Abstract
Immune cell function is critically dependent on precise control over transcriptional output from the genome. In this respect, integration of environmental signals that regulate gene expression, specifically by transcription factors, enhancer DNA elements, genome topography and non-coding RNAs (ncRNAs), are key components. The first three have been extensively investigated. Even though non-coding RNAs represent the vast majority of cellular RNA species, this class of RNA remains historically understudied. This is partly because of a lag in technological and bioinformatic innovations specifically capable of identifying and accurately measuring their expression. Nevertheless, recent progress in this domain has enabled a profusion of publications identifying novel sub-types of ncRNAs and studies directly addressing the function of ncRNAs in human health and disease. Many ncRNAs, including circular and enhancer RNAs, have now been demonstrated to play key functions in the regulation of immune cells and to show associations with immune-mediated diseases. Some ncRNAs may function as biomarkers of disease, aiding in diagnostics and in estimating response to treatment, while others may play a direct role in the pathogenesis of disease. Importantly, some are relatively stable and are amenable to therapeutic targeting, for example through gene therapy. Here, we provide an overview of ncRNAs and review technological advances that enable their study and hold substantial promise for the future. We provide context-specific examples by examining the associations of ncRNAs with four prototypical human autoimmune diseases, specifically rheumatoid arthritis, psoriasis, inflammatory bowel disease and multiple sclerosis. We anticipate that the utility and mechanistic roles of these ncRNAs in autoimmunity will be further elucidated in the near future.
Keywords: non-coding RNA, circular RNA, enhancer RNA, autoimmunity, single cell RNA-sequencing, rheumatoid arthritis, psoriasis
Introduction
Precise regulation of gene transcription is essential for the normal function of immune cells, balancing the need for appropriate responsiveness to pathogens and danger signals while maintaining tolerance to self-components. Broadly, transcriptional output from the genome can be divided into coding (messenger) ribonucleic acids (mRNAs) and non-coding RNAs (ncRNAs) based on the potential for translation to proteins. Although ncRNAs account for a large fraction of RNA species in most eukaryotic cells, relatively little is known about the roles played by ncRNAs in the context of immune cell differentiation and fate decisions. In fact, current understanding of genome biology in immune cells is mainly centered on the regulation of coding genes by transcription factors (TFs) and non-coding regulatory elements, such as enhancers. Therefore, a major knowledge gap remains in elucidating the functions of ncRNAs in human physiology and immune-related diseases. Nevertheless, recent primary studies have demonstrated a unique role for a variety of ncRNAs in the regulation of immune cell function at multiple levels, including lymphoid development, inflammatory pathway activation, cellular metabolism, antibody repertoire selection and cytokine secretion. Such functions indicate that ncRNAs participate in xeno-responses to provide immunity to pathogens. Moreover, they have also been implicated in anti-self responses, where components of the immune system are dysregulated and harm otherwise healthy tissues, evident as clinical autoimmunity. Importantly, while TFs and enhancers are typically challenging to target therapeutically, non-coding RNAs may provide better alternatives for gene therapy [1–3]. Thus, not only can ncRNAs act as potential biomarkers and enhance our understanding of the basic molecular mechanisms of disease, but they can also be therapeutically leveraged for the treatment of patients. Recent developments in technology required for the broad detection and discrete analysis of ncRNAs could also shed light on their roles in immune cells. Here, we provide an up-to-date review of ncRNAs, specifically discussing advances in technologies for their detection, mechanisms of action and putative roles orchestrated in human autoimmune diseases.
Classes of ncRNAs
Although ncRNAs make up the majority (~90%) [4,5] of transcriptional products, the functions of most ncRNAs remain poorly understood. Pioneering studies of transfer [6] and ribosomal [7] RNA in the 1950s described the function of the first ncRNAs, and were followed by the study of RNAse P [8], RNA granules [9], RNA methylation and processing [10], RNA structure prediction [11], and XIST-mediated X-inactivation [12]. With advancements in RNA sequencing [13], the number of ncRNAs detected amongst human cells has increased exponentially, with ~0.6 million ncRNAs now estimated to exist (Table 1). This compares to approximately 20,000 genes encoding mRNAs. In humans, the true number of ncRNAs and their expression in nature may vary more widely by cell-type and condition and, like mRNAs, alternative-splicing. Broadly, ncRNAs are divided into small (<200bp in length) and long ncRNAs [14] (variously defined as either >200bp with or without a putative open reading frame (ORF), or >300bp without an ORF) (Figure 1). Small ncRNAs include micro-RNAs (miRs) [15], transfer RNAs (tRNAs) [16], tRNA-derived small RNAs (tsRNAs) [17], piwi-interacting RNAs (piRNAs) [18] and small nucleolar RNAs (snoRNAs) [19]. The long ncRNAs include linear long non-coding RNAs (lncRNAs) [20], long intergenic non coding RNAs (lincRNAs – essentially lncRNAs that do not overlap with protein-coding genes) [21], enhancer RNAs (eRNAs) [22,23] and circular RNAs (circRNAs) [24,25]. The number of ncRNAs in each category is increasing exponentially. Table 1 lists the number of ncRNAs described to date in each category and the databases curated to enable browsing of all those described.
Table 1.
ncRNA types, detection, genomics, databases, and biogenesis.
| ncRNAs | Size (bp) | Detection method(s) | Known GRCh38 Coverage | Databases | Biogenesis | Roles in immune cell regulation | Ref | |
|---|---|---|---|---|---|---|---|---|
| Small to mid-size ncRNAs | miR | 19–25 | Microarray, RNA-seq, Northern blotting, qRT-PCR, ddPCR, SEXPAR, miRacles PCR In situ stain |
Total # of hairpin precursor: 1917 Total # of mature miRs: 2654 ---- Intronic: 56.44% Intergenic: 23.72% Exonic: 10.38% Other: 13.96% |
http://www.mirbase.org http://www.microrna.gr/tarbase (For miR-gene interaction) http://www.microrna.gr/LncBase (For miR-lncRNA interaction) |
RNA pol II, RNA pol III, For maturation: Drosha, DGCR8 and Dicer | Target mRNA degradation, translation repression T cells homeostasis, proliferation, plasticity, lineage determination, and functionality (promote and dampen). Regulates B cells development and antibody production |
[258–268] |
| tRNA | 76–90 | RNA-seq, Northern blotting PCR In situ stain |
Total # of tRNAs: 429 # of tRNAs with introns: 28 |
http://gtrnadb.ucsc.edu | RNA pol III | 1/3rd of tRNAs are associated with MHCI on chromosome 6 | [269–271] | |
| tsRNA | 14–40 | RNA-seq, Northern blotting PCR In situ stain |
Total # of tRNAs generate tRFs: 321 Total # of tRFs: 28824 |
http://genome.bioch.virginia.edu/trfdb/ http://cm.jefferson.edu/MINTbase/ (For mitochondrial and nuclear tRNA fragments) |
tRNAs cleavage by angiogenin (ANG), Rny1p, Dicer and RNaseZ | [17,272–277] | ||
| piRNA | 24–33 | RNA-seq followed by piRNApredi ctor, Piano, Pibomd, piRPred IpiRId PCR In situ stain |
Total # of piRNAs: 35356 Total 89 clusters |
http://resulatorvrna.org/database/piRNA/
http://pirnabank.ibab.ac.in/stats.html https://www.pirnadb.org |
Ping-pong amplification | piR30840 (A snoRNA derived piRNA): Binds with IL-4 pre-mRNA intron and inhibit Th2 cells development | [18,278–288] | |
| snoRNA | 60–300 | RNA-seq PCR In situ stain |
Total # of snoRNAs: 2064 Intergenic: 1320 Intronic: 744 |
http://scottgroup.med.usherbrooke.ca/snoDB/
http://snoatlas.bioinf.uni-leipzig.de http://snoopv.med.mivazaki-u.ac.jp https://www-snorna.biotoul.fr/ |
RNA pol I, II and III followed by exonuclease activity of RNase III | [288–290] | ||
| Long ncRNAs | rRNA | 121–5070 | RNA-seq PCR In situ stain |
https://www.arb-silva.de
http://bioinformatics.psb.ugent.be/webtools/rRNA/ |
RNA pol I, RNA pol III | [291,292] | ||
| eRNA | 200 – 4000 | RNA-seq PCR In situ stain |
Total # of enhancers: 2534123 |
http://www.enhanceratlas.org
https://hanlab.uth.edu/HeRA/ http://gong_lab.hzau.edu.cn/Animal-eRNAdb/ |
RNA pol II | Regulates chromatin modification, Promotes enhancer-promoter interaction, genes transcription. Releases paused RNA pol II by acting as decoy for NELF. Promotes monocytes activation, TCR recombination. | [23,39,293–296] | |
| circRNA | <100 - >4000 | Divergent primer PCR, Northern blotting, Gel trap, 2D Gel electrophore sis, RNaseH, Exonuclease In situ stain |
Total # of circRNAs: 413657 Exonic: 252494 Intronic: 51291 Intergenic: 22557 5’ UTR: 754 3’ UTR: 1686 Non-repeat: 24896 Antisense: 39060 |
http://circatlas.biols.ac.cn/
http://www.circbase.org https://mioncocirc.github.io https://github.com/Xinglab/isoCirc |
RNA pol II followed by back splicing | Decoy, promotes autoimmune diseases (SLE, allergic rhinitis, MS, colitis, T1D, RA), Biomarker | [297–302] | |
| lincRNA/lncRNA | >200 | RNA-seq, RNA-FISH, Microarray, qRT-PCR In situ stain |
Total # of lncRNAs: 172216 Total # of lncRNA genes: 96308 |
http://www.noncode.org
http://biocc.hrbmu.edu.cn/LNCat/ http://lncrnadb.org/ http://bigd.big.ac.cn/lncbook https://www.monocldb.org/ https://ngdc.cncb.ac.cn/lncrnawiki1/index.php/Main_Page |
RNA pol II | Upregulates and downregulates cellular activation and differentiation. Promote and impair immune responses |
[71,303–305] | |
RT-qPCR: Reverse transcription qPCR; ddPCR: droplet digital PCR; SEXPAR: symmetric exponential amplification reaction; miRacles: miR-activated conditional looping of engineered switches. Some databases cover more than one type of ncRNA and are listed here: https://rnacentral.org/; https://nrdr.ncrnadatabases.org/.
Figure 1. Types of non-coding RNAs and schematic of Circular (circRNAs) and enhancer (eRNAs) RNAs.
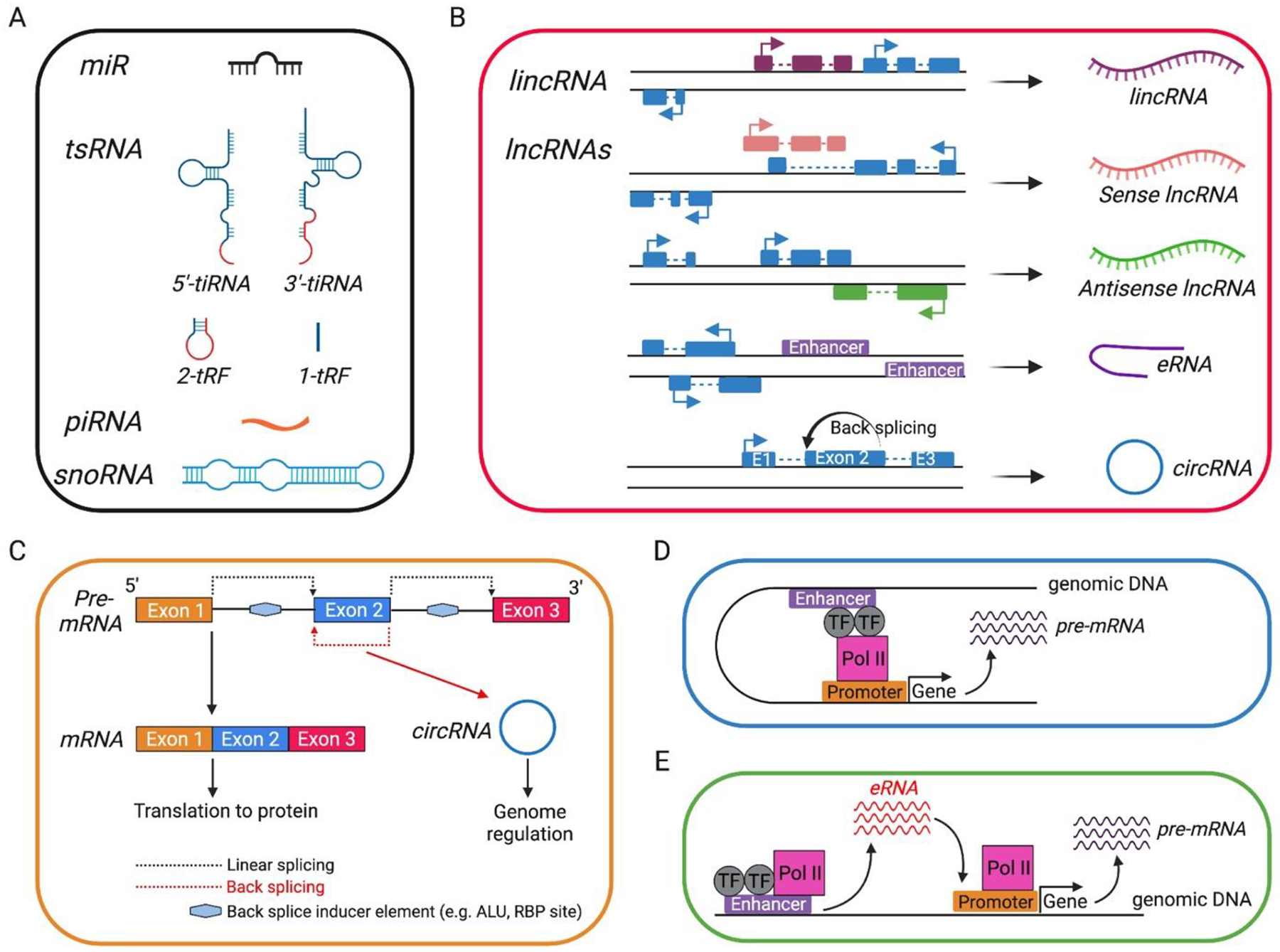
A, Small ncRNAs. B, Long ncRNAs. In B, coding exons are denoted as blue boxes. C, pre-mRNA (above) can be spliced in a linear fashion to generate mRNA (below left) or undergo back-splicing to generate circRNA (below right). D, conventional enhancers participate in DNA looping bringing transcription factors (TF) and polymerase II (Pol II) together with gene promoters to enhance gene transcription. E, some enhancers are themselves transcribed to non-coding RNAs (called eRNAs) that participate in loop formation to enhance gene transcription. For simplicity, loop formation has not been depicted in the cartoon in E. miR, micro-RNA; tsRNA, tRNA-derived small RNA (5’-tiRNA/3’-tiRNA, stress induced tRNA-derived RNAs from 5’ or 3’ ends of tRNA; 2-tRF, anti-codon loop cleaved by unknown ribonuclease; 1-tRF, generated by RNase Z in the 3’ trailer of tRNA); piRNA, piwi-interacting RNA, snoRNA, small nucleolar RNAs; lncRNA, linear long non-coding RNA; lincRNA, long intergenic non-coding RNA (lncRNAs that do not overlap with protein-coding genes); eRNA, enhancer RNA; circRNA, circular RNA.
Overall, many mechanisms of function have been ascribed to ncRNAs, including those that are specific to each class. Broadly, ncRNAs may regulate expression of other genes and basic cellular processes of cells. Some of these are generic to all cells, while others are specific to immune cells. These include regulation of cellular metabolism [26], polycomb repression [27], active-chromatin binding [28], gene transcription [29], alternative splicing [30], CTCF-associated class-switch recombination [31], ribosome activity [32,33], signal-transduction[34] and cytokine production [15,35]. Whereas miRs disrupt target molecule mRNA expression via the miR-induced silencing complex (miRISC) that includes argonaute-based nuclease machinery [36], long ncRNAs generally function differently, for example by acting as signals (i.e. transcriptional regulators), decoys (i.e. for miR binding and sponge activity), guides (i.e. directing localization of molecules) and molecular scaffolds (supporting multimeric complexes) [37]. Some ncRNAs integrate multiple functions. For example, HOTAIR, a regulator for anatomic specificity in anterior posterior differentiation, binds to the polycomb repressor complex (PRC) 2 and lysine-specific demethylase (LSD)1 complexes as a modular scaffold and targets PRC2 localization toward discrete genomic locales. Thus collectively HOTAIR regulates cellular position identity in complex tissues by regulating gene expression through mobilization of chromatin modifiers [37]. The most common mechanisms by which ncRNAs regulate cellular function are shown graphically in Figure 2.
Figure 2. Common mechanisms of action by which ncRNAs regulate cellular function.
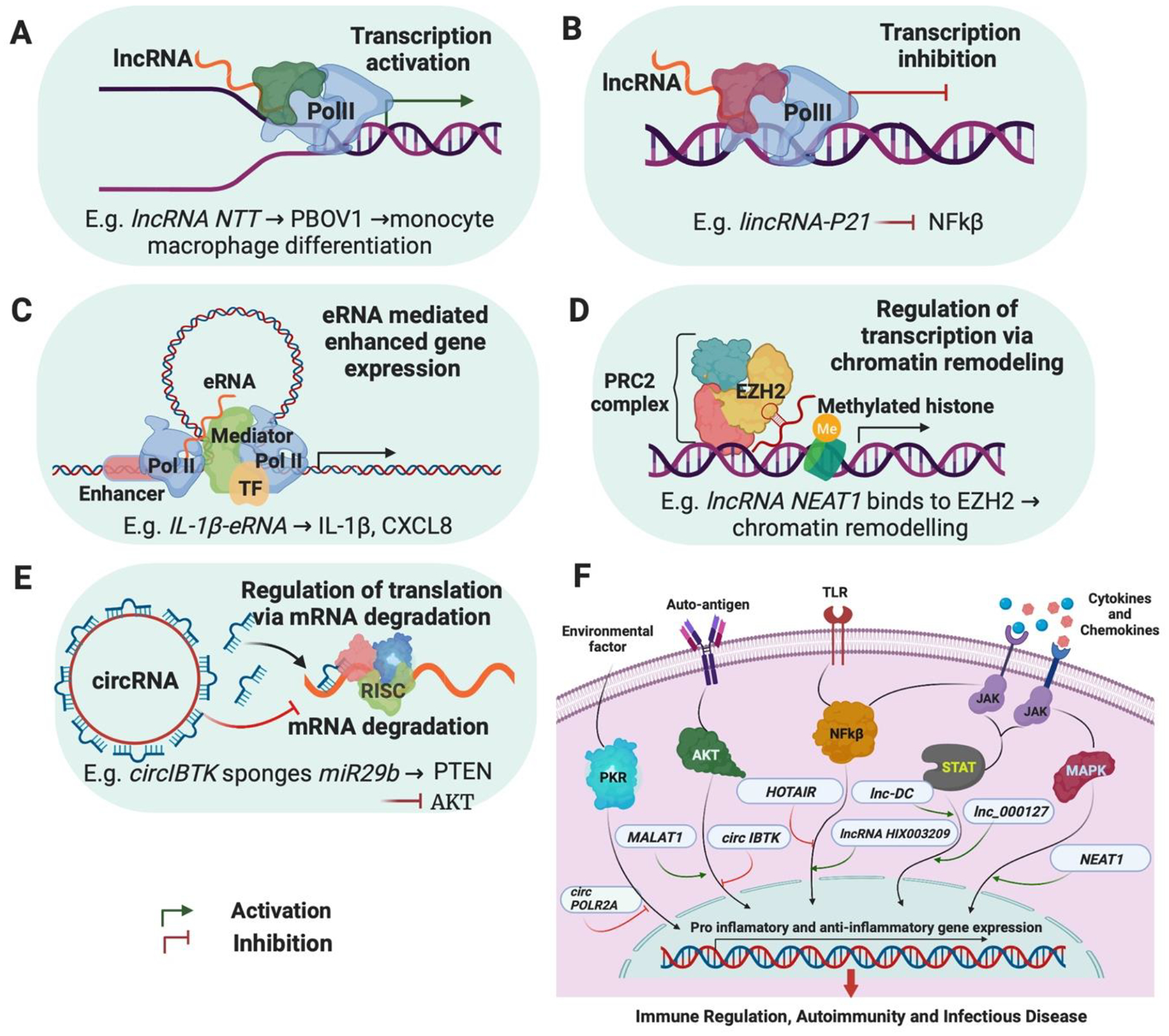
Shown in A-E are examples of how ncRNAs regulate cellular function, with exemplar genes included. A, activation of gene transcription. A common mechanism is physical interactions that facilitate recruitment of the transcriptional machinery to the promotors of target genes to enhance transcription. B, inhibition of gene transcription. Here, lncRNAs can act as decoys and bind to transcription factors to alter their recruitment or that of polymerase II to the promoter, resulting in transcriptional suppression. C, enhancement of gene transcription. eRNAs transcribed from enhancer elements commonly function to stabilize enhancer-promoter interactions via DNA looping and recruit transcription factors and polymerase II. D, chromatin remodeling. ncRNAs, for example lncRNAs, can interact with chromatin remodeling complex proteins, such as EZH2 and PRC2 (promoting polycomb repressor complexes), modulating histone modification and thereby gene expression. E, competing endogenous RNAs. Some ncRNAs, such as lncRNAs and circRNAs can act as sponges for miRs, thereby modulating the degradation of mRNAs that are the cognate targets of those miRs. F, many of these mechanisms can converge to regulate biological pathways in cells. Shown are just some of the cellular processes implicated in the pathogenesis of autoimmune diseases and how ncRNAs could impact pro- and anti-inflammatory gene expression programs in those pathways. TLR, Toll Like Receptor; PKR, Protein Kinase RNA; AKT, Protein Kinase B; NFκB, Nuclear Factor kappa-light-chain-enhancer of activated B cells; STAT, Signal Transducer and Activator of Transcription; MAPK, Mitogen-Activated Protein Kinase; JAK, Janus Kinase.
The focus of much of the literature has historically been on the study of lncRNAs and miRs. More recent attention has shifted to circRNAs and eRNAs. circRNAs and eRNAs are critically important ncRNAs increasingly recognized for regulating fundamental cellular processes. circRNAs are abundant and formed by back-splicing events distinguishing them from linear RNAs [38] (Figure 1C). eRNAs are non-coding RNAs transcribed from enhancer regions of DNA and regulate target gene expression [39–41] (Figure 1D–E). Both circRNAs and eRNAs are critical for mammalian gene regulation in cis and/or trans. Moreover, controlled over-expression or ablation of specific circRNAs and eRNAs can modulate biological functions of cells [42,43], providing a novel means to alter transcriptional activity of target genes. Because some of these, such as circRNAs, are relatively stable and long-lived, they also have the potential to act as biomarkers of disease or to be the targets of gene therapy.
Detection of ncRNAs in immune cells – old challenges and new
Conventionally, a significant portion of the knowledge gap that exists in the study of ncRNAs stemmed from limitations in ncRNA detection. At present, a variety of techniques exist to detect ncRNAs in cells and tissues under diverse immunological states. These range from prior knowledge-based techniques, such as qRT-PCR, to high-throughput methods that include bulk RNA-sequencing (RNAseq). While RNAseq is now standardized, it is important to point out that most experiments ‘capture’ RNAs that are 3’-polyadenylated. This is desirable to bias capture towards mRNAs and avoid reactions being saturated by rRNAs (the majority of cellular RNAs). However, because a large fraction of cellular ncRNAs is not poly-adenylated, this approach has low efficiency at capturing the non-coding transcriptome. An approach more suitable to whole transcriptome sequencing uses random priming combined with reagents that deplete ribosomal RNAs (‘ribo-depletion’) or enrich for RNA species of interest (e.g. RNase R treatment for circRNA enrichment). However, this approach is particularly susceptible to inefficiency if the quality of starting RNA is sub-optimal, as ribo-depletion of partially degraded RNA works poorly. It is also worth pointing out that data from whole transcriptome RNAseq still pose significant challenges when trying to discover novel ncRNAs, partly because many of the ncRNAs have low expression and discerning them from “transcriptional noise” requires large numbers of samples, increased sequencing depth and confirmation by orthogonal methods. An additional challenge is identifying the exact transcription start and end coordinates which would be needed for functional studies, such as overexpression or gene-editing. An additional computational challenge for circRNAs is that identifying exact back-splicing junctions requires sufficient sequencing reads through the junction to distinguish it from artifacts introduced during RNA-seq library preparation, such as random template switching [44]. Such depth of coverage can be difficult to achieve for low-expressed transcripts.
Other novel opportunities for identifying ncRNAs comes from the variety of methods now available for sequencing of nascent RNA molecules, such as methods that sequence transcripts without capture or library prep to enable high-throughput characterization of total nucleotides. These include uninterrupted long-range sequencing, global nuclear run-on sequencing (GRO-seq), precision run-on sequencing (PRO-seq), native elongating transcript sequencing (NET-seq), mammalian NET-seq (mNET-seq) and fastGRO [45–49]. Some of these approaches are gaining traction but are still dependent on input bulk RNA derived from multiple cells and generally measure actively transcribing genes rather than steady-state stable RNA levels.
Single cell RNA-seq (scRNAseq) is a contemporary approach to detect and quantify cell-state specific expression without the need of microdissection [50] or laser capture [51]. It theoretically enables the discrete analysis of cellular transcriptomes on a per cell basis and the determination of heterogeneity within a given population of cells. However, current scRNAseq methods are less than ideal for the study of ncRNAs principally for two reasons. Firstly, current scRNAseq platforms are susceptible to ‘drop-out’, a phenomenon by which a transcript is observed in one cell, often at low or moderate level of expression, but is not detected in another cell within the same population. Thus, because many ncRNA species are of low abundance (NeST, for example [see below] is estimated to be expressed at one molecule or less per T cell) or low expression, they are often subject to dropout within a population of identical cells. Secondly, many commercially available scRNAseq kits employ a reverse transcription (RT)-PCR reaction primed by polydT, which predominantly captures 3’-polyadenylated transcripts. This is fine for some lncRNAs that contain 3’polyA tails and can be captured by priming with polydT. For example, Zhou et al., recently conducted polydT-primed single-cell profiling of lncRNAs during hematopoiesis in vivo and produced an atlas of such ncRNAs during hematopoiesis [52]. Unfortunately, the lack of 3’polyA tails in many ncRNAs results in most of these species being ignored by the polydT-primed RT-PCR used in most routine scRNAseq techniques. Conversely, techniques that bias capture towards ncRNAs (e.g. RT-PCR primed with random hexamers) are limited by the opposite problem, namely that the majority of the PCR products are unwanted ribosomal RNAs, which saturate and dominate sequencing reactions, resulting in low depth of sequencing reads for detecting and quantifying desired ncRNAs. In theory, combining random priming with techniques that deplete unwanted rRNAs before or after RT-PCR could enable whole transcriptome assessment and more efficient measurement of ncRNAs at the single cell level, as they have in bulk RNAseq. However, protocols to achieve this aim and efficiently deplete rRNAs are not yet available. It is also conceivable that that innovation in methods for sequencing of nascent RNA molecules will capitalize on existing plate-based or oil-immersion approaches to perform nascent identification of RNAs and other molecules in singe cells at longer lengths.
Achieving single cell transcriptomic readout of cells is clearly an important advance in understanding biology in complex tissues. However, there are two clear limitations that need to be acknowledged. Firstly, many studies use cells from surrogate sites, such as cerebrospinal fluid or blood, to identify disturbed ncRNA expression. While hypothesis-generating, this approach most likely only hints at the exact ncRNAs regulating disease within affected tissues. Secondly, most immune reactions in complex diseases are spatially oriented and dependent on cell-to-cell interactions between cells. Examples include the processes occurring in the synovia of patients with rheumatoid arthritis, kidney tissues in lupus nephritis or brains of patients affected by neuro-immunological diseases, such as multiple sclerosis. Tissue disruption required for single cell methods, such as scRNAseq, by intention disrupts this information. This limitation necessitated the development of post-hoc computational methods, such as CellphoneDB [53], that infer and approximate the interactions that might have occurred in vivo in that tissue using existing receptor-ligand pair knowledge base. This approach is generally limited and somewhat tangential.
