Abstract
Nerve injury can induce aberrant changes in ion channels, enzymes, and cytokines/chemokines in the dorsal root ganglia (DRGs); these changes are due to or at least partly governed by transcription factors that contribute to the genesis of neuropathic pain. However, the involvement of transcription factors in neuropathic pain is poorly understood. In this study, we report that transcription factor (TF) ETS proto-oncogene 1 (ETS1) is required for the initiation and development of neuropathic pain. Sciatic nerve chronic constrictive injury (CCI, a clinical neuropathic pain model) increases ETS1 expression in the injured male mouse DRG. Blocking this upregulation alleviated CCI-induced mechanical allodynia and thermal hyperalgesia, with no apparent effect on locomotor function. Mimicking this upregulation results in the genesis of nociception hypersensitivity; mechanistically, nerve injury-induced ETS1 upregulation promotes the expression of histone deacetylase 1 (HDAC1, a key initiator of pain) via enhancing its binding activity to the HDAC1 promotor, leading to the elevation of spinal central sensitization, as evidenced by increased expression of p-ERK1/2 and GFAP in the dorsal spinal horn. It appears that the ETS1/HDAC1 axis in DRG may have a critical role in the development and maintenance of neuropathic pain, and ETS1 is a potential therapeutic target in neuropathic pain.
Keywords: Neuropathic pain, ETS proto-oncogene 1, histone deacetylase 1, chronic constriction injury, dorsal root ganglia
Introduction
Neuropathic pain, which is caused by disease or injury of the nervous system, is characterized by spontaneous and irritant-induced pain sensation. Neuropathic pain affects up 7-8% of people in worldwide and has become a public health problem, with its incidence increasing at a rate of 0.82% every year.1–4 Moreover, there is a lack of effective clinical drugs for treatment of neuropathic pain owing to side effects.5 Thus, exploration of the mechanisms underlying neuropathic pain could contribute to the development of new treatment approaches for this disorder. Peripheral nerve injury induces dysfunctional regulation of expression of intracellular kinases, channels, receptors, and TFs in the dorsal root ganglia (DRGs), contributing to the production of neuropathic pain.5–8 However, the causes of these aberrant changes remain elusive.
ETS1 is a member of the ETS TF family.9 As ETS1 contains an evolutionarily conserved ETS domain, it attributes to DNA recognition and binding, in addition to its function in protein–protein interactions.10,11 An increasing body of evidence has established strong links between ETS1 and cell proliferation and growth. ETS1 has also been implicated in diseases of various systems, including the central nervous system (for instance, malignant gliomas ischemic stroke and depression),12–14 and it is widely expressed in many tissues, including lymphatic and hematopoietic tissues, and in the vascular system.6,9,12,13 ETS1 has been documented to be associated with natural killer cell development, glial cell formation, and angiogenesis.14–16 Notably, ETS1 has a critical role in the central nervous system.12,15,16 Feng et al. (2016) reveals for the first time that ETS1 may be associated with nervous system inflammation and shows that it promotes neuronal apoptosis in neuroinflammation by upregulating nuclear factor κB activation.12 Later, Yu et al. finds that the expression of ETS1 in hippocampal neurons is significantly increased in a cerebral ischemia model, where it promotes κB activation and then leads to neuronal apoptosis in neuroinflammation, by contrast, inhibiting the expression of ETS1 and significantly reducing neuronal apoptosis.15 In addition, a large study by Luo et al. shows that overexpression of ETS1 in the ventral hippocampal brain region of rats increases GalR2 expression and counteracts the depression-like behavior.14 However, whether ETS1 participates in pain processes has remained unknown.
Acetylation modification of histones plays an important part in the regulation of chronic pain. HDAC1 has been demonstrated to have functions in various types of pain, including chronic neuropathic pain, inflammation pain and cancer-original pain.17–20 For example, chronic nerve injury or spared nerve injury (SNI) is found to significantly increase HDAC1 expression in the spinal cord, whereas administration of selective HDAC1 inhibitor LG325 can relieve pain in mice.17 In the SNL mouse model, the HDAC1 promotor region recruits Sp1 to promote SOX10 expression, inducing neuropathic pain-like behavior.21 Another study shows that HDAC1 has an important effect on pain, with evidence demonstrates that intrathecal administration of T10 and SAHA, inhibitors of HDAC1, relieves bone cancer pain in rats by inhibiting the activation of glial cells in the spinal dorsal horn and DRG.19 Thus, it is important to further explore the functions of HDAC1 in the regulation of neuropathic pain.
