-HA and anti-V5 antibodies. IP, immunoprecipitation; IgG, immunoglobulin G; IB, immunoblot. (C) The Y128F mutation in sun-1(ie143) disrupts interaction between MJL-1 and SUN-1. Transition zone nuclei stained with antibodies against SUN-1 (green in merged images) and HA (magenta) from wild-type and mutant hermaphrodites expressing HA::MJL-1. Scale bar, 5 μm. (D) Illustration of the inferred interaction between MJL-1 and SUN-1. INM, inner nucler membrane.
Our genetic screen also identified a separation-of-function mutation in sun-1 that resulted in meiotic defects similar to sun-1(jf18) without disrupting the mitotic functions of SUN-1. This missense mutation, sun-1(ie143), changes tyrosine-128 to phenylalanine (Y128F). This conserved tyrosine residue lies near the C terminus of the predicted transmembrane domain in SUN-1 and may contribute to anchoring the transmembrane domain through interactions with phospholipid head groups (49). In contrast to sun-1(jf18), sun-1(ie143) resulted in loss of MJL-1 protein from the NE (Fig. 4C and fig. S4). This suggests that SUN-1 and MJL-1 may interact through their transmembrane domains and/or adjacent regions in the perinuclear lumen (Fig. 4D). These regions are relatively well conserved within Caenorhabditis MJL-1 (fig. S5).
MJL-1 was also detected in apoptotic nuclei in the loop region of the germ line. These nuclei retain SUN-1 but not ZYG-12 at their NE (fig. S6), which may reflect disruption of the outer nuclear membrane during apoptosis, although this has not been directly demonstrated. The persistence of MJL-1 in these nuclei, together with our evidence that the protein requires SUN-1 for its localization to the NE (above) and connects PCs to SUN-1 (below), indicates that MJL-1 probably resides within the inner nuclear membrane with its N-terminal domain in the nucleoplasm, similar to mouse MAJIN (35).
MJL-1 interacts with meiotic PCs and LINC complex proteins
Sequence alignment of Caenorhabditis MJL-1 homologs revealed a short region of relatively high conservation containing several acidic residues (Fig. 5A). Using two synthetic CRIPSR RNAs (crRNAs) flanking this region, we generated an in-frame deletion of amino acids 34 to 49. The resulting MJL-1Δacidic protein was expressed and localized to the NE but did not colocalize with PCs (Fig. 5A and fig. S7). Animals homozygous for this deletion allele showed extensive nonhomologous synapsis, similar to mjl-1 null mutants (fig. S8). Together with evidence that SUN-1 is essential for localization and stability of MJL-1 (above), this suggests that the mutant protein retains the ability to interact with SUN-1 but does not link PC proteins to SUN-1. In contrast, an in-frame deletion of MJL-1 amino acids 9 to 26, which are also fairly well conserved within MJL-1 homologs, led to no apparent defects (fig. S9).
Fig. 5. A small domain of MJL-1 enriched in acidic amino acids is essential for its interaction with PC proteins.
(A) Sequence alignment of the N-terminal region of MJL-1 homologs within Caenorhabditis, generated using MAFFT. (B) Maximum intensity projection images showing transition zone nuclei stained with antibodies against HIM-8 (green in merged images) and HA (magenta) from hermaphrodites expressing HA::MJL-1 (top) or HA::MJL-1Δacidic (bottom). Scale bar, 5 μm. (C) PLK-2 activity is required for interaction between MJL-1 (magenta) and PC proteins (green). Scale bar, 5 μm. (D) Recruitment of PLK-2 by HIM-8 (green), which is abrogated in him-8(T64A) and him-8(me4) mutants, is required for the association of HIM-8 with MJL-1 (magenta). Scale bar, 5 μm.
The Polo-like kinase PLK-2 is recruited to PCs through Polo box–binding motifs in the PC proteins and is required for colocalization of PCs and LINC complexes (50, 51). In animals expressing only kinase-dead PLK-2K65M (52, 53), MJL-1 localized throughout the NE and did not concentrate with PC proteins (Fig. 5B). This indicates that PLK-2 activity is likely required for association between PC proteins and MJL-1/SUN-1 but presumably not for interaction between MJL-1 and SUN-1. Recruitment of PLK-2 to PCs depends on phosphorylation of the Polo box–interacting motifs in the HIM-8/ZIM proteins by Checkpoint kinase 2 homolog (CHK-2) (54). Consistent with this, we found that auxin-induced degradation of CHK-2 also abrogated the colocalization of MJL-1 and PCs (Fig. 5C).
We examined the colocalization of MJL-1 with two mutant versions of HIM-8, the zinc finger protein specific for the X chromosome PC: HIM-8T64A, which has a point mutation in its only Polo box–interacting motif (50), and HIM-8S85F [encoded by the him-8(me4) allele], which is not phosphorylated by CHK-2 and thus does not recruit PLK-2 (54). Neither mutant protein colocalized with patches of MJL-1 (Fig. 5C). Together, we conclude that recruitment of PLK-2 to HIM-8 is required for its association with MJL-1 and SUN-1. This is likely to be true for autosomal PCs as well, based on the failure of MJL-1 to form patches in plk-2(K65M) homozygotes.
