Abstract
Transterm is a database that facilitates studies of translation and the translational control of protein synthesis. It contains a curated collection of elements in mRNAs that control translation, and biologically relevant mRNA regions extracted from GenBank. It is organised largely on a taxonomic basis with files and summaries for each species. Global patterns that may affect translation in particular species, for example bias in the context of initiation codons (Kozak’s consensus or Shine–Dalgarno sequences) or termination codons, can be detected in the consensus and information content bias summaries. Several types of access are provided via a web browser interface. Transterm defined elements may be matched in a user’s sequence or in the database. Alternatively, elements can be entered by the user to search specific sections of the database (for example, coding regions or 3′ flanking regions or the 3′-UTRs) or the user’s sequence. Each Transterm defined element has an associated biological description with references. The database is accessible at http://uther.otago.ac.nz/Transterm.html.
INTRODUCTION
Transterm is a database of elements in mRNAs and mRNA sequences in which they may be found. Elements in particular mRNAs and translated viral RNAs have been shown to mediate many post-translational controls in cells (reviewed in 1–5). In the Transterm database, elements are now stored and classified in two ways, functionally and structurally.
ELEMENTS FOUND IN CELLULAR AND VIRAL RNAS
Functional classification of elements
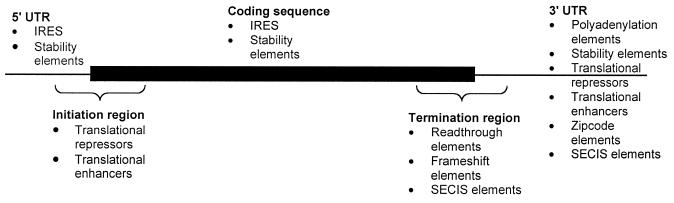
For the purposes of classification we divide the elements into the following classes: (i) mRNA localisation elements (zip code elements); (ii) stability elements (SE); (iii) translational repressors (TR); (iv) translation enhancers (TE) (6,7); (v) polyadenylation elements (PE) (8); (vi) elements that control the efficiency of translation initiation in abnormal contexts [e.g. Internal Ribosome Entry Sites (IRES)]; and (vii) elements which promote alternative reading of the genetic code [e.g. frameshifting elements (FSE), readthrough elements (RE), selenocysteine incorporation elements (SECIS)]. This is not an absolute classification but facilitates searching and comparison. At time of writing the database contained 47 such elements. For each element, information concerning its structure, functional description, location in the mRNA, confirmed phylogenetic distribution, example mRNA, place discovered, required elements or factors in cis or trans, structural classification and a short bibliography are provided. These elements are typically located in certain regions of the mRNA (Fig. 1).
Figure 1.
Typical locations of the functional classes of elements in a mRNA sequence.
Structural classification of elements
These elements can be classified into three broad classes based on sequence and structure: (i) sequence alone; (ii) secondary structure alone; and (iii) combination of sequence and structure. These classifications are a useful approximation of the biology but may not describe the biology fully; for example, elements defined as sequence alone may still require an ‘A’ helical mRNA structure (1,9). Also, some elements originally defined largely by core primary structure have subsequently been shown to require additional structure (10).
BIOLOGICALLY RELEVANT mRNA REGIONS EXTRACTED FROM GenBank
The Transterm database contains files of specific parts of mRNA sequences extracted from GenBank. Currently these are the 5′ flank, initiation region, CDS, termination region and 3′ flank. The regions are extracted based on GenBank annotation with a reduction in redundancy, several accuracy checks and constraints as previously described (11).
Species by species consensuses of initiation, termination and coding regions
As the database is organised by species using GenBank TAXIDs, global summaries for each species can be calculated (12). For the termination and initiation regions summary statistics are calculated for each species. These include the incidence of each base before and after the initiation codon, a consensus of this region and the information content in this region. For all the CDSs of a species the codon usage is calculated and provided in GCG format.
Parameters describing each mRNA
In addition to containing the sequence of regions for each CDS, Transterm contains several calculated parameters describing the CDS. These include measures of codon bias, GC% and length.
APPLICATIONS OF THE DATABASE
The database and associated tools provide users with an entry point to address several types of biological questions. FAQs and extensive online help can be found on the web site (http://uther.otago.ac.nz/Transterm.html). Examples of applications include: does my mRNA sequence contain any defined translational control elements? Is my newly discovered element found in any other 3′-UTRs? What is the bias around the context of the initiation codon in a particular species? What is the codon usage in a particular species?
Acknowledgments
ACKNOWLEDGEMENTS
The work is supported by a Marsden fund grant to C.M.B. and NZ Health Research Council grant to W.T.
REFERENCES
- 1.Richter J.D. and Theurkauf,W.E. (2001) Development. The message is in the translation. Science, 293, 60–62. [DOI] [PubMed] [Google Scholar]
- 2.Bakheet T., Frevel,M., Williams,B.R., Greer,W. and Khabar,K.S. (2001) ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res., 29, 246–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson J.R. (2000) Induced fit in RNA–protein recognition. Nature Struct. Biol., 7, 834–837. [DOI] [PubMed] [Google Scholar]
- 4.Pesole G., Liuni,S., Grillo,G., Licciulli,F., Larizza,A., Makalowski,W. and Saccone,C. (2000) UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Nucleic Acids Res., 28, 193–196. Updated article in this issue: Nucleic Acids Res. (2002), 30, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conne B., Stutz,A. and Vassalli,J.D. (2000) The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology. Nature Med., 6, 637–641. [DOI] [PubMed] [Google Scholar]
- 6.Miller W.A., Brown,C.M. and Wang,S.P. (1997) New punctuation in the genetic code: luteovirus gene expression. Seminars Virol., 8, 3–13. [Google Scholar]
- 7.Qu F. and Morris,T.J. (2000) Cap-independent translational enhancement of turnip crinkle virus genomic and subgenomic RNAs. J. Virol., 74, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Echevarria M.J. and Peltz,S.W. (2000) The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell, 101, 741–751. [DOI] [PubMed] [Google Scholar]
- 9.Wang X. and Tanaka Hall,T.M. (2001) Structural basis for recognition of AU-rich element RNA by the HuD protein. Nature Struct. Biol., 8, 141–145. [DOI] [PubMed] [Google Scholar]
- 10.Thisted T., Lyakhov,D.L. and Liebhaber,S.A. (2001) Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and αCP-2KL, suggest distinct modes of RNA recognition. J. Biol. Chem., 276, 17484–17496. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs G.H., Stockwell,P.A., Schrieber,M.J., Tate,W.P. and Brown,C.M. (2000) Transterm: a database of messenger RNA components and signals. Nucleic Acids Res., 28, 293–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. Updated article in this issue: Nucleic Acids Res. (2002), 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]