Abstract
Background
Low vitamin D status is often associated with systemic low-grade inflammation as reflected by elevated C-reactive protein (CRP) levels. We investigated the causality and direction of the association between vitamin D status and CRP using linear and non-linear Mendelian randomization (MR) analyses.
Methods
MR analyses were conducted using data from 294 970 unrelated participants of White-British ancestry from the UK Biobank. Serum 25-hydroxyvitamin D [25(OH)D] and CRP concentrations were instrumented using 35 and 46 genome-wide significant variants, respectively.
Results
In non-linear MR analysis, genetically predicted serum 25(OH)D had an L-shaped association with serum CRP, where CRP levels decreased sharply with increasing 25(OH)D concentration for participants within the deficiency range (<25 nmol/L) and levelled off at ∼50 nmol/L of 25(OH)D (Pnon-linear = 1.49E-4). Analyses using several pleiotropy-robust methods provided consistent results in stratified MR analyses, confirming the inverse association between 25(OH)D and CRP in the deficiency range (P = 1.10E-05) but not with higher concentrations. Neither linear or non-linear MR analysis supported a causal effect of serum CRP level on 25(OH)D concentration (Plinear = 0.32 and Pnon-linear = 0.76).
Conclusion
The observed association between 25(OH)D and CRP is likely to be caused by vitamin D deficiency. Correction of low vitamin D status may reduce chronic inflammation.
Keywords: Non-linear Mendelian randomization, vitamin D, serum 25-hydroxyvitamin D concentration, C-reactive protein, chronic inflammation
Key Messages.
Our bidirectional Mendelian randomization (MR) analysis confirmed that the association between serum 25-hydroxyvitamin D [25(OH)D] and C-reactive protein (CRP) is likely to be driven by an effect of 25(OH)D on CRP rather than vice versa.
Non-linear MR analyses showed that the effect of 25(OH)D on CRP is restricted to the vitamin D deficiency range, where higher genetically predicted 25(OH)D was associated with lower CRP concentrations.
There was no evidence for an effect of genetically predicted serum CRP concentration on 25(OH)D in linear or non-linear MR analyses.
Given that the serum CRP level is a widely used biomarker for chronic inflammation, these results suggest that improving vitamin D status may reduce chronic inflammation, but only for people with vitamin D deficiency.
Introduction
Systemic low-grade inflammation, characterized by prolonged release of inflammatory mediators and activation of harmful signal-transduction pathways, is associated with various complex somatic and neuropsychiatric diseases and disorders.1,2 It is considered that nutritional factors can influence many aspects of inflammation.3 Vitamin D is a pro-hormone and an essential micronutrient, and although its classical roles are related to the regulation of calcium homeostasis, various types of immune cells express both the vitamin D receptor and metabolizing enzymes,4 suggesting that hormonal vitamin D could also play a role in modulating inflammatory responses.5 This is supported by an inverse association between serum 25-hydroxyvitamin D [25(OH)D] concentrations and C-reactive protein (CRP), which is frequently reported in observational studies.6,7 Serum 25(OH)D is the best indicator for vitamin D status whereas CRP is one of the most widely used inflammatory biomarkers in clinical practice. However, there is an ongoing debate about the causal nature of the association between 25(OH)D and CRP, and the observational association has not been supported by randomized trials.8 Indeed, it has been suggested that the association between serum 25(OH)D and CRP simply reflects reverse causality or confounding, where a low 25(OH)D concentration is either a consequence of chronic inflammation or results from behaviours such as less time outdoors in people who are unwell.8
Sitting at the interface between observational studies and randomized–controlled trials (RCTs), Mendelian randomization (MR) has been increasingly used to strengthen causal evidence in observational studies.9 It uses genetic variants associated with the exposure of interest to approximate the exposure and, conditional on the key method assumptions being met, MR has the benefit of reducing bias due to confounding and reverse causation.9 The association of 25(OH)D and CRP has previously been investigated using the MR approach, with no evidence to support a causal effect.10,11 However, all previous studies have only used the standard linear MR method, which cannot rule out the possibility of a threshold effect restricted to vitamin D deficiency.12 Indeed, it is logical to expect that improving vitamin D status would be relevant only in the presence of vitamin D deficiency, whereas any further additions may be redundant and, in the high extreme of supplementation, might become toxic.13 These types of non-linear dose–response relationships can be tested by the non-linear MR approach, which allows us to interrogate the shape of the association.14 This method has been recently used to provide evidence for the adverse effect of vitamin D deficiency on cardiovascular disease (CVD) risk and mortality, which is not visible using the standard linear MR approach.15,16 In this study we set out to examine evidence for the direction and causality of the association between serum 25(OH)D and CRP, also allowing for possible threshold effects. We performed these analyses using data from 294 970 participants in UK Biobank, representing the largest cohort to date with measured serum 25(OH)D concentrations.
Methods
Study participants
UK Biobank is a large prospective cohort study with >500 000 participants aged 37–73 years recruited from 22 assessment centres across the UK between 13 March 2006 and 1 October 2009 with a goal to improve the prevention, diagnosis and treatment of diseases of middle and old age.17 Participants filled in questionnaires to provide broad information on health and lifestyles at baseline survey and provided blood samples for biomarker and genetic assays. We restricted the analyses to unrelated individuals who were identified as White-British ancestry based on self-report and genetic profiling18 and excluded participants with mismatched information between self-reported and genetic sex. Final genetic analyses were conducted among individuals with complete information on serum 25(OH)D and CRP concentrations and relevant covariates (N = 294 970) (Supplementary Figure S1, available as Supplementary data at IJE online). The present study was conducted under UK Biobank application number 20175. The UK Biobank study was approved by the National Information Governance Board for Health and Social Care and North West Multicentre Research Ethics Committee (11/NW/0382). All participants provided informed consent to participate.
Serum 25(OH)D and CRP concentrations
Blood samples of participants were collected at the time of recruitment. Serum 25(OH)D concentration (nmol/L) was measured using the LIAISON XL 25(OH)D assay (DiaSorin, Stillwater, USA) and serum CRP concentration (mg/L) was measured using high-sensitivity immunoturbidimetric assay on a Beckman Coulter AU580019,20 (see Supplementary methods, available as Supplementary data at IJE online, for details). Since the distribution of serum CRP concentration is highly skewed, we natural-log-transformed serum CRP concentrations to facilitate analyses.