Spatial transcriptomics approaches have overcome some of the limitations of scRNAseq by probing tissue-level events and cell-to-cell contacts experimentally but have their own limitations. “On-slide” first generation spatial transcriptomic methods showed promise in this domain but were generally of low resolution and provided regional, rather than single cell, information. One reason for the low resolution is exemplified by the 10X Genomics Visium platform [54] that integrates existing scRNAseq approaches to capture cellular transcripts by arrays of barcoded mRNA-binding oligonucleotides tiled across glass histology slides. Inefficient capture of transcripts from individual cells, physical limitations of the number of barcoded oligonucleotides that can be tiled and deadspace between capture spots, results in low resolution and loss of spatial information. On the other hand, this approach is suitable for discovery-led approach as the method is not dependent on a prior knowledge and will sequence all RNAs captured. Recent improvements in spatial transcriptomic methods have pushed the boundaries in this domain. These second-generation methods have largely eschewed RNA-capture and sequencing in favor of probe-based methods that enable higher resolution of capture. Some of these technologies, such as multiplexed error-robust fluorescence in situ hybridization (MERFISH) [55] or the 10X Genomics Xenium [56] platform, now enable mapping of transcript expression at even sub-cellular resolutions. This depth of resolution holds great promise for revealing how expression of ncRNAs in single cells or within sub-cellular compartments of single cells, can influence biological processes in individual cells and across tissues. In fact, a recent paper combined MERFISH with sub-cellular imaging to show enrichment of ~10,000 coding and non-coding RNAs in organelles and nuclei within a given cell [57]. The major drawback of such second-generation spatial approaches is that they currently rely on limited numbers of probes/genes, which requires a priori knowledge of the target transcripts. This reduces the ability to make novel discoveries, such as unexpected expression of a ncRNA. A second major limitation is that a given cell type may not be represented, or represented at a very low frequency, within the plane of the tissue slice selected for probing. Thus, spatial transcriptomics are currently especially useful when combined with data from single cell or bulk RNAseq approaches (which have their inherent drawbacks, as discussed above) and for confirmation of data previously obtained using other methods.
Some of the detection techniques could also shed light on ncRNA function. For example, as discussed, one of the mechanisms of function of ncRNAs is RNA-RNA interactions. A number of methods currently exist to detect RNA-RNA interactions (Table 2). Briefly, they can be classified into low-throughput methods that probe specific individual interactions (‘one to one’) or high-throughput methods that identify a compendium of all interactions (‘one to many’ or ‘many to many’). Moreover, based on the method, the detected interactions can be either directly or indirectly inferred, as delineated in Table 2. RNA-FISH (Fluorescent in situ hybridization) [58] is an enhanced resolution low-throughput method to study probed RNA-RNA interactions that relies of Förster resonance energy transfer (FRET) [59]. Some methods, such as hiCLIP (hybrid and individual-nucleotide resolution UV cross-linking and immunoprecipitation) [60], MS2-TRAP (MS2-tagged RNA affinity purification) [61] and CLASH (cross-linking, ligation and sequencing of hybrids) [62], can be utilized to identify many target RNAs interacting with a previously defined RNA molecule of interest. More recent to the field, high-throughput methods, such as RIC-seq (RNA in situ conformation sequencing) [63], RNA proximity sequencing, PARIS (psoralen analysis of RNA interactions and structures) [64], SPLASH (sequencing of psoralen crosslinked, ligated and selected hybrids) [65], Liger-seq (LIGation of interacting RNA followed by high-throughput sequencing) [66] and MARIO (Mapping RNA interactome in vivo) [67], have been devised to map global RNA-RNA interactions. RIC-seq, in particular, has high sensitivity, relatively lower background and better coverage among all available high-throughput methods. Nevertheless, the resolution remains low, and advancements in the computational dissection and high-resolution visualization of RNA interactions in vitro and in vivo is still required. A relatively new method on the market is MERFISH. Although ostensibly a spatial transcriptomics solution, the enhanced (sub-cellular) resolution or MERFISH [55,57] enables it to function as a spatial method to probe 100s to 1000s of targets and provide a visual confirmation of interactions or co-localization, even amongst other labeled molecules. Nonetheless all of these methods, with the exception of RNA-FISH, require millions of cells (or tissues) for input, which is a real limitation when studying rare cell populations, such as immune cells derived from tissues of patients. Thus, there is an unmet need for technology development that can utilize low(er) cell numbers.
Table 2. Novel and Well-Defined Methods to detect RNA to RNA interactions.
Please note that besides RNA-FISH, all methods here require millions of cells at least.
| Type | Method | Direct or indirect? | Advantages | Disadvantages |
|---|---|---|---|---|
| One to one | RNA-FISH [58,306–309] | Direct | Single cell approach, visualizes target RNA-RNA interactions, can detect nascent and mature transcripts simultaneously. | Low-throughput, fixation req., permeabilization required, difficult to reveal if target sequence is masked by RBPs or any structure. |
| One to many | hiCLIP (hybrid and individual-nucleotide resolution UV cross-linking and immunoprecipitation) [60,310] | Indirect | Linker addition to make chimeric RNAs improves efficiency and read quality, low background, provides secondary structure of RNAs interacting with target RBPs. | High cell input, antibodies req. against RBPs, two rounds of immunoprecipitation and radiolabeling steps add to complexity, relatively laborious. |
| MS2-TRAP (MS2-tagged RNA affinity purification) [61,311] | Indirect | RNA-RBP interactions can be explored, may be utilized to visualize RNA-RNA/RBP interactions in vivo by replacing GST tag with fluorescent protein. | Require high cell input, laborious approach, lower coverage, RNA-tagging may interfere with RNAs interactions. | |
| CLASH (cross-linking, ligation and sequencing of hybrids) [62] | Indirect | One of the earliest established methods for RNA-RNA interactions, UV-cross linking reduces background signal. | High cell input, lower efficiency and quality, limited RNA-RNA interactions, dependency on antibody availability. | |
| Many to many | RIC-seq (RNA in situ conformation sequencing) [63] | Direct | Global mapping of RNA-RNA interactions, pCp-biotin incorporation enhances proximity ligation and read quality, better coverage and accuracy over all existing methods. | High cell inputs needed, formaldehyde crosslinking introduces some background signal and lower nucleotide resolution. |
| RNA proximity sequencing [64] | Direct | Global 3D-spatial organization of RNAs in situ, can distinguish RNA-dense from sparse regions, reveals nascent and mature RNAs, groups colocalization in sub-nucleolar space, can monitor the speed of RNA-Pol elongation and limits of alternative splicing. | Higher cell inputs needed, requires optimal dilution of single-random barcoded beads and target cells, complex and time-consuming approach. | |
| PARIS (psoralen analysis of RNA interactions and structures) [65] | Direct | Uses psoralen a nucleotide crosslinker for interacting RNAs, can provide a global map of RNA-RNA interactions. | High cell inputs, complex and laborious method, precise and controlled RNase digestion required, lower coverage and resolution. | |
| SPLASH (sequencing of psoralen crosslinked, ligated and selected hybrids) [65,312] | Indirect | Biotin-psoralen intercalation in interacting RNAs provides combinatorial method for crosslinking and fragments enrichment, provides inter and intra molecular RNA-RNA interaction map. | High cell inputs, lower yield, relatively large background, low resolution. | |
| LIGR-seq (LIGation of interacting RNA followed by high-throughput sequencing) [66] | Direct | Crosslinking of interacting RNAs by intercalating psoralen derivative 4’-aminomethyltrioxalen (AMT), followed by interacting RNA circularization and RNaseR treatment enriches target RNAs, can provide global map of RNA interactome. | Requires large numbers of cells, lower coverage, low resolution. | |
| MARIO (Mapping RNA interactome in vivo) [67] | Indirect | UV-cross linking for reduced background signal, linker enhances proximity ligation, cross-linking can facilitate the study of closely associated RBP-RNA interactions. | High cell input, partially in vivo method, complex and laborious. | |
| MERFISH (Multiplexed error-robust fluorescence in situ hybridization) [55], Xenium [56] | Direct | Single cell resolution identifies broad range of RNAs with subcellular compartmentalization, able to identify cell-cycle stage-associated transcriptional changes. | Requires target RNAs information to design probes (non-discovery led approach), necessitates specific imaging platform, complex and laborious method (probe-design to imaging and decoding) |
Known functions of ncRNAs in immune cells
Among early examples of ncRNAs regulating mammalian immunity was the discovery by the Bartel group that miRs regulate mouse hematopoiesis [68]. ncRNAs have since been found to be involved in many immunological processes and participate in the development and function of both innate and adaptive immune cells and are particularly highly expressed in specific cell types. Examples include GAS5, which promotes macrophage and microglial polarization [69,70] and inhibits Th17-cell differentiation [71]; lnc-DC, which promotes monocyte to dendritic cell differentiation [72]; NeST, which regulates IFN-γ production in both Th1 and Natural Killer (NK) cells [73,74]; linc-MAF-4, which guides Th1 specification [75]; lincR-Ccr2-5′-AS, which regulates T cell development [76];-ncRNAs that direct CD8+ T cell specification [77]; the TAD-associated (topologically-associated domain) enhancer lncCSRIgA, which regulates IgA repertoire diversity in B cells [78]; and MALAT1, which sponges miR-155 for polarizing dendritic cells and Tregs [79]. Figure 3 shows some of the well-characterized ncRNAs associated with specificity for hematopoietic cells at different stages of lineage differentiation and maturation and Figure 4 shows some of the molecular functions controlled by these. A more comprehensive list of ncRNAs regulating function in different immune cell types is shown in Table 3. Although eRNAs and circRNAs have, in general, been less studied in immune cells, accumulating evidence indicates that both classes of RNA play a critical role in immune responses, particularly in cells of the myeloid and lymphoid lineages [3,80]. Some are transcribed especially in response to sensing of danger signals, including a number of eRNAs, transcribed from the IL1B, SOCS3, TNFSF8, SLC30A4, MARCKS, AZIN1 and ACSL1 loci in myeloid cells activated by lipopolysaccharide (via, for example, mitogen activated (MAP) kinase and NF-κB signaling)[81,82]. Some of these induce the expression of pro-inflammatory cytokines and chemokines, notably IL-1β and CXCL8 [81]. Likewise, in lymphoid cells, regulation of T cell receptor (TCR) recombination in both αβ- and γδ-T cell development are dependent on eRNAs derived from the TCRA and TCRB gene loci [83,84]. Similarly, expanding evidence suggests a diverse role played by circRNAs in regulating the differentiation of both adaptive and innate immune cells (circNUP214: Th17 cells; circKcnt2, circTmem241: ILC3; circSnx5: DCs; circHIPK3: macrophage) and polarization (M1 and M2 macrophage polarization by circCdyl and has_circ_0005, respectively) [85–89].
Figure 3. Well-characterized ncRNAs associated with specific hematopoietic lineage cells.
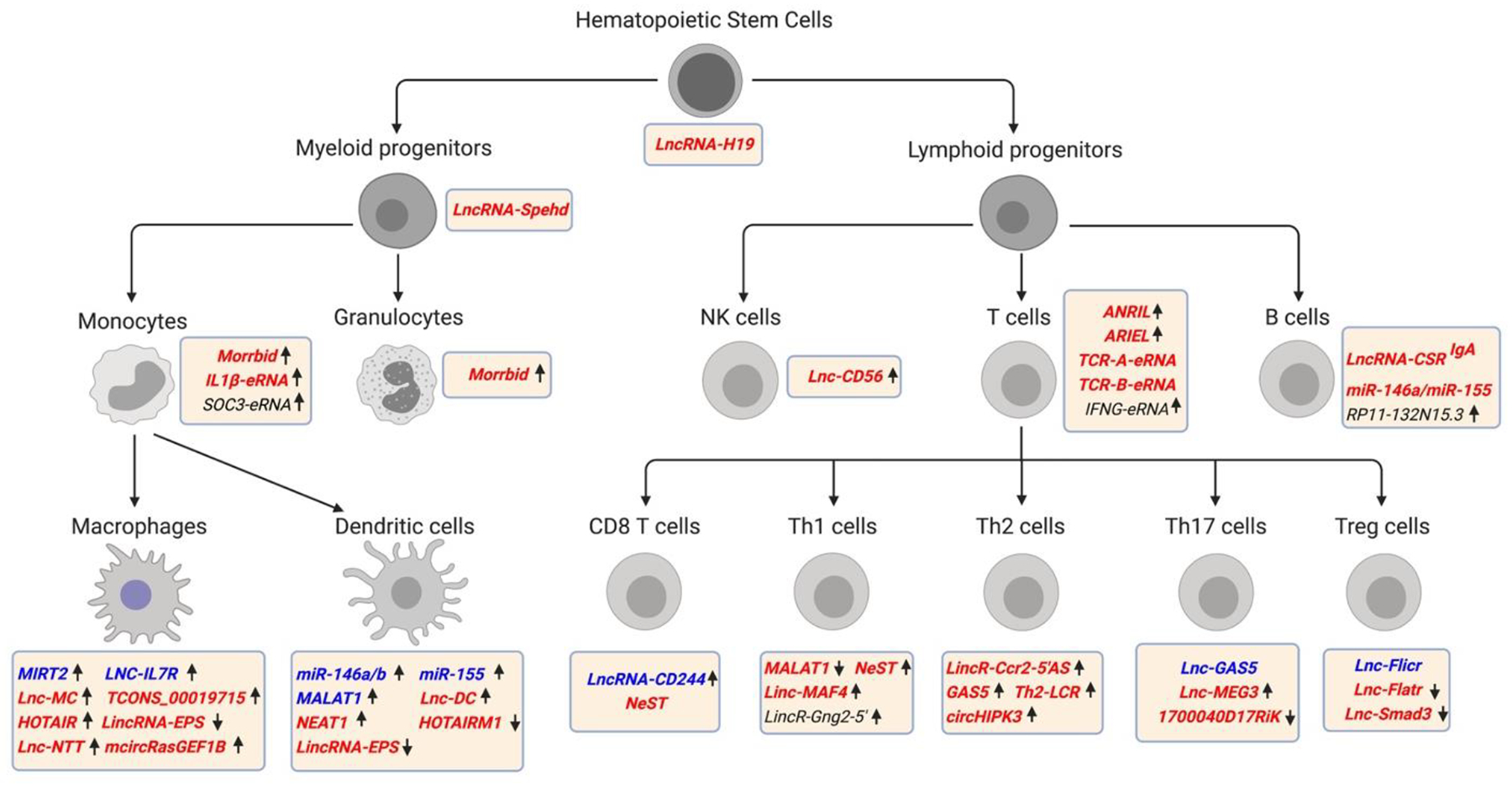
Expression of labeled ncRNAs in different immune cell-types are marked. Arrows indicate expression of ncRNAs in cells: up-arrow, ncRNA upregulated; down-arrow, ncRNA downregulated. Colors represent functions of the indicated ncRNA: blue, impairs cell differentiation and/or function; red, induces cell differentiation and/or function; black, inconclusive effect on differentiation and/or function.
Figure 4. Immune cell ncRNA regulatory circuits.
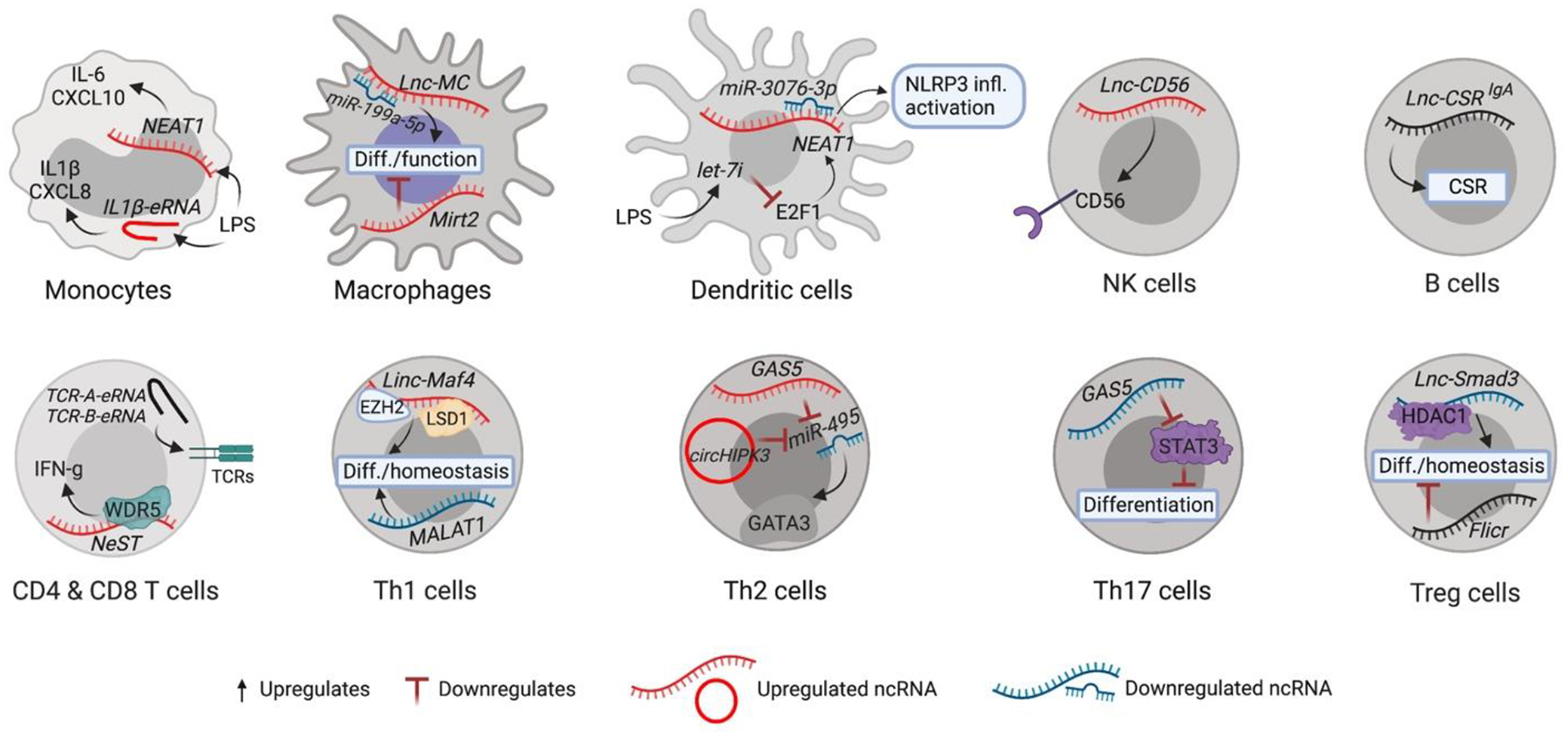
Examples of known effects of ncRNAs in diverse myeloid and lymphoid cell types. Putative mechanism of regulation is shown for each cell type indicated. Details of these mechanisms (and those of other ncRNAs) are included in Table 3. CSR, class switch recombination; LPS, lipopolysaccharide; Infl., inflammasome; LSD1, lysine-specific demethylase 1; EZH2, enhancer of zeste homolog 2; WDR, tryptophan-aspartic (WD) repeat subunits; Diff., differentiation.
Table 3.
Described functions of ncRNAs in immune cells.
| Immune cells | Type of ncRNA | ncRNA | Expression level | Role | Detection Method | Ref |
|---|---|---|---|---|---|---|
| DCs | miR | miR-146a, miR-146b | Upregulated | Promotes apoptosis, impairs pro-inflammatory cytokines production via diminished TRAF6 and IRAK1 expression | Microarray | [313] |
| lncRNA | HOTAIRM1 | Downregulated | Promotes monocytes to dendritic cells differentiation via competitive binding to miR-3960 | qRT-PCR, ChIP-chip assay | [314] | |
| Lnc-DC (LOC645638) | Upregulated | Promotes DCs differentiation via direct interaction with STAT3 through phosphorylation on Tyr-705, prevents SHP1 mediated STAT3 dephosphorylation | Microarray & RNA-seq | [72] | ||
| Malat1 | Upregulated | Promotes tolerogenic DCs (via lowered CD80, CD86 and MHCII) by inducing DC-SIGN expression through sponging miR-155, induces Treg expansion | Microarray, qRT-PCR and ChIP assay | [79] | ||
| NEAT1 | Upregulated | Promotes DCs maturation, enhances NLRP3 induced inflammasome via direct binding with miR-3076p (a negative regulator of NLRP3) | RNA-seq | [124] | ||
| circRNA | circSnx5 | N/A | Ectopic expression of circSxn5 induces tolerogenic DCs, impairs T cell response, acts as a sponge for miR-544 to inhibit SOCS1 expression as well as impairs the nuclear translocation of PU.1, negatively regulating DC activation | Overexpression and KO experiments | [315] | |
| DCs and Macrophages | lncRNA | lincRNA-EPS | Downregulated | Associated with enhanced pro-inflammatory cytokines (IL1B, IL6 and TNF), iNOS and nitric oxide production | RNA-seq | [316,317] |
| moDCs | miR | miR-155 | Upregulated | Dampens pro-inflammatory cytokine production | Microarray | [318] |
| Macrophages | lncRNA | HOTAIR | Upregulated | Promotes proinflammatory responses (enhanced IL-6 and iNOS expression) by NF-kB activation via IkBalpha degradation | qRT-PCR, ChIP assay | [319] |
| Lnc-IL7R | Upregulated | Dampens pro-inflammatory responses via association with increased H3K27me3 deposition at promoters of VCAM-1 and E-selectin | Microarray, qRT-PCR | [320] | ||
| Lnc-MC (lnc monocytic RNA) | Upregulated | Sequesters miR-199a-5p and enhances ACVR1B expression promoting monocyte/macrophage differentiation | RNA-seq and PU. 1 ChIP-seq | [321] | ||
| lncRNA-KCNQ1OT1 | Downregulated | Polarizes M1 macrophages to M2 macrophages, directly binds miR-21a-5p | qRT-PCR | [322] | ||
| Mirt2 | Upregulated | Impairs inflammatory responses, attenuates TRAF6 oligomerization and K63 mediated autoubiquitination inhibiting NF-kB and MAPK pathways to dampen inflammatory molecules (e.g. TNF, IL1B, IL6 and IL12) | Microarray, qRT-PCR | [323] | ||
| TCONS_00019715 | Up in M1 and down in M2 | Regulates M1/M2 switching, Promotes M1 macrophage differentiation | Microarray | [69] | ||
| PACER | Upregulated | Decoys p50 homodimer, a repressive subunit of NF-kB, promoting COX-2 expression | ChIP assay | [324] | ||
| circRNA | circCdyl | Upregulated | Induces the polarization of M1 macrophages by inhibiting the entry of interferon regulatory factor 4 (IRF4) into the nucleus. Sponge for let-7c | Microarray | [88] | |
| circHIPK3 | Upregulated | Induces Macrophage activation via sponging miR-192 and miR-561, in turn upregulating NLRP3 inflammasome and TLR4 expression. | qRT-PCR | [325] | ||
| circRNA-003424, circRNA-001489, circRNA-018127, circRNA-013630 | Downregulated | Down in M1 polarized compared to M2 macrophages, mechanism not established | Microarray | [326] | ||
| circRNA-010056, circRNA-003780, circRNA-010231 | Upregulated | Up in M1 polarized compared to M2 macrophages, mechanism not established | Microarray | [326] | ||
| circPPM1F | Upregulated | Found to be highly upregulated in type 1 diabetes mellitus patients, associated with LPS-induced M1 macrophage by inducing NF-κB pathway | Microarray | [327] | ||
| hsa_circ_0005567 | Downregulated | Ectopic expression of this circRNA induces M2 macrophage polarization, sponge for miR-492 to prevent SOCS2 loss, associated with inhibited osteoarthritis progression | qRT-PCR | [89] | ||
| mcircRasGEF1B | Upregulated | Promotes immune responses via stabilizing ICAM-1 mRNAs | RNA-seq | [328] | ||
| Monocytes | lncRNA | NEAT1 | Upregulated | Promote pro-inflammatory molecules (IL6, CXCL10, CXCL11, CCL2 and CCL8) expression | qRT-PCR | [99] |
| eRNA | IL1B-eRNA | Upregulated | Enhances pro-inflammatory responses, induces IL1B and CXCL8 expression | RNA-seq | [81] | |
| Monocytic cells (THP1) | eRNA | SOC3-eRNA, TNFSF8-eRNA, SLC30A4-eRNA, MARCKS-eRNA, AZIN1-eRNA, ACSL1-eRNA | Upregulated | Upregulated in LPS induced THP1 cells | qRT-PCR, ChIP assay | [82] |
| Monocytes, macrophages and THP1 cells | lncRNA | NTT | Upregulated in RA patients | Upregulates PBOV-1 expression via hnRNP-U binding, NTT overexpression promotes cell cycle arrest, macrophage differentiation and CXCL10 expression | ChIP-PCR, qRT-PCR | [171] |
| Neutrophils, eosinophils and Monocytes | lncRNA | Morrbid | Up: Neu>Eos>Mon | Required for myeloid cell survival by repressing Bcl2l11 (Bim) expression in cis | RNA-seq, Chip-seq, FISH and qRT-PCR studies | [329] |
| NK cells | lncRNA | Lnc-CD56 (AB128931) | Upregulated | Development, positively regulates CD56 expression in human NK cells | Microarray | [330] |
| ILC3 | circRNA | circKcnt2 | Upregulated | Associated with impaired ILC3 activation during colitis resolution, assembles the nucleosome remodeling deacetylase (NuRD) complex onto the Batf promoter to reduce Batf expression, suppressing IL17 expression and ILC3 activation | Microarray | [86] |
| circTmem241 | Upregulated | Induces ILC3 differentiation via recruiting histone methyltransferase ASH1 on Elk3 promoter to initiate Elk3 transcription | Microarray, qRT-PCR | [87] | ||
| B cells | miR | miR-146a | Basal level | Required for marginal zone (MZ) B cells development, directly interacts with Numb (a negative regulator of Notch2) to maintain MZ B cells | RNA-seq, qRT-PCR | [331] |
| miR-155 | N/A | Regulates IgG class-switched plasma cells development via direct inhibition of transcription factor Pu.1, maintains memory response | Northern blot, microarray and KO studies | [332] | ||
| lncRNA | lncRNA-CSR IgA | N/A | Required for class switch recombination (CSR) to IgA in B cells of the Peyer’s patches | High-throughput chromosome capture (Hi-C) | [78] | |
| RP11–132N15.3 | Upregulation | Mechanism and function not established | Microarray | [333] | ||
| T cells (Jurkat cells) | eRNA | ARIEL/XLOC 005968 | Upregulated | Essential in T-cell leukemia cells progression, recruits Mediator Complex proteins to the ARID5B enhancer, promotes enhancer-promoter looping and ARID5B expression thereby promotes MYC oncogene and TAL1-induced transcriptional machinery | RNA-seq, qRT-PCR and ChIP-seq | [115] |
| T cells | lncRNA | ANRIL | Upregulated | Promotes T-cell leukemia cells proliferation, activate NF-kB pathway via ANRIL/EZH2/p65 ternary complex | ChIP-qPCR and KD studies | [334] |
| GAS5 | N/A | Induces apoptosis and impedes cell cycle | KD and overexpression studies | [335] | ||
| NRON | N/A | Represses NFAT activity via sequestering phosphorylated NFAT in the cytoplasm of unactivated T cells | qRT-PCR, shRNA-based study | [336,337] | ||
| eRNA-IFNG | Upregulated | Promotes Uveitis | qRT-PCR | [114] | ||
| eRNA | TCR-A eRNA | Basal level | Required for alphabeta and gammadelta T cells development, promotes V alpha to J alpha rearrangement in alphabeta T cells, required for VdeltaDJdelta and J alpha transcript in gammadelta T cells | Mutation | [84] | |
| TCR-B eRNA | Basal level | Required for alphabeta T cells development, mediates V(D)J recombination during alphabeta T cells development | Deletion | [83] | ||
| CD8 T cells | lncRNA | lncRNA-CD244 (lncRNA-BC050410) | Upregulated | Inhibits CD8 T cell responses (reduces IFN-γ and TNF expression) in TB patients via EZH2 recruitment to ifng and tnfa promoter | Microarray, qRT-PCR, ChIP-qPCR, RIP-qPCR | [338] |
| NeST | N/A | Binds with WDR5, a component of H3K4 methyltransferase complex and promotes IFN-γ production | Transgenic expression studies | [339] | ||
| circRNA | circRNA100783 | Differential exp | Precise role unknown | Microarray | [340] | |
| CD4 T cells | miR | miR-126a-5p | Upregulated | Induces Th1 phenotype via promoting Notch-1 signaling in the Echinococcus granulosus (a cause of cystic echinococcosis disease) infected mice | qRT-PCR | [341] |
| miR-181a-5p | Downregulated | miR-181a-5p overexpression in allergic rhinitis (AR) model impedes the binding of RAGE and HMGB1, thereby alleviates Th17/Treg imbalance to block asthma development from AR | qRT-PCR | [342] | ||
| lncRNA | linc01882 | Downregulated | Downregulated in anti-CD3 and anti-CD28 mediated CD4 T cell activation | RNA-seq, qRT-PCR and KD studies | [343] | |
| lncSnhg7 | Upregulated | Upregulated in the offspring exposed to cadmium during gestation | RNAseq | [344] | ||
| XIST | Upregulated | Induces proliferation and differentiation of CD4 T cells in the PBMCs of primary biliary cholangitis (PBC) patients | qRT-PCR | [345] | ||
| lincRNA | lincRNA00892 | Upregulated | Promotes CD4 T cell differentiation by inducing CD40L protein expression (minimal impact on mRNA levels), augments B cell activation and IgG secretion | Microarray, qRT-PCR | [346] | |
| circRNA | circ003912 | Upregulated | Sponge of miR-31, miR-1231 and miR-647, associated with enhanced expression of miR-146a and Foxp3 | Microarray, qRT-PCR | [347] | |
| Th1 cells | lncRNA | linc-MAF-4 | Upregulated | Maintains Th1 cells homeostasis, recruits chromatin modifiers (EZH2 and LSD1) with MAF, dampens Th2 differentiation | RNA-seq, qRT-PCR | [96] |
| lincR-Gng2-5′ | Upregulated | Positively regulates STAT4 levels during Th1 differentiation | RNA-seq | [76] | ||
| NeST | Upregulated | Required for IFN-γ production | RNA-seq | [348] | ||
| RP11–552114.1, RP11–37L2.1, linc-MYNN-7, linc-C14orf-up-AS_1, RP11–489018.1, linc-CST7–2, linc-TMEM99–2, linc-LRRC49–4 | Upregulated | Precise role unknown | RNA-seq | [96] | ||
| RUNX1-IT1 | Upregulated | Promotes Th1 differentiation in the peripheral blood of Graves’ disease patients by regulating Tbet | qRT-PCR | [349] | ||
| Th1 cells, Th2 cells | lncRNA | MALAT1 | Downregulated | MALAT1 downregulation leads to reduced Maf4 expression (a required TF for IL-10 expression) and thereby promotes Th1 and Th2 differentiation by dampening anti-inflammatory cytokines | RNA-seq, qRT-PCR | [143] |
| Th2 cells | lncRNA | GAS5 | Upregulated | Promotes Th2 differentiation by inducing GATA3 expression via miR-495 sponging | qRT-PCR | [137] |
| GATA3-AS1 | Upregulated | Up in effector Th2 cells | qRT-PCR | [137,350] | ||
| lincR-Ccr2-5′AS | Upregulated | Promotes migration to the target site | RNA-seq | [76] | ||
| lncRNA FR215775 | Upregulated | Induces Th2 differentiation in murine allergic rhinitis | qRT-PCR | [351] | ||
| lincR-Epas1-3′AS | Upregulated | Positively regulated with STAT6 expression in Th2 differentiation | RNA-seq | [76] | ||
| LincRNA | Th2-LCR (AC004041.2) | Upregulated | Required for Th2 cytokine production. Interacts with WDR5 and hnRNPs | RNA-seq | [350] | |
| circRNA | circHIPK3 | Upregulated | Promotes differentiation by upregulating GATA3 expression via sponging miR-495 | qRT-PCR | [137] | |
| circPVT1 | Upregulated | Function and mechanism not established | qRT-PCR | [137] | ||
| Th17 cells | lncRNA | 1700040D17Rik | Downregulated | Promotes differentiation in SLE, biomarker | Microarray, qRT-PCR | [352] |
| GAS5 | Downregulated | Promotes differentiation in ITP, regulates TRAF6 mediated STAT3 ubiquitination thereby ROR-γt and IL17 production | qRT-PCR | [71] | ||
| GAS5 | Downregulated | Reduced expression of GAS5 (act as a sponge for miR-23a) in myasthenia gravis patients induces Th17 phenotype | qRT-PCR | [353] | ||
| MEG3 | Upregulated | Sponges miR-17 (a repressing molecule for ROR-γt) thereby promoting Th17 differentiation and function in asthma patients | Microarray | [354,355] | ||
| lincRNA | LINC01588 | Upregulated | Regulates Treg/Th17 cell balance via peroxisome proliferator-activated receptor (PPAR) signaling pathway | RNA-seq, FISH | [356] | |
| MIAT | Upregulated | During the development of human Th17 cells STAT3 binds at the promoter of MIAT to induce MIAT expression, ultimately enhanced expression of MIAT induces IL17A expression through PKCa | RNA-seq, qRT-PCR | [357] | ||
| circRNA | circINPP4B | Upregulated | Induces Th17 differentiation and EAE progression, sponges miR-30a | Microarray | [253] | |
| circNUP214 | Upregulated | Induces IL-23R and IL-17A expression in RA patients, sponge for miR-125a-3p | qRT-PCR | [85] | ||
| Treg cells | lncRNA | Flatr | Upregulated | No effect on thymic Treg cells generation, anticipates iTreg cells differentiation | RNA-seq | [358] |
| Flicr | N/A | Dampens Foxp3 expression and destabilizes Foxp3 expression, limiting IL-2 concentration | RNA-seq and KO studies | [21] | ||
| lnc-Smad3 | Downregulated | Reduced level associated with enhanced iTreg generation, associates with HDAC1 to reduce Smad3 (a required molecule for iTreg conversion) expression | KD and overexpression studies | [359] | ||
| HSC | lncRNA | H19 | N/A | Required for HSC development from endothelium, regulates demethylation of various critical TFs (Runx1 and Spi1) required for hematopoiesis | Single cell RNA-seq, qRT-PCR | [52] |
| Common myeloid progenitor | lncRNA | Spehd | N/A | Maintains oxidative phosphorylation and multi-lineage differentiation | RNA-seq, qRT-PCR, RNA, smRNA-FISH | [360] |
DC-SIGN: Dendritic cell-specific intercellular adhesion molecule-3 grabbing nonintegrin; NTT: Noncoding Transcript in T Cells; PACER: P50-Associated COX-2 Extragenic RNA; Lnc-MC: long non-coding; monocytic RNA; Morrbid: myeloid RNA regulator of Bim-induced death; Malat1: metastasis-associated lung adenocarcinoma; transcript 1; ACVR1B: activin A receptor type 1B (ACVR1B; ARIEL: ARID5B-inducing enhancer associated long noncoding RNA; NRON: Non-coding repressor of NFAT; NFAT: nuclear factor of activated T cells; TH2-LCR: Th2 locus control region; ITP: Immune thrombocytopenia; Flicr: Foxp3 long intergenic noncoding RNA.
