Abstract
The Prostate Expression Databases (PEDB and mPEDB) are online resources designed to allow researchers to access and analyze gene expression information derived from the human and murine prostate, respectively. Human PEDB archives more than 84 000 Expressed Sequence Tags (ESTs) from 38 prostate cDNA libraries in a curated relational database that provides detailed library information including tissue source, library construction methods, sequence diversity and sequence abundance. The differential expression of each EST species can be viewed across all libraries using a Virtual Expression Analysis Tool (VEAT), a graphical user interface written in Java for intra- and inter-library sequence comparisons. Recent enhancements to PEDB include (i) the development of a murine prostate expression database, mPEDB, that complements the human gene expression information in PEDB, (ii) the assembly of a non-redundant sequence set or ‘prostate unigene’ that represents the diversity of gene expression in the prostate, and (iii) an expanded search tool that supports both text-based and BLAST queries. PEDB and mPEDB are accessible via the World Wide Web at http://www.pedb.org and http://www.mpedb.org.
INTRODUCTION
Diseases of the prostate are among the most common pathologies to afflict aging men. Prostate carcinoma is the most frequently diagnosed non-cutaneous malignancy in the US with more than 180 000 new cases estimated for 2001 (1). In order to characterize molecular alterations that accompany prostate disease processes and provide resources for virtual and physical analyses, we have developed the Prostate Expression Database (PEDB) (2). PEDB serves as a centralized collection of gene expression information derived from the human prostate that is organized in a fashion suitable for sequence-based queries, assessment of gene expression diversity, and comparative expression analyses. Expressed Sequence Tags (ESTs) and full-length cDNA sequences derived from 38 human prostate cDNA libraries are archived and represent gene expression profiles reflecting a wide spectrum of normal, benign and malignant prostate disease states. Detailed library information including tissue source, library construction methods, sequence diversity and sequence abundance are maintained in a relational database management system (RDBMS). Prostate ESTs are assembled into distinct species groups using the sequence assembly program Phrap, and annotated with information from the GenBank, dbEST and Unigene public sequence databases.
In recognition of the emerging uses of the mouse as a model system for the study of normal and pathological prostate development, we have developed a database complementary to PEDB that serves to archive and analyze murine prostate gene expression information. The mouse Prostate Expression Database (mPEDB) currently comprises >6000 ESTs from five mouse prostate cDNA libraries constructed from distinct developmental stages and anatomical locations. A detailed description of the database development, data inventory and utilities is available online: www.pedb.org/OVERVIEW/ (Table 1).
Table 1. Table of contents for PEDB overview (http://www.pedb.org/OVERVIEW).
| 1. | Introduction to PEDB |
| 2. | PEDB Information |
| 2.a. Construction | |
| 2.b. Dataflow | |
| 2.c. Build Process | |
| 2.d. Current PEDB Build | |
| 3. | PEDB Utilities |
| 3.a. BLAST Queries | |
| 3.b. Search Engine | |
| 3.c. Prostate Unigene | |
| 3.d. Virtual Analysis Expression Tool (VEAT) | |
| 3.e. Prostate Transcriptome | |
| 3.f. Prostate Proteome | |
| 4. | mPEDB |
| 5. | PEDB References and Resources |
PEDB DATA AND ANALYSIS TOOLS
PEDB consists of archives of ESTs derived from 38 human prostate cDNA libraries. These ESTs are obtained from public sequence repositories such as GenBank (3), the database of ESTs (dbEST) (4), the Cancer Genome Anatomy Project (CGAP) (5), The Institute for Genome Research (TIGR) or from in-house EST sequencing projects. Sequence processing and curation involves a pipeline of sequence submission, sequence masking, sequence assembly and assembly annotation that now incorporates quality-based assemblies using Phred and Phrap base-calling and sequence assembly algorithms (6,7) (www.pedb.org/OVERVIEW/). Assembled consensus sequences are used for BLAST queries against the Unigene, GenBank and dbEST databases to provide cluster annotation and to further facilitate the assembly process.
The most recent build of PEDB ESTs was assembled starting with 84 832 prostate ESTs. Portions of EST sequences with homology to cloning vectors, Escherichia coli genomic DNA and human repetitive DNA sequences were masked. Sequences annotating to the mitochondrial genome were removed and the remaining ESTs with >300 bp of high quality sequence were admitted to the assembly process. A total of 68 426 high-quality ESTs were assembled using Phrap to produce 28 182 clusters. Each cluster was annotated by searching the Unigene, GenBank and dbEST databases using BLASTN. Clusters annotating to the same database sequence were joined to further reduce the number of distinct clusters to 20 187. These annotated assemblies represent the prostate transcriptome: that portion of the genome that is used or expressed in the prostate.
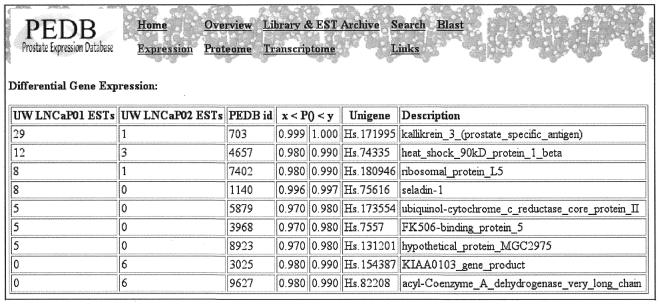
The primary work sites of PEDB involve text-based queries and a BLAST interface for sequence-based searches against PEDB and Unigene datasets. Dynamic gene expression profiles based upon EST assembly and annotation information can be generated using the Virtual Expression Analysis Tool (VEAT). The VEAT provides user-directed inter- and intra-library analysis of transcript abundance, diversity and differential expression. We have recently incorporated a statistical algorithm developed by Audic and Claverie (8) that can determine probabilities of differential transcript abundance levels in datasets comprised of varying numbers of sequences. We have used these tools to identify prostate genes regulated by androgens and genes differentially expressed between adenocarcinoma and small cell carcinoma of the prostate (Fig. 1).