Genetic instrument for serum 25(OH)D concentration
We constructed a weighted genetic score (vitaminD-GS) consisting of 35 single-nucleotide polymorphisms (SNPs) to instrument serum 25(OH)D concentration (Supplementary Table S1, available as Supplementary data at IJE online). All 35 SNPs are common genome-wide significant variants (minor allele frequency > 5%), discovered in a recent genome-wide association analysis (GWAS) for serum 25(OH)D concentration in UK Biobank21 and were replicated with a consistent direction and a P-value < 0.05 in the earlier GWAS by the SUNLIGHT consortium22 (Supplementary Figure S2, available as Supplementary data at IJE online). Benefits of replication in the SUNLIGHT consortium are 2-fold. It ensures the robustness of the GWAS signals and also allows us to take weights for vitaminD-GS from an independent sample, avoiding bias arising from using internal weights.23 VitaminD-GS was constructed by first computing the weighted average of the number of 25(OH)D-increasing alleles for an individual and then multiplying it by the number of available variants. The weight for each SNP was the effect estimate of the association of the SNP with serum 25(OH)D in the SUNLIGHT consortium.22 As a sensitivity analysis, we constructed an alternative instrument using a broader set of SNPs consisting of 122 autosomal variants (vitaminD-GS-122) (Supplementary methods, Supplementary Figure S2 and Supplementary Table S1, available as Supplementary data at IJE online). Further, to minimize any potential influence by horizontal pleiotropy (via variants with no clear biological links to vitamin D metabolism), in particular by metabolic traits (including lipids traits24), we included two additional instruments in the sensitivity analysis, which were constructed using only variants near/in genes related to vitamin D metabolism. A focused score16 consists of 21 variants from four loci, including GC, DHCR7, CYP2R1 and CYP24A1 (Supplementary methods, available as Supplementary data at IJE online). As GC and CYP24A1 [which are involved in transport and clearance of 25(OH)D, respectively] are downstream of the production of 25(OH)D, their associations with high serum 25(OH)D concentrations may not accurately reflect concentrations of bioavailable 25(OH)D.25 As an attempt to better represent the level of ‘effective’ 25(OH)D, we repeated analyses using a synthesis score26,27 that only included variants from DHCR7 and CYP2R1, which directly contribute to substrate availability and synthesis of 25(OH)D (Supplementary methods, available as Supplementary data at IJE online).
Genetic instrument for serum CRP concentration
The genetic score for serum CRP concentration (CRP-gwasGS) was constructed using 46 genome-wide significant variants associated with serum CRP concentration, which were discovered in a recent GWAS meta-analysis with no overlap with UK Biobank.28 Information on these 46 variants can be found in Supplementary Table S2 (available as Supplementary data at IJE online). CRP-gwasGS was constructed in the same way as vitaminD-GS with the weight for each SNP being the effect estimate of the association of the SNP with CRP in the aforementioned meta-analysis.28 As a sensitivity analysis, we constructed an alternative instrument using four cis-acting variants29 (Supplementary Table S2, available as Supplementary data at IJE online).
Statistical analysis
We performed linear and non-linear MR analyses to examine the genetic evidence for the bidirectional association between serum 25(OH)D and CRP concentrations. If a non-linear association was evident in the non-linear MR analysis, we also performed stratified MR analyses as a sensitivity analysis to gauge the robustness of the detected non-linearity. For the full analytical strategy, please see Supplementary Figure S3 (available as Supplementary data at IJE online). MR estimates of the linear, non-linear and stratified MR analyses were calculated using the genetic score-based approach (GS-based one-sample approach). For the linear and stratified analyses, where estimation via the SNP-based two-sample approach is also possible, we repeated the analyses applying five SNP-based two-sample methods as sensitivity analyses to gauge the robustness of the results to horizontal pleiotropy. Both GS-based one-sample and SNP-based two-sample approaches are described in detail below in the context of linear, stratified and non-linear MR analyses.
GS-based one-sample analysis for linear and stratified MR
In the GS-based one-sample analysis, the MR estimate is computed using the ratio-of-coefficients method,30 in which in the same sample (i.e. one-sample) GS–exposure and GS–outcome association estimates are computed and then taken as inputs to estimate the causal effect. For the linear MR analysis, the MR estimate is computed in the full sample, whereas in the stratified analysis, stratum-specific MR estimates are computed in the strata of residual exposure, which is defined as the exposure level minus the variation induced by the genetic score. Stratification is performed using residual exposure (rather than the raw exposure level) to avoid collider bias31,32 that could potentially induce spurious associations between GS and outcome, and bias stratum-specific MR estimates.