Broadly, ncRNAs have been described as accessory immunoregulatory nodes in the regulation of key processes, including regulation of interferons [90], cellular damage resolution [91], inflammasome regulation [92], viral interaction [93], cell growth [94], wound-healing [95] and tissue-repair [26]. While much informative work can be done by detecting and functionally assessing ncRNAs in sorted cells from non-inflammatory sites, such as understanding hematopoietic or immune cell differentiation [76,96], one of the challenges facing the ncRNA field in the context of immunity is that multiple cells work in co-operation and are often spatio-temporally organized. As most immunological responses are characterized by the interactions of many cell types and/or pathogens acting in tandem, experiments directly validating cell-type specific ncRNA expression and function amongst multiple cell types and states in the same reaction is limited in the current literature. Single cell identification of ncRNAs (as discussed earlier) or microscopic analysis of ncRNAs in hematopoiesis, thymic development, lymph nodes or in tissues, such as in spleen or in lungs, are largely absent in the literature. This creates a barrier for truly establishing cell-type specific roles of specific ncRNAs in most autoimmune and pathogen related datasets, which, as discussed, mostly opt for bulk tissue instead of isolated cells.
ncRNAs in immune related diseases
ncRNAs have been experimentally described in several autoimmune diseases, including Sjögren’s [97], inflammatory bowel disease (IBD) [98], systemic lupus erythematosus (SLE) [99,100], rheumatoid arthritis (RA) [101,102], psoriasis [103], psoriatic arthritis [104], uveitis [105,106], Behçet’s [107], multiple sclerosis (MS) [108], lupus nephritis (LN) [109], myasthenia gravis [110], autoimmune hemolytic anemia [111], immune thrombocytopenia [71] and in other immune-related conditions such as hyperinflammatory lung disease [112], and dyskeratosis congenita [113]. Indeed, the role of ncRNAs in human diseases may be much broader than currently thought and possibly context-dependent. For example, alongside their known roles in immune cell development, eRNAs such as ARIEL (ARID5B-inducing enhancer associated long noncoding RNA) and eRNA-IFNG are also associated with progression of T-cell leukemia in patients with T-ALL and uveitis (Table 3) [114,115]. Moreover, some pathogens produce ncRNAs that assist their propagation and/or evasion of host immunity, for example Epstein-Barr virus-encoded miRs [116]. Some of these studies are, however, primarily based on in silico observations, such as the identification of SARS-CoV-2 miRs that are predicted to interact with transcriptional co-activator subunits and STAT1 [117], rather than via validation experiments, such as those that are still required to detect and test these interactions between SARS-CoV-2 predicted ncRNAs and host cells.
Searching PubMed reveals a plethora of human ncRNAs associated with autoimmune diseases, several being associated with multiple diseases including small ncRNAs miR-21, miR-146a, miR-155, and lncRNAs NEAT1, GAS5, MEG3, MALAT1, TUG1, PRINS, HOTAIR, lnc-DC, hsa_circ_0044235, and circCAMSAP1. These are summarized in Tables 4–7. Of these autoimmune disease-associated ncRNAs, most are specifically expressed in immune cells, implying that they are likely to play a role in the etiology or pathogenesis of these diseases [118]. It is possible, of course, that the functions of ncRNAs differ from cell to cell (e.g. between T and B cells) or between cell states (e.g. resting vs activated T cells, or between Th1 and Th2 cells). Of note are three lncRNAs, NEAT1, GAS5, and MALAT1 that demonstrate the diversity of immunoregulatory ncRNA circuits acting amongst cell types in autoimmunity. These are further discussed below.
Table 4.
Discrete ncRNAs associated with rheumatoid arthritis.
| Class of ncRNA | ncRNA | Cells/Tissue | Expression (up or down) in disease | Role | Detection method | Refs |
|---|---|---|---|---|---|---|
| lncRNA |
ENST00000572491
ENST00000563752 ENST00000569543 |
PBMCs, CD4 | Up | Biomarkers | Microarray | [361] |
| FAM66C | PBMC | Associated in GWAS with RA | GWAS | [362] | ||
| GAS5 | T cells | Up | Biomarker | qRT-PCR | [363] | |
| HIX003209 | PBMCs Myeloid cells | Up | Overexpression of HIX003209 increased THP1 cell proliferation and proinflammatory cytokine secretion | qRT-PCR | [169] | |
| HOTAIR | PBMCs, Serum exosomes | Up | Forced upregulation of HOTAIR in synoviocytes and osteoclasts decreases MMP-2 and MMP-13 activation | Microarray | [167] | |
| Serum | Up | Biomarker, upregulated in serum of patients | qRT-PCR | [166] | ||
| HOXA3as | Synoviocytes and osteoclasts | Down | Biomarker | Microarray | [167] | |
| LINC00638 | PBMCs | Down | Activation of Nrf2/HO-1 pathway | qRT-PCR | [364] | |
| lincRNA-p21 | PBMCs | Down | Reduced expression of lincRNA-p21 increases NFκB activity | qRT-PCR | [365] | |
|
LOC100506036
LOC100652951 |
T cells | Up | Knockdown in Jurkat cells linked with decreased IFN-γ and NFAT expression during PMA/Ionomycin treatment | qRT-PCR | [172] | |
| Lnc-Cox2 | Serum | Up | Biomarker, upregulated in serum of patients | qRT-PCR | [166] | |
|
LUST
MEG9 SNHG4 |
Exosomes | Up | Biomarkers | Microarray | [167] | |
| MALAT1 | PBMCs | Down | MALAT1 SNPs rs619586 and rs3200401 were, interestingly, not associated with RA susceptibility | qRT-PCR | [366] | |
| MEG3 | Fibroblast like Synoviocytes (FLS) | Down | MEG3 knockdown resulted in activation of STAT3, PI3K/AKT pathway, FLS proliferation, IL-6 and IL-8 secretion | qRT-PCR | [367] | |
| NEAT1 | PBMCs | Up | Th17 differentiation | qRT-PCR | [125] | |
| NTT | PBMCs | Up | Monocyte macrophage differentiation, Cytokine secretion | qRT-PCR | [171] | |
| PRINS | Exosomes | Down | Biomarkers | Microarray | [167] | |
|
RNA143598
RNA143596 HIX003209 IGHCgamma1 XLOC_002730 |
Serum | Up | Correlation with disease parameters observed. Not functionally assessed | Microarray | [168] | |
| THRIL | T cells | Up | Biomarker | qRT-PCR | [363] | |
| TUG1 | Exosomes | Up | Biomarker | Microarray | [167] | |
| circRNA | circHIPK3 | Serum | Up | Sponging of miR-124a | qRT-PCR | [368] |
| circNUP214 | PBMCs | Up | Biomarker, promotes Th17 cells through IL23R | qRT-PCR | [85] | |
| circ_000836 | Fibroblast like Synoviocytes (FLS) | Down | Represses histone deacetylase 4 by sponging miR-135b-5p to limit proliferation and production of IL-1β, IL-6, and TNF | qRT-PCR | [181] | |
|
circRNA_104871
circRNA 003524 circRNA 101873 circRNA_103047 |
PBMCs | Up | Biomarkers | Microarray | [369] | |
| ciRS-7/CDR1as | PBMCs | Up | Biomarker, may inhibit miR-7 and upregulate mTOR | qRT-PCR | [174] | |
| hsa_circ_0001859 | Synovium SW982 cells | Not shown | Competing endogenous RNA for miR-204/211. Knockdown of hsa circ 0001859 decreased expression of ATF2, c-Jun, c-Fos | qRT-PCR | [177] | |
| hsa_circ_0044235 | Peripheral blood | Down | Biomarker | qRT-PCR | [370] | |
| hsa_circ_0092285 hsa_circ_0058794 hsa_circ_0088088 hsa_circ_0038644 | PBMC | Up Up Down Down |
miR sponge | Microarray | [101] |
Table 7.
Discrete ncRNAs associated with multiple sclerosis.
| Class of ncRNA | ncRNA | Cells/Tissue | Expression (up or down) in disease | Role | Detection method | Refs |
|---|---|---|---|---|---|---|
| lncRNA | ANRIL | PBMCs | Significant association between ANRIL polymorphisms and MS risk | PCR | [230] | |
|
ENSG00000231898.3
XLOC_009626 XLOC_010881 |
PBMCs | Up | Biomarker | Microarray | [235] | |
|
ENSG00000233392.1
ENSG00000259906.1 XLOC_010931 |
PBMCs | Down | Biomarker | Microarray | [235] | |
|
ENSG00000260302
ENSG00000270972 ENSG00000272512 ENSG00000223387 |
Whole Blood | Up | Biomarker | RNAseq | [236] | |
| FAM13A-AS1 | Brain | Central hubs in competing endogenous RNA networks in brain tissue | Microarray | [238] | ||
| GAS5 | Peripheral blood | Significant association between rs2067079 polymorphisms and MS risk | PCR | [225] | ||
| GAS5 | Peripheral blood | Significant association between rs2067079 polymorphisms and MS risk | PCR | [227] | ||
| GAS5 | Peripheral blood | Significant association between rs55829688 polymorphisms and MS risk | PCR | [228] | ||
|
GAS8
GAS8-AS1 |
PBMC | Up | Biomarker | qRT-PCR | [240] | |
| HOTAIR | PBMCs | Up | VitD supplementation to MS patients reduced expression of HOTAIR | qRT-PCR | [378] | |
| HOTAIR | PBMCs | Significant association between rs4759314 polymorphisms and MS risk | PCR | [229] | ||
|
IFNG-AS1
APOA1-AS |
PBMCs | Up | Biomarkers. Elevated during disease relapse. | qRT-PCR | [243] | |
|
IFNG-AS1
GSTT1-AS1 |
PBMCs | Down | Biomarkers. | qRT-PCR | [245] | |
| LncDDIT4 | PBMCs, CD4 cells | Up | Regulates DDIT4 expression in cis, to promote Th17 differentiation | Microarray | [246] | |
| Linc-MAF-4 | PBMCS, CD4 cells | Up | Knockdown of linc-MAF-4 promotes Th2 differentiation and overexpression increases Th1 differentiation | Microarray | [75] | |
|
MALAT1
Lnc-DC |
Serum | Up | Biomarker | qRT-PCR | [233] | |
| MALAT1 | PBMCs | Up | Biomarker. Contributes to splicing and backsplicing events in cell lines | Microarray | [234] | |
| MALAT1 | Peripheral Blood samples | Up | SNPs were associated with MS risk Biomarker | PCR | [226] | |
|
MALAT1
ANRIL TUG1 XIST |
PBMCs | Down | Biomarkers | qRT-PCR | [244] | |
| MEG3 | PBMCs | Down | Biomarker (AUC 0.91 to distinguish patients from healthy controls) | qRT-PCR | [241] | |
| NEAT1 | Serum | Up | Biomarker | qRT-PCR | [232] | |
|
NEAT1
TUG1 PANDA |
PBMCs | Up | Biomarkers | qRT-PCR | [122] | |
| PINK1-AS | PBMCs | Up | Biomarker | qRT-PCR | [242] | |
|
PVT1
FAS-AS1 |
PBMCs | Down | Biomarkers | qRT-PCR | [239] | |
| RN7SK | Serum | Up | Biomarker | qRT-PCR | [232] | |
| THRIL | PBMCs | Up | Biomarker | qRT-PCR | [239] | |
| TUG1 | Serum | Up | Biomarker | qRT-PCR | [232] | |
|
XIST
OIP5-AS1 CTB-89H12.4 |
PBMCs | Network | Central hubs in competing endogenous RNA networks in peripheral blood, e.g. XIST-miR-326-HNRNPA1 | Microarray | [237] | |
| circRNA | circINPP4B | Peripheral blood lymphocytes | Up | Induces Th17 differentiation in mouse CD4 model of EAE. Sponges miR-30a | Microarray | [253] |
| hsa_circ_0005402 has_circ_0035560 | PBMCs | Down | Biomarkers | Microarray | [247] | |
| circ_0000518 | CSF, PBMCs microglia cells | Up | Induces M1 phenotype and immune cell infiltration to CNS | qRT-PCR | [252] | |
|
hsa_circRNA_001896
hsa_circRNA_101145 |
PBMC | Down | Biomarkers | Microarray qRT-PCR | [249] | |
|
hsa_circRNA_101348
hsa_circRNA_102611 hsa_circRNA_104361 |
PBMCs | Up | Biomarkers Interacting miRs proposed |
Microarray qRT-PCR | [248] |
Nuclear enriched assembly transcript (NEAT1)
NEAT1 is associated with multiple autoimmune diseases (Tables 4, 6–7), as well as some human tumors [119] and neurological diseases (Parkinson [120] and Alzheimer [121] diseases). In fact, NEAT1 expression is reciprocally correlated with age of onset in female patients with MS, and overall NEAT1 expression is higher in the peripheral blood [122]. In human SLE, NEAT1 is overexpressed in circulating myeloid cells and is a regulator of TLR4-inflamatory pathways and cytokines, such as IL-6 and CXCL10 [99].
Table 6.
Discrete ncRNAs associated with inflammatory bowel disease.
| Class of ncRNA | ncRNA | Cells/Tissue | Expression (up or down) in disease | Role | Detection method | Refs |
|---|---|---|---|---|---|---|
| lncRNA | DIO3OS | Colon tissue and plasma | Down | Biomarkers | qRT-PCR | [212] |
|
DPP10-AS1
PDZK1P2 ANRIL |
Colon tissue | Down | Biomarkers | Microarray | [209] | |
| DQ786243 | PBMCs | Up | Positively corelated with CREB and Foxp3 expression in patient samples | Microarray | [214] | |
|
ENST00000555407
ENST00000427085 (eRNA) TCONS_00027621 (lincRNA) |
Plasma | Down | Biomarkers | Microarray | [371] | |
|
ENST00000569039
ENST00000422548 (eRNA) TCONS_00027580 (lincRNA) |
Plasma | Up | Biomarkers | Microarray | [371] | |
| GAS5 | PBMCs LoVo, HeLa, | No change in PBMC | Affects Glucocorticoid (GC) response | qRT-PCR | [372] | |
| GAS5 | Colon tissue, THP1 cells | Down | GAS regulated expression of matrix metalloprotease (MMP) 2 and MMP9 | qRT-PCR | [373] | |
| H19 | Colon tissue | Up | H19 was induced by IL-22 and enhanced proliferation H19 bound p53 and miRs that normally inhibit cell proliferation | qRT-PCR | [374] | |
| IFNG-AS1 (NeST or TMEVPG1) | Colon tissue, CD4 cells | Up | Increased expression of NeST/IFNG-AS1 leads to increased IFN-γ | Microarray | [211] | |
|
LINC01272
KIF9-AS1 |
Colon tissue and plasma | Up | Biomarkers | qRT-PCR | [212] | |
| lncRNA BC012900 | Colon tissue | Up | Apoptosis of colon epithelial cells, knockdown of lncRNA BC012900 leads to decreased caspase 3/7 activity | Microarray | [210] | |
| lncRNA Mirt2 | Plasma | Down | Positively regulates IL-22 expression | qRT-PCR | [213] | |
|
MMP12
FAM66D SAA2-SAA4 |
Colon tissue | Up | Biomarkers | Microarray | [209] | |
| NEAT1 | Colon tissue | Up | NEAT1 induced TNFRSF1B expression, a receptor for TNF, to induce inflammatory signaling. Mechanism not established | qRT-PCR | [375] | |
| THRIL | Serum | Up | Biomarker. | qRT-PCR | [215] | |
| circRNA | circCDKN2B-AS1 | Colon tissue | Down | Colonic epithelial cell proliferation | Microarray | [219] |
| circGMLC1 | Colon tissue | Down | Regulates Inflammation and Autophagy | Microarray | [376] | |
| circRNA_0001187 | Colon tissue | Up | Upregulates Myd88 and TNF | qRT-PCR | [377] | |
|
circRNA_092520
circRNA_102610 circRNA_004662 circRNA_103124 |
PBMCs | Up | Biomarkers. circRNA_004662 has binding potential for miRs related to mTOR | Microarray | [220] | |
| circRNA_103516 | PBMCs | Up | Biomarker. Possible sponge for miR-19b-1–5p | qRT-PCR | [222] | |
| circRNA_103765 | PBMC | Up | Biomarker. Induced by TNF. Functions as competitive endogenous RNA to sponge miR-30 family and impair negative regulation of delta-like ligand 4. Protects epithelial cells from apoptosis | qRT-PCR | [221] |
NEAT1 is expressed constitutively in many immune and non-immune cells. It directly binds active chromatin, suggesting a broad modulatory function on gene expression [28]. NEAT1 promotes the activation of the NLRP3 inflammasome, a key component of mature IL-1β production [123], in at least dendritic cells, monocytes, and lipopolysaccharide (LPS)-activated THP-1 cells [99,124] (Figure 4). Knockdown of NEAT1 suppresses Th17 polarization by modulation of STAT3-ubiquitination [125,126], which may partially also explain why NEAT1 is considered by some as a bridge between STAT3 and histone 3 lysine 27 acetylation (H3K27Ac) [121]. Collectively, elevated NEAT1 appears to promote inflammatory signatures and contribute to autoimmunity. NEAT1 is expressed in many cell types and is associated with several diseases, and thus there are shared and distinct mechanisms of action amongst cell types in the context of inflammation. Normalizing the expression of NEAT1 would be an attractive means of ameliorating some of these processes. However, the mechanisms by which NEAT1 is overexpressed, and can therefore be normalized, remain less clear.
Growth Arrest Specific 5 (GAS5)
GAS5 is a dynamic, alternatively-spliced, ‘inside-out’ lncRNA (i.e. one that contains processed introns that become intermediates for a variety of snoRNAs [126]). It can exert effects on gene transcription as a ncRNA decoy of the glucocorticoid receptor (GR) [127], or as a miR-sponge for a variety of miRs including miR-495 (32242002) and miR-21 [128]. GAS5 is associated with cellular proliferation, metastasis [129–131] and is downregulated in many cancers [128,132–134], and associated with multiple autoimmune diseases, including rheumatoid arthritis (RA), multiple sclerosis (MS), systemic lupus erythematosus (SLE), allergy, inflammatory bowel disease (IBD) and immune thrombocytopenia (IT) (Tables 4, 6–7). In fact, GAS5 expression in plasma can act as a biomarker of SLE [135]. In PBMCs from patients with IT, GAS5 expression is inversely associated with Th17 polarization via promoting STAT3 degradation by ubiquitin-mediated proteolysis. This is consistent with a direct interaction between GAS5 and STAT3 and miR-21-mediated downregulation of GAS5 driving Th17 differentiation [71]. However, others have reported that CD4+ T cells from patients with SLE have elevated expression of GAS5, as well as miR-21 [136], which suggests that the miR-21-GAS5 relationship may not hold true in every disease. GAS5 is also credited as a regulator of Th2 balance via the miR-495:circHIPK3 axis to upregulate GATA3 expression and promote allergic rhinitis [137], and as a putative driver of demyelination in MS, where it inhibits microglial M2 polarization [70].
Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1)
Besides cancer, MALAT1 is associated with multiple autoimmune diseases (Tables 4, 7) and widely expressed amongst immune cells. Expression of MALAT1 is detectable by conventional scRNAseq, as it contains a non-canonical 3’ RNAse P cleavage site, revealing a polyadenylation signal that directs the long MALAT1 (~7.4kb) cleavage product toward nuclear speckles, with the small cleavage product, termed mascRNA (that is a tRNA-like structure), shuttled to the cytoplasm [138]. Thus, for some ncRNA molecules, like MALAT1, this adenylation is positively regulatory, directing the molecule to gain functions, whereas adenylation of other ncRNAs, such as rRNA and tRNA influences their degradation [139,140]. MALAT1 also has a circularized form termed circ-MALAT1 that is regulated by GDF15 to induce tolerogenic DCs by dampening NFκB signaling during allograft heart transplantation [141]. In airway epithelial cells (AECs) co-cultured with DCs, MALAT1 levels are increased and modulate AEC cytokine secretion. Here, MALAT1 inhibition in AECs induces proinflammatory cytokines (IL-6, IFN-γ and TNF) and chemokines (CXCR2 and CXCR4) secreted by the co-cultured DCs and siRNA directed against MALAT1 in AECs drives IL-6 secretion [142].
In many contexts, MALAT1 is found to be regulatory. Notably, overexpression of MALAT1 in dendritic cells induces regulatory T cells by sponging of miR-155 [79]. Likewise, MALAT1 is responsible for regulating IL-10 expression by exerting control over expression of MAF [143]. Thus, mice with MALAT1 deficiency produce more aggressive responses to infections and are protected against experimental leishmaniasis and malaria [143]. This is concordant with human asthma, in which reduced MALAT1 expression is associated with loss of regulatory and enhanced inflammatory cytokine production [144]. Consistent with these observations, reduced MALAT1 expression is observed in activated B and T cells and macrophages, and is important for class-switch recombination, Th1 effector function, and M1 polarization [79,143,145]. In the experimental autoimmune encephalomyelitis (EAE) model, a murine model of human MS, MALAT1 is downregulated in the CNS and the magnitude of the change correlates inversely with the degree of inflammatory change [146].
Collectively, it is evident that dysregulated expression of NEAT1, GAS5, and MALAT1 are associated with multiple autoimmune diseases. While it is not possible to state definitively whether this association is cause or effect, the fact that all three ncRNAs are potent regulators of immune function suggests that they play an active role in the pathogenesis of these diseases. As ncRNAs commonly demonstrate a range of expression among immune cells, they might have shared and unique cell-type specific functions. However, many of these potential facets remain unexplored. By further analyzing specific ncRNAs associated with autoimmunity and dissecting mechanisms of function in studies of autoimmunity, we may be able to further explore common immunopathogenic mechanisms and the dynamic range under which various ncRNAs operate.
ncRNAs and Specific Autoimmune Diseases
Autoimmunity, the targeting of self-antigens by host, involves a heterogeneous set of processes, that are impacted by a number of cellular mechanisms and extrinsic factors. These include cell signaling events [147], soluble factors [148], gene transcription [149], cellular metabolism [150], regional microbiota [151], circadian rhythms [152], and genetic polymorphisms [153]. In-depth reviews of autoimmunity can be found elsewhere in the literature, for example [154]. According to the current central molecular dogma, all these cellular processes are mediated or modified in part by protein-coding (mRNA) and non-protein coding transcriptional intermediates. Autoimmune-associations with transcriptional products have been investigated since at least the 1960s beginning with the detection of anti-ribosome autoantibodies [155]. Taking cues from this early work, current studies are exploring whether RNA molecules can be useful in the diagnosis of autoimmunity and the design of precision therapies.