In the present study, we found that chronic constriction injury (CCI) of peripheral nerves — a model of neuropathic pain that mimics clinical symptoms — caused increases in ETS1 and HDAC1 expression in mouse DRGs. There was a predictive binding motif sequence in the HDAC1 promotor. Therefore, we hypothesize that ETS1 regulates neuropathic pain via targeting HDAC1. This ETS1/HDAC1 axis provides novel insight into the mechanisms underlying neuropathic pain.
Materials and methods
Animals
Adult KM male mice (provided by the Experimental Animal Center of Xuzhou Medical University) weighing 25–30 g was used for the experiments. All mice were fed in a central house under a standard 12 h light/dark cycle, with water and food pellets available ad libitum. The room was maintained at a humidity of 20–30% and temperature of 20 ± 4°C. The experimental manipulations were started after the mice had been allowed to acclimatize for about 1 week.
CCI model
Sciatic nerve CCI in mice was conducted as described previously.22 Briefly, animals were placed on the operation table under anesthesia with 3% isoflurane, the mouse’s left leg was disinfected with iodophor after hair removal, the skin was cut open with scissors, and the unilateral middle and upper sciatic trunk was exposed and loosely ligated with 4-0 silk thread at three sites at intervals of about 1 mm. Then, the surgical incision was sutured and disinfected. Sham surgery animals received the same operation but without the ligation or transection of the sciatic nerve. After they awoke, they were returned to the mouse room.
Behavioral testing
Behavioral testing including mechanical paw withdrawal frequency (PWF) and thermal paw withdrawal latency (PWL) was performed as described previously.23,24 Briefly, all animal behavioral testing was performed in a quiet environment and required 1-2 h of early adaptation to the environment; thus, behavioral testing was performed between 09:00 and 14:00.
For PWF, the mouse was placed in an organic glass hood with a metal mesh at the bottom. After the mouse had been allowed to adapt to the environment quietly, 0.07 g and 0.4 g von Frey filament (Stoelting Co Wood Dale, IL) stimuli were used to simulate touch-induced pain and hyperalgesia, respectively. Through the metal mesh at the bottom, the central plantar region of the hind limb of the mice was vertically stimulated with stable pressure each time to bend the filament in an “S” pattern, which was maintained for 1.5 s. The occurrence of paw withdrawal and paw licking in the mice was marked as a positive reaction. The time interval between two adjacent rounds of stimulation was no less than 5 min. After repeated testing 10 times, the numbers of positive responses of mice were recorded, and the frequency percentage of mouse paw withdrawal under the stimulation of the same mass of filaments was calculated as the mechanical PWL.
For PWL, thermal hyperalgesia was measured using an IITC Plantar Analgesia Meter (Model 336, Series 8; IITC Life Science, USA). The mouse was placed on a mm-thick glass plate housed a single compartment with a transparent plexiglass box and allowed to adapt for more than 30 min. Measurements were obtained after the mice had become quiet. The center of the sole of the foot of the mouse was irradiated with a thermal radiation stimulator. The time until the mouse lifted its foot or moved and licked the sole of the foot was recorded. To avoid thermal damage caused by non-lifting of feet, the instrument system was set to automatically end irradiation after 20 s. We performed 3–5 consecutive measurements at intervals of no less than 10 min. Taking the mean of the recorded results gave the PWL value for each mouse.
DRG microinjection
DRG microinjection was carried out as described previously.23,25 Briefly, each mouse was anesthetized and placed on an operating table, the dorsal hair was shaved and the area disinfected with iodophor, a small incision was made in the paravertebral region of the backs of mice with surgical scissors to expose the transverse process, and the corresponding articular was removed with forceps to expose the DRG. A glass micropipette, which had been drawn in advance, was connected to the needle of a 10 μL microsyringe to microinject virus or short interfering RNA (siRNA). We adjusted the manipulator under the microscope, a glaze needle was inserted into the DRG, and corresponding fluids were injected and then removed after about 10 min. The muscle and skin were sutured after anti-infection treatment. The sequence of the siRNA was as follows: ETS1 siRNA sense 5′-GCAGAUGUCCCGCUGUUAATT-3′, antisense: 5′-UUAACAGCGGGACAUCUGCTT-3′.