Non-linear MR analysis
The non-linear MR is also a GS-based one-sample approach and for our analysis we used the fractional polynomial method to capture the non-linearity of the exposure–outcome association.14 Briefly, the full UK Biobank sample was stratified into 100 strata using the residuals of exposure after regressing on the corresponding GS. Within each stratum, the localized average causal effect (LACE) was computed using the ratio-of-coefficients method, which is the ratio of the coefficient of the GS–outcome association estimate to that of the GS–exposure association estimate. Meta-regression of LACE against the stratum-specific mean exposure was then performed by fitting a range of fractional polynomial exposure–outcome models of Degrees 1 and 2, and the best-fitting model selected based on the likelihood ratio test. We report the fractional polynomial test for non-linearity in which the best-fitting fractional polynomial model of Degree 1 is compared against the linear model.14 Non-linear MR analyses assume that the GS–exposure association is constant over the entire distribution of exposure. To test this assumption, we examined the heterogeneity of GS–exposure associations across 100 strata using the Cochran’s Q test and trend test.14
SNP-based two-sample analysis for linear and stratified MR
We included five SNP-based two-sample methods in the linear and stratified MR analyses, including inverse-variance weighted (IVW), MR–Egger, weighted median, weighted mode and MR–Presso (Supplementary Figure S3, available as Supplementary data at IJE online). All these methods use individual SNPs (instead of an aggregate genetic score as in the GS-based approach) and take SNP–exposure and SNP–outcome association estimates as inputs. It is important to note that SNP–exposure and SNP–outcome association estimates need to be taken from independent samples (i.e. two-sample) otherwise bias can be introduced.33 The five SNP-based two-sample methods have largely independent assumptions on horizontal pleiotropy and a good agreement across these methods suggests a credible causal estimate (see Supplementary methods, available as Supplementary data at IJE online, for details). In the linear MR analysis, in which SNP-25(OH)D and SNP–CRP association estimates were all taken from UK Biobank, we applied the split-sample strategy to avoid bias due to overlapping samples.33 More specifically, the full UK Biobank sample is randomly split into two subsamples of equal size (Samples A and B); the MR estimate is calculated for each sample (MRA and MRB) and then combined to obtain the overall estimate (MRmeta) (Supplementary Figure S3, available as Supplementary data at IJE online). MRA is computed using SNP–outcome association estimates from Sample A and SNP–exposure association estimates from Sample B, whereas SNP–outcome association estimates from Sample B and SNP–exposure association estimates from Sample A are used to compute MRB. MRA and MRB are then combined in a fixed-effects meta-analysis to compute MRmeta. In the stratified analyses, the stratum-specific MR estimate is computed using SNP–outcome association estimates from the stratum under consideration with SNP–exposure association estimates taken from the full sample leaving out the stratum under consideration (Supplementary Figure S3, available as Supplementary data at IJE online).
Given that serum 25(OH)D and CRP concentrations are continuous, all GS/SNP–exposure/outcome association estimates required for the linear, stratified and non-linear analyses were computed by fitting linear regression models. All models were adjusted for age, sex, assessment centre, birth location, SNP array, top 40 genetic principal components and nuisance factors related to the measurement of serum 25(OH)D and/or CRP concentrations, including month in which blood sample was taken, fasting time before blood sample was taken and sample aliquots for measurement.20 Adjustment of birth location and 40 genetic components is recommended to account for latent population structure in UK Biobank.34 GS-based one-sample linear and stratified analyses were performed using STATA, version 17.0 (StataCorp LP, College Station, Texas, USA). SNP-based two-sample analyses and non-linear MR analysis were conducted in R (version 4.0.2) using the TwoSampleMR (version 0.5.6)35 and nlmr (version 2.0)14 package, respectively.
Results
In total, 294 970 participants were included in the analysis (Supplementary Figure S1, available as Supplementary data at IJE online). The average 25(OH)D concentration was 50.0 nmol/L (range 10–340 nmol/L) and 11.7% (n = 34 403) of the participants had concentrations of <25 nmol/L (Table 1). There were notable variations in serum 25(OH)D and CRP concentrations with respect to distributions of demographics, lifestyle, general health and socio-economic factors (Table 1). Serum 25(OH)D and CRP concentrations are inversely related in a dose-dependent manner (P < 1.0E-300) (Table 1).
Table 1.
Serum 25(OH)D and CRP concentration by baseline characteristics in UK Biobank
| 25(OH)D (nmol/L) | CRP (mg/L) | ||
|---|---|---|---|
| N (%) | Mean (SD) | Gmean (GSD) | |
| Age (years) | |||
| <65 | 236 445 (80.16) | 49.31 (21.01) | 1.32 (2.90) |
| ≥65 | 58 525 (19.84) | 52.03 (20.57) | 1.64 (2.78) |
| P a | 2.06E-188 | <1.0E-300 | |
| Sex | |||
| Male | 138 911 (47.09) | 49.92 (21.03) | 1.34 (2.78) |
| Female | 156 059 (52.91) | 49.80 (20.89) | 1.42 (2.98) |
| P a | 2.60E-03 | <1.0E-300 | |
| BMI (kg/m2) | |||
| <18.5 | 1443 (0.49) | 50.72 (24.42) | 0.56 (3.18) |
| (18.5–25) | 96 053 (32.56) | 53.01 (21.82) | 0.86 (2.78) |
| (25–30) | 125 559 (42.57) | 50.61 (20.56) | 1.41 (2.60) |
| ≥30 | 71 037 (24.08) | 44.32 (19.20) | 2.57 (2.56) |
| Missing | 878 (0.30) | 42.53 (21.28) | 2.23 (3.16) |
| P a | <1.0E-300 | 9.36E-88 | |
| Smoking | |||
| Non-smoker | 160 897 (54.55) | 50.07 (20.63) | 1.27 (2.86) |
| Ex-smoker | 103 600 35.12 | 50.81 (21.03) | 1.47 (2.86) |