MJL-1 directly promotes homolog pairing
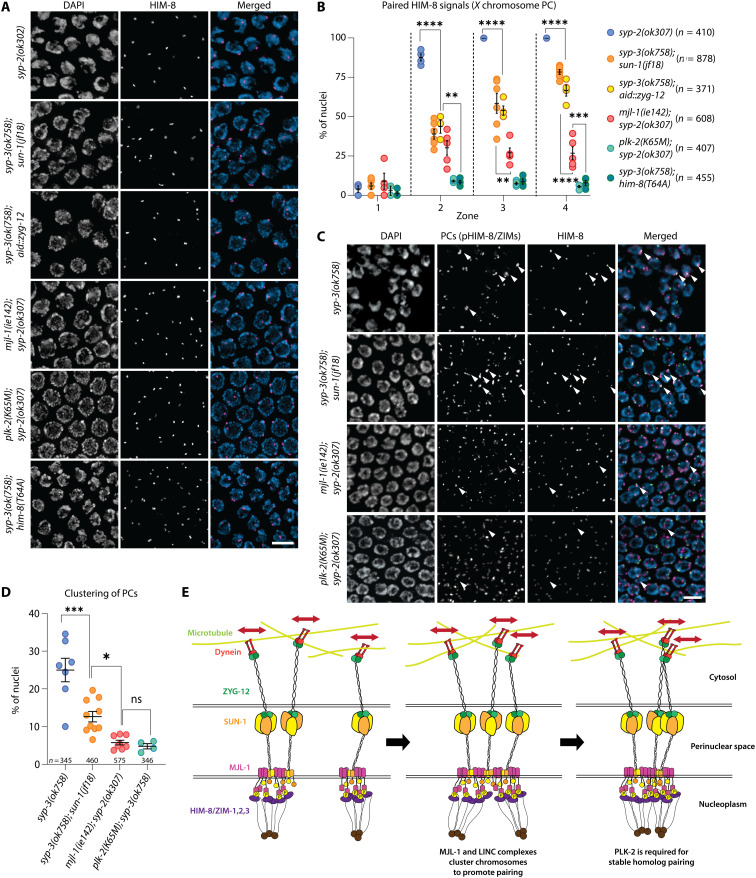
Mutations in SC proteins prevent nonhomologous synapsis and partially restore X chromosome pairing in sun-1(jf18) mutants (30). To assess the role of MJL-1 in pairing, we compared the extent of X chromosome pairing in aid::zyg-12 following auxin treatment, sun-1(jf18), and mjl-1(ie142), all in the absence of synapsis. Loss of MJL-1 resulted in more severe pairing defects than depletion of ZYG-12 or the sun-1(jf18) mutation, indicating that connection of chromosomes to MJL-1 and LINC complexes promotes limited pairing even in the absence of rapid chromosome movements (Fig. 6, A and B). We also observed a reduction in nonhomologous associations between HIM-8 and other PCs in the absence of MJL-1 compared to sun-1(jf18) mutants, suggesting that MJL-1 promotes clustering of PCs (Fig. 6, C and D), which may be required for efficient pairing of homologous PCs (Fig. 6E).
Fig. 6. MJL-1 promotes pairing even in the absence of chromosome movements.
(A) Blocking synapsis does not restore pairing of HIM-8 in mjl-1(ie142) mutants, in contrast to sun-1(jf18) or depletion of AID::ZYG-12 by treatment with auxin for 24 hours. Prior work has shown that syp-2 and syp-3 null mutants have indistinguishable effects on pairing and synapsis (72, 73). Nuclei display polarized morphology due to a cell cycle delay in the absence of synapsis (54). Scale bar, 5 μm. (B) Quantification of X chromosome pairing. The extended transition zone was divided into three equal regions (zones 2 to 4) by length (zone 1, premeiotic). Each point represents a single gonad. P values were calculated by one-way ANOVA with pairwise post hoc Bonferroni correction (**P < 0.01; ***P < 0.001; ****P < 0.0001). (C) In the absence of synapsis, clustering of HIM-8 with other PC proteins is lower in mjl-1(ie142) than in sun-1(jf18). Arrowheads indicate clusters of PCs. Scale bar, 5 μm. (D) Quantification of clustering between HIM-8 and other PC proteins in various mutants. Only nuclei in zone 2 were analyzed since pHIM-8/ZIM staining becomes undetectable in zones 3 and 4. P values were calculated using one-way ANOVA with post hoc pairwise Bonferroni correction (*P < 0.05; ***P < 0.001). ns, not significant. (E) Overview of homolog pairing. Upon meiotic entry, PCs recruit PC proteins (purple) and are connected to MJL-1 (magenta) and SUN-1 (yellow/orange). CHK-2 and PLK-2 are required for this association. MJL-1 interacts with SUN-1 through its transmembrane/perinuclear region. SUN-1 trimers interact with ZYG-12 dimers (green) that are connected to dynein (red) to generate processive chromosome movements (red arrows). These movements promote homolog searching (left). MJL-1 and SUN-1 cluster to promote homolog pairing (middle). PLK-2 phosphorylates unknown substrates to stabilize homolog pairing (right).
