Abstract
Transcription Regulatory Regions Database (TRRD) is an informational resource containing an integrated description of the gene transcription regulation. An entry of the database corresponds to a gene and contains the data on localization and functions of the transcription regulatory regions as well as gene expression patterns. TRRD contains only experimental data that are inputted into the database through annotating scientific publication. TRRD release 6.0 comprises the information on 1167 genes, 5537 transcription factor binding sites, 1714 regulatory regions, 14 locus control regions and 5335 expression patterns obtained through annotating 3898 scientific papers. This information is arranged in seven databases: TRRDGENES (general gene description), TRRDLCR (locus control regions); TRRDUNITS (regulatory regions: promoters, enhancers, silencers, etc.), TRRDSITES (transcription factor binding sites), TRRDFACTORS (transcription factors), TRRDEXP (expression patterns) and TRRDBIB (experimental publications). Sequence Retrieval System (SRS) is used as a basic tool for navigating and searching TRRD and integrating it with external informational and software resources. The visualization tool, TRRD Viewer, provides the information representation in a form of maps of gene regulatory regions. The option allowing nucleotide sequences to be searched for according to their homology using BLAST is also included. TRRD is available at http://www.bionet.nsc.ru/trrd/.
DESCRIPTION OF TRRD
Transcription Regulatory Regions Database (TRRD) has been developed and supported at the Institute of Cytology and Genetics SB RAS (Novosibirsk, Russia) since 1993. The main goal while developing TRRD was to provide a most complete and adequate description of the structure–function organization of transcription regulatory regions of eukaryotic genes. Both the TRRD structure and format were formed to achieve this goal and are still developing. The current TRRD release (6.0) comprises seven databases linked with cross-references: TRRDGENES (general gene description), TRRDLCR (locus control regions); TRRDUNITS (regulatory regions: promoters, enhancers, silencers, etc.), TRRDSITES (transcription factor binding sites), TRRDFACTORS (transcription factors), TRRDEXP (expression patterns) and TRRDBIB (bibliography).
The format of TRRD allows the transcription regulation of the eukaryotic genes transcribed by RNA polymerase II to be described in an integrated manner in all the organs, tissues and cell types of the organism as well as in cell lines. First, TRRD contains the data on structural organization of transcription regulatory regions of the following hierarchical levels: (i) transcription factor binding sites (TRRDSITES); (ii) regulatory units, including promoters, enhancers and silencers (TRRDUNITS); (iii) regulatory regions, including 5′- and 3′-regulatory regions, exons and introns (TRRDGENES); and (iv) locus control regions (TRRDLCR). Secondly, TRRD accumulates functional characteristics of regulatory elements of all the levels, such as the effect of the gene on transcriptional activity; specific function at a certain stage of the cell cycle or ontogenesis, in particular cell types, tissues or organs; and involvement of a regulatory element in regulation of gene expression in response to various intracellular and external stimuli or influences. Thirdly, TRRD contains the data on patterns of gene expression (TRRDEXP). The informational fields RegUnitLink (RP) and SiteLink (RS) of this database are hyperlinked to textual descriptions of regulatory units (promoters, enhancers and silencers, described in TRRDUNITS) and transcription factor binding sites (TRRDSITES) that realize specific expression features typical of each pattern.
A distinguishing feature of the TRRD database is accumulation of the information confirmed experimentally. The data are inputted into TRRD through annotating publications describing results of experiments of different types, exemplified in Table 1. These experiments may be aimed at (i) detection and primary analysis of extended regulatory regions of genes; (ii) detection of transcription factor binding sites; (iii) confirming the functional importance of the site; and (iv) identification of DNA-binding proteins. Each type of experiment has its own code (second column of the table). Digital codes together with the information on cell types involved in experiments are given in the informational fields ExperimentCodes (AG) of the databases TRRDGENES, TRRDSITES and TRRDUNITS, thereby allowing the types of experiments underlying the description of particular site or regulatory unit in the corresponding entry of TRRDGENES, TRRDSITES and TRRDUNITS to be indicated.