| Smokerb | 7561 (2.56) | 49.09 (21.57) | 1.40 (2.94) |
| Cigars/pipe | 1682 (0.57) | 45.49 (21.08) | 1.83 (2.71) |
| <1 to 15 cigs/day | 12 056 (4.09) | 45.36 (21.63) | 1.73 (2.95) |
| >15 cigs/day | 8158 (2.77) | 41.55 (21.77) | 2.35 (2.84) |
| Missing | 1016 (0.34) | 49.92 (21.76) | 1.74 (2.90) |
| P a | <1.0E-300 | <1.0E-300 | |
| Alcohol intake | |||
| Non-drinker | 19 178 (6.50) | 45.96 (20.95) | 1.69 (3.06) |
| Special occasions or 1–3 times/month | 63 647 (21.58) | 47.17 (20.36) | 1.62 (2.94) |
| 1 or 2 times/week | 78 199 (26.51) | 50.44 (20.74) | 1.38 (2.86) |
| 3 or 4 times/week | 71 296 (24.17) | 51.41 (20.87) | 1.23 (2.82) |
| Daily or almost daily | 62 445 (21.17) | 51.28 (21.51) | 1.27 (2.82) |
| Missing | 205 (0.07) | 45.08 (21.27) | 1.81 (2.98) |
| P a | <1.0E-300 | <1.0E-300 | |
| Physical activity | |||
| Light | 88 245 (29.92) | 46.35 (20.20) | 1.61 (2.92) |
| Moderate | 142 803 (48.41) | 50.64 (20.80) | 1.28 (2.85) |
| Vigorous | 57 373 (19.45) | 54.02 (21.42) | 1.24 (2.79) |
| Missing | 6549 (2.22) | 43.40 (21.30) | 2.24 (3.09) |
| P a | <1.0E-300 | <1.0E-300 | |
| Education | |||
| None | 50 706 (17.19) | 50.42 (21.42) | 1.84 (2.82) |
| NVQ/CSE/A-levels | 105 433 (35.74) | 50.56 (21.18) | 1.43 (2.87) |
| Degree/professional | 136 357 (46.23) | 49.08 (20.58) | 1.21 (2.86) |
| Missing | 2474 (0.84) | 50.56 (20.96) | 1.65 (2.88) |
| P a | 7.65E-87 | <1.0E-300 | |
| TDI quartiles (min–max) | |||
| Q1 (–6.26 to –3.76) | 73 533 (24.93) | 51.94 (20.72) | 1.27 (2.81) |
| Q2 (–3.76 to –2.37) | 73 780 (25.01) | 51.53 (20.71) | 1.33 (2.83) |
| Q3 (–2.37 to 0.030) | 73 655 (24.97) | 49.95 (20.77) | 1.37 (2.88) |
| Q4 (0.030 to 10.82) | 73 658 (24.97) | 45.99 (21.08) | 1.58 (3.0) |
| Missing | 344 (0.12) | 49.99 (20.46) | 1.47 (3.14) |
| P a | <1.0E-300 | <1.0E-300 | |
| Self-rated health | |||
| Excellent | 49 237 (16.69) | 53.06 (21.17) | 0.98 (2.69) |
| Good | 172 566 (58.50) | 50.68 (20.71) | 1.31 (2.78) |
| Fair | 60 087 (20.37) | 46.46 (20.63) | 1.88 (2.91) |
| Poor | 12 080 (4.10) | 42.27 (21.18) | 2.65 (3.17) |
| Missing | 1000 (0.34) | 43.70 (20.08) | 1.91 (3.04) |
| P a | <1.0E-300 | <1.0E-300 | |
| Long-term illness | |||
| No | 194 251 (65.85) | 50.76 (20.75) | 1.23 (2.79) |
| Yes | 94 129 (31.91) | 48.11 (21.27) | 1.74 (2.99) |
| missing | 6590 (2.23) | 47.96 (20.63) | 1.58 (2.92) |
| P a | <1.0E-300 | <1.0E-300 | |
| 25(OH)D (nmol/L) | |||
| <25 | 34 403 (11.66) | – | 1.64 (3.03) |
| 25–50 | 121 812 (41.30) | – | 1.44 (2.88) |
| 50–75 | 103 400 (35.05) | – | 1.30 (2.82) |
| >75 | 35 355 (11.99) | – | 1.21 (2.91) |
| P a | – | <1.0E-300 | |
| CRP quartiles (mg/L, min–max) | |||
| Q1 (0.08–0.64) | 72 663 (24.63) | 51.91 (21.30) | – |
| Q2 (0.65–1.3) | 74 111 (25.12) | 50.97 (20.81) | – |
| Q3 (1.31–2.72) | 74 359 (25.21) | 49.33 (20.56) | – |
| Q4 (2.73–79.95) | 73 837 (25.03) | 47.24 (20.86) | – |
| P a | <1.0E-300 | – |
CRP, C-reactive protein; 25(OH)D, 25-hydroxyvitamin D; BMI, body mass index; NVQ, National Vocational Qualification; CSE, Certificate of Secondary Education; A-levels, Advanced levels; SD, standard deviation; Gmean: geometric mean; GSD: geometric standard deviation; Q, quartiles; cig, cigarette; TDI, Townsend deprivation index.
P-values have been adjusted for age, sex, assessment centre and nuisance factors that could affect serum 25(OH)D measurements, including month in which blood sample was taken, fasting time before blood sample was taken and sample aliquots for measurement.
Current smokers without information on types of tobacco that they smoke.
Instrument validation for vitaminD-GS
VitaminD-GS was robustly associated with serum 25(OH)D concentration in UK Biobank, explaining 2.8% of variation (F-statistic = 8646, P < 1.0E-300) (Supplementary Figure S4, available as Supplementary data at IJE online). We examined the association of vitaminD-GS with serum 25(OH)D across 100 strata of residuals of serum 25(OH)D. We detected evidence for heterogeneity (PCochran’s Q < 1.0E-300; Ptrend = 0.23), where vitaminD-GS–25(OH)D association in the 1st and 100th strata appeared to be outliers (Supplementary Figure S5, available as Supplementary data at IJE online). We found an association between the genetic instrument with serum low-density lipoprotein (LDL) and triglyceride concentrations (P < 0.001 for both). In contrast there was no evidence that vitaminD-GS was associated with other potential confounders in UK Biobank, including body mass index (BMI), smoking, alcohol intake, physical activity, education and Townsend deprivation index (uncorrected P 0.051 for all) (Supplementary Table S3, available as Supplementary data at IJE online). We also examined vitaminD-GS–confounder associations across 100 strata of residuals of serum 25(OH)D and found no evidence of association after accounting for multiple testing (Supplementary Figure S6, available as Supplementary data at IJE online).
Instrument validation for CRP-gwasGS
CRP-gwasGS explained 6.0% of variation in the serum log CRP concentration with a F-statistic of 18 687 (P < 1.0E-300) (Supplementary Figure S7, available as Supplementary data at IJE online). There was evidence of heterogeneity in the CRP-gwasGS–CRP association across 100 strata of residuals of serum CRP, with the 1st and 100th strata appearing to be the outliers (PCochran’sQ = 2.66E-13, Ptrend = 0.28) (Supplementary Figure S8, available as Supplementary data at IJE online). CRP-gwasGS was not associated with the selected confounders, except for BMI (uncorrected P = 1.066E-13) (Supplementary Table S3, available as Supplementary data at IJE online) and physical activity (uncorrected P = 0.004) (Supplementary Table S3, available as Supplementary data at IJE online). There was no evidence that CRP-gwasGS was associated with selected confounders across 100 strata of residuals of serum CRP concentrations after accounting for multiple testing (Supplementary Figure S9, available as Supplementary data at IJE online).