As of today, many studies have described expression of autoimmune-associated ncRNAs using high-throughput techniques (such as microarray and RNAseq) in peripheral blood mononuclear cells [101,156], plasma [100], primary immune cell subsets [3,157], tissue biopsies [103,109,158], and cultured human cell lines [157]. The accumulating data suggest that ncRNAs play important roles in the regulation of pathways contributing to autoimmunity and inflammation (Tables 4–7).
Moreover, ncRNAs that are consistently differentially expressed could potentially be used as diagnostic or prognostic biomarkers of specific autoimmune diseases, especially as some ncRNAs (e.g. circRNAs) are far less susceptible to degradation than others (e.g. linear mRNA). Here we review some of the known functions of noncoding ncRNAs in specific named autoimmune diseases and their putative functional significance towards the pathogenesis of the disease.
Rheumatoid Arthritis (RA)
RA is a systemic autoimmune disease exemplified by inflammation of the synovia leading to joint, cartilage and bone erosion [159]. This is a well-studied disease and, not surprisingly, miRs have been a major component of the research effort in ncRNAs. A variety of miRs are dysregulated in RA and affect expression of target mRNAs encoding cytokines, chemokines, and inflammation-related signaling pathways including TLR signaling [160,161]. For example, in the plasma of patients with RA, 33 miRs are differentially expressed compared to healthy controls, amongst which a single one, miR-9–5p, positively correlates with plasma IFN-γ, TNF, IL-17A, IL-4 and CXCL9 levels [162]. A comprehensive review about miRs in RA can be found elsewhere [163,164].
Apart from miRs, many lncRNAs have also been suggested as potential biomarkers of RA and some have been shown to regulate inflammatory processes and tissue damage in both patients and mouse models [165]. These are summarized in Table 4. Owing to unbiased high-throughput approaches, the list of disease-associated lncRNAs is growing. For example, one study identified upregulation of HOTAIR, together with 83 other lncRNAs, in the PBMCs of RA patients, as well as in serum exosomes. Although this was corroborated by an independent, targeted study, using qRT-PCR from serum [166], the value of HOTAIR as a biomarker of RA remains unestablished [167]. Likewise, microarray screening of serum from patients with RA identified 73 up-regulated and 61 down-regulated lncRNAs, including LncHIX003209, which correlated with a number of clinical outcomes [168]. A follow-up study from the same group confirmed elevated expression of LncHIX003209 in both PBMCs and circulating myeloid cells and correlation with parameters of disease activity [169]. Further functional assessment identified LncHIX003209 as a facilitator of macrophage activation via the TLR4/NF-κB pathway [169]. Specifically, LncHIX003209 acts as a competing endogenous RNA sponge for miR-6089 [169], a miR that targets TLR4 directly to limit IL-6, IL-29, and TNF generation [170]. Although exhibiting some promise, the utility of LncHIX003209 as a clinical biomarker remains unknown at present. Other studies in RA have taken a more hypothesis-driven approach aimed at testing the role of specific lncRNAs in disease. For example, one group reported that lncNTT expression is upregulated in PBMCs of RA patients compared to healthy controls and regulates differentiation of monocytes to macrophages [171]. This is achieved through interaction with the promoter of PBOV1, which is required for maturation and secretion of mediators, such as CXCL10, that recruit inflammatory cells [171]. Consequently, a direct correlation is observed between lncNTT expression and parameters of disease severity in RA [171]. Another study noted abnormal expression of two out of ten hypothesized lncRNAs, LOC100652951 and LOC100506036, in T cells of patients with RA and demonstrated some correlation with clinical parameters [172]. Although the functionality of these lncRNAs were not fully investigated, silencing of LOC100506036 in Jurkat cells was associated with decreased IFN-γ expression [172].
With the growing evidence of the importance of circRNAs in immunobiology, attention in RA has recently also shifted to this class of ncRNAs (Table 4). More than 500 differentially expressed circRNAs have been noted in PBMCs of patients from RA from microarray studies, with some (limited) computational prediction of miRs that may be sponged by these circRNAs [101]. This approach is limited because no experimental validation of target miRs was attempted, and because the methodology, in principle, does not consider the possibility that samples from patients and controls could have different cellularities, which may give the impression of differential circRNA usage. Other studies taking a more targeted approach have assessed individual circRNAs and assessed expression of their target miRs directly. For example, Tang et. al., demonstrated increased expression of hsa-circRNA-ciRS-7 in PBMCs of patients with RA. In this setting, expression of its target, miR-7 (ciRS-7 has more than 70 binding sites for miR-7), was repressed, which correlated inversely with elevated mRNA expression of the mammalian target of Rapamycin (mTOR) that harbors miR-7 binding site in its 3’ UTR [173,174]. A similar targeted approach focused on circNUP214, which functions as a competitive endogenous RNA sponging miRs and has previously been implicated in Hashimoto thyroiditis [175]. Here, the investigators noted elevated expression of circNUP214 in PMBCs from patients with RA, although no correlation could be identified between its expression and parameters of disease severity or activity [85]. The authors did, however, demonstrate that circNUP214 indirectly induces expression of IL23R mRNA (via sponging of miR-125a-3p [176]), which encodes the IL-23 receptor required for efficient generation of pathogenic Th17 subsets [85]. Although the direct relevance of ciRS-7 and circNUP214 to RA is still only tangentially demonstrated, both studies did attempt to make a case for their utility as diagnostic biomarkers. Area under the receptor operator characteristic (ROC) curves (AUC) for the expression of these two circRNAs in PBMCs to distinguish patients with RA from healthy controls were remarkably similar, 0.76 for both [85,174], which represents a moderate diagnostic test. Their utility when used together or in combination with other metrics has not been directly tested.
Despite being a systemic autoimmune disease, one could argue that the predominant site of inflammation, synovial tissues, may offer greater insights into the roles played by ncRNAs in directing inflammation. To our knowledge, there are only a few published high-throughput approaches studying ncRNAs in synovium from RA [177–179]. This report suggests that expression of activating transcription factor 2 (ATF2) and inflammatory cytokines are enhanced at this site and that this transcription factor is targeted for repression by miR-204/211, which can in turn be sponged by hsa-circ-0001859 [177]. There are a number of key limitations that should be acknowledged in this report. Firstly, this study is based on historical expression profiling by array (GSE2053) [180] in which multiple samples were pooled from different donors before RNA extraction and expression profiling. Thus, the provenance of dysregulated ATF2 expression with respect to donor and cell type is not known. Secondly, neither actual expression nor abnormal expression of hsa-circ-0001859 was demonstrated, so its role in the local pathogenesis of RA is circumspect and inferential at best. The insights gained from this publication remain unconfirmed by others. However, there are some instances of more targeted, hypothesis-driven studies. For example, fibroblast-like synoviocytes derived from patients with RA demonstrate decreased expression of circ_0008360, a circRNA that represses synoviocyte proliferation and production of inflammatory cytokines, including IL-1β, IL-6, and TNF, attributable to sponging of miR-135b-5p, a miR that represses expression of histone deacetylase 4 [181].
Psoriasis
Psoriasis is a chronic inflammatory skin disease with rapid proliferation of skin cells that leads to the development of red thick scaly patches on the skin. Similar to other autoimmune diseases, psoriasis is mediated by both innate and adaptive arms of the immune system [182]. Several studies have investigated the expression of ncRNAs in psoriatic skin (Table 5).
Table 5.
Discrete ncRNAs associated with psoriasis.
| Class of ncRNA | ncRNA | Cells/Tissue | Expression (up or down) in disease | Role | Detection method | Refs |
|---|---|---|---|---|---|---|
| lncRNA |
AL035425.3
PWAR6 |
Skin Biopsies | Down | Competing endogenous RNA, candidate regulator of a miR network associated with psoriasis | High-throughput sequencing data from Gene Expression Omnibus (GEO) | [184] |
| AGXT2L1–2:2 | Keratinocytes | Up | Inhibits apoptosis and promotes proliferation of keratinocytes | qRT-PCR | [188] | |
|
CARD14
CYP4Z2P HINT1 RPSAP58 TRHDE-AS1 |
Skin Biopsies | Up | Biomarkers | RNAseq | [103] | |
|
lnc-RP11–701H24.7
lnc-RNU12 |
PBMCs | Up | Biomarkers | RNAseq | [187] | |
|
LUCAT1
TRIM55–1 LINC00657 LINC00909 |
PBMCs | Up | Decoded to interplay with immune response via pathway analysis | Microarray | [104] | |
| MEG3 | Skin Biopsies | Down | Interacts with miR-21 to regulate caspase-8 levels, implicated in cell proliferation and apoptosis | qRT-PCR | [190] | |
| NORAD | Keratinocytes | Up | Promotes proliferation of keratinocytes | qRT-PCR | [189] | |
| PRINS | Keratinocytes | Down in inflammato ry cytokine treated cells | PRINS overexpression decreased IL-6 and IL-8 expression | qRT-PCR | [191] | |
| PRINS | Plasma | Down | Biomarker | qRT-PCR | [192] | |
| SPRR2C | Keratinocytes and psoriatic lesion | Up | Promotes Inflammation and cell viability, sponges miR-330 to enhance STAT1 and S1007 | RNAseq and qRT-PCR | [186] | |
| XIST | Serum | Up | Biomarker and miR sponge for miR-338–5p, which targets IL6 mRNA for silencing | qRT-PCR | [193] | |
| Many | Skin biopsies | Up and Down | 1214 lncRNAs up and down-regulated in lesional psoriasis | RNAseq | [183] | |
| circRNA |
CDR1as
hsa_circ_0109327 hsa_skin_05227 |
Skin Biopsies | Down | Computationally predicted to act as competitive endogenous RNAs sponging cellular miRs | RNAseq | [206] |
|
circCAMSAP1
circTRIM35 circTULP4 circARAP2 ciRS-7/CDR1as |
Skin Biopsies | Down | Genome-wide profiling of circRNAs from paired lesional vs non-lesional skin | RNAseq NanoString CISH |
[195] | |
| circEIF5 | Skin Biopsies | Up | Knockdown and overexpression in HaCaT cells inhibited and enhanced proliferation, respectively Activates NF-κB and STAT3 | qRT-PCR | [201] | |
| circOAS3 | Skin Biopsies | Up | Interacting with Hsp70 and regulates signaling through the JNK MAPK, NF-κB and STAT3 pathways to enhance proliferation of keratinocytes | Microarray | [200] | |
| circRAB3B | Skin Biopsies | Down | Downregulated by IL-22 (in HaCaT cells) Inhibits cell proliferation by sponging miR-1228–3p, which targets PTEN |
qRT-PCR | [198] | |
|
circRNA
chr2:206992521|206994966 |
Skin Biopsies | Down | Regulation of IL-11, IL-6 and hepatocyte growth factor in skin MSCs; regulation of co-cultured T cell proliferation | RNAseq | [203] | |
|
circTULP4
circPARD3 circPTPRA ciRS-7 |
Skin Biopsies | Down | Repressed circRNAs that normalized after treatment with anti-IL-17 | Targeted NanoString panel |
[197] | |
|
ciRS-7
circZRANB1 |
Skin Biopsies | Down Up |
Biomarkers of disease | RNAseq | [196] | |
|
hsa_circ_0003689
chr4:121675708|121732604 hsa_circ_0003718 |
Skin Biopsies Plasma | Up | Biomarkers | RNAseq | [204] | |
| hsa_circ_0056856 | Serum samples | Up | Biomarker, predicted to function as a competitive endogenous RNA | Microarray qRT-PCR | [205] | |
| hsa_circ_0061012 | Skin Biopsies | Up | miR sponge | Microarray | [199] | |
| hsa_circ_0061012 | Skin Biopsies | Up | Upregulated in HaCaT cells by IL-22. Silencing reduced proliferation of HaCat cells. Upregulates GAB2 by Sponging miR-194–5p | qRT-PCR | [202] |
High throughput and discovery-led methods have identified a number of differentially expressed lncRNAs in psoriatic skin compared to healthy control skin. One relatively sizeable study performed RNA-seq on 99 lesional psoriatic, 27 uninvolved psoriatic and 90 normal skin biopsies and identified 1214 differentially expressed lncRNAs between normal and lesional psoriatic skin samples. Intriguingly, 505 of these were novel lncRNAs [183]. Although the study did not focus on any particular lncRNA, the authors did note that 26 differentially expressed lncRNAs were from known psoriasis susceptibility regions and that 336 of the differentially expressed lncRNAs could be induced or repressed by IL-17 and TNF, cytokines that play key roles in the pathogenesis of psoriasis [183]. Another study found 971 differentially expressed lncRNAs between lesional skin compared to normal healthy donor samples and confirmed 4 of these (CYP4Z2P, HINT1, RPSAP58 and TRHDE-AS1) by qRT-PCR. Like the previous study, a large proportion of these lncRNAs were normalized after treatment with anti-TNF therapy, indicating that many lncRNAs are responsive to inflammatory signals and may be the effect rather than cause of disease [103]. Collectively, these data implicate lncRNAs as both potential mediators of psoriasis and as potential bystanders induced by local cytokines. To help resolve these possibilities, a third study analyzed publicly available transcriptomes deposited in the gene expression omnibus (GEO) database and constructed a psoriasis-associated lncRNA-miR-mRNA network based on the hypothesis that lncRNAs act as competitive endogenous RNAs for miRs. This intriguing study identified two key lncRNAs, named AL035425.3 and PWAR6, that interacted with at least 5 different miRs, were downregulated and hypothesized to play a part in a network that may be pathogenic. Although neither of these were experimentally validated, the analysis does provide a hypothetical pathogenic and targetable node for further exploration in psoriatic skin [184], especially since at least PWAR6 harbors binding sites for miR-155, a driver of NLRP3-dependent inflammation in psoriasis [185]. Interestingly, another high-throughput analysis of psoriatic lesions (12 patients and 12 controls) employing a different computational methodology (weighted gene co-expression network analysis, WGNA) did not identify either of these lncRNAs as key components of a pathogenic causative network, but instead suggested lnc-SPRR2C as a candidate repressing miR-300 by sponging, leading to enhanced inflammatory STAT1 and S100A7 signaling [186]. Similarly, potential biomarkers of psoriatic arthritis (a variant of psoriasis that also affects the joints) have been identified using data analyzed from high-throughput methods deployed on PBMCs. Although only 4 subjects with disease were compared to 4 healthy subjects by RNA-seq, confirmation by qRT-PCR in a larger test cohort of >90 subjects per group suggested that some candidates lncRNAs (lnc-RP11–701H24.7 and lnc-RNU12) were upregulated in disease, correlated with clinical parameters and could distinguish patients from healthy subjects with moderate accuracy (AUCs of 0.76 and 0.84, respectively) [187]. These potential biomarkers have yet to be confirmed in a larger, independent cohort as part of a follow-up study.
Hypothesis-led methods have also identified abnormal expression of several lncRNAs in psoriatic skin, such as AGXT2L1–2:2 [188], NORAD [189], MEG3 [190], PRINS [191,192] and XIST [193] and experimental work has revealed some of their functions (summarized in Table 5). For example, MEG3 is downregulated in psoriasis samples compared to healthy controls and regulates the proliferation and apoptosis of keratinocytes via binding to miR-21, which in turn regulates Caspase 8[190]. Over-expression of MEG3 in keratinocytes, on the other hand, suppresses the PI3K-AKT-mTOR pathway and decreases inflammation on TNF treatment [194]. It is interesting to note that few of these studies include lncRNAs identified in the discovery-led approaches discussed above.
Review articles on the subject are largely in agreement about the disposition and diversity of circRNAs in psoriatic skin, indicating that lesional psoriatic skin mostly demonstrates repression of expressed circRNAs. This position, however, may reflect a bias or tendency to investigate the well-known sponging mechanism of circRNAs (Table 5). For example, one (small) study reported that circRNAs are largely depleted, demonstrating that circiRS-7, circCAMSAP1, circTRIM35, circTULP4, circARAP2 are all decreased, without a correlative change in miRs, in lesional skin compared to paired non-lesional skin using RNAseq [195]. The same group subsequently provided additional evidence of this phenomenon and demonstrated that ciRS-7 is downregulated and could act as a disease biomarker distinguishing psoriasis from atopic dermatitis (AUC of 0.92), a distinction that can sometimes be difficult to make using current clinical tools [196]. The same publication, however, also noted that circZRANB1 is actually upregulated and that its performance as a biomarker is comparable to that of ciRS-7 (AUC of 0.89) [196]. The top 50 most abundant circRNAs annotated in skin of psoriasis patients by these papers [195] were adopted in a subsequent publication from an independent group for designing a targeted NanoString panel to study circRNAs in lesional skin [197]. As expected, the majority of these were found to be repressed in lesional skin and subsequently be normalized following treatment with secukinumab (anti-IL-17 antibodies), suggesting that IL-17, a known driver of the disease, may be a driver of circRNA repression in psoriasis [197]. Hypothesis-led studies also largely follow similar patterns. Notably, circRAB3B has been shown to be repressed in psoriatic skin and to regulate PTEN by sponging miR-1228–3p. Thus, low circRAB3B expression enhances cell proliferation and migration [198].
Other studies, in contrast, suggest that circRNAs can be both up- and down-regulated in psoriatic lesional skin. For example, one publication reported 3016 upregulated and 1940 downregulated circRNAs in psoriatic lesions compared to healthy control samples, with many of these, such as hsa_circ_0061012, predicted to sponge miRs that regulate T cell function and development [199]. Additional experiments by the same group demonstrated that circOAS3, a psoriatic skin induced circRNA, physically interacts with Hsp70 and regulates signaling through the JNK-MAPK, NF-κB and STAT3 pathways to enhance proliferation of keratinocytes [200]. In a further follow-up study, the authors of the original publication [199], found that one of the circRNAs upregulated in lesional skin, termed circEIF5, is an activator of the NF-κB and STAT3 pathways (through undetermined mechanisms) and regulates cellular proliferation and chemokine production [201]. Of these circRNAs, hsa_circ_0061012 has since been independently verified by others as upregulated in psoriatic skin and to regulate proliferation of HaCaT cells, an immortalized human keratinocyte cell line, by upregulating GAB2 via sponging of miR-194–5p [202]. Meanwhile, two other high-throughput analyses reported differentially expressed circRNAs in skin mesenchymal stromal cells (sMSCs) of patients compared to healthy donors [203,204]. The first identified 123 upregulated and only 6 downregulated circRNAs in sMSCs [203]. One of the downregulated circRNAs labeled chr2:206992521|206994966 was functionally analyzed and its knockdown in sMSCs found to support enhanced T cell proliferation and altered cytokines secretion (higher IL-11 and lower IL-6 and hepatocyte growth factor secretion)[203]. In the second study, 129 circRNAs were differentially expressed; although it is clear that more circRNAs were up rather than down in terms of detected level, the exact number of detected circRNA were not reported [204]. Of these, expressions of three circRNAs, hsa_circ_0003689, chr4:121675708|121732604 and hsa_circ_0003718, were also elevated in plasma and correlated with clinical severity index [204]. The performance of these circRNAs as predictors of disease was not evaluated, nor were functional analyses conducted, but a link to the MAPK pathway was computationally suggested [204]. A number of circRNAs in serum from psoriatic patients detected by microarray are differentially expressed (123 upregulated, 9 nine downregulated) compared to healthy donors, of which the topmost induced, hsa_circ_005685, shows some promise as a disease biomarker (AUC of 0.853 to distinguish patients over controls) and iss predicted to act as a miR sponge [205].
Psoriasis is a complex disease, with many immune and non-immune cells participating in the pathology. The major drawback of the available data is due to their being generated from bulk RNA inputs derived from biopsy or blood-derived samples. This means that at least some of the highlighted changes in ncRNAs might reflect changes in cellularity rather than changes in transcriptional output in cells that express transcripts of interest. What is striking from reviewing publications using high-throughput methods is the lack of agreement among them. This may represent an inherent problem with the approach (bulk cells instead of single cells), differences in populations, sample size or the effects of treatments on patients in different cohorts. Part of the disparity in the numbers and identities of circRNAs reported in different publications also relates to computational challenges in identifying back-spliced circularized RNA species. There is room for computational innovation in this area and recent evidence points to novel methods that have been deployed in psoriasis to identify additional sets of previously unrecognized differentially expressed circRNAs, including those that may act as competitive endogenous RNAs sponging cellular miRs [206]. The lack of data on eRNAs is also evident and may reflect similar difficulties in correctly identifying eRNA species using simple methods. Coupled with these limitations, the paucity of proof-of-concept experiments renders it difficult to assess whether highlighted transcripts are abnormally expressed as cause or effect of the disease. These studies, are however, hypothesis-generating and are a reasonable starting point for more in-depth studies that address causation and mechanism.
Inflammatory Bowel Diseases (IBD)
IBD, including ulcerative colitis and Crohn disease, characteristically causes inflammation of the gastrointestinal mucosa. These are another group of diseases in which host genetics and cell-intrinsic mediators (e.g. CXCL10 [207]) interact with cell-extrinsic factors, such as the microbiome. As a result, there is considerable heterogeneity in these diseases, resulting in variable presentation to the clinic. Consensus on the role of the roles of ncRNAs may therefore prove to be difficult to achieve in this sphere.
Aberrantly expressed ncRNAs in Crohn disease are of broad groups and including lncRNAs, lincRNAs and circRNAs (Table 6). miRs in IBD are reviewed elsewhere [208]. Multiple groups have assessed ncRNA expression in mucosal biopsies from patients [209–212] and reported dysregulation of a number of lncRNAs. As anticipated, and similar to other diseases reviewed in this article, there is generally low agreement amongst studies, which may reflect biopsies taken at different sites or differences in cellularity or clinical activity between collected samples used in bulk RNAseq. Briefly, one study identified 329 lncRNAs upregulated and 126 downregulated in colitis using a microarray approach on sigmoid colon biopsies [210]. Here, the authors focused on enhanced expression of lncRNA BC012900, which could also be induced by treatment of colorectal adenocarcinoma cells (HT29) with pro-inflammatory agents, including TNF, IL1-β, LPS, Flagelin or CpG oligodeoxynucleotide [210]. The increased expression of BC012900 was positively correlated with apoptosis in HT29 cells and could be reversed using siRNA mediated knockdown of BC012900 [210]. In another study, lncRNAs KIF9-AS1, linc01272, and DIO3OS were found to be differentially expressed in colitis and were suggested as potential biomarkers for the disease, with some of them demonstrating reasonable AUCs (generally 0.8–0.9) in differentiating patients from healthy controls [212]. In a third study, differentially expressed lncRNAs identified in Crohn disease and ulcerative colitis were associated with antigen processing and presentation, immune system processes, and NK cell activation [209]. Finally, a fourth group identified between ~300–2000 lncRNAs as differentially expressed between healthy mucosa and mucosa from patients with uncreative colitis [211], amongst which upregulation of NeST, a key regulator of IFN-γ expression, was one of the most statistically significant [211]. In blood, targeted approaches (e.g., qRT-PCR) have identified downregulated lncMIRT2, which regulates IL-22 production and apoptosis of colonic epithelial cells [213], and upregulated DQ786243, a driver of CREB and FOXP3 expression, in patients with IBD [214]. Perhaps importantly, elevated expression of the lncRNA THRIL in serum has very high predictive power for distinguishing patients with IBD from healthy individuals (AUC 1.0) [215]. The interested reader is directed to a number of review articles for a more in-depth discussion of the association between many lncRNAs and IBD [98,216].
Relatively less is known about the role of other ncRNAs in IBD. CircRNAs are no exception in this regard. Although circRNAs are known to play an important role in epithelial cell function in the bowels [217,218] and to immune cell function more generally, little is known about their role in inflammatory bowel diseases. In colonic tissues, circCDKN2B-AS1 has been reported to be downregulated, and to be a negative regulator of colonic epithelial cell proliferation [219]. In blood, a study of PBMCs from Crohn disease identified 155 upregulated and 229 downregulated circRNAs, amongst which circRNA_0004662 was predicted to sponge mTOR pathway-related miR [220], hinting at dynamic regulation of cellular immunometabolism. The same group proposed another circRNA, named circRNA_103765, as a candidate biomarker of IBD. Although circRNA_103765 was not a strong biomarker (AUC 0.65–0.7), this circRNA was induced by TNF and normalized after therapy with anti-TNF drugs. Functional experiments indicated that circRNA_103765 functions to protect epithelial cells from TNF-associated apoptosis, possibly through sponging of members of the miR-30 family, which negatively regulate expression of delta-like ligand 4 [221]. In other studies, expression of another circRNA, circRNA_103516, in PBMC has been shown to be elevated in patients and to correlate with disease severity [222] however the utility of this circRNA as a single biomarker appears to be modest (AUC between 0.6 and 0.79). To our knowledge, eRNAs have not been the subject of intense study in IBD, potentially for the same reasons as their understudy in other diseases, namely difficulties in correctly identifying eRNA species using simple methods.
Multiple Sclerosis (MS)
Multiple sclerosis is an inflammatory immune-mediated demyelinating disease of the central nervous system. Inflammation and the loss of myelin disrupt neural signal conduction and lead to a broad spectrum of mild to severe relapsing-remitting and/or progressive symptoms including pain, motor impairment, cognitive deficit and sensory disturbances [223]. Many miRs are associated with MS, and these are reviewed elsewhere [224]. Of the long ncRNAs, the existing data is currently light on mechanistic insights as most published articles have sought to identify lncRNAs that are surrogates or biomarkers of disease occurring in the central nervous system (CNS), a site that is difficult to access routinely in live humans. The lack of concerted follow-up or overlapping publications from different groups also makes it difficult to obtain consensus on the disposition of ncRNAs in MS. The main findings are summarized in Table 7.
Genomic variants in several lncRNAs including GAS5, MALAT1, HOTAIR and ANRIL are associated with MS [225–230] with several other susceptibility loci close to or within lncRNA-encoding regions [231]. Although it is known that GAS5 can inhibit microglial M2 polarization and exacerbate myelin loss in EAE [70], the mechanisms of these associations remain largely unknown. In the serum, initial studies reported that lncTUG1, NEAT1 and RN7SK (7SK small nuclear RNA) [232], plus MALAT1 and lnc-DC [233], are upregulated in MS patients. The ability of these lncRNAs as biomarkers of disease were either not reported, or in the case of MALAT1 and lnc-DC they had modest performance (AUC ~0.65 alone and 0.75 in combination) [233]. There is, however, some biological plausibility to the functional relevance of MALAT1 in MS as this lncRNA is a regulator of gene splicing and backsplicing events, including those of MS-associated genes [234]. Microarrays conducted on PBMCs have revealed 2353 upregulated and 389 downregulated lncRNAs in patients with MS and enrichment of these in pathways including tight junctions, regulation of axon guidance, axon guidance receptor activity, and regulation of endothelial cell chemotaxis [235]. Separately, RNAseq of PBMCs using a discovery and validation cohort of patients identified four novel lncRNAs that might potentially serve as biomarkers for the disease [236]. However, the strength of these lncRNAs as biomarkers is still not established and their functional relevance to MS, if any, remains unknown. The associations of transcriptional profiles derived from blood with MS is certainly interesting as these cells are remote from the site of CNS injury. One method to systematically classify peripheral blood transcriptional changes involves creation of an MS-associated competing endogenous RNA network, in which lncRNAs are considered as central hubs. Such a network was constructed by Ding et al., who identified three discrete lncRNAs, XIST, OIP5-AS1, and CTB-89H12.4 as hub lncRNAs. Of these, XIST was proposed as part of an XIST-miR-326-HNRNPA1 regulatory module that could be pharmacologically targeted [237]. Interestingly, another method to computationally delineate an MS-associated competing endogenous RNA network by weighted gene co-expression network analysis (WGCNA) identified none of these as a key node and instead focused on FAM13A-AS1 as the central hub [238]. Unfortunately, the validity and utility of the insights from either analysis have not yet been tested.