Table 1. Examples of experiments underlying the information inputted into TRRD.
| Type of experiment | Assay code in TRRD |
|---|---|
| Delineation and initial analysis of large regulatory regions | |
| Insertion of the promoter region upstream of reporter gene | 6.8 |
| Attachment of DNA fragment of interest to homologous or heterologous promoter and reporter gene | 6.3.1 |
| Deletion analysis | 6.1.1 |
| Assessing appropriate regulation by different agents in transient transfection assay | 6.5 |
| Detection of transcription factor binding sites | |
| DNase I footprinting with nuclear extract | 1.1.1 |
| OP-Cu footprinting with nuclear extract | 1.6 |
| DNase I footprinting with purified or recombinant protein | 1.1.5 |
| Genomic footprinting | 1.5 |
| Methylation protection assay | 4.1 |
| Methylation interference assay | 4.2 |
| Electrophoretic mobility shift assay (EMSA) with nuclear extract | 3.1 |
| EMSA performed in the presence of competitive oligonucleotides | 3.2 |
| EMSA performed with mutant probes or competitors | 3.3 |
| Confirming the functional importance of the site | |
| Insertion of isolated site 5′ of homologous or heterologous promoter | 6.3.2 |
| Comprehensive mutant analysis | 6.2 |
| Trans-activation of a reporter gene by overexpression of a distinct transcription factor | 6.6 |
| Genomic footprinting | 1.5 |
| Identification of DNA-binding proteins | |
| DNase I footprinting with purified or recombinant protein | 1.1.5 |
| DNase I footprinting with nuclear extract and specific antibodies | 1.1.6 |
| EMSA with purified or recombinant protein | 3.5 |
| EMSA with nuclear extract and specific antibodies | 3.6 |
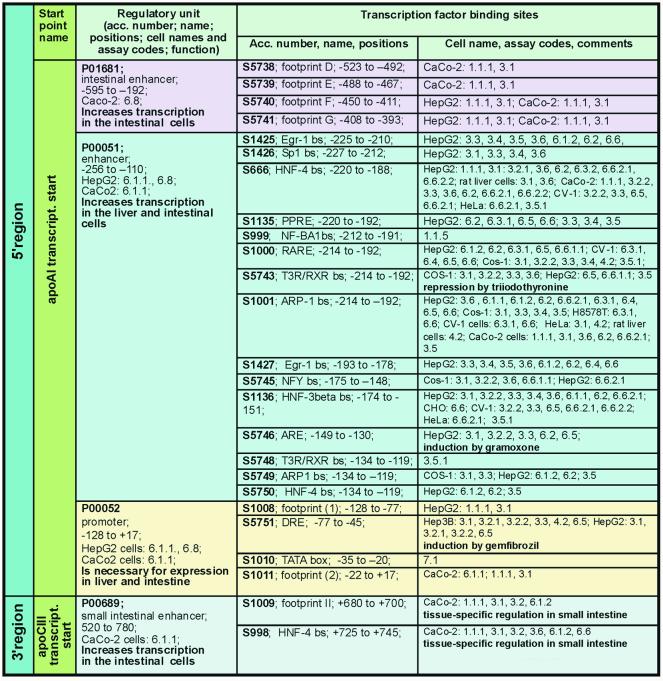
A brief characterization of the regulatory elements (regulatory regions, regulatory units and transcription factor binding sites) of human apolipoprotein A1 gene was composed (Fig. 1) using the data compiled in TRRD (the entry with accession no. A00264 in TRRDGENES). This gene contains four regulatory units. Three of them [promoter (–128/+17) and two enhancers (–256/–110 and –595/–192)] are localized to 5′-region. The fourth regulatory unit [enhancer (+520/+780)] is localized to 3′-region; its positions are indicated with reference to the transcription start of apolipoprotein C3 gene, which is located ∼9 kb from the transcription start of apolipoprotein A1 gene. In the regulatory regions of this gene, 25 transcription factor binding sites were identified. There are data indicating that the sites S1009 and S998 provide a tissue-specific action of the enhancer P00689, while several sites mediate the regulation of the gene expression by triiodothyronine (S5743); gramoxone, an inducer of oxidative stress (S5746), and gemfibrozil, a hypolipidemic drug (S5751).
Figure 1.
The structure–function characterization of regulatory regions of human apolipoprotein A1 based on the TRRD data (accession no. a00264 in TRRDGENES).
TRRD FORMAT
The formats of TRRD releases 4.1 and 4.2 are described in detail by Kolchanov et al. (1,2). Both the format and structure of TRRD release 6.0 are essentially expanded and modified.