MR of 25(OH)D on CRP
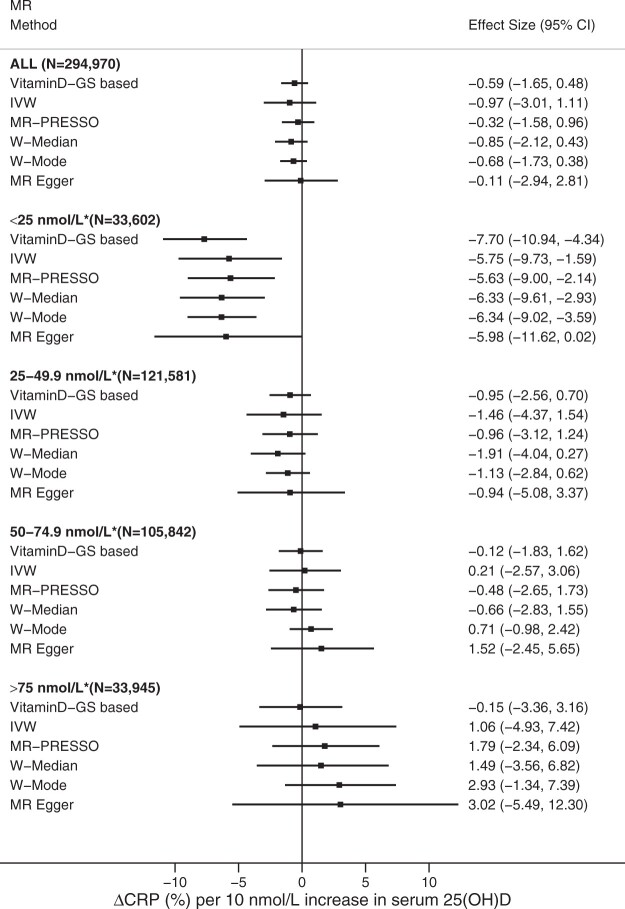
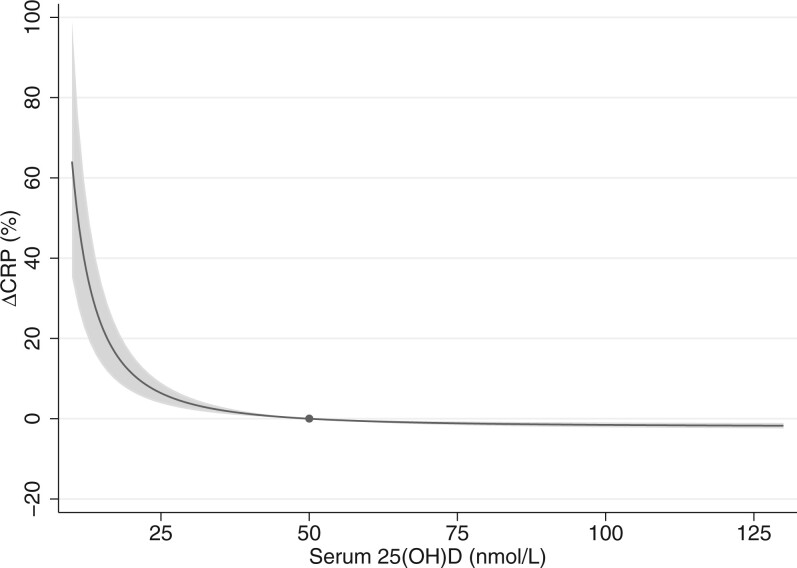
Linear MR analysis did not support a causal association of serum 25(OH)D with CRP concentration [–0.59% (95% CI, –1.65, 0.48) per 10-nmol/L increase in 25(OH)D, P = 0.28) (Figure 1). The non-linear MR analysis suggested an ‘L-shaped’ association in which the serum CRP level peaked at the lowest 25(OH)D concentration, dropping steeply with increasing 25(OH)D concentration and levelling off at ∼50 nmol/L (Pnon-linear = 1.49E-4) (Figure 2). Compared with those with 50 nmol/L, individuals with serum 25(OH)D concentration at 25 nmol/L have a 6.4% (95% CI, 3.85–8.98) higher serum CRP concentration.
Figure 1.
Linear and stratified MR analyses of serum 25(OH)D with CRP concentration. Adjustment includes age, sex, assessment centre, birth location, SNP array, top 40 genetic principal components and nuisance factors that could affect serum 25(OH)D and/or CRP measurements, including month in which blood sample was taken, fasting time before blood sample was taken and sample aliquots for measurement. CRP, C-reactive protein; 25(OH)D, 25-hydroxyvitamin D; IVW, inverse-variance weighted MR; W-Median, weighted median MR; W-Mode, weighted mode MR. *Residuals of serum 25(OH)D concentrations.
Figure 2.
Non-linear MR analysis of serum 25(OH)D with CRP concentration. The dot represents the reference point of serum 25(OH)D of 50 nmol/L. The shaded areas represent the 95% confidence intervals. Adjustment includes age, sex, assessment centre, birth location, SNP array, top 40 genetic principal components and nuisance factors that could affect serum 25(OH)D and/or CRP measurement, including month in which blood sample was taken, fasting time before blood sample was taken and sample aliquots for measurement. CRP, C-reactive protein; 25(OH)D, 25-hydroxyvitamin D.