Western blotting
The DRG tissue was removed as described previously,26 minced with scissors, homogenized by contact ultrasound in RIPA lysis buffer (P0013B, Beyotime) with 130 μL of 1 mM PMSF (ST506, Beyotime) and 1 mM protease inhibitor (p8340, Sigma Aldrich), and then centrifuged at 12,000 g for 20 min. The supernatant was collected, and protein concentrations were determined using a BCA Protein Assay Kit (P0010, Beyotime). Proteins were denatured with sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer at 100°C for 8 min and separated on a glycine-SDS-PAGE gel. The proteins were electrophoretically transferred onto a polyvinylidene fluoride membrane (IPVH00010, Millipore). After blocking for 2 h by 5% bovine serum albumin in Tris-buffered saline (TBS) containing 0.1% Tween-20 (P9416, Sigma Aldrich), the membrane was incubated with rabbit anti-ETS1 antibody (1:1000; A13302, ABclonal), rabbit anti-HDAC1 antibody (1:1000; 10,197-1-AP, Proteintech), rabbit anti-HDAC2 antibody (1:1000; 12,922-3-AP, Proteintech), rabbit anti-HDAC3 antibody (1:1000; 10,255-1-AP, Proteintech), rabbit anti-p-ERK antibody (1:1000; 28,733-1-AP, Proteintech), mouse anti-ERK antibody (1:1000; 11,257-1-AP, Proteintech), mouse anti-GFAP antibody (1:1000; 60,190-1-Ig, Proteintech), tubulin antibody (1:2000; 10,094-1-AP, Proteintech), or rabbit anti-GAPDH antibody (1:5000; ET1601-4, HuaBio) at 4°C overnight. The membranes were washed three times in 0.1% TBS-Tween 20 and then incubated with HRP-labeled goat anti-rabbit IgG (1:2000; A0208, Beyotime) or HRP-labeled goat anti-mouse IgG (1:2000; A0216, Beyotime) for 2 h at room temperature and washed three times. The immune complexes were detected with an ECL chemiluminescent assay kit (P0018S, Beyotime). The optical density of each band was then measured with a computer-assisted imaging analysis system (ImageJ) and normalized with tubulin or GAPDH.
Cell line culture and transfection
HEK293T (ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle medium (DMEM; D0821, Sigma Aldrich) containing 10% fetal bovine serum (FBS; F8318, Sigma Aldrich) at 37°C in a humidified cell incubator with 5% CO2 and 95% O2. The plasmids were transfected into the HEK293 T cells using ExFect Transfection Reagent (T101-01/02, Vazyme) according to the manufacturer’s instructions.
Luciferase reporter assay
To construct HDAC1 gene reporter plasmids, the 799-base-pair (bp) fragment from the HDAC1 gene promotor region (including the ETS1-binding motif) was amplified by polymerase chain reaction (PCR) from genomic DNA with the following primer pairs: forward: 5′-GCCGGTACCGCTAGCCTCGAGTAGGTGAGATGGTTGCTGCCA-3′; reverse: 5′-TCTACGCGTGAGCTCCTCGAGGAGTCTGCGCCATCTTGCTC-3′. The PCR products were ligated into the pGL6-Basic vector (containing the firefly luciferase reporter gene, Promega, Madison, WI) using the Xhol (R-146, NEB) restriction sites. DNA sequencing was carried out to verify the sequence of the reconstruction vector. HEK293T cells were cultured as described above. One day after plating on a 24-well plate, the cells were transfected with 200 ng of pGL6-Basic vectors as a control using ExFect Transfection Reagent (T101-01/02, Vazyme) according to the manufacturer’s instructions. The wells were divided into groups as needed. After transfection for 48 h, the cells were collected in passive lysis buffer. Approximately 100 μL of supernatant was used to measure the luciferase activity with a Dual-Luciferase Reporter Assay System (E1910, Promega). Independent transfection experiments were repeated three times. The relative reporter activity was calculated after normalization of the ratio of firefly to Renilla luciferase activity.