PLK-2 plays multiple roles in homolog pairing
Hermaphrodites expressing kinase-dead PLK-2K65M or the mutant HIM-8T64A protein displayed virtually no X chromosome pairing in the absence of synapsis, in contrast to mjl-1 mutants (Fig. 6, A and B). This indicates that the role of PLK-2 in homolog pairing goes beyond promoting interactions between PCs and the LINC complexes.
Phosphorylation of the nuclear lamin protein LMN-1 by PLK-2 promotes chromosome mobility along the NE during meiosis, likely by partially disrupting the lamina (52). To test whether this might account for the essential role of PLK-2 in homolog pairing, we depleted LMN-1 using RNA interference (RNAi). This depletion did not rescue X chromosome pairing in syp-3(ok758); him-8(T64A) mutants, revealing that PLK-2 activity at PCs contributes to pairing through a lamin-independent mechanism (fig. S10).
In SC-deficient mutants, homologous PCs pair shortly after meiotic entry and dissociate during late prophase (25, 55). We found that dissociation of paired PCs occurs concomitantly with the disappearance of PLK-2, indicating that PLK-2 activity is important to maintain synapsis-independent interactions between homologous PCs (fig. S11).
DISCUSSION
Similarities and differences between MJL-1 and MAJIN
We were able to identify homologs of MJL-1 only within Caenorhabditis. Homologs of HIM-8 and ZIM proteins can been detected in related nematode genera (47) but show rapid divergence, particularly in their N-terminal domains, which act as scaffolds to recruit kinases and may also directly interact with NE proteins. If the zinc finger proteins interact with MJL-1, as suggested by our findings, then rapid coevolution of these proteins may partially account for our inability to detect MJL-1 homologs in other nematode species.
Our evidence indicates that MJL-1 connects the PC proteins to LINC complexes in C. elegans. Similarly, in mice, MAJIN connects shelterin-binding proteins TERB1 and TERB2 to LINC complexes (35, 56). MAJIN interacts with SUN-1 through its nucleoplasmic domain (57), while MJL-1 and SUN-1 may interact through their transmembrane domains and/or perinuclear regions. In fission yeast, a direct interaction between NE proteins Bqt3 and Bqt4 and LINC complexes has not been detected (33). Whereas MJL-1 requires PLK-2 activity to interact with PC proteins, cyclin-dependent kinase 2 (CDK2) activity is required for interaction between MAJIN and SUN-1 in mice (Fig. 7) (57, 58).
Fig. 7. Similarities in molecular architecture of meiotic chromosome-LINC complex attachments in fission yeast, mouse, and C. elegans.
In fission yeast, Bqt4 (magenta) connects the telomere shelterin complex (red) to the NE in vegetative cells. Bqt4 interacts with a multi-pass NE protein, Bqt3 (pink) (33). Meiosis-specific proteins Bqt1 and Bqt2 (blue) are also required to connect telomeres to LINC complexes (yellow/orange) (left) (33, 34). In mice, TERB2 (blue) interacts with MAJIN (magenta), and TERB1 (blue) interacts with SUN-1 (35, 56). MAJIN also associates with SUN-1 (yellow/orange). This requires CDK2 activity (light blue) (middle) (57). In C. elegans, MJL-1 (magenta) connects PC proteins (purple) to SUN-1 (yellow/orange). This requires CHK-2 and PLK-2 activities (light blue), which are recruited by the PC proteins. MJL-1 and SUN-1 cluster to promote homolog pairing (right).
MAJIN and Bqt4 contain N-terminal DNA binding motifs that are required for recruitment of telomeres to the NE (35, 37). The DNA binding motif of MAJIN has been implicated in “telomere cap exchange,” whereby telomeres release shelterin and directly interact with TERB1, TERB2, and MAJIN (35). In contrast, MJL-1 lacks an apparent DNA binding motif, and PCs in C. elegans appear to associate with the NE even in the absence of MJL-1 or SUN-1. Thus, while MJL-1 shares similarity with MAJIN and Bqt4, the way it connects to chromosomes is likely distinct from vertebrate MAJIN proteins. Together with the lack of apparent sequence homology, this makes it unclear whether MJL-1 is evolutionarily related to other meiotic NE proteins.
Roles of MJL-1 and LINC complexes in regulation of pairing and synapsis
Connections between PCs and LINC complexes inhibit inappropriate synapsis in C. elegans (29, 30). This is consistent with observations that these associations and resulting chromosome movements persist even after pairing of homologous PCs, which occurs soon after entry into meiosis. Loss of telomere-LINC complex connections and/or chromosome movements in mice also leads to some nonhomologous synapsis, albeit not as extensive as in C. elegans (11, 35, 56, 59). Moreover, loss of CDK2, which is associated with LINC complexes during early prophase in mouse spermatocytes, results in extensive nonhomologous synapsis (58, 60–62), suggesting that regulation of synapsis may be a general role of chromosome-LINC complex attachments despite differences in the details of this regulation between organisms. Intriguingly, telomeric attachments in mammals and PC attachments in C. elegans each require a kinase (CDK2 and PLK-2, respectively) that is also involved in crossover regulation, suggesting that coordination between chromosome attachment sites and crossover (CO) intermediates may be a conserved feature of meiosis (63–65).