(i) A new database, TRRDUNITS, compiling the information on regulatory regions (promoters, enhancers, silencers, etc.), was formed. The data are distributed between 13 informational fields: RegUnitAC (AP), the accession number of regulatory unit; GeneID (ID), identifier of the entry in TRRD; RegRegion (RG), transcription regulatory region; RegUnit (PR), name and localization of the regulatory unit, its start point, and the list of accession numbers of the sites localized to this regulatory unit; DNA_BankLink (AQ), the first and last nucleotide positions of the sequence according to the EMBL/GenBank and the hyperlink to this database; LeftTrunc (LQ), the number of nucleotides of the regulatory unit from the left end that are absent in the field Sequence, because it was not found in corresponding sequence in EMBL/GenBank; RightTrunc (RQ), the number of nucleotides of the regulatory unit from the right end that are absent in the field Sequence, because it was not found in corresponding sequence in EMBL/GenBank; SeqLength (SL), the number of nucleotides in the sequence contained in the field Sequence; Sequence (SE), DNA sequence; PromotTisSp (PT), promoter tissue-specific characteristics; PromotInd (PI), promoter induction pattern; MultipleStarts (MS), positions of alternative transcription starts (if applicable); and ExperimentCodes (AG), cells, assay codes and reference to the paper annotated.
(ii) A new database, TRRDLCR, was formed. Its format allows the specific structure–function features of these regulatory regions to be described. Each entry of this database describes an individual locus control region (LCR). The information compiled in TRRDLCR is distributed between 40 informational fields, containing the data on the structure of the regulated gene locus, structure of the LCR itself and the functional characterization of its elements. TRRDLCR is hyperlinked to two databases. The first database, TRRDGENES, contains the description of regulatory regions of the genes forming the locus under the control of particular LCR. The second database, TRRDUNITS, has hyperlinks to detailed descriptions of the transcription factor binding sites detected in the LCR functional elements. In addition, links to the EMBL, SWISS-PROT and MEDLINE databases are provided.
(iii) The format of TRRDSITES is expanded. Three new fields are added to TRRDSITES. The field ImportPos (IP) is designed to compile the information on the nucleotides within transcription factor binding sites that are important for interactions with the corresponding proteins. The field IP is filled based on the data of methylation interference assays, transient transfection assays and gel mobility shift assays using DNA fragment carrying mutations.
Two other new fields, SeqContradiction (SC) and PosContradiction (PC), are introduced for adequate representation of information in the case when the annotator finds a discrepancy either in the site sequence or its positions between the paper annotated and the corresponding data from EMBL/GenBank. The site sequence and positions corresponding to EMBL/GenBank are indicated in the fields Sequence (SQ) and SequencePosition (PQ) of the database TRRDSITES, while the sequence and positions from the paper annotated are given in the fields SeqContradiction (SC) PosContradiction (PC).
INFORMATIONAL CONTENT OF THE CURRENT TRRD RELEASE 6.0
The information system TRRD is supplemented monthly with new information. The number of entries in TRRD release 4.2.5 (2) and the current release are shown in Table 2.
Table 2. Informational content of TRRD.
| Name of database | Number of entries in the release 4.2.5 | Number of entries in the release 6.0 |
|---|---|---|
| TRRDGENES | 760 | 1167 |
| TRRDUNITS | – | 1714 |
| TRRDEXP | 3403 | 5335 |
| TRRDSITES | 3604 | 5537 |
| TRRDFACTORS | 2862 | 4600 |
| TRRDLCR | – | 14 |
| TRRDBIB | 2537 | 3898 |
While developing TRRD, we focused our attention on descriptions of genes belonging to certain functional systems, represented in nine subject sections. The sections Interferon-Inducible Genes (IIG-TRRD), Heat Shock-Induced Genes (HS-TRRD), Glucocorticoid-Regulated Genes (GR-TRRD) and Redox-Sensitive Genes (ROS-TRRD) comprise the genes whose expression depends on the corresponding inducer. The section Erythroid-Specific Regulated Genes (ESRG-TRRD) contains the genes that are regulated specifically in the erythroid cells. The sections Genes of Lipid Metabolism (LM-TRRD), Endocrine System Transcription Regulatory Regions Database (ES-TRRD) and Cell Cycle-Dependent Genes (CYCLE-TRRD) include the genes that are involved in the regulation of lipid metabolism, endocrine system and cell cycle, respectively. The section Plant Genes (PLANT-TRRD) describes the genes of plants.