Lentivirus production
Lentivirus production was carried out as described previously.27 Briefly, the constructed core plasmid (8 μg) and two envelope plasmids, the PSPAX2 (6 μg) vector and PMD2G (4 μg) vector, were co-transfected into 293T cells in a 10 cm dish containing DMEM without FBS. After transfection for 6 h, complete medium containing 10% FBS was used according to the manufacturer’s instructions provided with the ExFect Transfection Reagent (T101-01/02, Vazyme). The supernatant was collected at 48–72 h after transfection and concentrated using a Centricon Plus-70 filter unit (UFC910096, Millipore, MA, USA). Lentivirus with titer 108 TU/ml was used in the experiments.
Quantitative PCR (qPCR)
qPCR was performed as previously described.27 Briefly, total RNA extraction was performed according to the instructions provided with TRIzol reagent (9190, Takara), and the quality and quantity of total RNA were evaluated with a Nanodrop ND-2000 (Thermo Scientific, USA). RNA (400 ng) was reverse transcribed into single-stranded cDNA using 4 × gDNA wiper MIX and 5× HiScript II Select Qrt SuperMIX II (R233-01, Vazyme). Reverse transcription of RNA was performed at 42°C for 2 min, 50°C for 5 min, and 85°C for 5 s. The cDNA template (1.5 μL) was amplified by real-time qPCR with the primers listed in Table 1 The signal for each sample was determined in triplicate using SYBR Green Master Mix (Q111-02, Vazyme). Reactions were carried out in an ABI system. Data were normalized to Gapdh (cycle threshold [CT]) using the 2-(ΔΔCT) method. Any value among triplicates that showed a marked difference (≥1.00) compared with the average of the other two was discarded. A total of 1.5 μL cDNA was used for real-time quantification, in which 1.2 μL forward and reverse primers (5 μM) each, 7.5 μL TB Green Premix, and 0.3 μL Rox Reference Dye II were used to configure a 15 μL system. Each sample was repeated in duplicate, mixed in eight tubes, and measured with an ABI quantitative PCR instrument (Thermo Scientific, USA). The reaction conditions were as follows. After pre-denaturation at 95°C for 1 min, 40 cycles of 95°C for 10 s, 58°C for 20 s, and 72°C for 30 s were performed. The primer sequences were as follows: ETS1 forward 5′-ACAGACTACTTTCGGATCAAGCA-3′, reverse 5′- ACGCTCTCAAAAGAGTCCTGG-3′; Gapdh forward 5′-GGTGAAGG TCGGTGTGAACG-3′, reverse 5′-CTCGCTCCTGGAAGATGG TG-3′.
Table 1.
All primers and siRNA used.
| Names | Sequences | Names | Sequences |
|---|---|---|---|
| RT-qPCR | ChIP-PCR | ||
| ETS1 F | 5′-ACAGACTACTTTCGGATCAAGCA-3′ | ChIP-Hdac1 F1 | 5′-GTAACACTGAATAAGCTCTTCA-3′ |
| ETS1 R | 5′-ACGCTCTCAAAAGAGTCCTGG-3′ | ChIP-Hdac1 R11 | 5′-AGGACTCAGGAGGCAGAGTC-3′ |
| Gapdh F | 5′-GGTGAAGGTCGGTGTGAACG-3′ | ChIP-Hdac1 R12 | 5′-TGGGCGTGGTTGCTATGTCT-3′ |
| Gapdh R | 5′-CTCGCTCCTGGAAGATGGTG-3′ | ChIP-Hdac1 F2 | 5′-CTCTGATCTTTGCACACGC-3′ |
| siRNA | ChIP-Hdac1 R21 | 5′-GCCGTACCATCGAGTCCC-3′ | |
| Ets1-siRNA-441S | 5′-GCAGAUGUCCCGCUGUUAATT-3′ | ChIP-Hdac1 R22 | 5′-GGGCAGCTTAGAGAAGCCA-3′ |
| Ets1-siRNA-441AS | 5′-UUAACAGCGGGACAUCUGCTT-3′ | ChIP-Hdac1 F | 5′-AGACATGCTTTCACTACGGA-3′ |
| HDAC1-siRNA-S | 5′-CCGGUCAUGUCCAAAGUAA-3′ | ChIP-Hdac1 R | 5′-AGAGTCAGGAAGATCTCTGT-3′ |
| HDAC1-siRNA-AS | 5′-UUACUUUGGACAUGACCGG-3 | ChIP-Hdac1 F3 | 5′-TGCAGAGTTTACGGAGCCGA-3′ |
| luciferase assay | ChIP-Hdac1 R3 | 5′-TCACTCTAACCAATCGGCAT-3′ | |
| pGL6-Hdac1 F1 | 5′-GCCGGTACCGCTAGCCTCGAGTAGGTGAGATGGTTGCTGCCA-3′ | ||
| pGL6- Hdac1 F2 | 5′-GCCGGTACCGCTAGCCTCGAGGTAACACTGAATAAGCTCTTCA-3′ | ||
| pGL6- Hdac1 R | TCTACGCGTGAGCTCCTCGAG GAGTCTGCGCCATCTTGCTC-3′ |
RT: Reverse-transcription. F: Forward. R: Reverse.