MATERIALS AND METHODS
C. elegans strains
N2 Bristol was used as the wild-type C. elegans strain; all mutations described here were generated in this background. The Hawaiian isolate CB4856 was used for genetic mapping. All strains were maintained at 20°C under standard laboratory conditions. The following mutations and balancers were used: mjl-1(ie142) (I:9419844; C to T), mjl-1(tm1651), mjl-1(Δacidic), sun-1(jf18), sun-1(ok1282), sun-1(ie143) (V:13193099; T to A), syp-2(ok307), syp-3(ok758), him-8(me4), him-8(T64A) (50), plk-2(K65M)::3xflag (53), nT1[qIs51] (IV;V), hT2 [bli-4(e937) let-?(q782) qIs48] (I;III). The following constructs were used for auxin-inducible degradation: aid::ha::zyg-12 and aid::v5::sun-1, and ha::aid::chk-2, where “aid” designates a DNA sequence encoding a 44–amino acid degron (66).
To generate ha::mjl-1 strains, single-stranded DNA (ssDNA) templates were designed to insert one or two copies of the HA tag (YPYDVPDYA) at the N terminus of MJL-1, separated by a flexible linker (GGGGS). These were coinjected with Cas9-Nuclear Localization Signal (NLS) prebound to duplexed guide RNAs (gRNAs), as well as a gRNA and ssDNA template for co-CRISPR of dpy-10 (67). Final concentrations are as follows: dpy-10 crRNA, 20 μM; mjl-1 crRNA, 50 μM; trans-activating CRISPR RNA (tracrRNA), 40 μM; Cas9-NLS protein, 20 μM; dpy-10 repair template, 1 μM; mjl-1 repair template, 1 μM. To label MJL-1 using the split-GFP system and V5 tag, a template to insert GFP11 and V5 was coinjected with the Cas9-gRNA ribonucleoprotein complex into DUP223 glh-1{sam129[glh-1::T2A::sGFP2 (1-10)]} (68). Essentially the same procedure was used to generate in-frame deletions in mjl-1, except that two gRNAs were used.
Auxin-induced degradation
A stock solution containing 250 mM indole acetic acid (auxin) in EtOH was diluted to 1 mM in Nematode Growth Medium) (NGM) agar just before pouring plates. After drying overnight, plates were seeded with Escherichia coli: OP50 freshly cultured overnight to saturation in 50 ml of LB was pelleted by centrifugation at 3000g for 5 min and resuspended in 500 μl of M9 buffer containing 1 mM auxin. This concentrated bacteria + auxin was spread on the plates and allowed to grow at room temperature for 1 to 2 days. To deplete degron-tagged proteins, young adult animals aged 24 to 48 hours from L4 were picked onto these plates and analyzed 4 to 24 hours later.
RNA interference
Carbenicillin and isopropyl-β-d-thiogalactopyranoside were added to cooled NGM agar to 200 μg/ml and 1 mM final concentration, respectively, just before pouring plates. Clones from the Ahringer laboratory (69) were freshly cultured overnight to saturation in 10 ml of LB containing carbenicillin (200 μg/ml). The culture was pelleted by centrifugation at 3000g for 5 min and resuspended in 50 μl of M9 buffer. This concentrated E. coli was spread on the plates and allowed to grow at 37°C for 1 day. For feeding RNAi, young adult animals aged 24 to 48 hours from L4 were picked onto these plates and analyzed 24 to 48 hours later. LMN-1 depletion was confirmed by LMN-1 immunofluorescence.
Cytological methods
Immunofluorescence and in situ hybridization were performed essentially as described previously (70). In brief, young adult worms were cut with a scalpel blade in egg buffer containing 0.05% tetramisole to release their gonads on slides, fixed in 1% formaldehyde in egg buffer for 2 min, transferred to tubes, and incubated with methanol prechilled to −30°C for 5 min. The tissue was then washed three times in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBST) at room temperature. Tissues were blocked using 1× Roche blocking reagent in PBST for 20 min. Primary antibodies were diluted into the same blocking solution and incubated with the tissues overnight at 4°C. Secondary antibodies were prepared in the blocking solution (1:200), mixed with samples, and incubated 1 to 2 hours at room temperature. Samples were mounted in Prolong Diamond mounting medium containing DAPI (Invitrogen).
For fluorescence in situ hybridization, dissected gonads were fixed in 2% formaldehyde in egg buffer for 5 min, incubated with methanol prechilled to −30°C for 5 min, and washed three times in 2× SSC containing 0.1% Tween 20 (2× SSCT) at room temperature. The tissue was then incubated in 50% formamide in 2× SSCT overnight at 37°C. Fluorophore-conjugated oligonucleotide probes (0.5 to 1 μl of 100 μM; IDT) “IV-2” (71) were added to 100 μl of hybridization buffer (50% formamide and 10% dextran sulfate in 2× SSCT), and tissues were moved into this mix and incubated for 2 to 3 min at 91°C and then overnight at 37°C. The tissues were washed three times in 2× SSCT at room temperature and mounted as for immunofluorescence (above).
Images were acquired using a DeltaVision Elite wide-field microscope system (GE) with a 100× 1.40 or 1.45 numerical aperture (NA) oil-immersion objective, or a CSU-W1 SoRa confocal microscope system equipped with a 100×, 1.49 NA oil-immersion objective [Intelligent Imaging Innovations Inc. (3i)]. Deconvolution, projection, and analysis were performed using the softWoRx package or Slidebook 6 (3i).