QUALITY MANAGEMENT WHILE INPUTTING THE DATA INTO TRRD
The information is input into TRRD by expert biologists through annotation of experimental papers. While inputting the data, both syntactic and semantic analyses are performed using original software. The program TRRD-INPUT (2) allows both the new data to be inputted specifying the structure of the text file in advance and the text files created earlier to be checked and edited. TRRD-INPUT checks the words from the informational fields of the databases TRRDGENES, TRRDEXP, TRRDSITES and TRRDFACTORS for their compliance with the terms of the corresponding controlled vocabularies. So far, 22 vocabularies with a totality of over 3200 terms have been created and are maintained. Vocabularies of morphological terms (organs, tissue types and cell types) are organized hierarchically. Another program checks the compliance of the information on positions of transcription factor binding sites and transcription start sites of genes with the DNA sequence from the corresponding EMBL/GenBank entry. If any discrepancies between the transcription start positions calculated from the information on positions of several sites are lacking, the program extracts the nucleotide sequence of the regulatory unit from the sequence of the corresponding EMBL/GenBank entry. Prior to launching each new TRRD release, both the cross-references within the system itself and hyperlinks to external databases, such as EMBL/GenBank, SWISS-PROT, TRANSFAC, GeneNet, ACTIVITY, SAMPLES, EPD, GDB, IMGT/LIGM, are checked. A specialized program allows the TRRD flat file to be converted into an XML format.
VISUALIZATION TOOL
TRRD 6.0 has a new visualization tool, TRRD Viewer, realized as a Java applet using jdk 1.6, which possesses additional options compared with the version described previously (1).
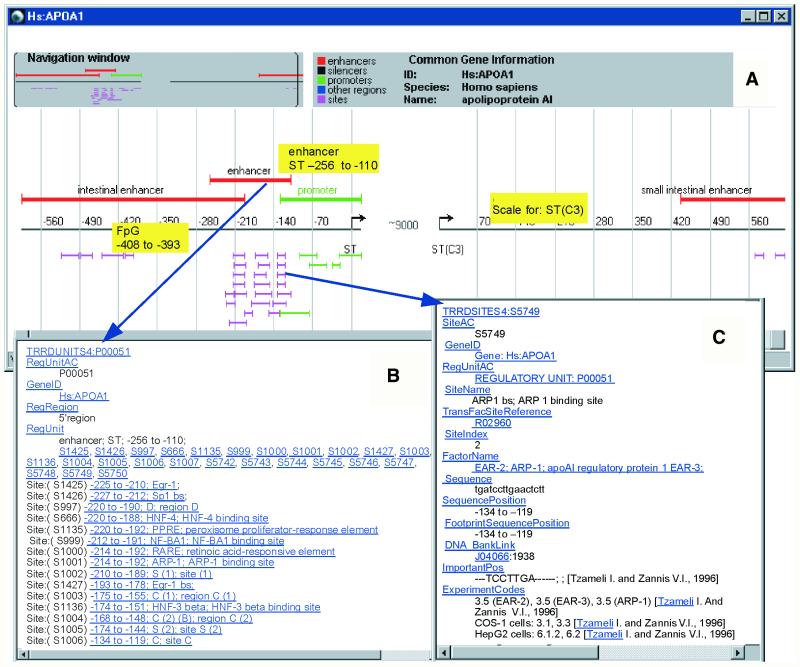
The data on structural organization of the regulatory regions are represented in TRRD Viewer as maps of gene regulatory regions, as exemplified in Figure 2A. The interface of TRRD Viewer includes three windows: (i) navigation window; (ii) the window with textual description and designations; and (iii) the window with map of gene regulatory regions. Moving brackets in the navigation window allows the left and right limits of the region to be shown in the lower window to be specified. The lower window in Figure 2A demonstrates the map of human apolipoprotein A1 gene regulatory regions, including two enhancers in 5′-region (red line segments above the reference scale), promoter (green line segment) and one enhancer in 3′-region (red line segment above the reference scale); 4, 21, 4 and 2 transcription factor binding sites are localized to these regions, respectively (short line segments below the scale). Clicking on the images of regulatory units (promoters, enhancers and silencers) or transcription factor binding sites pops up a tool tip with brief textual descriptions of the corresponding elements (yellow text boxes). Single clicking of the left mouse button on the image of a regulatory unit paints this image and the images of the related transcription factor binding sites with the same color according to the legend (in Fig. 2A, the sites related to promoter are colored green). The images of regulatory regions and sites are hyperlinks to their complete textual descriptions in the databases TRRDUNITS and TRRDSITES (Fig. 2B and C) accessible through double clicking the left button.