Double-labeling immunofluorescence
Immunofluorescence was performed as described in previous reports.28 Briefly, L3-L4 DRGs were rapidly detached from mice perfused with 4% paraformaldehyde (BL539 A, Biosharp) and post-fixed with 4% paraformaldehyde for 2 h, followed by dehydration in 5%, 15%, 30% sucrose gradients, respectively, and sectioned at a thickness of 12 mm on a cryostat. After blocking at room temperature for 1 h and washing with PBST (containing 0.04% Triton), the slices were incubated with anti-rabbit ETS1 antibody (1:1000; A13302, ABclonal), mouse anti-glutamine synthetase (GS; 1:800; EM1902-39, Hua Bio), mouse anti-β-tubulin (1:800; M0805-8, Hua Bio), mouse anti-CGRP (1:50; SC57053, Santa Cruz), mouse anti-P2X3 (1:100; SC390572, Santa Cruz), or mouse anti-NF200 (1:100; 60,331-1-Ig, Proteintech) at 4°C overnight. After washing with PBST, slices were finally incubated with fluorescent-conjugated secondary antibodies Alexa FluorTM 488 goat anti-rabbit IgG (A21206, Molecular Probes) and Alexa Fluor™ 594 goat anti-mouse IgG (A10036, Molecular Probes). The sections were mounted on a mounting medium with DAPI (0100-20, Southern Biotech) and scanned using a high-resolution digital confocal microscope (Olympus FV1000, Japan).
Chromatin immunoprecipitation assays
The ChIP assays were performed according to the instructions of the Beyotime CHIP Assay kit (P2078, Beyotime). Briefly, DRG minced with scissors was cross-linked with 1% formaldehyde at room temperature for 8 min. The process of cross-linking was terminated by adding glycine (0.25 M); then, after centrifugation at 12,000 g for 6 min, the pellet was collected and lysed in SDS lysis buffer containing protease inhibitor PMSF. The lysis was performed by sonication at medium intensity until the DNA was broken into fragments with a length of 200–1000 bp. Each sample was divided into two parts, with 50 μL used as an input (appositive control) and the remainder (1.8 mL) pre-treated with 60 μL protein A + G agarose beads rotation for 1 h at 4°C. After centrifugation, the supernatant was divided into two tubes; subsequently, 2 μg anti-rabbit ETS1 was added to one tube, followed by incubation with rotation overnight at 4°C. To the other tube, 2 μg Normal Rabbit IgG (A7058, Beyotime) was added as a negative control, 70 μL protein A + G agarose beads was added to the mixture, followed by incubation with rotation for 2 h at 4°C, and the chromatin was pulled down using beads. After purification, the DNA fragments were amplified using teal-time PCR with the primers listed in Table 1.
Locomotor function
Locomotor function, including placing, grasping, and righting reflexes, was assessed as previously described.29 Briefly, for the placing reflex, we placed the hindlimb of the mouse slightly below the forelimb with back of the hind paw in contact with the edge of the table and recorded whether the hind paws were placed on the table surface reflexively. For the righting reflex, we placed the mouse supine on a flat surface and observed whether it immediately returned to its normal upright position. For the grasping reflex, the mouse was placed on a wire mesh perpendicular to the ground to observe whether it could grasp the wire mesh with its hind paws. Scores for placing, grasping, and righting reflexes were determined according to the counts for each normal reflex in five trials.
Statistical analyses
All data were statistically analyzed using GraphPad Prism 8.0 software. All measurement data are expressed as mean ± SEM. Comparisons between two groups were performed by Student’s t-test, comparisons among three or more groups were performed by one-way or two-way analysis of variance (ANOVA), and pairwise comparisons between groups were performed by q-test. The data collected from mouse behavioral tests at different time points were analyzed by repeated measurements using two-way ANOVA. Differences were considered statistically significant at p < 0.05.