Antibodies
All antibodies used in this study were obtained from commercial sources or have been previously described. They include the following antibodies and dilutions: mouse monoclonal anti-HA (Invitrogen 2-2.2.14) (1:250), mouse monoclonal anti-V5 (Invitrogen 46-0705) (1:250), and rabbit polyclonal anti-V5 (Sigma-Aldrich, V8137) (1:250). Custom polyclonal antibodies included rabbit anti–SUN-1 (1:250) (30), rat anti–HIM-8 (1:500) (26), rabbit anti–SYP-2 (1:500) (72), rabbit anti–phospho-HIM-8/ZIMs (1:1000) (54), and guinea pig anti–ZIM-2 (1:2000, affinity purified). Fluorophore-conjugated secondary antibodies were purchased from Jackson ImmunoResearch and used at 1:200 dilution.
Quantification of homolog pairing
Three-dimensional (3D) distances between HIM-8 or fluorescence in situ hybridization (FISH) signals were measured using the “Measure Distance” tool in softWoRx. Foci closer than 0.6 μm were scored as paired. We defined this threshold on the basis of the maximum width of PC protein patches associated with paired chromosomes in wild-type oocytes. For analysis of pairing of FISH signals, we included only nuclei displaying two clear foci.
In vivo imaging and quantification of chromosome movement
The 3D confocal image acquisition was performed essentially as described (21) using a Marianas spinning-disc confocal microscope system equipped with a 100×, 1.46 NA oil-immersion objective [Intelligent Imaging Innovations Inc. (3i)]. Exposure time was set to 100 to 150 ms depending on the brightness of foci. Stacks of 10 to 20 optical sections at 0.5-μm z spacing were acquired every 5 s for a total of 5 to 10 min. The 3D time-lapse images were analyzed using Imaris 9.2.0 (Bitplane). Background drift was corrected using the “Reference Frame” tool. Foci were detected using the “Spots” tool with an estimated XY diameter of 1.33 μm and filtered with “Quality” and “Intensity Sum” (Imaris). Tracks were obtained with a max distance of 1.75 μm and a max gap of 3. Tracks from the background noise were manually removed. P values were calculated using the pairwise t test with post hoc Bonferroni correction.
Western blots
Two hundred young adult animals aged 48 hours from the L4 stage were picked into tris-buffered saline (TBS) with 0.1% Tween 20 buffer, washed three times, and then incubated in 1× SDS sample buffer at 50°C for 10 min. Samples were vortexed for 2 to 3 min until no visible solid material remained. Proteins were separated using SDS–polyacrylamide gel electrophoresis gradient gels (Invitrogen, NuPAGE, 4 to 12%, bis-tris, 1.0 mm, 10-well, MES SDS running buffer) and transferred to Amersham Hybond P 0.45 polyvinylidene difluoride membranes. The membrane was cut into slices and probed with mouse anti-HA (Invitrogen, 2-2.2.14) (1:1000), mouse anti-V5 (Invitrogen) (1:1000), or mouse anti–α-tubulin (Sigma-Aldrich, DM1A) (1:5000) antibodies overnight at 4°C. Horseradish peroxidase–conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch) (1:10,000) was incubated with membranes for 1 hour at room temperature. SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) was used for detection. ImageJ mean gray value was used for quantification.
Immunoprecipitation
Approximately 200,000 young adult animals grown in liquid culture, aged 48 hours from the L4 stage, were collected by centrifugation and homogenized with a Douncer in 1× egg buffer with 250 mM sucrose until 90% of adults were broken. Debris was precipitated by centrifugation at 50g for 2 min. The supernatant was filtered using 40-μm filters followed by 20-μm filters to remove additional debris. Nuclei were pelleted by centrifugation at 2000g for 10 min and resuspended in 800 μl of lysis buffer [130 mM NaCl, 2.5 mM MgCl2, 25 mM Hepes (pH 7.4), 2 mM EGTA, and 1% Triton X-100, with Roche cOmplete Mini EDTA-free protease inhibitor cocktail]. Nuclei were sonicated in a Bioruptor Twin sonication bath (Diagenode, Denville, NJ) at 4°C for 30-min periods of alternating 30 s on high power and 30 s off. The resulting lysate was centrifuged at 10,000g for 2 min, and supernatant was incubated with antibody-coated Dynabeads Protein A (Invitrogen) at 4°C for 2 hours and eluted in urea solution (6 M urea, 6% SDS, and 5% 2-mercaptoethanol).
Acknowledgments
We thank M. Colaiácovo and V. Jantsch for providing antibodies, the laboratory of S. Mitani for the mjl-1(tm1651) mutant, and Y. Kim for the plk-2(K65M) mutant. We thank members of the Dernburg laboratory for helpful comments on the manuscript and F. Wu for strains. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding: This work was supported by the Investigator award from HHMI (A.F.D.), Fellowship from the Korea Foundation for Advanced Studies (KFAS) (H.J.K.), and Life Sciences Research Foundation (C.L.)