We conducted several sensitivity analyses to gauge the robustness of the detected non-linear association. First, we performed a stratified MR analysis using four categories of residual 25(OH)D concentration; <25.0, 25–49.9, 50.0–74.9 and 75.0 nmol/L. Consistently with the L-shaped association seen in the non-linear analysis, we only observed a genetic association of serum 25(OH)D with CRP concentration among individuals with residual 25(OH)D < 25 nmol/L, where each 10-nmol/L higher serum 25(OH)D concentration was associated with 7.70% lower CRP (95% CI, –10.9, –4.34, P = 1.1E-05) (Figure 1). There was a good agreement in MR estimates across all five SNP-based two-sample methods within the <25-nmol/L strata (Figure 1). Further, as heterogeneity of the vitaminD-GS–25(OH)D association was detected (Supplementary Figure S5, available as Supplementary data at IJE online), we reanalysed the data excluding local causal estimates from the outlying strata of residuals of serum 25(OH)D (i.e. the 1st and 100th strata, after exclusion PCochran’s Q = 0.30; Ptrend = 0.28), and this did not affect our findings (Supplementary Figure S10, available as Supplementary data at IJE online). Given the association between the vitamin-GS and blood lipids, we reanalysed the association adjusting for LDL and triglycerides, and restricting the instrument to variants that were not associated with any metabolic traits (a non-metabolic score) (Supplementary methods, available as Supplementary data at IJE online). Both approaches replicated the L-shaped association between 25(OH)D and CRP (Supplementary Figure S11, available as Supplementary data at IJE online) (Pnon-linear = 1.4E-03 for LDL adjustment, Pnon-linear = 0.057 for triglyceride adjustment and Pnon-linear = 1.6E-03 for the non-metabolic score). We also conducted reanalyses of non-linear MR using alternative instruments including vitaminD-GS-122 (Pnon-linear = 7.7E-07) (Supplementary Figure S12, available as Supplementary data at IJE online), focused score (Pnon-linear = 6.8E-10) (Supplementary Figure S13, available as Supplementary data at IJE online) and synthesis score (Pnon-linear = 3.3E-07) (Supplementary Figure S14, available as Supplementary data at IJE online) and found similar results.
MR of CRP on 25(OH)D
Neither linear nor non-linear MR analyses provided any support for a causal effect of serum CRP on 25(OH)D concentration (Plinear = 0.32 and Pnon-linear = 0.76) (Supplementary Table S4, available as Supplementary data at IJE online).
We performed several sensitivity analyses to gauge whether the null association in the linear and non-linear MR analyses could be related to the study design. First, as there is some evidence that CRP-gwasGS is associated with BMI and physical activity in UK Biobank (Supplementary Table S3, available as Supplementary data at IJE online), we re-performed linear and non-linear analyses adjusting for BMI, physical activity or both BMI and physical activity, and found similar results (Supplementary Table S4, available as Supplementary data at IJE online). Further, for the non-linear analysis, exclusion of local causal estimates from the outlying strata of residuals of serum CRP concentrations (i.e. the 1st and 100th strata, after exclusion PCochran’s Q = 0.51; Ptrend = 0.28) did not affect our findings (Pnon-linear = 0.79) (Supplementary Table S4, available as Supplementary data at IJE online). Reanalyses using instrument with four cis-acting variants also provided similar results (Supplementary Table S4, available as Supplementary data at IJE online).
Discussion
In this large-scale genetic analysis, we observed evidence for a causal effect of vitamin D status on CRP with no support for CRP as a determinant of 25(OH)D concentrations. The association between 25(OH)D and CRP was largely restricted to the deficiency range, where only individuals with low serum 25(OH)D concentrations have elevated serum CRP. The shape of the observed association supports the previously proposed threshold effect,12 suggesting that correction of vitamin D deficiency in the affected individuals is likely to reduce systemic low-grade inflammation and potentially mitigate the risk or severity of chronic illnesses with inflammatory components.
The earlier RCTs or MR studies have failed to provide evidence for an effect of vitamin D on CRP,8,10,11 which seemingly contradicts our finding. However, if the causal effect of vitamin D is truly L-shaped and restricted to concentrations within the deficiency range as seen in our study, it would have been overlooked both by the existing supplementation trials and linear MR studies. Severe deficiency is relatively rare36 and as it is unethical to subject participants to undue harm, supplementation trials often can only include individuals who are already vitamin D replete,12 rendering the health effect of supplementation in the deficiency range largely unassessed. Previous linear MR studies10,11 would have had limited statistical power to capture the threshold effect restricted to the deficiency range, which is indeed what was reflected in our linear MR analysis. We also found no evidence for an effect by genetically instrumented CRP on serum 25(OH)D concentration. This provides evidence against the notion that both molecules serve as acute phase reactants and that their association arises merely from confounding by inflammation.37 Indeed, if inflammation truly drives low serum 25(OH)D concentrations, we would have expected genetically instrumented CRP values to be associated with serum 25(OH)D concentration. Overall, our bidirectional MR analyses suggest that rather than vitamin D acting as a bystander [where 25(OH)D-CRP association arises merely from confounding by inflammation37], increasing 25(OH)D concentrations to alleviate a state of severe deficiency may help to mitigate the severity of inflammation. This said, it is important to note that these findings provide no support for a need to use high-dose vitamin D supplementation, as the observed benefits appeared to become largely saturated by the time 25(OH)D concentrations reach 50 nmol/L. It should also be noted that a higher vitamin D status may benefit some subpopulations or with respect to other disease outcomes,38 which is an area warranting further investigation.