Figure 2.
Graphical map of human apolipoprotein A1 produced by TRRD Viewer.
DATA RETRIEVAL FROM TRRD
Sequence Retrieval System (SRS) is used as a basic software tool for accessing TRRD via the Internet; this provides an efficient search not only of TRRD, but also of the linked databases. A system of hyperlinks integrates seven databases of the informational system TRRD with one another and informational and software modules of GeneExpress-2 (3), underlying a quick access to both the experimental information on regulation of expression of a particular gene and the programs for computer analyses of its regulatory sequences.
TRRD also provides the possibility of searching for genes via browsing lists of gene names and species.
The genes from functionally significant groups listed above can be quickly accessed via the TRRD subject sections.
POSSIBILITIES OF DATA ANALYSIS USING THE INFORMATION COMPILED IN TRRD
The user is provided with the possibility of searching for the regulatory regions described in TRRD and homologous to the sequence of his own interest using the program BLAST (4).
Another type of query is the search for regions homologous to the transcription factor binding sites described in TRRD in the sequence of user’s interest. The program BinomSite performs the search by binomial probability estimation of the similarity between fragments of user’s sequence and each of the transcription factor binding sites described in TRRD.
AVAILABILITY
TRRD 6.0 is available via the World Wide Web at http://www.bionet.nsc.ru/trrd/. TRRD flat and XML files are available on a collaborative basis. TRRD cannot be included into other databases without explicit permission of the authors. All rights reserved. The TRRD scientific supervisor Nikolay A. Kolchanov can be contacted by Email: kol@bionet.nsc.ru. Users are welcome with any comments, corrections and requests for additional information via Email, or Fax: +7 3832 331278. Users are asked to refer to this paper and the 2000 publication (2) when reporting results obtained through TRRD application.
CONCLUSION
Numerous databases containing diverse information on regulation of eukaryotic gene expression are available now. TRRD is a unique database as it contains simultaneously the information obtained while studying extended regulatory regions, transcription factor binding sites and specific expression patterns of various eukaryotic genes. This information is input into the database through annotation of scientific papers describing various types of experiments and further standardization and coordination of the data. TRRD may be useful for a wide range of researchers involved in a diversity of branches of molecular biology, genetics, pharmacology, biotechnology and biomedicine.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to I. V. Lokhova and L. V. Katokhina for bibliographic support; to D. A. Grigorovich and E. V. Maksakov, for development of the necessary software; to expert biologists O. E. Belova, T. V. Busygina, V. M. Merkulov, T. N. Goryachkovskaya, V. V. Suslov, T. M. Khlebodarova, C. A. Fedorova, S. S. Ibragimova, O. G. Smirnova and A. L. Proskura for annotating publications; and to G. B. Chirikova for translation of the paper into English. The work was supported by the National Human Genome Program, State Committee on Science and Technology of the Russian Federation, Integrative Project of the Siberian Branch of the Russian Academy of Sciences, and Russian Foundation for Basic Research (grant nos 01-07-376, 00-04-49229, 00-04-49225, 00-07-90337 and 99-07-90203).
REFERENCES
- 1.Kolchanov N.A., Ananko,E.A., Podkolodnaya,O.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N., Busygina,T.V. et al. (1999) Transcription Regulatory Regions Database (TRRD): its status in 1999. Nucleic Acids Res., 27, 303–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolchanov N.A., Podkolodnaya,O.A., Ananko,E.A., Ignatieva,E.V., Stepanenko,I.L., Kel-Margoulis,O.V., Kel,A.E., Merkulova,T.I., Goryachkovskaya,T.N. et al. (2000) Transcription Regulatory Regions Database (TRRD): its status in 2000. Nucleic Acids Res., 28, 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolchanov N.A., Ponomarenko,M.P., Frolov,A.S., Ananko,E.A., Kolpakov,F.A., Ignatieva,E.V., Podkolodnaya,O.A., Goryachkovskaya,T.N., Stepanenko,I.L., Merkulova,T.I. et al. (1999) Integrated databases and computer systems for studying eukaryotic gene expression. Bioinformatics, 15, 669–686. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]