Results
Peripheral nerve injury upregulates ETS1 in DRG neurons
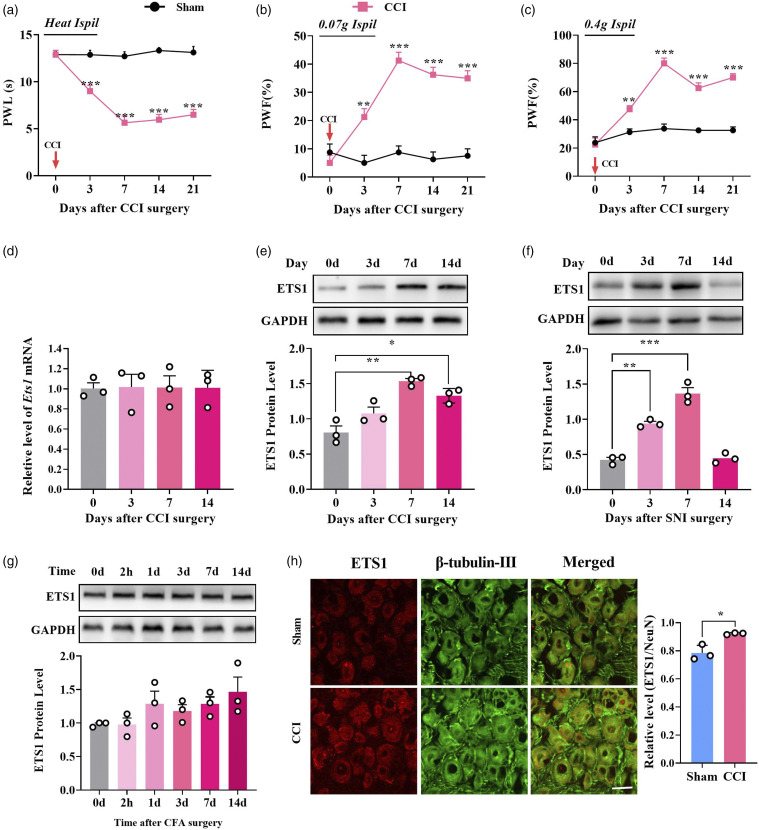
To determine whether DRG ETS1 was associated with neuropathic pain, we first examined the expression of ETS1 in DRG of mice subjected to unilateral sciatic nerve CCI (a clinical neuropathic pain model). As previously described, mice with CCI surgery showed significant pain hypersensitivity in response to mechanical and thermal stimulus starting from day 3 after surgery; however, sham surgery did not change the basal pain threshold (Figures 1(a) to (c)). CCI-induced nerve injury did not change the level of Ets1 mRNA in the unilateral injured DRGs from day 3–14 after surgery (Figure 1(d)), but ETS1 protein expression was markedly upregulated, by 90.6% on day 7 and 64.8% on day 14, in the unilateral injured DRGs after CCI surgery (Figure 1(e)). A similar increase was observed in the ipsilateral DRGs of SNI mice, another neuropathic pain model, on day 7 and day 14 after surgery (Figure 1(f)). To examine whether peripheral chronic inflammation pain also changed ETS1 expression in the DRGs, we injected complete Freund’s adjuvant (CFA)30 unilaterally into the plantar side of the hind paw. Unexpectedly, no ETS1 change was seen in the DRGs from 2 h to 14 days post-CFA injection (Figure 1(g)). Immunofluorescence staining further confirmed the increase in ETS1 in DRGs on day 7 after CCI, compared with the sham group (Figure 1(h)). Thus, it is likely that Ets1 is associated with chronic neuropathic pain but not chronic inflammation pain.
Figure 1.