Vitamin D is a pro-hormone. Its anti-inflammatory property captured by our analysis could be mediated through its hormonal effect on vitamin D receptor-expressing immune cells, such as monocytes, B cells, T cells and antigen-presenting cells.4 Indeed, cell experiments have shown that active vitamin D can inhibit the production of pro-inflammatory cytokines, including TNF-, IL-1, IL-6, IL-8 and IL-12, and promote the production of IL-10, an anti-inflammatory cytokine.4,5 Further, the anti-inflammatory effect also raises the possibility that having adequate vitamin D concentrations may mitigate complications arising from obesity and reduce the risk or severity of chronic illnesses with an inflammatory component, such as CVDs, diabetes, autoimmune diseases and neurodegenerative conditions, among others.1 If the related effects are indeed true, given the high prevalence of serum 25(OH)D levels of <50 nmol/L across the world (≤40% in some European countries),36,39–42 population-wide correction of low vitamin D status (e.g. by food fortification) could potentially be a cost-effective measure to reduce the burden of chronic disease. In fact, in linear MR analyses higher 25(OH)D concentrations have been associated with a lower risk of type 2 diabetes43 and multiple sclerosis (a chronic inflammatory disease of the central nervous system),44 with recent non-linear MR analyses providing evidence that correction of vitamin D deficiency can decrease the risk for CVDs15 and all-cause mortality.16
To the best of our knowledge, this is the first non-linear MR study to explore the bidirectional association between serum 25(OH)D and CRP concentrations. We used several strategies to ensure that our MR analyses are not affected by horizontal pleiotropy, where variants may influence the outcome through pathways other than through the exposure of interest.9 First, we restricted our vitaminD-GS to 35 variants with robust replicated evidence for an association with serum 25(OH)D concentrations. Second, we confirmed that the L-shaped association of serum 25(OH)D and CRP was consistent between the non-linear and stratified MR analyses, with the inverse association of serum 25(OH)D and CRP in the deficiency range consistently observed across pleiotropy-robust methods with largely independent assumptions on the pattern of pleiotropy. Third, the L-shaped association of serum 25(OH)D and CRP was confirmed to be robust using a spectrum of alternative instruments, including when using a broader set of variants, when excluding variants related to lipids and other metabolic traits and when restricting the set of variants to those directly related to vitamin D metabolism. Despite these strengths, our study also has some limitations. Although CRP is a widely used inflammatory biomarker, it certainly cannot capture the full complexity of the immune system and hence investigation of more specific biomarkers (such as TNF- and IL-6) is required to provide a more detailed understanding on the anti-inflammatory effects of hormonal vitamin D. We restricted our analysis to participants of White-British descent to minimize bias due to population stratification; however, this may limit the transferability of our findings to other ethnic groups. As with all MR studies, genetic instruments approximate the average effects over the life course and the true biological association between serum 25(OH)D and CRP may be more complex than that indexed in our study. With only a 5% response rate at the recruitment, UK Biobank is not representative of the general public in the UK45 despite its large sample size. It is uncertain to what extent this selection could affect the non-linear MR analysis. However, given that risk factor–disease associations show close agreement between UK Biobank and nationally representative studies46 and that an earlier publication from UK Biobank using the same non-linear MR approach has replicated the expected J-shaped association between BMI and mortality,47 this lack of representativeness may not be affecting our findings.
Conclusions
In conclusion, using a large population-based cohort, we provide genetic evidence for an L-shaped association of serum 25(OH)D with CRP, suggesting that the benefit of increasing 25(OH)D is restricted to individuals with low vitamin D status. Our finding suggests that improving vitamin D status in the deficiency range could reduce systemic low-grade inflammation and potentially mitigate the risk or severity of chronic illnesses with an inflammatory component.
Ethics approval
The present study was conducted under UK Biobank application number 20175. The UK Biobank study was approved by the National Information Governance Board for Health and Social Care and North West Multicentre Research Ethics Committee (11/NW/0382).
Supplementary Material
Contributor Information
Ang Zhou, Australian Center for Precision Health, University of South Australia Cancer Research Institute, Adelaide, Australia; South Australian Health and Medical Research Institute, Adelaide, Australia.
Elina Hyppönen, Australian Center for Precision Health, University of South Australia Cancer Research Institute, Adelaide, Australia; South Australian Health and Medical Research Institute, Adelaide, Australia; Population, Policy and Practice, UCL Institute of Child Health, London, UK.
Data availability
Data are available from UK Biobank for researchers who meet the criteria and gain approvals to access the research database from the UK Biobank access management committee at the University of Oxford.
Supplementary data
Supplementary data are available at IJE online.
Author contributions
E.H. conceived of the study and designed the research question; A.Z. analysed the data and wrote the first draft. Both authors interpreted the results, drafted and revised the manuscript critically for important intellectual content, and read and approved the final manuscript.
Funding
This study was financially supported by the National Health and Medical Research Council, Australia (grant number 1123603). The funder had no role in the design, implementation, analysis, interpretation of the data, approval of the manuscript and decision to submit the manuscript for publication.
Conflict of interest
None declared.
References
- 1. Furman D, Campisi J, Verdin E. et al. Chronic inflammation in the etiology of disease across the life span. Nat Med 2019;25:1822–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu CH, Abrams ND, Carrick DM. et al. Biomarkers of chronic inflammation in disease development and prevention: challenges and opportunities. Nat Immunol 2017;18:1175–80. [DOI] [PubMed] [Google Scholar]
- 3. Calder PC, Ahluwalia N, Albers R. et al. A consideration of biomarkers to be used for evaluation of inflammation in human nutritional studies. Br J Nutr 2013;109(Suppl 1):S1–34. [DOI] [PubMed] [Google Scholar]
- 4. Charoenngam N, Holick MF.. Immunologic effects of vitamin D on human health and disease. Nutrients 2020;12:2097.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calton EK, Keane KN, Newsholme P, Soares MJ.. The impact of vitamin D levels on inflammatory status: a systematic review of immune cell studies. PLoS One 2015;10:e0141770.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mellenthin L, Wallaschofski H, Grotevendt A, Völzke H, Nauck M, Hannemann A.. Association between serum vitamin D concentrations and inflammatory markers in the general adult population. Metabolism 2014;63:1056–62. [DOI] [PubMed] [Google Scholar]
- 7. Murr C, Pilz S, Grammer TB. et al. Vitamin D deficiency parallels inflammation and immune activation, the Ludwigshafen Risk and Cardiovascular Health (LURIC) study. Clin Chem Lab Med 2012;50:2205–12. [DOI] [PubMed] [Google Scholar]
- 8. Autier P, Mullie P, Macacu A. et al. Effect of vitamin D supplementation on non-skeletal disorders: a systematic review of meta-analyses and randomised trials. Lancet Diabetes Endocrinol 2017;5:986–1004. [DOI] [PubMed] [Google Scholar]
- 9. Davies NM, Holmes MV, Smith GD.. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liefaard MC, Ligthart S, Vitezova A. et al. Vitamin D and C-reactive protein: a Mendelian randomization study. PLoS One 2015;10:e0131740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palaniswamy S, Gill D, De Silva NM. et al. Could Vitamin D reduce obesity-associated inflammation? Observational and Mendelian randomization study. Am J Clin Nutr 2020;111:1036–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scragg R. Emerging evidence of thresholds for beneficial effects from vitamin D supplementation. Nutrients 2018;10:561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heaney RP. Guidelines for optimizing design and analysis of clinical studies of nutrient effects. Nutr Rev 2014;72:48–54. [DOI] [PubMed] [Google Scholar]
- 14. Staley JR, Burgess S.. Semiparametric methods for estimation of a nonlinear exposure-outcome relationship using instrumental variables with application to Mendelian randomization. Genet Epidemiol 2017;41:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou A, Selvanayagam JB, Hyppönen E.. Non-linear Mendelian randomization analyses support a role for vitamin D deficiency in cardiovascular disease risk. Eur Heart J 2021;ehab809. [DOI] [PubMed] [Google Scholar]
- 16. Sofianopoulou E, Kaptoge SK, Afzal S. et al. Estimating dose-response relationships for vitamin D with coronary heart disease, stroke, and all-cause mortality: observational and Mendelian randomisation analyses. Lancet Diabetes Endocrinol 2021;9:837–46. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Sudlow C, Gallacher J, Allen N. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bycroft C, Freeman C, Petkova D. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fry D, Almond R, Moffat S, Gordon M, Singh P. Companion Document to Accompany Serum Biomarker Data. Version 1.0 (11/03/2019). https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/serum_biochemistry.pdf (30 November 2021, date last accessed).