Author contributions: Conceptualization: H.J.K. and A.F.D. Methodology: H.J.K. and A.F.D. Investigation: H.J.K., C.L., and L.Z. Supervision: A.F.D. Writing—original draft: H.J.K. and A.F.D.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The mjl-1(tm1651) mutant allele can be provided upon request with written permission from S. Mitani or requested from the National Bioresource Project (https://shigen.nig.ac.jp/c.elegans/).
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
REFERENCES AND NOTES
- 1.D. Zickler, N. Kleckner, Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Y. Blat, R. U. Protacio, N. Hunter, N. Kleckner, Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 111, 791–802 (2002). [DOI] [PubMed] [Google Scholar]
- 3.S. L. Page, R. S. Hawley, The genetics and molecular biology of the synaptonemal complex. Annu. Rev. Cell Dev. Biol. 20, 525–558 (2004). [DOI] [PubMed] [Google Scholar]
- 4.C. K. Cahoon, R. S. Hawley, Regulating the construction and demolition of the synaptonemal complex. Nat. Struct. Mol. Biol. 23, 369–377 (2016). [DOI] [PubMed] [Google Scholar]
- 5.D. Zickler, N. Kleckner, Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. Kleckner, Chiasma formation: Chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194 (2006). [DOI] [PubMed] [Google Scholar]
- 7.M. N. Conrad, A. M. Dominguez, M. E. Dresser, Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276, 1252–1255 (1997). [DOI] [PubMed] [Google Scholar]
- 8.P. R. Chua, G. S. Roeder, Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11, 1786–1800 (1997). [DOI] [PubMed] [Google Scholar]
- 9.J. P. Cooper, Y. Watanabe, P. Nurse, Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392, 828–831 (1998). [DOI] [PubMed] [Google Scholar]
- 10.D.-Q. Ding, A. Yamamoto, T. Haraguchi, Y. Hiraoka, Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell 6, 329–341 (2004). [DOI] [PubMed] [Google Scholar]
- 11.X. Ding, R. Xu, J. Yu, T. Xu, Y. Zhuang, M. Han, SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev. Cell 12, 863–872 (2007). [DOI] [PubMed] [Google Scholar]
- 12.M. N. Conrad, C.-Y. Lee, G. Chao, M. Shinohara, H. Kosaka, A. Shinohara, J.-A. Conchello, M. E. Dresser, Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133, 1175–1187 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Y. Hiraoka, A. F. Dernburg, The SUN rises on meiotic chromosome dynamics. Dev. Cell 17, 598–605 (2009). [DOI] [PubMed] [Google Scholar]
- 14.J. Link, V. Jantsch, Meiotic chromosomes in motion: A perspective from Mus musculus and Caenorhabditis elegans. Chromosoma 128, 317–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Hiraoka, Meiotic telomeres: A matchmaker for homologous chromosomes. Genes Cells 3, 405–413 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Y. Chikashige, D. Q. Ding, H. Funabiki, T. Haraguchi, S. Mashiko, M. Yanagida, Y. Hiraoka, Telomere-led premeiotic chromosome movement in fission yeast. Science 264, 270–273 (1994). [DOI] [PubMed] [Google Scholar]
- 17.H. Scherthan, S. Weich, H. Schwegler, C. Heyting, M. Härle, T. Cremer, Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 134, 1109–1125 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D. Zickler, N. Kleckner, The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32, 619–697 (1998). [DOI] [PubMed] [Google Scholar]
- 19.H. Scherthan, A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 2, 621–627 (2001). [DOI] [PubMed] [Google Scholar]
- 20.O. Rog, A. F. Dernburg, Chromosome pairing and synapsis during Caenorhabditis elegans meiosis. Curr. Opin. Cell Biol. 25, 349–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. J. Wynne, O. Rog, P. M. Carlton, A. F. Dernburg, Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J. Cell Biol. 196, 47–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R. E. Rosenbluth, D. L. Baillie, The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics 99, 415–428 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.K. S. McKim, A. M. Howell, A. M. Rose, The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120, 987–1001 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. M. Villeneuve, A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136, 887–902 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A. J. MacQueen, C. M. Phillips, N. Bhalla, P. Weiser, A. M. Villeneuve, A. F. Dernburg, Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123, 1037–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.C. M. Phillips, C. Wong, N. Bhalla, P. M. Carlton, P. Weiser, P. M. Meneely, A. F. Dernburg, HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123, 1051–1063 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. M. Phillips, A. F. Dernburg, A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11, 817–829 (2006). [DOI] [PubMed] [Google Scholar]
- 28.C. M. Phillips, X. Meng, L. Zhang, J. H. Chretien, F. D. Urnov, A. F. Dernburg, Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat. Cell Biol. 11, 934–942 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Penkner, L. Tang, M. Novatchkova, M. Ladurner, A. Fridkin, Y. Gruenbaum, D. Schweizer, J. Loidl, V. Jantsch, The nuclear envelope protein matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell 12, 873–885 (2007). [DOI] [PubMed] [Google Scholar]