Hypothesis-driven studies using qRT-PCR have identified that lncRNA-PVT1, FAS-AS1 and MEG3 expression are downregulated and THRIL, NEAT1, lncTUG1, PANDA, lncGAS8, GAS8-AS1 and PINK1-AS expression are upregulated in PBMCs from MS patients compared to healthy controls [122,239–242]. Likewise, APOA1-AS and IFNG-AS1 are elevated in PBMCs, especially during relapses [243]. Because the number of subjects in many of these studies are small and there is heterogeneity in severity, disease stage and treatment, it is not surprising that opposing results for some have also been reported. For example, others have reported that IFNG-AS1, MALAT1, ANRIL, lncTUG1 and XIST are actually repressed in PBMCs of patients with MS [244,245]. Furthermore, despite these associations, almost no functional experiments have been conducted to help substantiate a plausible biological link. One exception is the case of LncDDIT4, which is more highly expressed in PBMCs and CD4+ T cells of patients and acts to promote DDIT4 mRNA in cis, thus enhancing differentiation of naïve CD4+ T cells to Th17 cells [246]. Similarly, another group identified enhanced expression of linc-MAF-4 in patients with MS and, in a series of functional experiments, showed that linc-MAF-4 regulates MAF expression to promote encephalitogenic Th1 responses [75].
Comparatively fewer data exist with respect to other forms of ncRNAs. Some studies have described a broad distribution of differentially expressed circRNAs in PBMC samples of patients with MS [247–249], including downregulated circRNAs originated from ANXA2 (circ_0005402 and circ_0035560) as putative markers of disease [247] and circRNA species that potentially sponge interacting miRs [248,249]. Some differentially expressed circRNAs in blood are detectable within extracellular vesicles [250]. Another study examining exosomes isolated from the cerebrospinal fluid of patients with immune-mediated demyelinating disease (MS and Guillain-Barré syndrome) agreed with broadly dysregulated circRNA dynamics (2364 upregulated and 2730 downregulated), suggesting that exo-hsa_circ_0087862 and exo-hsa_circ_0012077 in CSF can function as diagnostic biomarkers for immune-mediated demyelinating diseases [251]. Both these circRNAs performed admirably to distinguish patients from controls (AUCs of 1.0 for both when used alone), although it has to be acknowledged that this was a very small study (n=5 patients and controls each) that has not yet been validated in an independent cohort [251]. A recent report indicates the increased expression of circ_0000518 at multiple sites, including PBMCs and CSF, of patients with MS [252]. Here, experimental work in microglial cell lines showed that knockdown of this circRNA increased M1 and decreased M2 phenotype [252]. Mechanistically, circ_000518 interacted with the RNA Binding Protein FUS and its knockdown decreased immune cell infiltration in the CNS of mice with EAE [252]. A significant development in obtaining mechanistic insights into the roles of circRNAs in MS was recently published. Here, the authors showed that circINPP4B is upregulated in the peripheral blood CD4+ T cells of EAE and that it positively correlates with the clinical score [253]. Knockdown of circINPP4B ameliorated clinical severity and decreased Th17 cells in vivo and decreased Th17 differentiation in vitro. Mechanistically, circINPP4B sponged miR-30a, although the targets of miR-30a were not elucidated. Finally, the authors showed upregulation of circINPP4B in peripheral blood lymphocytes of 18 patients with MS compared to 20 healthy controls [253]. Little is currently known about the disposition of enhancer RNAs in MS.
To summarize, there is a broad collection of dysregulated ncRNAs detected in many autoimmune diseases (Tables 4–7). In most autoimmune workflows, the activities, functions, and utility of many of these ncRNAs remain mostly unknown. There are also many other caveats to these studies, such as the collection of plasma, bulk tissue or circulating PBMCs in lieu of affected tissue or single cells. Consistency is lacking, biased by detection techniques, sample handling, and or biological variability in human samples amongst diverse genomic backgrounds. Nevertheless, emerging studies have genuinely expanded the mechanism of pathogenesis of these various autoimmune diseases and are beginning to show how ncRNAs underpin some of the basic molecular mechanisms of autoimmunity.
Exploiting ncRNAs as diagnostic and therapeutic targets
The study of ncRNAs may offer three potential benefits. Firstly, strong associations between disease and a given ncRNA or sets of ncRNAs could point to clinical utility as biomarkers for diagnostic or prognostic purposes. Autoimmunity, especially if sub-clinical, can be difficult to diagnose in patients and to track. Biomarkers for diagnosis, especially early disease detection, and disease activity represent some of the unmet needs in this domain. The accumulating evidence suggests that most autoimmune diseases are associated with abnormal expression of at least some ncRNAs in accessible sites. Indeed, most human tissues and fluids, such as peripheral blood, plasma, cerebrospinal fluid, vitreous and aqueous humors, saliva, urine, nasal washes, tears, sperm, synovial fluid, vomitus, skin, biliary brushings, feces, cerumen, amniotic fluid, and sputum contain ncRNA signatures and can be sampled. Secondly, a strong association between transcriptional products and a given ailment can form the basis for a more detailed mechanistic understanding of the basic processes of disease. This supposition is likely to depend on precise delineation of the site of expression, the cells in which expression occurs and how expression is switched on and off. Finally, greater understanding of how a transcriptional product relates to disease opens the possibility for therapeutic intervention, either by directly targeting the RNA species of interest, a key node in its regulatory pathway or its mechanism of action. RNA is amenable to the design of discretely targeted therapies, using antisense and related technologies targeted toward the exact RNA of interest. For example, antagonizing miR activity can be accomplished with antisense oligonucleotides to decrease active levels of mature miRs or overexpression of miR sponges (synthetically designed or native), or miR activity can be enhanced using miR mimics. Silencing of long ncRNAs can be achieved with antisense (e.g siRNA, shRNA [254]) or CRISPR technologies (e.g. CRISPR-Cas13 [255]) and disruption of context-specific promoter-associated ncRNAs to modulate gene activation or silencing. Some ncRNAs are candidates for targeted therapies, such as anti- miR-21, MALAT1, MEG3, GAS5, and NEAT1. Expression of these molecules can theoretically be normalized or modulated, however delivery of the intervention to the correct cellular and sub-cellular compartment is likely to be key [256]. Indeed, delivery is the major limiting factor given most tissues are composed of orthogonally stacked cells that form intricate layers. It is likely that a technological evolution will be needed for the optimal delivery of anti-RNA or RNA-decoy lncRNAs to specific subsets of cells. Care must be taken not to alter putatively healthy cells, and to limit off-target behaviors. Some tissues lend themselves to particular routes of local administration, such as lungs (inhaled delivery), skin (topical delivery), eyes (eyedrops and topical ointments) and joints (direct injection), but off-target effects on “healthy” cells in these tissues is a real possibility. Other tissues will most likely require development of novel delivery methods.
There are some clear limitations with respect to the current state of the field that should be acknowledged. Most of the studies reviewed here are limited by small sample size and geographically isolated populations (most are of European descent). Markers associated with disease in such small studies need first to be confirmed in larger, more heterogeneous populations. Second, poor reproducibility across different studies could indicate either differences in cellularity of samples, or in the case of RNAs that track with disease activity, differences in disease activity/stage at the time of sampling and/or computational challenges. Processing of samples before capture of ncRNAs should also be standardized, given that sample handling and experimental isolation and necrosis of material can alter the expression of RNA and possibly result in a thanatotranscriptome [257].
Concluding Remarks
As this collection of studies demonstrates, ncRNAs are dynamically associated with and/or regulate a plethora of autoimmune diseases. High-throughput methods reveal a complex network of tissue-level and disease associations. Implementation of single cell methods and advances in spatial transcriptomics are enabling finer molecular dissection of the roles of ncRNAs within individual organelles, individual cells and within tissues as a whole. Rigorous computational network modeling is most likely needed to accurately quantify ncRNAs and correlate respective datasets with one another, given the propensity for autoimmune diseases to share common and unique signatures. An in silico machine learning approach of all data may well also define unbiased novel ncRNA regulator associations with immune cell function, which can then be experimentally validated. The curation of larger datasets is critical as part of this endeavor. It is possible, of course, that some ncRNAs may be patient-specific and dependent on individual genetics, so coupling of transcriptomes to patient-specific genomes will probably be an important resource in linking ncRNAs to disease across populations. Likewise, ncRNA studies should be performed in the context of mRNAs, proteins, lipids, organelles and other omics and functional assays, establishing gene-set relationships that are integrated between the library of all available molecules. The majority of studies have focused on highly detected ncRNA molecules. It is likely that low expressed, or harder to detect ncRNAs, such as NeST (reported as one ncRNA molecule per IFNG allele in some conditions) [73], and their subtle differences in expression, are likely critical for properly assessing the regulatory nature of all ncRNAs in the context of immunological function and disease. Nevertheless, by using and regularly curating community-driven ncRNA resources, it is reasonable to be confident that future studies will directly test and find novelty in many of these hypotheses. Finally, because there are too many ncRNAs with unknown function and/or importance to disease, screening of ncRNAs, for example using a CRISPR library or RNAi, may help to uncover potential functions.
Highlights.
Precise regulation over transcriptional output is key for appropriate immune function
ncRNAs represent the majority of cellular RNAs and perform indispensable functions
ncRNAs act as biomarkers of autoimmune diseases and are therapeutic targets
We review novel methods for their detection and known roles in autoimmunity
Acknowledgments
This work was supported by extramural research programs of the NIH (R35GM138283) and the Showalter Trust (research award to MK). This research was supported (in part) by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases (project number ZIA/DK075149 to BA). The authors also gratefully acknowledge the support from the Purdue University Center for Cancer Research, P30CA023168.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to declare.
References
- [1].Roberts TC, Wood MJA, Therapeutic targeting of non-coding RNAs, Essays Biochem. 54 (2013) 127–145. 10.1042/bse0540127. [DOI] [PubMed] [Google Scholar]
- [2].Zhong X, Zhang D, Xiong M, Zhang L, Noncoding RNA for Cancer Gene Therapy, Recent Results Cancer Res. Fortschritte Krebsforsch. Progres Dans Rech. Sur Cancer 209 (2016) 51–60. 10.1007/978-3-319-42934-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu C-X, Li X, Nan F, Jiang S, Gao X, Guo S-K, Xue W, Cui Y, Dong K, Ding H, Qu B, Zhou Z, Shen N, Yang L, Chen L-L, Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity, Cell. 177 (2019) 865–880.e21. 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- [4].Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Stamatoyannopoulos JA, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SCJ, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Dutta A, Guigó R, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Flicek P, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermüller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung W-K, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Dermitzakis ET, Margulies EH, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei C-L, Ruan Y, Snyder M, Birney E, Struhl K, Gerstein M, Antonarakis SE, Gingeras TR, Brown JB, Flicek P, Fu Y, Keefe D, Birney E, Denoeud F, Gerstein M, Green ED, Kapranov P, Karaöz U, Myers RM, Noble WS, Reymond A, Rozowsky J, Struhl K, Siepel A, Stamatoyannopoulos JA, Taylor CM, Taylor J, Thurman RE, Tullius TD, Washietl S, Zheng D, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Collins FS, Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Hou M, Taylor J, Nikolaev S, Montoya-Burgos JI, Löytynoja A, Whelan S, Pardi F, Massingham T, Brown JB, Huang H, Zhang NR, Bickel P, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Gerstein M, Antonarakis SE, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Pachter L, Green ED, Sidow A, Weng Z, Trinklein ND, Fu Y, Zhang ZD, Karaöz U, Barrera L, Stuart R, Zheng D, Ghosh S, Flicek P, King DC, Taylor J, Ameur A, Enroth S, Bieda MC, Koch CM, Hirsch HA, Wei C-L, Cheng J, Kim J, Bhinge AA, Giresi PG, Jiang N, Liu J, Yao F, Sung W-K, Chiu KP, Vega VB, Lee CWH, Ng P, Shahab A, Sekinger EA, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Clelland GK, Wilcox S, Dillon SC, Andrews RM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Carter NP, Vetrie D, Kapranov P, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project, Nature. 447 (2007) 799–816. 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ponting CP, Belgard TG, Transcribed dark matter: meaning or myth?, Hum. Mol. Genet 19 (2010) R162–168. 10.1093/hmg/ddq362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC, A soluble ribonucleic acid intermediate in protein synthesis, J. Biol. Chem 231 (1958) 241–257. [PubMed] [Google Scholar]
- [7].Danon D, Marikovsky Y, Littauer UZ, A comparative electron microscopical study of RNA from different sources, J. Biophys. Biochem. Cytol 9 (1961) 253–261. 10.1083/jcb.9.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schaefer KP, Altman S, Söll D, Nucleotide modification in vitro of the precursor of transfer RNA of Escherichia coli, Proc. Natl. Acad. Sci. U. S. A 70 (1973) 3626–3630. 10.1073/pnas.70.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gall JG, Small granules in the amphibian oocyte nucleus and their relationship to RNA, J. Biophys. Biochem. Cytol 2 (1956) 393–396. 10.1083/jcb.2.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Greenberg H, Penman S, Methylation and processing of ribosomal RNA in HeLa cells, J. Mol. Biol 21 (1966) 527–535. 10.1016/0022-2836(66)90025-8. [DOI] [PubMed] [Google Scholar]
- [11].Delisi C, Crothers DM, Prediction of RNA secondary structure, Proc. Natl. Acad. Sci. U. S. A 68 (1971) 2682–2685. 10.1073/pnas.68.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].McCarrey JR, Dilworth DD, Expression of Xist in mouse germ cells correlates with X-chromosome inactivation, Nat. Genet 2 (1992) 200–203. 10.1038/ng1192-200. [DOI] [PubMed] [Google Scholar]
- [13].Stark R, Grzelak M, Hadfield J, RNA sequencing: the teenage years, Nat. Rev. Genet 20 (2019) 631–656. 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- [14].Chen YG, Satpathy AT, Chang HY, Gene regulation in the immune system by long noncoding RNAs, Nat. Immunol 18 (2017) 962–972. 10.1038/ni.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Salvi V, Gianello V, Tiberio L, Sozzani S, Bosisio D, Cytokine Targeting by miRNAs in Autoimmune Diseases, Front. Immunol 10 (2019). 10.3389/fimmu.2019.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berg MD, Brandl CJ, Transfer RNAs: diversity in form and function, RNA Biol. 18 (2021) 316–339. 10.1080/15476286.2020.1809197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li S, Xu Z, Sheng J, tRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA, Genes. 9 (2018) E246. 10.3390/genes9050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhong F, Zhou N, Wu K, Guo Y, Tan W, Zhang H, Zhang X, Geng G, Pan T, Luo H, Zhang Y, Xu Z, Liu J, Liu B, Gao W, Liu C, Ren L, Li J, Zhou J, Zhang H, A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes, Nucleic Acids Res. 43 (2015) 10474–10491. 10.1093/nar/gkv954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Negishi H, Endo N, Nakajima Y, Nishiyama T, Tabunoki Y, Nishio J, Koshiba R, Matsuda A, Matsuki K, Okamura T, Negishi-Koga T, Ichinohe T, Takemura S, Ishiwata H, Iemura S-I, Natsume T, Abe T, Kiyonari H, Doi T, Hangai S, Yanai H, Fujio K, Yamamoto K, Taniguchi T, Identification of U11snRNA as an endogenous agonist of TLR7-mediated immune pathogenesis, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 23653–23661. 10.1073/pnas.1915326116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yao R-W, Wang Y, Chen L-L, Cellular functions of long noncoding RNAs, Nat. Cell Biol 21 (2019) 542–551. 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- [21].Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C, Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E3472–E3480. 10.1073/pnas.1700946114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Aune TM, Crooke PS, Patrick AE, Tossberg JT, Olsen NJ, Spurlock CF, Expression of long non-coding RNAs in autoimmunity and linkage to enhancer function and autoimmune disease risk genetic variants, J. Autoimmun 81 (2017) 99–109. 10.1016/j.jaut.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang Z, Hong W, Ruan H, Jing Y, Li S, Liu Y, Wang J, Li W, Diao L, Han L, HeRA: an atlas of enhancer RNAs across human tissues, Nucleic Acids Res. 49 (2021) D932–D938. 10.1093/nar/gkaa940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhou Z, Sun B, Huang S, Zhao L, Roles of circular RNAs in immune regulation and autoimmune diseases, Cell Death Dis. 10 (2019) 1–13. 10.1038/s41419-019-1744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Patop IL, Wüst S, Kadener S, Past, present, and future of circRNAs, EMBO J. 38 (2019) e100836. 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ, Lin28 enhances tissue repair by reprogramming cellular metabolism, Cell. 155 (2013) 778–792. 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brockdorff N, Noncoding RNA and Polycomb recruitment, RNA N. Y. N 19 (2013) 429–442. 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE, The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites, Mol. Cell 55 (2014) 791–802. 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R, Activating RNAs associate with Mediator to enhance chromatin architecture and transcription, Nature. 494 (2013) 497–501. 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M, Splicing regulation by long noncoding RNAs, Nucleic Acids Res. 46 (2018) 2169–2184. 10.1093/nar/gky095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rothschild G, Zhang W, Lim J, Giri PK, Laffleur B, Chen Y, Fang M, Chen Y, Nair L, Liu Z-P, Deng H, Hammarström L, Wang J, Basu U, Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class switch recombination, Sci. Immunol 5 (2020). 10.1126/sciimmunol.aay5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N, An mRNA-Derived Noncoding RNA Targets and Regulates the Ribosome, Mol. Cell 54 (2014) 147–155. 10.1016/j.molcel.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pircher A, Gebetsberger J, Polacek N, Ribosome-associated ncRNAs: An emerging class of translation regulators, RNA Biol. 11 (2015) 1335–1339. 10.1080/15476286.2014.996459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao M, Zhu N, Hao F, Song Y, Wang Z, Ni Y, Ding L, The Regulatory Role of Non-coding RNAs on Programmed Cell Death Four in Inflammation and Cancer, Front. Oncol 9 (2019) 919. 10.3389/fonc.2019.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Carpenter S, Fitzgerald KA, Cytokines and Long Noncoding RNAs, Cold Spring Harb. Perspect. Biol 10 (2018). 10.1101/cshperspect.a028589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jonas S, Izaurralde E, Towards a molecular understanding of microRNA-mediated gene silencing, Nat. Rev. Genet 16 (2015) 421–433. 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- [37].Wang KC, Chang HY, Molecular mechanisms of long noncoding RNAs, Mol. Cell 43 (2011) 904–914. 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen L-L, Yang L, Regulation of circRNA biogenesis, RNA Biol. 12 (2015) 381–388. 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME, Widespread transcription at neuronal activity-regulated enhancers, Nature. 465 (2010) 182–187. 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei C-L, Natoli G, A large fraction of extragenic RNA pol II transcription sites overlap enhancers, PLoS Biol. 8 (2010) e1000384. 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kim T-K, Hemberg M, Gray JM, Enhancer RNAs: a class of long noncoding RNAs synthesized at enhancers, Cold Spring Harb. Perspect. Biol 7 (2015) a018622. 10.1101/cshperspect.a018622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bose DA, Berger SL, eRNA binding produces tailored CBP activity profiles to regulate gene expression, RNA Biol. 14 (2017) 1655–1659. 10.1080/15476286.2017.1353862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Meng H, Bartholomew B, Emerging roles of transcriptional enhancers in chromatin looping and promoter-proximal pausing of RNA polymerase II, J. Biol. Chem 293 (2018) 13786–13794. 10.1074/jbc.R117.813485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yan B, Chakravorty S, Mirabelli C, Wang L, Trujillo-Ochoa JL, Chauss D, Kumar D, Lionakis MS, Olson MR, Wobus CE, Afzali B, Kazemian M, Host-Virus Chimeric Events in SARS-CoV-2-Infected Cells Are Infrequent and Artifactual, J. Virol 95 (2021) e0029421. 10.1128/JVI.00294-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Core LJ, Waterfall JJ, Lis JT, Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters, Science. 322 (2008) 1845–1848. 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS, Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution, Cell. 161 (2015) 541–554. 10.1016/j.cell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ, Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing, Cell. 161 (2015) 526–540. 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gardini A, Global Run-On Sequencing (GRO-Seq), Methods Mol. Biol. Clifton NJ 1468 (2017) 111–120. 10.1007/978-1-4939-4035-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Barbieri E, Hill C, Quesnel-Vallières M, Zucco AJ, Barash Y, Gardini A, Rapid and Scalable Profiling of Nascent RNA with fastGRO, Cell Rep. 33 (2020) 108373. 10.1016/j.celrep.2020.108373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chauss D, Basu S, Rajakaruna S, Ma Z, Gau V, Anastas S, Brennan LA, Hejtmancik JF, Menko AS, Kantorow M, Differentiation state-specific mitochondrial dynamic regulatory networks are revealed by global transcriptional analysis of the developing chicken lens, G3 Bethesda Md. 4 (2014) 1515–1527. 10.1534/g3.114.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Atkins N, Miller CM, Owens JR, Turek FW, Non-laser capture microscopy approach for the microdissection of discrete mouse brain regions for total RNA isolation and downstream next-generation sequencing and gene expression profiling, J. Vis. Exp. JoVE (2011) 3125. 10.3791/3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhou J, Xu J, Zhang L, Liu S, Ma Y, Wen X, Hao J, Li Z, Ni Y, Li X, Zhou F, Li Q, Wang F, Wang X, Si Y, Zhang P, Liu C, Bartolomei M, Tang F, Liu B, Yu J, Lan Y, Combined Single-Cell Profiling of lncRNAs and Functional Screening Reveals that H19 Is Pivotal for Embryonic Hematopoietic Stem Cell Development, Cell Stem Cell. 24 (2019) 285–298.e5. 10.1016/j.stem.2018.11.023. [DOI] [PubMed] [Google Scholar]
- [53].Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R, CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes, Nat. Protoc 15 (2020) 1484–1506. 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- [54].Maynard KR, Collado-Torres L, Weber LM, Uytingco C, Barry BK, Williams SR, Catallini JL, Tran MN, Besich Z, Tippani M, Chew J, Yin Y, Kleinman JE, Hyde TM, Rao N, Hicks SC, Martinowich K, Jaffe AE, Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex, Nat. Neurosci 24 (2021) 425–436. 10.1038/s41593-020-00787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X, RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells, Science. 348 (2015) aaa6090. 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Janesick A, Shelansky R, Gottscho A, Wagner F, Rouault M, Beliakoff G, de Oliveira MF, Kohlway A, Abousoud J, Morrison C, Drennon TY, Mohabbat S, Williams S, 10x Development Teams, Taylor S, High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and in situ analysis of FFPE tissue, (2022) 2022.10.06.510405. 10.1101/2022.10.06.510405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xia C, Fan J, Emanuel G, Hao J, Zhuang X, Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 19490–19499. 10.1073/pnas.1912459116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yan Z, Huang N, Wu W, Chen W, Jiang Y, Chen J, Huang X, Wen X, Xu J, Jin Q, Zhang K, Chen Z, Chien S, Zhong S, Genome-wide colocalization of RNA–DNA interactions and fusion RNA pairs, Proc. Natl. Acad. Sci 116 (2019) 3328–3337. 10.1073/pnas.1819788116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lorenz M, Visualizing protein–RNA interactions inside cells by fluorescence resonance energy transfer, RNA. 15 (2009) 97–103. 10.1261/rna.1307809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, Luscombe NM, Ule J, hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1, Nature. 519 (2015) 491–494. 10.1038/nature14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yoon J-H, Srikantan S, Gorospe M, MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs, Methods San Diego Calif. 58 (2012) 81–87. 10.1016/j.ymeth.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D, Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 10010–10015. 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cai Z, Cao C, Ji L, Ye R, Wang D, Xia C, Wang S, Du Z, Hu N, Yu X, Chen J, Wang L, Yang X, He S, Xue Y, RIC-seq for global in situ profiling of RNA-RNA spatial interactions, Nature. 582 (2020) 432–437. 10.1038/s41586-020-2249-1. [DOI] [PubMed] [Google Scholar]
- [64].Morf J, Wingett SW, Farabella I, Cairns J, Furlan-Magaril M, Jiménez-García LF, Liu X, Craig FF, Walker S, Segonds-Pichon A, Andrews S, Marti-Renom MA, Fraser P, RNA proximity sequencing reveals the spatial organization of the transcriptome in the nucleus, Nat. Biotechnol 37 (2019) 793–802. 10.1038/s41587-019-0166-3. [DOI] [PubMed] [Google Scholar]