- 20. UK Biobank. Biomarker assay quality procedures: approaches used to minimise systematic and random errors (and the wider epidemiological implications). Version 1.2 (02/04/2019). https://biobank.ndph.ox.ac.uk/showcase/ukb/docs/biomarker_issues.pdf (30 November 2021, date last accessed).
- 21. Revez JA, Lin T, Qiao Z. et al. Genome-wide association study identifies 143 loci associated with 25 hydroxyvitamin D concentration. Nat Commun 2020;11:1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang X, O'Reilly PF, Aschard H. et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nat Commun 2018;9:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burgess S, Thompson SG.. Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 2013;42:1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burgess S, Gill D.. Genetic evidence for vitamin D and cardiovascular disease: choice of variants is critical. Eur Heart J 2021;ehab870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berry DJ, Vimaleswaran KS, Whittaker JC, Hingorani AD, Hyppönen E.. Evaluation of genetic markers as instruments for Mendelian randomization studies on vitamin D. PLoS One 2012;7:e37465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vimaleswaran KS, Cavadino A, Berry DJ. et al. ; Caroline Hayward. Association of vitamin D status with arterial blood pressure and hypertension risk: a Mendelian randomisation study. Lancet Diabetes Endocrinol 2014;2:719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vimaleswaran KS, Berry DJ, Lu C. et al. ; Genetic Investigation of Anthropometric Traits-GIANT Consortium. Causal relationship between obesity and vitamin D status: bi-directional Mendelian randomization analysis of multiple cohorts. PLoS Med 2013;10:e1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ligthart S, Vaez A, Võsa U. et al. Genome analyses of >200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am J Hum Genet 2018;103:691–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson T, Martin RM, Yarmolinsky J.. Mendelian randomisation analysis of circulating adipokines and C-reactive protein on breast cancer risk. Int J Cancer 2020;147:1597–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palmer TM, Sterne JA, Harbord RM. et al. Instrumental variable estimation of causal risk ratios and causal odds ratios in Mendelian randomization analyses. Am J Epidemiol 2011;173:1392–403. [DOI] [PubMed] [Google Scholar]
- 31. Cole SR, Platt RW, Schisterman EF. et al. Illustrating bias due to conditioning on a collider. Int J Epidemiol 2010;39:417–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dudbridge F, Allen RJ, Sheehan NA. et al. Adjustment for index event bias in genome-wide association studies of subsequent events. Nat Commun 2019;10:15611561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burgess S, Davies NM, Thompson SG.. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016;40:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Haworth S, Mitchell R, Corbin L. et al. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat Commun 2019;10:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hemani G, Zheng J, Elsworth B. et al. The MR-Base platform supports systematic causal inference across the human phenome. Loos R, editor. eLife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amrein K, Scherkl M, Hoffmann M. et al. Vitamin D deficiency 2.0: an update on the current status worldwide. Eur J Clin Nutr 2020;74:1498–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Antonelli M, Kushner I.. Low serum levels of 25-hydroxyvitamin D accompany severe COVID-19 because it is a negative acute phase reactant. Am J Med Sci 2021;362:333–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dawson-Hughes B, Staten MA, Knowler WC. et al. Intratrial exposure to vitamin D and new-onset diabetes among adults with prediabetes: a secondary analysis from the vitamin D and type 2 diabetes (D2d) study. Diabetes Care 2020;43:2916–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Umar M, Sastry KS, Chouchane AI.. Role of vitamin D beyond the skeletal function: a review of the molecular and clinical studies. Int J Mol Sci 2018;19:1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nimitphong H, Holick MF.. Vitamin D status and sun exposure in Southeast Asia. Dermatoendocrinology 2013;5:34–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Australian Government Department of Health. Vitamin D status. Aust. Gov. Dep. Health, 2018. https://www.health.gov.au/resources/pregnancy-care-guidelines/part-g-targeted-maternal-health-tests/vitamin-d-status (28 September 2021, date last accessed).
- 42. Lips P, Cashman KD, Lamberg-Allardt C. et al. Current vitamin D status in European and Middle East countries and strategies to prevent vitamin D deficiency: a position statement of the European Calcified Tissue Society. Eur J Endocrinol 2019;180:P23–54. [DOI] [PubMed] [Google Scholar]
- 43. Lu L, Bennett DA, Millwood IY. et al. Association of vitamin D with risk of type 2 diabetes: a Mendelian randomisation study in European and Chinese adults. PLoS Med 2018;15:e1002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mokry LE, Ross S, Ahmad OS. et al. Vitamin D and risk of multiple sclerosis: a Mendelian randomization study. PLoS Med 2015;12:e1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fry A, Littlejohns TJ, Sudlow C. et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol 2017;186:1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Batty GD, Gale CR, Kivimäki M, Deary IJ, Bell S.. Comparison of risk factor associations in UK Biobank against representative, general population based studies with conventional response rates: prospective cohort study and individual participant meta-analysis. BMJ 2020;368:m131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun Y-Q, Burgess S, Staley JR. et al. Body mass index and all cause mortality in HUNT and UK Biobank studies: linear and non-linear Mendelian randomisation analyses. BMJ 2019;364:l1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from UK Biobank for researchers who meet the criteria and gain approvals to access the research database from the UK Biobank access management committee at the University of Oxford.