- 30.A. Sato, B. Isaac, C. M. Phillips, R. Rillo, P. M. Carlton, D. J. Wynne, R. A. Kasad, A. F. Dernburg, Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139, 907–919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.A. M. Penkner, A. Fridkin, J. Gloggnitzer, A. Baudrimont, T. Machacek, A. Woglar, E. Csaszar, P. Pasierbek, G. Ammerer, Y. Gruenbaum, V. Jantsch, Meiotic chromosome homology search involves modifications of the nuclear envelope protein matefin/SUN-1. Cell 139, 920–933 (2009). [DOI] [PubMed] [Google Scholar]
- 32.A. Baudrimont, A. Penkner, A. Woglar, T. Machacek, C. Wegrostek, J. Gloggnitzer, A. Fridkin, F. Klein, Y. Gruenbaum, P. Pasierbek, V. Jantsch, Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLOS Genet. 6, e1001219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Y. Chikashige, M. Yamane, K. Okamasa, C. Tsutsumi, T. Kojidani, M. Sato, T. Haraguchi, Y. Hiraoka, Membrane proteins Bqt3 and −4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 187, 413–427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Y. Chikashige, C. Tsutsumi, M. Yamane, K. Okamasa, T. Haraguchi, Y. Hiraoka, Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125, 59–69 (2006). [DOI] [PubMed] [Google Scholar]
- 35.H. Shibuya, A. Hernández-Hernández, A. Morimoto, L. Negishi, C. Höög, Y. Watanabe, MAJIN links telomeric DNA to the nuclear membrane by exchanging telomere cap. Cell 163, 1252–1266 (2015). [DOI] [PubMed] [Google Scholar]
- 36.H. Shibuya, K. Ishiguro, Y. Watanabe, The TRF1-binding protein TERB1 promotes chromosome movement and telomere rigidity in meiosis. Nat. Cell Biol. 16, 145–156 (2014). [DOI] [PubMed] [Google Scholar]
- 37.C. Hu, H. Inoue, W. Sun, Y. Takeshita, Y. Huang, Y. Xu, J. Kanoh, Y. Chen, The inner nuclear membrane protein Bqt4 in fission yeast contains a DNA-binding domain essential for telomere association with the nuclear envelope. Structure 27, 335–343.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 38.I. da Cruz, C. Brochier-Armanet, R. Benavente, The TERB1-TERB2-MAJIN complex of mouse meiotic telomeres dates back to the common ancestor of metazoans. BMC Evol. Biol. 20, 55 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Hodgkin, H. R. Horvitz, S. Brenner, Nondisjunction mutants of the nematode CAENORHABDITIS ELEGANS. Genetics 91, 67–94 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.K. O. Kelly, A. F. Dernburg, G. M. Stanfield, A. M. Villeneuve, Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156, 617–630 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.M. Doitsidou, R. J. Poole, S. Sarin, H. Bigelow, O. Hobert, C. elegans Mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLOS ONE 5, e15435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. R. Wicks, R. T. Yeh, W. R. Gish, R. H. Waterston, R. H. A. Plasterk, Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28, 160–164 (2001). [DOI] [PubMed] [Google Scholar]
- 43.K. A. Swan, D. E. Curtis, K. B. McKusick, A. V. Voinov, F. A. Mapa, M. R. Cancilla, High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 12, 1100–1105 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.W. C. Spencer, G. Zeller, J. D. Watson, S. R. Henz, K. L. Watkins, R. D. McWhirter, S. Petersen, V. T. Sreedharan, C. Widmer, J. Jo, V. Reinke, L. Petrella, S. Strome, S. E. V. Stetina, M. Katz, S. Shaham, G. Rätsch, D. M. Miller III, A spatial and temporal map of C. elegans gene expression. Genome Res. 21, 325–341 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. Han, E. A. Schroeder, C. G. Silva-García, K. Hebestreit, W. B. Mair, A. Brunet, Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185–190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D. Grün, M. Kirchner, N. Thierfelder, M. Stoeckius, M. Selbach, N. Rajewsky, Conservation of mRNA and protein expression during development of C. elegans. Cell Rep. 6, 565–577 (2014). [DOI] [PubMed] [Google Scholar]
- 47.R. Rillo-Bohn, R. Adilardi, T. Mitros, B. Avşaroğlu, L. Stevens, S. Köhler, J. Bayes, C. Wang, S. Lin, K. A. Baskevitch, D. S. Rokhsar, A. F. Dernburg, Analysis of meiosis in Pristionchus pacificus reveals plasticity in homolog pairing and synapsis in the nematode lineage. eLife 10, e70990 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.A. Woglar, A. Daryabeigi, A. Adamo, C. Habacher, T. Machacek, A. L. Volpe, V. Jantsch, Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLOS Genet. 9, e1003335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. K. Chamberlain, Y. Lee, S. Kim, J. U. Bowie, Snorkeling preferences foster an amino acid composition bias in transmembrane helices. J. Mol. Biol. 339, 471–479 (2004). [DOI] [PubMed] [Google Scholar]
- 50.N. C. Harper, R. Rillo, S. Jover-Gil, Z. J. Assaf, N. Bhalla, A. F. Dernburg, Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell 21, 934–947 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. Labella, A. Woglar, V. Jantsch, M. Zetka, Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev. Cell 21, 948–958 (2011). [DOI] [PubMed] [Google Scholar]