- [65].Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR, Chang HY, RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure, Cell. 165 (2016) 1267–1279. 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ, Global Mapping of Human RNA-RNA Interactions, Mol. Cell 62 (2016) 618–626. 10.1016/j.molcel.2016.04.030. [DOI] [PubMed] [Google Scholar]
- [67].Nguyen TC, Cao X, Yu P, Xiao S, Lu J, Biase FH, Sridhar B, Huang N, Zhang K, Zhong S, Mapping RNA-RNA interactome and RNA structure in vivo by MARIO, Nat. Commun 7 (2016) 12023. 10.1038/ncomms12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chen C-Z, Li L, Lodish HF, Bartel DP, MicroRNAs modulate hematopoietic lineage differentiation, Science. 303 (2004) 83–86. 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- [69].Huang Z, Luo Q, Yao F, Qing C, Ye J, Deng Y, Li J, Identification of Differentially Expressed Long Non-coding RNAs in Polarized Macrophages, Sci. Rep 6 (2016) 19705. 10.1038/srep19705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Sun D, Yu Z, Fang X, Liu M, Pu Y, Shao Q, Wang D, Zhao X, Huang A, Xiang Z, Zhao C, Franklin RJ, Cao L, He C, LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination, EMBO Rep. 18 (2017) 1801–1816. 10.15252/embr.201643668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Li J, Tian J, Lu J, Wang Z, Ling J, Wu X, Yang F, Xia Y, LncRNA GAS5 inhibits Th17 differentiation and alleviates immune thrombocytopenia via promoting the ubiquitination of STAT3, Int. Immunopharmacol 80 (2020) 106127. 10.1016/j.intimp.2019.106127. [DOI] [PubMed] [Google Scholar]
- [72].Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X, The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation, Science. 344 (2014) 310–313. 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- [73].Petermann F, Pękowska A, Johnson CA, Jankovic D, Shih H-Y, Jiang K, Hudson WH, Brooks SR, Sun H-W, Villarino AV, Yao C, Singleton K, Akondy RS, Kanno Y, Sher A, Casellas R, Ahmed R, O’Shea JJ, The Magnitude of IFN-γ Responses Is Fine-Tuned by DNA Architecture and the Non-coding Transcript of Ifng-as1, Mol. Cell 75 (2019) 1229–1242.e5. 10.1016/j.molcel.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Stein N, Berhani O, Schmiedel D, Duev-Cohen A, Seidel E, Kol I, Tsukerman P, Hecht M, Reches A, Gamliel M, Obeidat A, Charpak-Amikam Y, Yamin R, Mandelboim O, IFNG-AS1 Enhances Interferon Gamma Production in Human Natural Killer Cells, IScience. 11 (2019) 466–473. 10.1016/j.isci.2018.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhang F, Liu G, Wei C, Gao C, Hao J, Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 31 (2017) 519–525. 10.1096/fj.201600838R. [DOI] [PubMed] [Google Scholar]
- [76].Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K, Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation, Nat. Immunol 14 (2013) 1190–1198. 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wells AC, Pobezinskaya EL, Pobezinsky LA, Non-coding RNAs in CD8 T cell biology, Mol. Immunol 120 (2020) 67–73. 10.1016/j.molimm.2020.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Noncoding RNA transcription alters chromosomal topology to promote isotype-specific class switch recombination | Science Immunology, (n.d.). https://immunology.sciencemag.org/content/5/44/eaay5864 (accessed June 20, 2020). [DOI] [PMC free article] [PubMed]
- [79].Wu J, Zhang H, Zheng Y, Jin X, Liu M, Li S, Zhao Q, Liu X, Wang Y, Shi M, Zhang S, Tian J, Sun Y, Zhang M, Yu B, The Long Noncoding RNA MALAT1 Induces Tolerogenic Dendritic Cells and Regulatory T Cells via miR155/Dendritic Cell-Specific Intercellular Adhesion Molecule-3 Grabbing Nonintegrin/IL10 Axis, Front. Immunol 9 (2018) 1847. 10.3389/fimmu.2018.01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zurawska A, Mycko MP, Selmaj KW, Circular RNAs as a novel layer of regulatory mechanism in multiple sclerosis, J. Neuroimmunol 334 (2019) 576971. 10.1016/j.jneuroim.2019.576971. [DOI] [PubMed] [Google Scholar]
- [81].IIott NE, Heward JA, Roux B, Tsitsiou E, Fenwick PS, Lenzi L, Goodhead I, Hertz-Fowler C, Heger A, Hall N, Donnelly LE, Sims D, Lindsay MA, Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes, Nat. Commun 5 (2014) 3979. 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Heward JA, Roux BT, Lindsay MA, Divergent signalling pathways regulate lipopolysaccharide-induced eRNA expression in human monocytic THP1 cells, FEBS Lett. 589 (2015) 396–406. 10.1016/j.febslet.2014.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bouvier G, Watrin F, Naspetti M, Verthuy C, Naquet P, Ferrier P, Deletion of the mouse T-cell receptor beta gene enhancer blocks alphabeta T-cell development, Proc. Natl. Acad. Sci. U. S. A 93 (1996) 7877–7881. 10.1073/pnas.93.15.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Sleckman BP, Bardon CG, Ferrini R, Davidson L, Alt FW, Function of the TCR alpha enhancer in alphabeta and gammadelta T cells, Immunity. 7 (1997) 505–515. 10.1016/s1074-7613(00)80372-6. [DOI] [PubMed] [Google Scholar]
- [85].Peng H, Xing J, Wang X, Ding X, Tang X, Zou J, Wang S, Liu Y, Circular RNA circNUP214 Modulates the T Helper 17 Cell Response in Patients With Rheumatoid Arthritis, Front. Immunol 13 (2022) 885896. 10.3389/fimmu.2022.885896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Liu B, Ye B, Zhu X, Yang L, Li H, Liu N, Zhu P, Lu T, He L, Tian Y, Fan Z, An inducible circular RNA circKcnt2 inhibits ILC3 activation to facilitate colitis resolution, Nat. Commun 11 (2020) 4076. 10.1038/s41467-020-17944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Liu N, He J, Fan D, Gu Y, Wang J, Li H, Zhu X, Du Y, Tian Y, Liu B, Fan Z, Circular RNA circTmem241 drives group III innate lymphoid cell differentiation via initiation of Elk3 transcription, Nat. Commun 13 (2022) 4711. 10.1038/s41467-022-32322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Song H, Yang Y, Sun Y, Wei G, Zheng H, Chen Y, Cai D, Li C, Ma Y, Lin Z, Shi X, Liao W, Liao Y, Zhong L, Bin J, Circular RNA Cdyl promotes abdominal aortic aneurysm formation by inducing M1 macrophage polarization and M1-type inflammation, Mol. Ther. J. Am. Soc. Gene Ther 30 (2022) 915–931. 10.1016/j.ymthe.2021.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Zhang J, Cheng F, Rong G, Tang Z, Gui B, Circular RNA hsa_circ_0005567 overexpression promotes M2 type macrophage polarization through miR-492/SOCS2 axis to inhibit osteoarthritis progression, Bioengineered. 12 (2021) 8920–8930. 10.1080/21655979.2021.1989999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, Wang W, Liu S, Liu W, Ma Y, Zhang L, Cao X, Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response, Cell. 173 (2018) 906–919.e13. 10.1016/j.cell.2018.03.064. [DOI] [PubMed] [Google Scholar]
- [91].Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, Kelly GP, Lobley A, Walker J, Spencer-Dene B, Howell M, Stewart A, Svejstrup JQ, UV Irradiation Induces a Non-coding RNA that Functionally Opposes the Protein Encoded by the Same Gene, Cell. 168 (2017) 843–855.e13. 10.1016/j.cell.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhang P, Cao L, Zhou R, Yang X, Wu M, The lncRNA Neat1 promotes activation of inflammasomes in macrophages, Nat. Commun 10 (2019) 1495. 10.1038/s41467-019-09482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li Y, Wang C, Miao Z, Bi X, Wu D, Jin N, Wang L, Wu H, Qian K, Li C, Zhang T, Zhang C, Yi Y, Lai H, Hu Y, Cheng L, Leung K-S, Li X, Zhang F, Li K, Li X, Wang D, ViRBase: a resource for virus-host ncRNA-associated interactions, Nucleic Acids Res. 43 (2015) D578–582. 10.1093/nar/gku903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chung JY, Ain QU, Song Y, Yong S-B, Kim Y-H, Targeted delivery of CRISPR interference system against Fabp4 to white adipocytes ameliorates obesity, inflammation, hepatic steatosis, and insulin resistance, Genome Res. 29 (2019) 1442–1452. 10.1101/gr.246900.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhang Y, Yuan F, Liu L, Chen Z, Ma X, Lin Z, Zou J, The Role of the miR-21/SPRY2 Axis in Modulating Proangiogenic Factors, Epithelial Phenotypes, and Wound Healing in Corneal Epithelial Cells, Invest. Ophthalmol. Vis. Sci 60 (2019) 3854–3862. 10.1167/iovs.19-27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ranzani V, Rossetti G, Panzeri I, Arrigoni A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M, De Francesco R, Geginat J, Bodega B, Abrignani S, Pagani M, The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4, Nat. Immunol 16 (2015) 318–325. 10.1038/ni.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dolcino M, Tinazzi E, Vitali C, Papa ND, Puccetti A, Lunardi C, Long Non-Coding RNAs Modulate Sjögren’s Syndrome Associated Gene Expression and Are Involved in the Pathogenesis of the Disease, J. Clin. Med 8 (2019) 1349. 10.3390/jcm8091349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Yarani R, Mirza AH, Kaur S, Pociot F, The emerging role of lncRNAs in inflammatory bowel disease, Exp. Mol. Med 50 (2018) 1–14. 10.1038/s12276-018-0188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zhang F, Wu L, Qian J, Qu B, Xia S, La T, Wu Y, Ma J, Zeng J, Guo Q, Cui Y, Yang W, Huang J, Zhu W, Yao Y, Shen N, Tang Y, Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus, J. Autoimmun 75 (2016) 96–104. 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- [100].Li H, Li K, Lai W, Li X, Wang H, Yang J, Chu S, Wang H, Kang C, Qiu Y, Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus, Clin. Chim. Acta Int. J. Clin. Chem 480 (2018) 17–25. 10.1016/j.cca.2018.01.026. [DOI] [PubMed] [Google Scholar]
- [101].Zheng F, Yu X, Huang J, Dai Y, Circular RNA expression profiles of peripheral blood mononuclear cells in rheumatoid arthritis patients, based on microarray chip technology, Mol. Med. Rep 16 (2017) 8029–8036. 10.3892/mmr.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Li Z, Li X, Jiang C, Qian W, Tse G, Chan MTV, Wu WKK, Long non-coding RNAs in rheumatoid arthritis, Cell Prolif. 51 (2018). 10.1111/cpr.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Gupta R, Ahn R, Lai K, Mullins E, Debbaneh M, Dimon M, Arron S, Liao W, Landscape of Long Noncoding RNAs in Psoriatic and Healthy Skin, J. Invest. Dermatol 136 (2016) 603–609. 10.1016/j.jid.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Dolcino M, Pelosi A, Fiore PF, Patuzzo G, Tinazzi E, Lunardi C, Puccetti A, Long Non-Coding RNAs Play a Role in the Pathogenesis of Psoriatic Arthritis by Regulating MicroRNAs and Genes Involved in Inflammation and Metabolic Syndrome, Front. Immunol 9 (2018) 1533. 10.3389/fimmu.2018.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Pockar S, Globocnik Petrovic M, Peterlin B, Vidovic Valentincic N, MiRNA as biomarker for uveitis - A systematic review of the literature, Gene. 696 (2019) 162–175. 10.1016/j.gene.2019.02.004. [DOI] [PubMed] [Google Scholar]
- [106].Hou S, Li N, Liao X, Kijlstra A, Yang P, Uveitis genetics, Exp. Eye Res 190 (2020) 107853. 10.1016/j.exer.2019.107853. [DOI] [PubMed] [Google Scholar]
- [107].Yue Y, Zhang J, Yang L, Liu S, Qi J, Cao Q, Zhou C, Wang Y, Kijlstra A, Yang P, Hou S, Association of Long Noncoding RNAs Polymorphisms With Ankylosing Spondylitis, Vogt-Koyanagi-Harada Disease, and Behcet’s Disease, Invest. Ophthalmol. Vis. Sci 59 (2018) 1158–1166. 10.1167/iovs.17-23247. [DOI] [PubMed] [Google Scholar]
- [108].Yang X, Wu Y, Zhang B, Ni B, Noncoding RNAs in multiple sclerosis, Clin. Epigenetics 10 (2018) 149. 10.1186/s13148-018-0586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Liao Z, Ye Z, Xue Z, Wu L, Ouyang Y, Yao C, Cui C, Xu N, Ma J, Hou G, Wang J, Meng Y, Yin Z, Liu Y, Qian J, Zhang C, Ding H, Guo Q, Qu B, Shen N, Identification of Renal Long Non-coding RNA RP11–2B6.2 as a Positive Regulator of Type I Interferon Signaling Pathway in Lupus Nephritis, Front. Immunol 10 (2019). 10.3389/fimmu.2019.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Kong X, Wang J, Cao Y, Zhang H, Lu X, Wang Y, Bo C, Wang T, Li S, Tian K, Liu Z, Wang L, The long noncoding RNA MALAT-1 functions as a competing endogenous RNA to regulate MSL2 expression by sponging miR-338–3p in myasthenia gravis, J. Cell. Biochem 120 (2019) 5542–5550. 10.1002/jcb.27838. [DOI] [PubMed] [Google Scholar]
- [111].Xing L, Xu W, Qu Y, Zhao M, Zhu H, Liu H, Wang H, Su X, Shao Z, miR-150 regulates B lymphocyte in autoimmune hemolytic anemia/Evans syndrome by c-Myb, Int. J. Hematol 107 (2018) 666–672. 10.1007/s12185-018-2429-z. [DOI] [PubMed] [Google Scholar]
- [112].Xie N, Liu G, ncRNA-regulated immune response and its role in inflammatory lung diseases, Am. J. Physiol. - Lung Cell. Mol. Physiol 309 (2015) L1076–L1087. 10.1152/ajplung.00286.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I, The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita, Nature. 413 (2001) 432–435. 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- [114].Hertweck A, Evans CM, Eskandarpour M, Lau JCH, Oleinika K, Jackson I, Kelly A, Ambrose J, Adamson P, Cousins DJ, Lavender P, Calder VL, Lord GM, Jenner RG, T-bet Activates Th1 Genes through Mediator and the Super Elongation Complex, Cell Rep. 15 (2016) 2756–2770. 10.1016/j.celrep.2016.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Tan SH, Leong WZ, Ngoc PCT, Tan TK, Bertulfo FC, Lim MC, An O, Li Z, Yeoh AEJ, Fullwood MJ, Tenen DG, Sanda T, The enhancer RNA ARIEL activates the oncogenic transcriptional program in T-cell acute lymphoblastic leukemia, Blood. 134 (2019) 239–251. 10.1182/blood.2018874503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Chakravorty S, Afzali B, Kazemian M, EBV-associated diseases: Current therapeutics and emerging technologies, Front. Immunol 13 (2022). https://www.frontiersin.org/articles/10.3389/fimmu.2022.1059133 (accessed October 28, 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Saçar Demirci MD, Adan A, Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection, PeerJ. 8 (2020). 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hrdlickova B, Kumar V, Kanduri K, Zhernakova DV, Tripathi S, Karjalainen J, Lund RJ, Li Y, Ullah U, Modderman R, Abdulahad W, Lähdesmäki H, Franke L, Lahesmaa R, Wijmenga C, Withoff S, Expression profiles of long non-coding RNAs located in autoimmune disease-associated regions reveal immune cell-type specificity, Genome Med. 6 (2014). 10.1186/s13073-014-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Dong P, Xiong Y, Yue J, Hanley SJB, Kobayashi N, Todo Y, Watari H, Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors, Front. Genet 9 (2018). 10.3389/fgene.2018.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Simchovitz A, Hanan M, Niederhoffer N, Madrer N, Yayon N, Bennett ER, Greenberg DS, Kadener S, Soreq H, NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 33 (2019) 11223–11234. 10.1096/fj.201900830R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang Z, Zhao Y, Xu N, Zhang S, Wang S, Mao Y, Zhu Y, Li B, Jiang Y, Tan Y, Xie W, Yang BB, Zhang Y, NEAT1 regulates neuroglial cell mediating Aβ clearance via the epigenetic regulation of endocytosis-related genes expression, Cell. Mol. Life Sci. CMLS 76 (2019) 3005–3018. 10.1007/s00018-019-03074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Dastmalchi R, Ghafouri-Fard S, Omrani MD, Mazdeh M, Sayad A, Taheri M, Dysregulation of long non-coding RNA profile in peripheral blood of multiple sclerosis patients, Mult. Scler. Relat. Disord 25 (2018) 219–226. 10.1016/j.msard.2018.07.044. [DOI] [PubMed] [Google Scholar]
- [123].Kelley N, Jeltema D, Duan Y, He Y, The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation, Int. J. Mol. Sci 20 (2019) E3328. 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zhang M, Zheng Y, Sun Y, Li S, Chen L, Jin X, Hou X, Liu X, Chen Q, Li J, Liu M, Zheng X, Zhang Y, Wu J, Yu B, Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome, Theranostics. 9 (2019) 3425–3442. 10.7150/thno.33178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shui X, Chen S, Lin J, Kong J, Zhou C, Wu J, Knockdown of lncRNA NEAT1 inhibits Th17/CD4+ T cell differentiation through reducing the STAT3 protein level, J. Cell. Physiol 234 (2019) 22477–22484. 10.1002/jcp.28811. [DOI] [PubMed] [Google Scholar]
- [126].Goustin AS, Thepsuwan P, Kosir MA, Lipovich L, The Growth-Arrest-Specific (GAS)-5 Long Non-Coding RNA: A Fascinating lncRNA Widely Expressed in Cancers, Non-Coding RNA. 5 (2019). 10.3390/ncrna5030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP, Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor, Sci. Signal 3 (2010) ra8. 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ji J, Dai X, Yeung S-CJ, He X, The role of long non-coding RNA GAS5 in cancers, Cancer Manag. Res 11 (2019) 2729–2737. 10.2147/CMAR.S189052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Jin F, Wang N, Zhu Y, You L, Wang L, De W, Tang W, Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells, Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 43 (2017) 2062–2073. 10.1159/000484191. [DOI] [PubMed] [Google Scholar]
- [130].Li J, Wang Y, Zhang C-G, Xiao H-J, Xiao H-J, Hu J-M, Hou J-M, He J-D, Effect of long non-coding RNA Gas5 on proliferation, migration, invasion and apoptosis of colorectal cancer HT-29 cell line, Cancer Cell Int. 18 (2018) 4. 10.1186/s12935-017-0478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Liu W, Zhan J, Zhong R, Li R, Sheng X, Xu M, Lu Z, Zhang S, Upregulation of Long Noncoding RNA_GAS5 Suppresses Cell Proliferation and Metastasis in Laryngeal Cancer via Regulating PI3K/AKT/mTOR Signaling Pathway, Technol. Cancer Res. Treat 20 (2021) 1533033821990074. 10.1177/1533033821990074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT, GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer, Oncogene. 28 (2009) 195–208. 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- [133].Sun M, Jin F, Xia R, Kong R, Li J, Xu T, Liu Y, Zhang E, Liu X, De W, Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer, BMC Cancer. 14 (2014) 319. 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Bian D, Shi W, Shao Y, Li P, Song G, Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in melanoma, Am. J. Transl. Res 9 (2017) 1509–1520. [PMC free article] [PubMed] [Google Scholar]
- [135].Wu G-C, Li J, Leng R-X, Li X-P, Li X-M, Wang D-G, Pan H-F, Ye D-Q, Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus, Oncotarget. 8 (2017) 23650–23663. 10.18632/oncotarget.15569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Suo Q-F, Sheng J, Qiang F-Y, Tang Z-S, Yang Y-Y, Association of long non-coding RNA GAS5 and miR-21 levels in CD4+ T cells with clinical features of systemic lupus erythematosus, Exp. Ther. Med 15 (2018) 345–350. 10.3892/etm.2017.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhu X, Wang X, Wang Y, Zhao Y, The regulatory network among CircHIPK3, LncGAS5, and miR-495 promotes Th2 differentiation in allergic rhinitis, Cell Death Dis. 11 (2020) 216. 10.1038/s41419-020-2394-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wilusz JE, Freier SM, Spector DL, 3’ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA, Cell. 135 (2008) 919–932. 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Slomovic S, Laufer D, Geiger D, Schuster G, Polyadenylation of ribosomal RNA in human cells, Nucleic Acids Res. 34 (2006) 2966–2975. 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Toompuu M, Tuomela T, Laine P, Paulin L, Dufour E, Jacobs HT, Polyadenylation and degradation of structurally abnormal mitochondrial tRNAs in human cells, Nucleic Acids Res. 46 (2018) 5209–5226. 10.1093/nar/gky159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zhang Y, Zhang G, Liu Y, Chen R, Zhao D, McAlister V, Mele T, Liu K, Zheng X, GDF15 Regulates Malat-1 Circular RNA and Inactivates NFκB Signaling Leading to Immune Tolerogenic DCs for Preventing Alloimmune Rejection in Heart Transplantation, Front. Immunol 9 (2018) 2407. 10.3389/fimmu.2018.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Li Z, Zhang Q, Wu Y, Hu F, Gu L, Chen T, Wang W, lncRNA Malat1 modulates the maturation process, cytokine secretion and apoptosis in airway epithelial cell-conditioned dendritic cells, Exp. Ther. Med 16 (2018) 3951–3958. 10.3892/etm.2018.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Hewitson JP, West KA, James KR, Rani GF, Dey N, Romano A, Brown N, Teichmann SA, Kaye PM, Lagos D, Malat1 Suppresses Immunity to Infection through Promoting Expression of Maf and IL-10 in Th Cells, J. Immunol (2020). 10.4049/jimmunol.1900940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Liang Z, Tang F, The potency of lncRNA MALAT1/miR-155/CTLA4 axis in altering Th1/Th2 balance of asthma, Biosci. Rep 40 (2020). 10.1042/BSR20190397. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [145].Cui H, Banerjee S, Guo S, Xie N, Ge J, Jiang D, Zörnig M, Thannickal VJ, Liu G, Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury, JCI Insight. 4 (2019). 10.1172/jci.insight.124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Masoumi F, Ghorbani S, Talebi F, Branton WG, Rajaei S, Power C, Noorbakhsh F, Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis, J. Neuroimmunol 328 (2019) 50–59. 10.1016/j.jneuroim.2018.11.013. [DOI] [PubMed] [Google Scholar]
- [147].Rawlings DJ, Metzler G, Wray-Dutra M, Jackson SW, Altered B cell signalling in autoimmunity, Nat. Rev. Immunol 17 (2017) 421–436. 10.1038/nri.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Barrat FJ, Crow MK, Ivashkiv LB, Interferon target-gene expression and epigenomic signatures in health and disease, Nat. Immunol 20 (2019) 1574–1583. 10.1038/s41590-019-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Harley JB, Chen X, Pujato M, Miller D, Maddox A, Forney C, Magnusen AF, Lynch A, Chetal K, Yukawa M, Barski A, Salomonis N, Kaufman KM, Kottyan LC, Weirauch MT, Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity, Nat. Genet 50 (2018) 699–707. 10.1038/s41588-018-0102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Patel CH, Leone RD, Horton MR, Powell JD, Targeting metabolism to regulate immune responses in autoimmunity and cancer, Nat. Rev. Drug Discov 18 (2019) 669–688. 10.1038/s41573-019-0032-5. [DOI] [PubMed] [Google Scholar]
- [151].Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, Cook S, Tankou S, Stuart F, Melo K, Nejad P, Smith K, Topçuolu BD, Holden J, Kivisäkk P, Chitnis T, De Jager PL, Quintana FJ, Gerber GK, Bry L, Weiner HL, Alterations of the human gut microbiome in multiple sclerosis, Nat. Commun 7 (2016) 12015. 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Scheiermann C, Kunisaki Y, Frenette PS, Circadian control of the immune system, Nat. Rev. Immunol 13 (2013) 190–198. 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Schmidt RE, Grimbacher B, Witte T, Autoimmunity and primary immunodeficiency: two sides of the same coin?, Nat. Rev. Rheumatol 14 (2017) 7–18. 10.1038/nrrheum.2017.198. [DOI] [PubMed] [Google Scholar]
- [154].Rosenblum MD, Remedios KA, Abbas AK, Mechanisms of human autoimmunity, J. Clin. Invest 125 (2015) 2228–2233. 10.1172/JCI78088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Dodd MC, Bigley NJ, Geyer VB, McCOY FW, Wilson HE, Autoimmune response in rabbits injected with rat and rabbit liver ribosomes, Science. 137 (1962) 688–689. 10.1126/science.137.3531.688. [DOI] [PubMed] [Google Scholar]
- [156].Ahmadi M, Yousefi M, Abbaspour-Aghdam S, Dolati S, Aghebati-Maleki L, Eghbal-Fard S, Khabbazi A, Rostamzadeh D, Alipour S, Shabani M, Nouri M, Babaloo Z, Disturbed Th17/Treg balance, cytokines, and miRNAs in peripheral blood of patients with Behcet’s disease, J. Cell. Physiol 234 (2019) 3985–3994. 10.1002/jcp.27207. [DOI] [PubMed] [Google Scholar]
- [157].Jennewein C, von Knethen A, Schmid T, Brüne B, MicroRNA-27b Contributes to Lipopolysaccharide-mediated Peroxisome Proliferator-activated Receptor γ (PPARγ) mRNA Destabilization, J. Biol. Chem 285 (2010) 11846–11853. 10.1074/jbc.M109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Xu G, Thielen LA, Chen J, Grayson TB, Grimes T, Bridges SL, Tse HM, Smith B, Patel R, Li P, Evans-Molina C, Ovalle F, Shalev A, Serum miR-204 is an early biomarker of type 1 diabetes-associated pancreatic beta-cell loss, Am. J. Physiol. Endocrinol. Metab 317 (2019) E723–E730. 10.1152/ajpendo.00122.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Aletaha D, Smolen JS, Diagnosis and Management of Rheumatoid Arthritis: A Review, JAMA. 320 (2018) 1360–1372. 10.1001/jama.2018.13103. [DOI] [PubMed] [Google Scholar]
- [160].Churov AV, Oleinik EK, Knip M, MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential, Autoimmun. Rev 14 (2015) 1029–1037. 10.1016/j.autrev.2015.07.005. [DOI] [PubMed] [Google Scholar]
- [161].Sujitha S, Rasool M, MicroRNAs and bioactive compounds on TLR/MAPK signaling in rheumatoid arthritis, Clin. Chim. Acta Int. J. Clin. Chem 473 (2017) 106–115. 10.1016/j.cca.2017.08.021. [DOI] [PubMed] [Google Scholar]
- [162].Wang W, Zhang Y, Zhu B, Duan T, Xu Q, Wang R, Lu L, Jiao Z, Plasma microRNA expression profiles in Chinese patients with rheumatoid arthritis, Oncotarget. 6 (2015) 42557–42568. 10.18632/oncotarget.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Evangelatos G, Fragoulis GE, Koulouri V, Lambrou GI, MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact, Autoimmun. Rev 18 (2019) 102391. 10.1016/j.autrev.2019.102391. [DOI] [PubMed] [Google Scholar]
- [164].Chang C, Xu L, Zhang R, Jin Y, Jiang P, Wei K, Xu L, Shi Y, Zhao J, Xiong M, Guo S, He D, MicroRNA-Mediated Epigenetic Regulation of Rheumatoid Arthritis Susceptibility and Pathogenesis, Front. Immunol 13 (2022) 838884. 10.3389/fimmu.2022.838884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Guo T, Xing Y, Chen Z, Zhu H, Yang L, Xiao Y, Xu J, Long Non-Coding RNA NEAT1 Knockdown Alleviates Rheumatoid Arthritis by Reducing IL-18 through p300/CBP Repression, Inflammation. 45 (2022) 100–115. 10.1007/s10753-021-01531-x. [DOI] [PubMed] [Google Scholar]
- [166].Shaker OG, Mahmoud RH, Abdelaleem OO, Ahmed TI, Fouad NA, Hussein HA, Nassr MH, Zaki OM, Abdelghaffar NK, Hefzy EM, Expression Profile of Long Noncoding RNAs, lnc-Cox2, and HOTAIR in Rheumatoid Arthritis Patients, J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res 39 (2019) 174–180. 10.1089/jir.2018.0117. [DOI] [PubMed] [Google Scholar]
- [167].Song J, Kim D, Han J, Kim Y, Lee M, Jin E-J, PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis, Clin. Exp. Med 15 (2015) 121–126. 10.1007/s10238-013-0271-4. [DOI] [PubMed] [Google Scholar]
- [168].Xu D, Jiang Y, Yang L, Hou X, Wang J, Gu W, Wang X, Liu L, Zhang J, Lu H, Long noncoding RNAs expression profile and functional networks in rheumatoid arthritis, Oncotarget. 8 (2017) 95280–95292. 10.18632/oncotarget.20036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Yan S, Wang P, Wang J, Yang J, Lu H, Jin C, Cheng M, Xu D, Long Non-coding RNA HIX003209 Promotes Inflammation by Sponging miR-6089 via TLR4/NF-κB Signaling Pathway in Rheumatoid Arthritis, Front. Immunol 10 (2019) 2218. 10.3389/fimmu.2019.02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Xu D, Song M, Chai C, Wang J, Jin C, Wang X, Cheng M, Yan S, Exosome-encapsulated miR-6089 regulates inflammatory response via targeting TLR4, J. Cell. Physiol 234 (2019) 1502–1511. 10.1002/jcp.27014. [DOI] [PubMed] [Google Scholar]