Figure 1.
Output of differential expression analysis with statistical filtering. The annotated ESTs in two prostate cDNA libraries were compared for relative abundance levels. The output of the analysis provides (i) the number of ESTs in each library corresponding to a specific transcript, (ii) the PEDB identification number, (iii) the statistical probability, P, of differential expression between the two library datasets, (iv) the Unigene database accession number, and (v) a description of the gene based upon GenBank or Unigene annotation.
MOUSE PEDB (mPEDB)
The mouse represents a versatile model organism for studying development, genetics, behavior and disease. Several murine models of prostate carcinogenesis have recently been reported (9,10), and the mouse has been used to study the effects of genes hypothesized to be important in the normal and neoplastic development of the human prostate (11). Recognizing the great utility of EST sequences for characterizing organ-specific gene expression, cloning novel genes and developing microarray reagent sets, we have initiated efforts to define the mouse prostate transcriptome by constructing and sequencing mouse prostate cDNA libraries. Interestingly, the extensive list of cDNA libraries provided at the Cancer Genome Anatomy Project web site lists more than 400 murine cDNA libraries, but none are derived from the prostate gland (http://www.ncbi.nlm.nih.gov/ncicgap/).
To date we have made five mouse prostate cDNA libraries, which are derived from microdissected anterior, dorsolateral and ventral prostatic lobes of mature mice, and from the urogenital sinus of E16 embryos. A total of 6145 ESTs have been sequenced, assembled, annotated and loaded into mPEDB in a fashion analogous to that described for processing human prostate sequence in PEDB. Virtual comparisons of transcriptomes derived from these distinct anatomical regions of the prostate suggest that the prostate lobes have specific functional attributes. Library summaries, text- and sequence-based queries, and virtual expression analyses tools are provided.
SUMMARY AND FUTURE DEVELOPMENTS
The human and mouse Prostate Expression Databases serve as centralized archives of gene expression information derived from the human and murine prostate that can be utilized by investigators studying normal and neoplastic prostate development. The assembled human prostate transcriptome currently comprises 20 187 distinct transcripts. Ongoing work involves the characterization of additional cDNA libraries representing specific prostate cell types and early developmental stages, the virtual comparative analyses of human and mouse prostate gene expression, and a database extension for archiving and analyzing cDNA microarray data derived from PEDB and mPEDB sequence resources. PEDB is accessible via the World Wide Web at http://www.pedb.org. mPEDB is accessible at http://www.mpedb.org or through a link from PEDB.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Alec Mooreman, Stacy Boness and Josh Epstein for sequencing assistance. We thank Burak Eroglu for database administration. This work was supported by the CaPCURE Foundation and grants to P.S.N. from the NCI (K08 CA75173-01A1), NIDDK (RO1 DK59125-02) and the USAMRMC Program in Prostate Cancer Research (DAMD17-98-1-8499).
REFERENCES
- 1.Greenlee R.T., Murray,T., Bolden,S. and Wingo,P.A. (2000) Cancer statistics. CA Cancer J. Clin., 50, 7–33. [DOI] [PubMed] [Google Scholar]
- 2.Hawkins V., Doll,D., Bumgarner,R., Smith,T., Abajian,C., Hood,L. and Nelson,P.S. (1999) PEDB: the Prostate Expression Database. Nucleic Acids Res., 27, 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D.A., Boguski,M.S., Lipman,D.J., Ostell,J. and Ouellette,B.F.F. (1998) GenBank. Nucleic Acids Res., 26, 1–7. Updated article in this issue: Nucleic Acids Res. (2002), 30, 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boguski M.S., Lowe,T.M.J. and Tolstoshev,C.M. (1993) dbEST—database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 5.Schaefer C., Grouse,L., Buetow,K. and Strausberg,R.L. (2001) A new cancer genome anatomy project web resource for the community. Cancer J., 7, 52–60. [PubMed] [Google Scholar]
- 6.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 7.Gordon D., Abajian,C., Green,P. (1998) Consed: a graphical tool for sequence finishing. Genome Res., 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 8.Audic S. and Claverie,J.M. (1997) The significance of digital gene expression profiles. Genome Res., 7, 986–995. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg N.M., DeMayo,F., Finegold,M.J., Medina,D., Tilley,W.D., Aspinall,J.O., Cunha,G.R., Donjacour,A.A., Matusik,R.J. and Rosen,J.M. (1995) Prostate cancer in a transgenic mouse. Proc. Natl Acad. Sci. USA, 92, 3439–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Cristofano A., De Acetis,M., Koff,A., Cordon-Cardo,C. and Pandolfi,P.P. (2001) Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nature Genet., 27, 222–224. [DOI] [PubMed] [Google Scholar]
- 11.Bhatia-Gaur R., Donjacour,A.A., Sciavolino,P.J., Kim,M., Desai,N., Young,P., Norton,C.R., Gridley,T., Cardiff,R.D., Cunha,G.R., Abate-Shen,C. and Shen,M.M. (1999) Roles for Nkx3.1 in prostate development and cancer. Genes Dev., 13, 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]