- 52.J. Link, D. Paouneskou, M. Velkova, A. Daryabeigi, T. Laos, S. Labella, C. Barroso, S. P. Piñol, A. Montoya, H. Kramer, A. Woglar, A. Baudrimont, S. M. Markert, C. Stigloher, E. Martinez-Perez, A. Dammermann, M. Alsheimer, M. Zetka, V. Jantsch, Transient and partial nuclear lamina disruption promotes chromosome movement in early meiotic prophase. Dev. Cell 45, 212–225.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J. N. Brandt, K. A. Hussey, Y. Kim, Spatial and temporal control of targeting Polo-like kinase during meiotic prophase. J. Cell Biol. 219, e202006094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Y. Kim, N. Kostow, A. F. Dernburg, The chromosome axis mediates feedback control of CHK-2 to ensure crossover formation in C. elegans. Dev. Cell 35, 247–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. J. MacQueen, M. P. Colaiácovo, K. McDonald, A. M. Villeneuve, Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Y. Wang, Y. Chen, J. Chen, L. Wang, L. Nie, J. Long, H. Chang, J. Wu, C. Huang, M. Lei, The meiotic TERB1-TERB2-MAJIN complex tethers telomeres to the nuclear envelope. Nat. Commun. 10, 564 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.G. Wang, X. Wu, L. Zhou, S. Gao, D. Yun, A. Liang, F. Sun, Tethering of telomeres to the nuclear envelope is mediated by SUN1-MAJIN and possibly promoted by SPDYA-CDK2 during meiosis. Front. Cell Dev. Biol. 8, 845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Z. Tu, M. B. Bayazit, H. Liu, J. Zhang, K. Busayavalasa, S. Risal, J. Shao, A. Satyanarayana, V. Coppola, L. Tessarollo, M. Singh, C. Zheng, C. Han, Z. Chen, P. Kaldis, J.-Å. Gustafsson, K. Liu, Speedy A-Cdk2 binding mediates initial telomere-nuclear envelope attachment during meiotic prophase I independent of Cdk2 activation. Proc. Natl. Acad. Sci. U.S.A. 114, 592–597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.H. F. Horn, D. I. Kim, G. D. Wright, E. S. M. Wong, C. L. Stewart, B. Burke, K. J. Roux, A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell Biol. 202, 1023–1039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.P. Mikolcevic, M. Isoda, H. Shibuya, I. del Barco Barrantes, A. Igea, J. A. Suja, S. Shackleton, Y. Watanabe, A. R. Nebreda, Essential role of the Cdk2 activator RingoA in meiotic telomere tethering to the nuclear envelope. Nat. Commun. 7, 11084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. Viera, J. S. Rufas, I. Martínez, J. L. Barbero, S. Ortega, J. A. Suja, CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J. Cell Sci. 122, 2149–2159 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Y. Chen, Y. Wang, J. Chen, W. Zuo, Y. Fan, S. Huang, Y. Liu, G. Chen, Q. Li, J. Li, J. Wu, Q. Bian, C. Huang, M. Lei, The SUN1-SPDYA interaction plays an essential role in meiosis prophase I. Nat. Commun. 12, 3176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.A. Woglar, A. M. Villeneuve, Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell 173, 1678–1691.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.T. Ashley, D. Walpita, D. G. de Rooij, Localization of two mammalian cyclin dependent kinases during mammalian meiosis. J. Cell Sci. 114, 685–693 (2001). [DOI] [PubMed] [Google Scholar]
- 65.N. Palmer, S. Z. A. Talib, P. Singh, C. M. F. Goh, K. Liu, J. C. Schimenti, P. Kaldis, A novel function for CDK2 activity at meiotic crossover sites. PLoS Biol. 18, e3000903 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.L. Zhang, J. D. Ward, Z. Cheng, A. F. Dernburg, The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.J. A. Arribere, R. T. Bell, B. X. H. Fu, K. L. Artiles, P. S. Hartman, A. Z. Fire, Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. Goudeau, C. S. Sharp, J. Paw, L. Savy, M. D. Leonetti, A. G. York, D. L. Updike, C. Kenyon, M. Ingaramo, Split-wrmScarlet and split-sfGFP: Tools for faster, easier fluorescent labeling of endogenous proteins in Caenorhabditis elegans. Genetics 217, iyab014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.A. G. Fraser, R. S. Kamath, P. Zipperlen, M. Martinez-Campos, M. Sohrmann, J. Ahringer, Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330 (2000). [DOI] [PubMed] [Google Scholar]
- 70.A. F. Dernburg, K. McDonald, G. Moulder, R. Barstead, M. Dresser, A. M. Villeneuve, Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998). [DOI] [PubMed] [Google Scholar]
- 71.R. S. Adilardi, A. F. Dernburg, Robust, versatile DNA FISH probes for chromosome-specific repeats in Caenorhabditis elegans and Pristionchus pacificus. G3 6, jkac121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. P. Colaiácovo, A. J. MacQueen, E. Martinez-Perez, K. McDonald, A. Adamo, A. L. Volpe, A. M. Villeneuve, Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5, 463–474 (2003). [DOI] [PubMed] [Google Scholar]
- 73.S. Smolikov, A. Eizinger, K. Schild-Prufert, A. Hurlburt, K. McDonald, J. Engebrecht, A. M. Villeneuve, M. P. Colaiácovo, SYP-3 restricts synaptonemal complex assembly to bridge paired chromosome axes during meiosis in Caenorhabditis elegans. Genetics 176, 2015–2025 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1