- [171].Yang C-A, Li J-P, Yen J-C, Lai I-L, Ho Y-C, Chen Y-C, Lan J-L, Chang J-G, lncRNA NTT/PBOV1 Axis Promotes Monocyte Differentiation and Is Elevated in Rheumatoid Arthritis, Int. J. Mol. Sci 19 (2018). 10.3390/ijms19092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Lu M-C, Yu H-C, Yu C-L, Huang H-B, Koo M, Tung C-H, Lai N-S, Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses, Immunol. Res 64 (2016) 576–583. 10.1007/s12026-015-8756-8. [DOI] [PubMed] [Google Scholar]
- [173].Wang Y, Liu J, Liu C, Naji A, Stoffers DA, MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells, Diabetes. 62 (2013) 887–895. 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Tang X, Wang J, Xia X, Tian J, Rui K, Xu H, Wang S, Elevated expression of ciRS-7 in peripheral blood mononuclear cells from rheumatoid arthritis patients, Diagn. Pathol 14 (2019) 11. 10.1186/s13000-019-0783-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Xiong S, Peng H, Ding X, Wang X, Wang L, Wu C, Wang S, Xu H, Liu Y, Circular RNA Expression Profiling and the Potential Role of hsa_circ_0089172 in Hashimoto’s Thyroiditis via Sponging miR125a-3p, Mol. Ther. Nucleic Acids 17 (2019) 38–48. 10.1016/j.omtn.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Peng H, Liu Y, Tian J, Ma J, Tang X, Yang J, Rui K, Zhang Y, Mao C, Lu L, Xu H, Wang S, Decreased expression of microRNA-125a-3p upregulates interleukin-23 receptor in patients with Hashimoto’s thyroiditis, Immunol. Res 62 (2015) 129–136. 10.1007/s12026-015-8643-3. [DOI] [PubMed] [Google Scholar]
- [177].Li B, Li N, Zhang L, Li K, Xie Y, Xue M, Zheng Z, Hsa_circ_0001859 Regulates ATF2 Expression by Functioning as an MiR-204/211 Sponge in Human Rheumatoid Arthritis, J. Immunol. Res 2018 (2018) 9412387. 10.1155/2018/9412387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Piao X, Zhou J, Hu J, Role of RP11–83J16.1, a novel long non-coding RNA, in rheumatoid arthritis, Am. J. Transl. Res 12 (2020) 1397–1414. [PMC free article] [PubMed] [Google Scholar]
- [179].He C, Wang Y, Wen Y, Li T, Hu E, Zeng S, Yang B, Xiong X, An intersectional analysis of LncRNAs and mRNAs reveals the potential therapeutic targets of Bi Zhong Xiao Decoction in collagen-induced arthritis rats, Chin. Med 17 (2022) 110. 10.1186/s13020-022-00670-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Ungethuem U, Haeupl T, Witt H, Koczan D, Krenn V, Huber H, von Helversen TM, Drungowski M, Seyfert C, Zacher J, Pruss A, Neidel J, Lehrach H, Thiesen HJ, Ruiz P, Bläss S, Molecular signatures and new candidates to target the pathogenesis of rheumatoid arthritis, Physiol. Genomics 42A (2010) 267–282. 10.1152/physiolgenomics.00004.2010. [DOI] [PubMed] [Google Scholar]
- [181].Hao J, Chen Y, Yu Y, Circular RNA circ_0008360 Inhibits the Proliferation, Migration, and Inflammation and Promotes Apoptosis of Fibroblast-Like Synoviocytes by Regulating miR-135b-5p/HDAC4 Axis in Rheumatoid Arthritis, Inflammation. 45 (2022) 196–211. 10.1007/s10753-021-01538-4. [DOI] [PubMed] [Google Scholar]
- [182].Gaspari AA, Innate and adaptive immunity and the pathophysiology of psoriasis, J. Am. Acad. Dermatol 54 (2006) S67–80. 10.1016/j.jaad.2005.10.057. [DOI] [PubMed] [Google Scholar]
- [183].Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, Kang HM, Nair RP, Chinnaiyan AM, Abecasis GR, Elder JT, Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin, Genome Biol. 16 (2015) 24. 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Zhou Q, Yu Q, Gong Y, Liu Z, Xu H, Wang Y, Shi Y, Construction of a lncRNA-miRNA-mRNA network to determine the regulatory roles of lncRNAs in psoriasis, Exp. Ther. Med 18 (2019) 4011–4021. 10.3892/etm.2019.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Luo Q, Zeng J, Li W, Lin L, Zhou X, Tian X, Liu W, Zhang L, Zhang X, Silencing of miR-155 suppresses inflammatory responses in psoriasis through inflammasome NLRP3 regulation, Int. J. Mol. Med 42 (2018) 1086–1095. 10.3892/ijmm.2018.3677. [DOI] [PubMed] [Google Scholar]
- [186].Luo M, Huang P, Pan Y, Zhu Z, Zhou R, Yang Z, Wang C, Weighted gene coexpression network and experimental analyses identify lncRNA SPRR2C as a regulator of the IL-22-stimulated HaCaT cell phenotype through the miR-330/STAT1/S100A7 axis, Cell Death Dis. 12 (2021) 86. 10.1038/s41419-020-03305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Yue T, Ji M, Qu H, Guo M, Bai F, Zhang Z, Wang W, Gong X, Zhang Z, Comprehensive analyses of long non-coding RNA expression profiles by RNA sequencing and exploration of their potency as biomarkers in psoriatic arthritis patients, BMC Immunol. 20 (2019) 28. 10.1186/s12865-019-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Wang R, Lin L, Lu X, Du J, Xu J, LncRNA AGXT2L1–2:2 facilitates keratinocytes proliferation and inhibits apoptosis by interacting with estrogen-related receptor alpha in psoriasis, Mol. Cell. Probes 62 (2022) 101803. 10.1016/j.mcp.2022.101803. [DOI] [PubMed] [Google Scholar]
- [189].Li S, Zhu X, Zhang N, Cao R, Zhao L, Li X, Zhang J, Yu J, LncRNA NORAD engages in psoriasis by binding to miR-26a to regulate keratinocyte proliferation, Autoimmunity. 54 (2021) 129–137. 10.1080/08916934.2021.1897976. [DOI] [PubMed] [Google Scholar]
- [190].Jia H-Y, Zhang K, Lu W-J, Xu G-W, Zhang J-F, Tang Z-L, LncRNA MEG3 influences the proliferation and apoptosis of psoriasis epidermal cells by targeting miR-21/caspase-8, BMC Mol. Cell Biol 20 (2019) 46. 10.1186/s12860-019-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Danis J, Göblös A, Bata-Csörgő Z, Kemény L, Széll M, PRINS Non-Coding RNA Regulates Nucleic Acid-Induced Innate Immune Responses of Human Keratinocytes, Front. Immunol 8 (2017). 10.3389/fimmu.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Abdallah HY, Tawfik NZ, Soliman NH, Eldeen LAT, The lncRNA PRINS-miRNA-mRNA Axis Gene Expression Profile as a Circulating Biomarker Panel in Psoriasis, Mol. Diagn. Ther 26 (2022) 451–465. 10.1007/s40291-022-00598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Wang Y, Jiang F, Chen F, Zhang D, Wang J, LncRNA XIST Engages in Psoriasis via Sponging miR-338–5p to Regulate Keratinocyte Proliferation and Inflammation, Skin Pharmacol. Physiol 35 (2022) 196–205. 10.1159/000523781. [DOI] [PubMed] [Google Scholar]
- [194].Tang Z-L, Zhang K, Lv S-C, Xu G-W, Zhang J-F, Jia H-Y, LncRNA MEG3 suppresses PI3K/AKT/mTOR signalling pathway to enhance autophagy and inhibit inflammation in TNF-α-treated keratinocytes and psoriatic mice, Cytokine. 148 (2021) 155657. 10.1016/j.cyto.2021.155657. [DOI] [PubMed] [Google Scholar]
- [195].Moldovan L-I, Hansen TB, Venø MT, Okholm TLH, Andersen TL, Hager H, Iversen L, Kjems J, Johansen C, Kristensen LS, High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome, BMC Med. Genomics 12 (2019) 174. 10.1186/s12920-019-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Moldovan LI, Tsoi LC, Ranjitha U, Hager H, Weidinger S, Gudjonsson JE, Kjems J, Kristensen LS, Characterization of circular RNA transcriptomes in psoriasis and atopic dermatitis reveals disease-specific expression profiles, Exp. Dermatol 30 (2021) 1187–1196. 10.1111/exd.14227. [DOI] [PubMed] [Google Scholar]
- [197].Seeler S, Moldovan L-I, Bertelsen T, Hager H, Iversen L, Johansen C, Kjems J, Sommer Kristensen L, Global circRNA expression changes predate clinical and histological improvements of psoriasis patients upon secukinumab treatment, PloS One. 17 (2022) e0275219. 10.1371/journal.pone.0275219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Lu J, Xu X, Li Y, Yu N, Ding Y, Shi Y, CircRAB3B suppresses proliferation, motility, cell cycle progression and promotes the apoptosis of IL-22-induced keratinocytes depending on the regulation of miR-1228–3p/PTEN axis in psoriasis, Autoimmunity. 54 (2021) 303–312. 10.1080/08916934.2021.1934825. [DOI] [PubMed] [Google Scholar]
- [199].Qiao M, Ding J, Yan J, Li R, Jiao J, Sun Q, Circular RNA Expression Profile and Analysis of Their Potential Function in Psoriasis, Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 50 (2018) 15–27. 10.1159/000493952. [DOI] [PubMed] [Google Scholar]
- [200].Yang Z, Yin X, Chen C, Huang S, Li X, Yan J, Sun Q, CircOAS3 Regulates Keratinocyte Proliferation and Psoriatic Inflammation by Interacting with Hsc70 via the JNK/STAT3/NF-κB Signaling Pathway, Inflammation. 45 (2022) 1924–1935. 10.1007/s10753-022-01664-7. [DOI] [PubMed] [Google Scholar]
- [201].Chen C, Yang Z, Yin X, Huang S, Yan J, Sun Q, CircEIF5 contributes to hyperproliferation and inflammation of keratinocytes in psoriasis via p-NFκB and p-STAT3 signalling pathway, Exp. Dermatol 31 (2022) 1145–1153. 10.1111/exd.14565. [DOI] [PubMed] [Google Scholar]
- [202].He Q, Liu N, Hu F, Shi Q, Pi X, Chen H, Li J, Zhang B, Circ_0061012 contributes to IL-22-induced proliferation, migration and invasion in keratinocytes through miR-194–5p/GAB1 axis in psoriasis, Biosci. Rep 41 (2021) BSR20203130. 10.1042/BSR20203130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Liu R, Chang W, Li J, Cheng Y, Dang E, Yang X, Wang Q, Wang G, Li X, Zhang K, Mesenchymal stem cells in psoriatic lesions affect the skin microenvironment through circular RNA, Exp. Dermatol 28 (2019) 292–299. 10.1111/exd.13890. [DOI] [PubMed] [Google Scholar]
- [204].Liu R, Wang Q, Chang W, Zhou L, Li J, Zhang K, Characterisation of the circular RNA landscape in mesenchymal stem cells from psoriatic skin lesions, Eur. J. Dermatol. EJD 29 (2019) 29–38. 10.1684/ejd.2018.3483. [DOI] [PubMed] [Google Scholar]
- [205].Zhang M, Han M, Li J, Tao K, Yang X, Zheng H, Yang R, Zhai Z, hsa_circ_0056856 in the serum serves as a potential novel biomarker for disease activity in psoriasis, Chin. Med. J. (Engl.) 135 (2022) 1759–1761. 10.1097/CM9.0000000000002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Liu X, Frost J, Bowcock A, Zhang W, Canonical and Interior Circular RNAs Function as Competing Endogenous RNAs in Psoriatic Skin, Int. J. Mol. Sci 22 (2021) 5182. 10.3390/ijms22105182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Peters LA, Perrigoue J, Mortha A, Iuga A, Song W, Neiman EM, Llewellyn SR, Di Narzo A, Kidd BA, Telesco SE, Zhao Y, Stojmirovic A, Sendecki J, Shameer K, Miotto R, Losic B, Shah H, Lee E, Wang M, Faith JJ, Kasarskis A, Brodmerkel C, Curran M, Das A, Friedman JR, Fukui Y, Humphrey MB, Iritani BM, Sibinga N, Tarrant TK, Argmann C, Hao K, Roussos P, Zhu J, Zhang B, Dobrin R, Mayer LF, Schadt EE, A functional genomics predictive network model identifies regulators of inflammatory bowel disease, Nat. Genet 49 (2017) 1437–1449. 10.1038/ng.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Krishnachaitanya SS, Liu M, Fujise K, Li Q, MicroRNAs in Inflammatory Bowel Disease and Its Complications, Int. J. Mol. Sci 23 (2022) 8751. 10.3390/ijms23158751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, Gorodkin J, Pociot F, Transcriptomic landscape of lncRNAs in inflammatory bowel disease, Genome Med. 7 (2015) 39. 10.1186/s13073-015-0162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Wu F, Huang Y, Dong F, Kwon JH, Ulcerative Colitis-Associated Long Noncoding RNA, BC012900, Regulates Intestinal Epithelial Cell Apoptosis, Inflamm. Bowel Dis 22 (2016) 782–795. 10.1097/MIB.0000000000000691. [DOI] [PubMed] [Google Scholar]
- [211].Padua D, Mahurkar-Joshi S, Law IKM, Polytarchou C, Vu JP, Pisegna JR, Shih D, Iliopoulos D, Pothoulakis C, A long noncoding RNA signature for ulcerative colitis identifies IFNG-AS1 as an enhancer of inflammation, Am. J. Physiol. Gastrointest. Liver Physiol 311 (2016) G446–457. 10.1152/ajpgi.00212.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Wang S, Hou Y, Chen W, Wang J, Xie W, Zhang X, Zeng L, KIF9-AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease, Mol. Med. Rep 17 (2018) 2195–2202. 10.3892/mmr.2017.8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Ding G, Ming Y, Zhang Y, lncRNA Mirt2 Is Downregulated in Ulcerative Colitis and Regulates IL-22 Expression and Apoptosis in Colonic Epithelial Cells, Gastroenterol. Res. Pract 2019 (2019) 8154692. 10.1155/2019/8154692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Qiao YQ, Huang ML, Xu AT, Zhao D, Ran ZH, Shen J, LncRNA DQ786243 affects Treg related CREB and Foxp3 expression in Crohn’s disease, J. Biomed. Sci 20 (2013) 87. 10.1186/1423-0127-20-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Elamir A, Shaker O, Kamal M, Khalefa A, Abdelwahed M, Abd El Reheem F, Ahmed T, Hassan E, Ayoub S, Expression profile of serum LncRNA THRIL and MiR-125b in inflammatory bowel disease, PloS One. 17 (2022) e0275267. 10.1371/journal.pone.0275267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Lin L, Zhou G, Chen P, Wang Y, Han J, Chen M, He Y, Zhang S, Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease?, Cell Death Dis. 11 (2020) 456. 10.1038/s41419-020-2657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Zhu P, Zhu X, Wu J, He L, Lu T, Wang Y, Liu B, Ye B, Sun L, Fan D, Wang J, Yang L, Qin X, Du Y, Li C, He L, Ren W, Wu X, Tian Y, Fan Z, IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3, Nat. Immunol 20 (2019) 183–194. 10.1038/s41590-018-0297-6. [DOI] [PubMed] [Google Scholar]
- [218].Li X-X, Xiao L, Chung HK, Ma X-X, Liu X, Song J-L, Jin CZ, Rao JN, Gorospe M, Wang J-Y, Interaction between HuR and circPABPN1 Modulates Autophagy in the Intestinal Epithelium by Altering ATG16L1 Translation, Mol. Cell. Biol 40 (2020) e00492–19. 10.1128/MCB.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Rankin CR, Lokhandwala ZA, Huang R, Pekow J, Pothoulakis C, Padua D, Linear and circular CDKN2B-AS1 expression is associated with Inflammatory Bowel Disease and participates in intestinal barrier formation, Life Sci. 231 (2019) 116571. 10.1016/j.lfs.2019.116571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Yin J, Hu T, Xu L, Li P, Li M, Ye Y, Pang Z, Circular RNA expression profile in peripheral blood mononuclear cells from Crohn disease patients, Medicine (Baltimore). 98 (2019) e16072. 10.1097/MD.0000000000016072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Ye Y, Zhang L, Hu T, Yin J, Xu L, Pang Z, Chen W, CircRNA_103765 acts as a proinflammatory factor via sponging miR-30 family in Crohn’s disease, Sci. Rep 11 (2021) 565. 10.1038/s41598-020-80663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Ye Y-L, Yin J, Hu T, Zhang L-P, Wu L-Y, Pang Z, Increased circulating circular RNA_103516 is a novel biomarker for inflammatory bowel disease in adult patients, World J. Gastroenterol 25 (2019) 6273–6288. 10.3748/wjg.v25.i41.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Faissner S, Plemel JR, Gold R, Yong VW, Progressive multiple sclerosis: from pathophysiology to therapeutic strategies, Nat. Rev. Drug Discov 18 (2019) 905–922. 10.1038/s41573-019-0035-2. [DOI] [PubMed] [Google Scholar]
- [224].Ma X, Zhou J, Zhong Y, Jiang L, Mu P, Li Y, Singh N, Nagarkatti M, Nagarkatti P, Expression, regulation and function of microRNAs in multiple sclerosis, Int. J. Med. Sci 11 (2014) 810–818. 10.7150/ijms.8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Mahdi Eftekharian M, Noroozi R, Komaki A, Mazdeh M, Taheri M, Ghafouri-Fard S, GAS5 genomic variants and risk of multiple sclerosis, Neurosci. Lett 701 (2019) 54–57. 10.1016/j.neulet.2019.02.028. [DOI] [PubMed] [Google Scholar]
- [226].Eftekharian MM, Noroozi R, Komaki A, Mazdeh M, Ghafouri-Fard S, Taheri M, MALAT1 Genomic Variants and Risk of Multiple Sclerosis, Immunol. Invest 48 (2019) 549–554. 10.1080/08820139.2019.1576728. [DOI] [PubMed] [Google Scholar]
- [227].Senousy MA, Shaker OG, Sayed NH, Fathy N, Kortam MA, LncRNA GAS5 and miR-137 Polymorphisms and Expression are Associated with Multiple Sclerosis Risk: Mechanistic Insights and Potential Clinical Impact, ACS Chem. Neurosci 11 (2020) 1651–1660. 10.1021/acschemneuro.0c00150. [DOI] [PubMed] [Google Scholar]
- [228].Moradi M, Gharesouran J, Ghafouri-Fard S, Noroozi R, Talebian S, Taheri M, Rezazadeh M, Role of NR3C1 and GAS5 genes polymorphisms in multiple sclerosis, Int. J. Neurosci 130 (2020) 407–412. 10.1080/00207454.2019.1694019. [DOI] [PubMed] [Google Scholar]
- [229].Taheri M, Noroozi R, Sadeghpour S, Omrani MD, Ghafouri-Fard S, The rs4759314 SNP within Hotair lncRNA is associated with risk of multiple sclerosis, Mult. Scler. Relat. Disord 40 (2020) 101986. 10.1016/j.msard.2020.101986. [DOI] [PubMed] [Google Scholar]
- [230].Rezazadeh M, Gharesouran J, Moradi M, Noroozi R, Omrani MD, Taheri M, Ghafouri-Fard S, Association Study of ANRIL Genetic Variants and Multiple Sclerosis, J. Mol. Neurosci. MN 65 (2018) 54–59. 10.1007/s12031-018-1069-3. [DOI] [PubMed] [Google Scholar]
- [231].Bahrami T, Taheri M, Omrani MD, Karimipoor M, Associations Between Genomic Variants in lncRNA-TRPM2-AS and lncRNA-HNF1A-AS1 Genes and Risk of Multiple Sclerosis, J. Mol. Neurosci. MN 70 (2020) 1050–1055. 10.1007/s1203-1020-01504-z. [DOI] [PubMed] [Google Scholar]
- [232].Santoro M, Nociti V, Lucchini M, De Fino C, Losavio FA, Mirabella M, Expression Profile of Long Non-Coding RNAs in Serum of Patients with Multiple Sclerosis, J. Mol. Neurosci. MN 59 (2016) 18–23. 10.1007/s12031-016-0741-8. [DOI] [PubMed] [Google Scholar]
- [233].Shaker OG, Mahmoud RH, Abdelaleem OO, Ibrahem EG, Mohamed AA, Zaki OM, Abdelghaffar NK, Ahmed TI, Hemeda NF, Ahmed NA, Mansour DF, LncRNAs, MALAT1 and lnc-DC as potential biomarkers for multiple sclerosis diagnosis, Biosci. Rep 39 (2019) BSR20181335. 10.1042/BSR20181335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Cardamone G, Paraboschi EM, Soldà G, Cantoni C, Supino D, Piccio L, Duga S, Asselta R, Not only cancer: the long non-coding RNA MALAT1 affects the repertoire of alternatively spliced transcripts and circular RNAs in multiple sclerosis, Hum. Mol. Genet 28 (2019) 1414–1428. 10.1093/hmg/ddy438. [DOI] [PubMed] [Google Scholar]
- [235].Zhang F, Gao C, Ma X-F, Peng X-L, Zhang R-X, Kong D-X, Simard AR, Hao J-W, Expression Profile of Long Noncoding RNAs in Peripheral Blood Mononuclear Cells from Multiple Sclerosis Patients, CNS Neurosci. Ther 22 (2016) 298–305. 10.1111/cns.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Gupta M, Martens K, Metz LM, de Koning AJ, Pfeffer G, Long noncoding RNAs associated with phenotypic severity in multiple sclerosis, Mult. Scler. Relat. Disord 36 (2019) 101407. 10.1016/j.msard.2019.101407. [DOI] [PubMed] [Google Scholar]
- [237].Ding Y, Li T, Yan X, Cui M, Wang C, Wang S, Zhang F, Zhang R, Identification of hub lncRNA ceRNAs in multiple sclerosis based on ceRNA mechanisms, Mol. Genet. Genomics MGG 296 (2021) 423–435. 10.1007/s00438-020-01750-1. [DOI] [PubMed] [Google Scholar]
- [238].Hao Y, He M, Fu Y, Zhao C, Xiong S, Xu X, Identification of Novel Key Genes and Pathways in Multiple Sclerosis Based on Weighted Gene Coexpression Network Analysis and Long Noncoding RNA-Associated Competing Endogenous RNA Network, Oxid. Med. Cell. Longev 2022 (2022) 9328160. 10.1155/2022/9328160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Eftekharian MM, Ghafouri-Fard S, Soudyab M, Omrani MD, Rahimi M, Sayad A, Komaki A, Mazdeh M, Taheri M, Expression Analysis of Long Non-coding RNAs in the Blood of Multiple Sclerosis Patients, J. Mol. Neurosci. MN 63 (2017) 333–341. 10.1007/s12031-017-0982-1. [DOI] [PubMed] [Google Scholar]
- [240].Patoughi M, Ghafouri-Fard S, Arsang-Jang S, Taheri M, GAS8 and its naturally occurring antisense RNA as biomarkers in multiple sclerosis, Immunobiology. 224 (2019) 560–564. 10.1016/j.imbio.2019.04.005. [DOI] [PubMed] [Google Scholar]
- [241].Moradi A, Rahimi Naiini M, Yazdanpanahi N, Tabatabaeian H, Nabatchian F, Baghi M, Azadeh M, Ghaedi K, Evaluation of The Expression Levels of Three Long Non-Coding RNAs in Multiple Sclerosis, Cell J. 22 (2020) 165–170. 10.22074/cellj.2020.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Patoughi M, Ghafouri-Fard S, Arsang-Jang S, Taheri M, Expression analysis of PINK1 and PINK1-AS in multiple sclerosis patients versus healthy subjects, Nucleosides Nucleotides Nucleic Acids. 40 (2021) 157–165. 10.1080/15257770.2020.1844229. [DOI] [PubMed] [Google Scholar]
- [243].Ghaiad HR, Elmazny AN, Nooh MM, El-Sawalhi MM, Shaheen AA, Long noncoding RNAs APOA1-AS, IFNG-AS1, RMRP and their related biomolecules in Egyptian patients with relapsing-remitting multiple sclerosis: Relation to disease activity and patient disability, J. Adv. Res 21 (2020) 141–150. 10.1016/j.jare.2019.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Fenoglio C, Oldoni E, Serpente M, De Riz MA, Arcaro M, D’Anca M, Pietroboni AM, Calvi A, Lecchi E, Goris A, Mallants K, Dubois B, Comi C, Cantello R, Scarpini E, Galimberti D, LncRNAs expression profile in peripheral blood mononuclear cells from multiple sclerosis patients, J. Neuroimmunol 324 (2018) 129–135. 10.1016/j.jneuroim.2018.08.008. [DOI] [PubMed] [Google Scholar]
- [245].Ganji M, Sayad A, Omrani MD, Arsang-Jang S, Mazdeh M, Taheri M, Expression analysis of long non-coding RNAs and their target genes in multiple sclerosis patients, Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neurophysiol 40 (2019) 801–811. 10.1007/s10072-019-3720-3. [DOI] [PubMed] [Google Scholar]
- [246].Zhang F, Liu G, Li D, Wei C, Hao J, DDIT4 and Associated lncDDIT4 Modulate Th17 Differentiation through the DDIT4/TSC/mTOR Pathway, J. Immunol. Baltim. Md 1950 200 (2018) 1618–1626. 10.4049/jimmunol.1601689. [DOI] [PubMed] [Google Scholar]
- [247].Iparraguirre L, Muñoz-Culla M, Prada-Luengo I, Castillo-Triviño T, Olascoaga J, Otaegui D, Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis, Hum. Mol. Genet 26 (2017) 3564–3572. 10.1093/hmg/ddx243. [DOI] [PubMed] [Google Scholar]
- [248].Zurawska AE, Mycko MP, Selmaj I, Raine CS, Selmaj KW, Multiple Sclerosis: circRNA Profile Defined Reveals Links to B-Cell Function, Neurol. Neuroimmunol. Neuroinflammation 8 (2021) e1041. 10.1212/NXI.0000000000001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Mycko MP, Zurawska AE, Selmaj I, Selmaj KW, Impact of Diminished Expression of circRNA on Multiple Sclerosis Pathomechanisms, Front. Immunol 13 (2022) 875994. 10.3389/fimmu.2022.875994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Iparraguirre L, Alberro A, Hansen TB, Castillo-Triviño T, Muñoz-Culla M, Otaegui D, Profiling of Plasma Extracellular Vesicle Transcriptome Reveals That circRNAs Are Prevalent and Differ between Multiple Sclerosis Patients and Healthy Controls, Biomedicines. 9 (2021) 1850. 10.3390/biomedicines9121850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].He J, Ren M, Li H, Yang L, Wang X, Yang Q, Exosomal Circular RNA as a Biomarker Platform for the Early Diagnosis of Immune-Mediated Demyelinating Disease, Front. Genet 10 (2019) 860. 10.3389/fgene.2019.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Jiang F, Liu X, Cui X, Hu J, Wang L, Xue F, Guo S, Wang X, Circ_0000518 Promotes Macrophage/Microglia M1 Polarization via the FUS/CaMKKβ/AMPK Pathway to Aggravate Multiple Sclerosis, Neuroscience. 490 (2022) 131–143. 10.1016/j.neuroscience.2021.12.012. [DOI] [PubMed] [Google Scholar]
- [253].Han J, Zhuang W, Feng W, Dong F, Hua F, Yao R, Qu X, The circular RNA circINPP4B acts as a sponge of miR-30a to regulate Th17 cell differentiation during progression of experimental autoimmune encephalomyelitis, Cell. Mol. Immunol 18 (2021) 2177–2187. 10.1038/s41423-021-00748-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Weinberg MS, Morris KV, Long Non-Coding RNA Targeting and Transcriptional De-Repression, Nucleic Acid Ther. 23 (2013) 9–14. 10.1089/nat.2012.0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, Lander ES, Voytas DF, Ting AY, Zhang F, RNA targeting with CRISPR-Cas13a, Nature. 550 (2017) 280–284. 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Lennox KA, Behlke MA, Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides, Nucleic Acids Res. 44 (2016) 863–877. 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Scott L, Finley SJ, Watson C, Javan GT, Life and death: A systematic comparison of antemortem and postmortem gene expression, Gene. 731 (2020) 144349. 10.1016/j.gene.2020.144349. [DOI] [PubMed] [Google Scholar]
- [258].Lee Y, Kim M, Han J, Yeom K-H, Lee S, Baek SH, Kim VN, MicroRNA genes are transcribed by RNA polymerase II, EMBO J. 23 (2004) 4051–4060. 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Borchert GM, Lanier W, Davidson BL, RNA polymerase III transcribes human microRNAs, Nat. Struct. Mol. Biol 13 (2006) 1097–1101. 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- [260].Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A, Identification of mammalian microRNA host genes and transcription units, Genome Res. 14 (2004) 1902–1910. 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Huntzinger E, Izaurralde E, Gene silencing by microRNAs: contributions of translational repression and mRNA decay, Nat. Rev. Genet 12 (2011) 99–110. 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- [262].Chen J, An T, Ma Y, Situ B, Chen D, Xu Y, Zhang L, Dai Z, Zou X, Isothermal Amplification on a Structure-Switchable Symmetric Toehold Dumbbell-Template: A Strategy Enabling MicroRNA Analysis at the Single-Cell Level with Ultrahigh Specificity and Accuracy, Anal. Chem 90 (2018) 859–865. 10.1021/acs.analchem.7b03713. [DOI] [PubMed] [Google Scholar]
- [263].Tian H, Sun Y, Liu C, Duan X, Tang W, Li Z, Precise Quantitation of MicroRNA in a Single Cell with Droplet Digital PCR Based on Ligation Reaction, Anal. Chem 88 (2016) 11384–11389. 10.1021/acs.analchem.6b01225. [DOI] [PubMed] [Google Scholar]
- [264].Ouyang T, Liu Z, Han Z, Ge Q, MicroRNA Detection Specificity: Recent Advances and Future Perspective, Anal. Chem 91 (2019) 3179–3186. 10.1021/acs.analchem.8b05909. [DOI] [PubMed] [Google Scholar]
- [265].Chandrasekaran AR, MacIsaac M, Dey P, Levchenko O, Zhou L, Andres M, Dey BK, Halvorsen K, Cellular microRNA detection with miRacles: microRNA- activated conditional looping of engineered switches, Sci. Adv 5 (2019) eaau9443. 10.1126/sciadv.aau9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF, miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 7080–7085. 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Ghorai A, Ghosh U, miRNA gene counts in chromosomes vary widely in a species and biogenesis of miRNA largely depends on transcription or post-transcriptional processing of coding genes, Front. Genet 5 (2014) 100. 10.3389/fgene.2014.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Kozomara A, Birgaoanu M, Griffiths-Jones S, miRBase: from microRNA sequences to function, Nucleic Acids Res. 47 (2019) D155–D162. 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Sharp SJ, Schaack J, Cooley L, Burke DJ, Söll D, Structure and transcription of eukaryotic tRNA genes, CRC Crit. Rev. Biochem 19 (1985) 107–144. 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- [270].Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S, Gene map of the extended human MHC, Nat. Rev. Genet 5 (2004) 889–899. 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- [271].Köhrer C, Srinivasan G, Mandal D, Mallick B, Ghosh Z, Chakrabarti J, Rajbhandary UL, Identification and characterization of a tRNA decoding the rare AUA codon in Haloarcula marismortui, RNA N. Y. N 14 (2008) 117–126. 10.1261/rna.795508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Thompson DM, Parker R, The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae, J. Cell Biol 185 (2009) 43–50. 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R, Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs, Genes Dev. 22 (2008) 2773–2785. 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Saikia M, Hatzoglou M, The Many Virtues of tRNA-derived Stress-induced RNAs (tiRNAs): Discovering Novel Mechanisms of Stress Response and Effect on Human Health, J. Biol. Chem 290 (2015) 29761–29768. 10.1074/jbc.R115.694661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Kumar P, Anaya J, Mudunuri SB, Dutta A, Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets, BMC Biol. 12 (2014) 78. 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Kumar P, Mudunuri SB, Anaya J, Dutta A, tRFdb: a database for transfer RNA fragments, Nucleic Acids Res. 43 (2015) D141–145. 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Pliatsika V, Loher P, Magee R, Telonis AG, Londin E, Shigematsu M, Kirino Y, Rigoutsos I, MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all The Cancer Genome Atlas projects, Nucleic Acids Res. 46 (2018) D152–D159. 10.1093/nar/gkx1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Zhang Y, Wang X, Kang L, A k-mer scheme to predict piRNAs and characterize locust piRNAs, Bioinforma. Oxf. Engl 27 (2011) 771–776. 10.1093/bioinformatics/btr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Wang K, Liang C, Liu J, Xiao H, Huang S, Xu J, Li F, Prediction of piRNAs using transposon interaction and a support vector machine, BMC Bioinformatics. 15 (2014) 419. 10.1186/s12859-014-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Liu X, Ding J, Gong F, piRNA identification based on motif discovery, Mol. Biosyst 10 (2014) 3075–3080. 10.1039/c4mb00447g. [DOI] [PubMed] [Google Scholar]
- [281].Brayet J, Zehraoui F, Jeanson-Leh L, Israeli D, Tahi F, Towards a piRNA prediction using multiple kernel fusion and support vector machine, Bioinforma. Oxf. Engl 30 (2014) i364–370. 10.1093/bioinformatics/btu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Boucheham A, Sommard V, Zehraoui F, Boualem A, Batouche M, Bendahmane A, Israeli D, Tahi F, IpiRId: Integrative approach for piRNA prediction using genomic and epigenomic data, PloS One. 12 (2017) e0179787. 10.1371/journal.pone.0179787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Wang J, Zhang P, Lu Y, Li Y, Zheng Y, Kan Y, Chen R, He S, piRBase: a comprehensive database of piRNA sequences, Nucleic Acids Res. 47 (2019) D175–D180. 10.1093/nar/gky1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Sai Lakshmi S, Agrawal S, piRNABank: a web resource on classified and clustered Piwi-interacting RNAs, Nucleic Acids Res. 36 (2008) D173–177. 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Lau NC, Robine N, Martin R, Chung W-J, Niki Y, Berezikov E, Lai EC, Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line, Genome Res. 19 (2009) 1776–1785. 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Ernst C, Odom DT, Kutter C, The emergence of piRNAs against transposon invasion to preserve mammalian genome integrity, Nat. Commun 8 (2017) 1411. 10.1038/s41467-017-01049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Weick E-M, Miska EA, piRNAs: from biogenesis to function, Dev. Camb. Engl 141 (2014) 3458–3471. 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- [288].Filipowicz W, Pogacić V, Biogenesis of small nucleolar ribonucleoproteins, Curr. Opin. Cell Biol 14 (2002) 319–327. 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- [289].Krishnan P, Ghosh S, Wang B, Heyns M, Graham K, Mackey JR, Kovalchuk O, Damaraju S, Profiling of Small Nucleolar RNAs by Next Generation Sequencing: Potential New Players for Breast Cancer Prognosis, PloS One. 11 (2016) e0162622. 10.1371/journal.pone.0162622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Bouchard-Bourelle P, Desjardins-Henri C, Mathurin-St-Pierre D, Deschamps-Francoeur G, Fafard-Couture É, Garant J-M, Elela SA, Scott MS, snoDB: an interactive database of human snoRNA sequences, abundance and interactions, Nucleic Acids Res. 48 (2020) D220–D225. 10.1093/nar/gkz884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A, The expanding RNA polymerase III transcriptome, Trends Genet. TIG 23 (2007) 614–622. 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- [292].Malinovskaya EM, Ershova ES, Golimbet VE, Porokhovnik LN, Lyapunova NA, Kutsev SI, Veiko NN, Kostyuk SV, Copy Number of Human Ribosomal Genes With Aging: Unchanged Mean, but Narrowed Range and Decreased Variance in Elderly Group, Front. Genet 9 (2018) 306. 10.3389/fgene.2018.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Koch F, Fenouil R, Gut M, Cauchy P, Albert TK, Zacarias-Cabeza J, Spicuglia S, de la Chapelle AL, Heidemann M, Hintermair C, Eick D, Gut I, Ferrier P, Andrau J-C, Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters, Nat. Struct. Mol. Biol 18 (2011) 956–963. 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- [294].Hsieh C-L, Fei T, Chen Y, Li T, Gao Y, Wang X, Sun T, Sweeney CJ, Lee G-SM, Chen S, Balk SP, Liu XS, Brown M, Kantoff PW, Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation, Proc. Natl. Acad. Sci. U. S. A 111 (2014) 7319–7324. 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Gao T, He B, Liu S, Zhu H, Tan K, Qian J, EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types, Bioinforma. Oxf. Engl 32 (2016) 3543–3551. 10.1093/bioinformatics/btw495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Schaukowitch K, Joo J-Y, Liu X, Watts JK, Martinez C, Kim T-K, Enhancer RNA facilitates NELF release from immediate early genes, Mol. Cell 56 (2014) 29–42. 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Wu H, Yang L, Chen L-L, The Diversity of Long Noncoding RNAs and Their Generation, Trends Genet. TIG 33 (2017) 540–552. 10.1016/j.tig.2017.05.004. [DOI] [PubMed] [Google Scholar]
- [298].Li X, Yang L, Chen L-L, The Biogenesis, Functions, and Challenges of Circular RNAs, Mol. Cell 71 (2018) 428–442. 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- [299].Jeck WR, Sharpless NE, Detecting and characterizing circular RNAs, Nat. Biotechnol 32 (2014) 453–461. 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, Cheng H, Yan J, Zhang S, Yang P, Zhao F, Expanded Expression Landscape and Prioritization of Circular RNAs in Mammals, Cell Rep. 26 (2019) 3444–3460.e5. 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- [301].Wu W, Ji P, Zhao F, CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes, Genome Biol. 21 (2020) 101. 10.1186/s13059-020-02018-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Xin R, Gao Y, Gao Y, Wang R, Kadash-Edmondson KE, Liu B, Wang Y, Lin L, Xing Y, isoCirc catalogs full-length circular RNA isoforms in human transcriptomes, Nat. Commun 12 (2021) 266. 10.1038/s41467-020-20459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Ransohoff JD, Wei Y, Khavari PA, The functions and unique features of long intergenic non-coding RNA, Nat. Rev. Mol. Cell Biol 19 (2018) 143–157. 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Schein A, Zucchelli S, Kauppinen S, Gustincich S, Carninci P, Identification of antisense long noncoding RNAs that function as SINEUPs in human cells, Sci. Rep 6 (2016) 33605. 10.1038/srep33605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Xu H, Jiang Y, Xu X, Su X, Liu Y, Ma Y, Zhao Y, Shen Z, Huang B, Cao X, Inducible degradation of lncRNA Sros1 promotes IFN-γ-mediated activation of innate immune responses by stabilizing Stat1 mRNA, Nat. Immunol 20 (2019) 1621–1630. 10.1038/s41590-019-0542-7. [DOI] [PubMed] [Google Scholar]
- [306].Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S, Imaging individual mRNA molecules using multiple singly labeled probes, Nat. Methods 5 (2008) 877–879. 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Shaffer SM, Wu M-T, Levesque MJ, Raj A, Turbo FISH: a method for rapid single molecule RNA FISH, PloS One. 8 (2013) e75120. 10.1371/journal.pone.0075120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Orjalo AV, Johansson HE, Stellaris® RNA Fluorescence In Situ Hybridization for the Simultaneous Detection of Immature and Mature Long Noncoding RNAs in Adherent Cells, Methods Mol. Biol. Clifton NJ 1402 (2016) 119–134. 10.1007/978-1-4939-3378-5_10. [DOI] [PubMed] [Google Scholar]
- [309].Querido E, Dekakra-Bellili L, Chartrand P, RNA fluorescence in situ hybridization for high-content screening, Methods San Diego Calif. 126 (2017) 149–155. 10.1016/j.ymeth.2017.07.005. [DOI] [PubMed] [Google Scholar]
- [310].Sugimoto Y, Chakrabarti AM, Luscombe NM, Ule J, Using hiCLIP to identify RNA duplexes that interact with a specific RNA-binding protein, Nat. Protoc 12 (2017) 611–637. 10.1038/nprot.2016.188. [DOI] [PubMed] [Google Scholar]
- [311].Chi SW, Zang JB, Mele A, Darnell RB, Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps, Nature. 460 (2009) 479–486. 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Aw JGA, Shen Y, Wilm A, Sun M, Lim XN, Boon K-L, Tapsin S, Chan Y-S, Tan C-P, Sim AYL, Zhang T, Susanto TT, Fu Z, Nagarajan N, Wan Y, In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation, Mol. Cell 62 (2016) 603–617. 10.1016/j.molcel.2016.04.028. [DOI] [PubMed] [Google Scholar]
- [313].Park H, Huang X, Lu C, Cairo MS, Zhou X, MicroRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins, J. Biol. Chem 290 (2015) 2831–2841. 10.1074/jbc.M114.591420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Xin J, Li J, Feng Y, Wang L, Zhang Y, Yang R, Downregulation of long noncoding RNA HOTAIRM1 promotes monocyte/dendritic cell differentiation through competitively binding to endogenous miR-3960, OncoTargets Ther. 10 (2017) 1307–1315. 10.2147/OTT.S124201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Chen Q, Mang G, Wu J, Sun P, Li T, Zhang H, Wang N, Tong Z, Wang W, Zheng Y, Tian J, E M, Zhang M, Yu B, Circular RNA circSnx5 Controls Immunogenicity of Dendritic Cells through the miR-544/SOCS1 Axis and PU.1 Activity Regulation, Mol. Ther. J. Am. Soc. Gene Ther 28 (2020) 2503–2518. 10.1016/j.ymthe.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD, Blin J, Braun JE, Gandhi P, Moore MJ, Chang HY, Lodish HF, Caffrey DR, Fitzgerald KA, A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation, Cell. 165 (2016) 1672–1685. 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Agliano F, Fitzgerald KA, Vella AT, Rathinam VA, Medvedev AE, Long Non-coding RNA LincRNA-EPS Inhibits Host Defense Against Listeria monocytogenes Infection, Front. Cell. Infect. Microbiol 9 (2019) 481. 10.3389/fcimb.2019.00481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, Pierre P, MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 2735–2740. 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Obaid M, Udden SMN, Deb P, Shihabeddin N, Zaki MH, Mandal SS, LncRNA HOTAIR regulates lipopolysaccharide-induced cytokine expression and inflammatory response in macrophages, Sci. Rep 8 (2018) 15670. 10.1038/s41598-018-33722-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Cui H, Xie N, Tan Z, Banerjee S, Thannickal VJ, Abraham E, Liu G, The human long noncoding RNA lnc-IL7R regulates the inflammatory response, Eur. J. Immunol 44 (2014) 2085–2095. 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Chen M-T, Lin H-S, Shen C, Ma Y-N, Wang F, Zhao H-L, Yu J, Zhang J-W, PU.1-Regulated Long Noncoding RNA lnc-MC Controls Human Monocyte/Macrophage Differentiation through Interaction with MicroRNA 199a-5p, Mol. Cell. Biol 35 (2015) 3212–3224. 10.1128/MCB.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Gao X, Ge J, Li W, Zhou W, Xu L, LncRNA KCNQ1OT1 ameliorates particle-induced osteolysis through inducing macrophage polarization by inhibiting miR-21a-5p, Biol. Chem 399 (2018) 375–386. 10.1515/hsz-2017-0215. [DOI] [PubMed] [Google Scholar]
- [323].Du M, Yuan L, Tan X, Huang D, Wang X, Zheng Z, Mao X, Li X, Yang L, Huang K, Zhang F, Wang Y, Luo X, Huang D, Huang K, The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation, Nat. Commun 8 (2017) 2049. 10.1038/s41467-017-02229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Krawczyk M, Emerson BM, p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes, ELife. 3 (2014) e01776. 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [325].Lian C, Sun J, Guan W, Zhang L, Zhang X, Yang L, Hu W, Circular RNA circHIPK3 Activates Macrophage NLRP3 Inflammasome and TLR4 Pathway in Gouty Arthritis via Sponging miR-561 and miR-192, Inflammation. 44 (2021) 2065–2077. 10.1007/s10753-021-01483-2. [DOI] [PubMed] [Google Scholar]
- [326].Zhang Y, Zhang Y, Li X, Zhang M, Lv K, Microarray analysis of circular RNA expression patterns in polarized macrophages, Int. J. Mol. Med 39 (2017) 373–379. 10.3892/ijmm.2017.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Zhang C, Han X, Yang L, Fu J, Sun C, Huang S, Xiao W, Gao Y, Liang Q, Wang X, Luo F, Lu W, Zhou Y, Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus, Theranostics. 10 (2020) 10908–10924. 10.7150/thno.48264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Ng WL, Marinov GK, Liau ES, Lam YL, Lim Y-Y, Ea C-K, Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway, RNA Biol. 13 (2016) 861–871. 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Kotzin JJ, Spencer SP, McCright SJ, Kumar DBU, Collet MA, Mowel WK, Elliott EN, Uyar A, Makiya MA, Dunagin MC, Harman CCD, Virtue AT, Zhu S, Bailis W, Stein J, Hughes C, Raj A, Wherry EJ, Goff LA, Klion AD, Rinn JL, Williams A, Flavell RA, Henao-Mejia J, The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan, Nature. 537 (2016) 239–243. 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Zhang R, Ni F, Fu B, Wu Y, Sun R, Tian Z, Wei H, A long noncoding RNA positively regulates CD56 in human natural killer cells, Oncotarget. 7 (2016) 72546–72558. 10.18632/oncotarget.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].King JK, Ung NM, Paing MH, Contreras JR, Alberti MO, Fernando TR, Zhang K, Pellegrini M, Rao DS, Regulation of Marginal Zone B-Cell Differentiation by MicroRNA-146a, Front. Immunol 7 (2016) 670. 10.3389/fimmu.2016.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KGC, Rada C, Enright AJ, Toellner K-M, Maclennan ICM, Turner M, microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells, Immunity. 27 (2007) 847–859. 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Petri A, Dybkær K, Bøgsted M, Thrue CA, Hagedorn PH, Schmitz A, Bødker JS, Johnsen HE, Kauppinen S, Long Noncoding RNA Expression during Human B-Cell Development, PloS One. 10 (2015) e0138236. 10.1371/journal.pone.0138236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Song Z, Wu W, Chen M, Cheng W, Yu J, Fang J, Xu L, Yasunaga J-I, Matsuoka M, Zhao T, Long Noncoding RNA ANRIL Supports Proliferation of Adult T-Cell Leukemia Cells through Cooperation with EZH2, J. Virol 92 (2018) e00909–18. 10.1128/JVI.00909-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [335].Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT, Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5), J. Cell Sci 121 (2008) 939–946. 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- [336].Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG, A strategy for probing the function of noncoding RNAs finds a repressor of NFAT, Science. 309 (2005) 1570–1573. 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- [337].Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A, Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 11381–11386. 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, Zhang H, Chen ZW, Zeng G, Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection, Proc. Natl. Acad. Sci. U. S. A 112 (2015) E3883–3892. 10.1073/pnas.1501662112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Gomez JA, Wapinski OL, Yang YW, Bureau J-F, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K, The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus, Cell. 152 (2013) 743–754. 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Wang Y-H, Yu X-H, Luo S-S, Han H, Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing, Immun. Ageing A 12 (2015) 17. 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [341].Du X, Zhu M, Zhang T, Wang C, Tao J, Yang S, Zhu Y, Zhao W, The Recombinant Eg.P29-Mediated miR-126a-5p Promotes the Differentiation of Mouse Naive CD4+ T Cells via DLK1-Mediated Notch1 Signal Pathway, Front. Immunol 13 (2022) 773276. 10.3389/fimmu.2022.773276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].He R, Chen Y, Chen X, Yuan B, Mechanism of miR-181a-5p in Regulatory T/T-Helper 17 Immune Imbalance and Asthma Development in Mice with Allergic Rhinitis, Int. Arch. Allergy Immunol 183 (2022) 375–388. 10.1159/000519703. [DOI] [PubMed] [Google Scholar]
- [343].Houtman M, Shchetynsky K, Chemin K, Hensvold AH, Ramsköld D, Tandre K, Eloranta M-L, Rönnblom L, Uebe S, Catrina AI, Malmström V, Padyukov L, T cells are influenced by a long non-coding RNA in the autoimmune associated PTPN2 locus, J. Autoimmun 90 (2018) 28–38. 10.1016/j.jaut.2018.01.003. [DOI] [PubMed] [Google Scholar]
- [344].McCall JL, Varney ME, Rice E, Dziadowicz SA, Hall C, Blethen KE, Hu G, Barnett JB, Martinez I, Prenatal Cadmium Exposure Alters Proliferation in Mouse CD4+ T Cells via LncRNA Snhg7, Front. Immunol 12 (2021) 720635. 10.3389/fimmu.2021.720635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].She C, Yang Y, Zang B, Yao Y, Liu Q, Leung PSC, Liu B, Effect of LncRNA XIST on Immune Cells of Primary Biliary Cholangitis, Front. Immunol 13 (2022) 816433. 10.3389/fimmu.2022.816433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Liu X, Lin J, Wu H, Wang Y, Xie L, Wu J, Qin H, Xu J, A Novel Long Noncoding RNA lincRNA00892 Activates CD4+ T Cells in Systemic Lupus Erythematosus by Regulating CD40L, Front. Pharmacol 12 (2021) 733902. 10.3389/fphar.2021.733902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Huang Z, Liu F, Wang W, Ouyang S, Sang T, Huang Z, Liao L, Wu J, Deregulation of circ_003912 contributes to pathogenesis of erosive oral lichen planus by via sponging microRNA-123, −647 and −31 and upregulating FOXP3, Mol. Med. Camb. Mass 27 (2021) 132. 10.1186/s10020-021-00382-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [348].Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM, Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells, J. Immunol. Baltim. Md 1950 189 (2012) 2084–2088. 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Huang F-J, Liu Y-L, Wang J, Zhou Y-Y, Zhao S-Y, Qin G-J, LncRNA RUNX1-IT1 affects the differentiation of Th1 cells by regulating NrCAM transcription in Graves’ disease, Cell Cycle Georget. Tex 21 (2022) 921–933. 10.1080/15384101.2022.2034431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Spurlock CF, Tossberg JT, Guo Y, Collier SP, Crooke PS, Aune TM, Expression and functions of long noncoding RNAs during human T helper cell differentiation, Nat. Commun 6 (2015) 6932. 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Ma Y, Shi L, Zhao K, Zheng C, lncRNA FR215775 Regulates Th2 Differentiation in Murine Allergic Rhinitis, J. Immunol. Res 2022 (2022) 7783481. 10.1155/2022/7783481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Guo W, Lei W, Yu D, Ge Y, Chen Y, Xue W, Li Q, Li S, Gao X, Yao W, Involvement of lncRNA-1700040D17Rik in Th17 cell differentiation and the pathogenesis of EAE, Int. Immunopharmacol 47 (2017) 141–149. 10.1016/j.intimp.2017.03.014. [DOI] [PubMed] [Google Scholar]
- [353].Xu Y, Ouyang Y, Long non-coding RNA growth arrest specific 5 regulates the T helper 17/regulatory T balance by targeting miR-23a in myasthenia gravis, J. Int. Med. Res 50 (2022) 3000605211053703. 10.1177/03000605211053703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [354].Qiu Y-Y, Wu Y, Lin M-J, Bian T, Xiao Y-L, Qin C, LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/RORγt, Biomed. Pharmacother. Biomedecine Pharmacother 111 (2019) 386–394. 10.1016/j.biopha.2018.12.080. [DOI] [PubMed] [Google Scholar]
- [355].Li J-Q, Hu S-Y, Wang Z-Y, Lin J, Jian S, Dong Y-C, Wu X-F, null Dai-Lan L-J Cao, Long non-coding RNA MEG3 inhibits microRNA-125a-5p expression and induces immune imbalance of Treg/Th17 in immune thrombocytopenic purpura, Biomed. Pharmacother. Biomedecine Pharmacother 83 (2016) 905–911. 10.1016/j.biopha.2016.07.057. [DOI] [PubMed] [Google Scholar]
- [356].Huang Z-X, Qu P, Wang K-K, Zheng J, Pan M, Zhu H-Q, Transcriptomic profiling of pemphigus lesion infiltrating mononuclear cells reveals a distinct local immune microenvironment and novel lncRNA regulators, J. Transl. Med 20 (2022) 182. 10.1186/s12967-022-03387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [357].Khan MM, Khan MH, Kalim UU, Khan S, Junttila S, Paulin N, Kong L, Rasool O, Elo LL, Lahesmaa R, Long Intergenic Noncoding RNA MIAT as a Regulator of Human Th17 Cell Differentiation, Front. Immunol 13 (2022) 856762. 10.3389/fimmu.2022.856762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Brajic A, Franckaert D, Burton O, Bornschein S, Calvanese AL, Demeyer S, Cools J, Dooley J, Schlenner S, Liston A, The Long Non-coding RNA Flatr Anticipates Foxp3 Expression in Regulatory T Cells, Front. Immunol 9 (2018) 1989. 10.3389/fimmu.2018.01989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Xia M, Liu J, Liu S, Chen K, Lin H, Jiang M, Xu X, Xue Y, Liu W, Gu Y, Zhang X, Li Z, Yi L, Qian Y, Zhou C, Li R, Zhang X, Li Z, Cao X, Ash1l and lnc-Smad3 coordinate Smad3 locus accessibility to modulate iTreg polarization and T cell autoimmunity, Nat. Commun 8 (2017) 15818. 10.1038/ncomms15818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [360].Delás MJ, Jackson BT, Kovacevic T, Vangelisti S, Munera Maravilla E, Wild SA, Stork EM, Erard N, Knott SRV, Hannon GJ, lncRNA Spehd Regulates Hematopoietic Stem and Progenitor Cells and Is Required for Multilineage Differentiation, Cell Rep. 27 (2019) 719–729.e6. 10.1016/j.celrep.2019.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].Li M, Ma K, Feng Z, Wang J, Zhou X, Zhou L, Differential long non-coding RNA expression profiles in the peripheral blood and CD4+ T cells of patients with active rheumatoid arthritis, Exp. Ther. Med 20 (2020) 461–471. 10.3892/etm.2020.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Aterido A, Palacio C, Marsal S, Avila G, Julià A, Novel insights into the regulatory architecture of CD4+ T cells in rheumatoid arthritis, PloS One. 9 (2014) e100690. 10.1371/journal.pone.0100690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].Moharamoghli M, Hassan-Zadeh V, Dolatshahi E, Alizadeh Z, Farazmand A, The expression of GAS5, THRIL, and RMRP lncRNAs is increased in T cells of patients with rheumatoid arthritis, Clin. Rheumatol 38 (2019) 3073–3080. 10.1007/s10067-019-04694-z. [DOI] [PubMed] [Google Scholar]
- [364].Sun Y, Liu J, Wen J, Huang D, Zhou Q, Zhang X, Ding X, Chen X, Overexpression of long noncoding RNA LINC00638 inhibits inflammation and oxidative stress in rheumatoid arthritis fibroblast-like synoviocytes by regulating the Nrf2/HO-1 pathway, Immun. Inflamm. Dis 10 (2022) e663. 10.1002/iid3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [365].Spurlock CF, Tossberg JT, Matlock BK, Olsen NJ, Aune TM, Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction, Arthritis Rheumatol. Hoboken NJ 66 (2014) 2947–2957. 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Zhang T-P, Zhu B-Q, Tao S-S, Fan Y-G, Li X-M, Pan H-F, Ye D-Q, Long Non-coding RNAs Genes Polymorphisms and Their Expression Levels in Patients With Rheumatoid Arthritis, Front. Immunol 10 (2019) 2529. 10.3389/fimmu.2019.02529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Lu X, Qian J, Downregulated MEG3 participates in rheumatoid arthritis via promoting proliferation of fibroblast-like synoviocytes, Exp. Ther. Med 17 (2019) 1637–1642. 10.3892/etm.2018.7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Saad El-Din S, Ahmed Rashed L, Eissa M, Eldemery AB, Abdelkareem Mohammed O, Abdelgwad M, Potential Role of circRNA-HIPK3/microRNA-124a Crosstalk in the Pathogenesis of Rheumatoid Arthritis, Rep. Biochem. Mol. Biol 10 (2022) 527–536. 10.52547/rbmb.10.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [369].Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, Zhu D, Shi G-P, Yang M, Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Rheumatoid Arthritis Patients, Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol 42 (2017) 651–659. 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- [370].Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing C, Su R, Xu J, Guo Y, Huang Z, Li J, Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis, Clin. Exp. Immunol 194 (2018) 118–124. 10.1111/cei.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [371].Chen D, Liu J, Zhao H-Y, Chen Y-P, Xiang Z, Jin X, Plasma long noncoding RNA expression profile identified by microarray in patients with Crohn’s disease, World J. Gastroenterol 22 (2016) 4716–4731. 10.3748/wjg.v22.i19.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Lucafò M, Di Silvestre A, Romano M, Avian A, Antonelli R, Martelossi S, Naviglio S, Tommasini A, Stocco G, Ventura A, Decorti G, De Iudicibus S, Role of the Long Non-Coding RNA Growth Arrest-Specific 5 in Glucocorticoid Response in Children with Inflammatory Bowel Disease, Basic Clin. Pharmacol. Toxicol 122 (2018) 87–93. 10.1111/bcpt.12851. [DOI] [PubMed] [Google Scholar]
- [373].Lucafò M, Pugnetti L, Bramuzzo M, Curci D, Di Silvestre A, Marcuzzi A, Bergamo A, Martelossi S, Villanacci V, Bozzola A, Cadei M, De Iudicibus S, Decorti G, Stocco G, Long Non-Coding RNA GAS5 and Intestinal MMP2 and MMP9 Expression: A Translational Study in Pediatric Patients with IBD, Int. J. Mol. Sci 20 (2019) E5280. 10.3390/ijms20215280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [374].Geng H, Bu H-F, Liu F, Wu L, Pfeifer K, Chou PM, Wang X, Sun J, Lu L, Pandey A, Bartolomei MS, De Plaen IG, Wang P, Yu J, Qian J, Tan X-D, In Inflamed Intestinal Tissues and Epithelial Cells, Interleukin 22 Signaling Increases Expression of H19 Long Noncoding RNA, Which Promotes Mucosal Regeneration, Gastroenterology. 155 (2018) 144–155. 10.1053/j.gastro.2018.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [375].Pan S, Liu R, Wu X, Ma K, Luo W, Nie K, Zhang C, Meng X, Tong T, Chen X, Wang X, Deng M, LncRNA NEAT1 mediates intestinal inflammation by regulating TNFRSF1B, Ann. Transl. Med 9 (2021) 773. 10.21037/atm-21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [376].J Z, Y S, H Y, J Q, Y Z, Y G, Y D, Y J, W Z, H W, Z L, L T, PLGA-microspheres-carried circGMCL1 protects against Crohn’s colitis through alleviating NLRP3 inflammasome-induced pyroptosis by promoting autophagy, Cell Death Dis. 13 (2022). 10.1038/s41419-022-05226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [377].Ouyang W, Wu M, Wu A, Xiao H, Circular RNA_0001187 participates in the regulation of ulcerative colitis development via upregulating myeloid differentiation factor 88, Bioengineered. 13 (2022) 12863–12875. 10.1080/21655979.2022.2077572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [378].Pahlevan Kakhki M, Nikravesh A, Shirvani Farsani Z, Sahraian MA, Behmanesh M, HOTAIR but not ANRIL long non-coding RNA contributes to the pathogenesis of multiple sclerosis, Immunology. 153 (2018) 479–487. 10.1111/imm.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]