Peripheral nerve injury-induced increased expression of ETS1 protein in mouse L3/4 injured DRGs. (a–c) CCI of unilateral sciatic nerve caused hypersensitivity to heat (a) and mechanical (b, c) stimuli. **p < 0.01, ***p < 0.001 versus sham; n = 8. Data were analyzed by two-way repeated-measures ANOVA followed by post hoc Tukey test. (d) Expression of Ets1 mRNA in the ipsilateral L3/4 DRGs on days 0, 3, 7, and 14 after CCI-induced chronic neuropathic pain. No significance versus day 0; n = 6. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (e) ETS1 protein expression in the ipsilateral L3/L4 DRGs on days 0, 3, 7, and 14 after CCI or sham surgery; n = 6 mice per time point. *p < 0.05, **p < 0.01 versus day 0. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (f) ETS1 protein expression in the ipsilateral L3/L4 DRGs on days 0, 3, 7, and 14 after spared nerve injury (SNI) or sham surgery; n = 6 mice per time point. **p < 0.01 versus day 0. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (g) ETS1 expression in the ipsilateral L3/L4 DRGs at 0 d, 2 h, 1 d, 3 d, 7 d, and 14 d after Freund’s adjuvant (CFA)-induced inflammation pain or sham surgery; n = 6 mice per time point. No significance versus day 0. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (h) Co-staining of ETS1 (red) and β-tubulin III (a neuronal marker, green) immunofluorescence in the ipsilateral L3/4 DRGs on day 7 after CCI or sham surgery; n = 5. Scale bar, 30 μm. *p < 0.05 versus the corresponding sham group. Data were analyzed by Student’s t-test. PWF: Paw withdrawal frequency; PWL: Paw withdrawal latency.
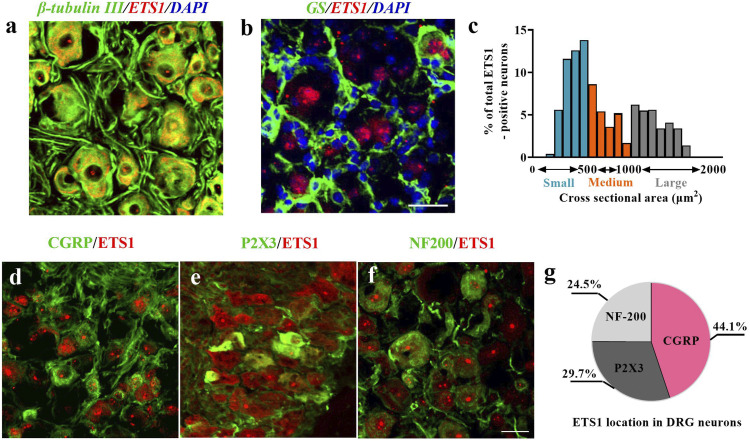
Next, to examine the cellular localization of ETS1 in DRGs, we performed ETS1 double-labeling staining with different DRG cellular marker proteins. Approximately 100% of ETS1-positive cells were labeled by β-tubulin III (a neuron marker) (Figure 2(a)), but almost no ETS1-positive cells by GS (a microsatellite glia cell marker) (Figure 2(b)). Notably, ETS1 was found in both cytoplasm and nuclei of neurons (Figures 2(a) and (b)). To further examine the subcellular distribution of ETS1 in DRG neurons, we analyzed its expression in DRG neurons of different sizes. ETS1 was expressed in all three types of neurons with different abundance. A cross-sectional area analysis showed that 24.5% of ETS1-positive neurons were co-labeled with NF200 (large cell type neurons; Figure 2(f)), 29.7% with P2X3 (small- and medium-sized cell type neurons; Figure 2(e)), and 44.1% with CGRP (small cell type peptidergic neurons; Figure 2(d)). Owing to the close linkage of medium- and small-sized neurons with pain sensation, the above results suggest that ETS1 may be involved in the pain process.
Figure 2.
Distribution of ETS1 protein in L3/4 DRGs of mouse. (a, b) ETS1 (red) was co-expressed exclusively with β-tubulin III (a, green) in cellular nuclei and undetected in cellular nuclei (DAPI, blue) of cells labelled with glutamine synthetase (GS, b, green). (c) Analysis of ETS1 in positive neuronal somata of cross-sectional area: large, 29.7%; medium, 24.5%; small, 44.1%. (d–f) Distribution of ETS1-positive neurons. ETS1-positive neurons were labelled by calcitonin gene-related peptide (CGRP; d, green), P2X3 (e, green), or neurofilament 200 (NF200; f, green); n = 4. Scale bar, 30 mm. (g) Schematic representation of ETS1 positive neurons.
Blockade of ETS1 upregulation alleviates CCI-induced nociception hypersensitivity
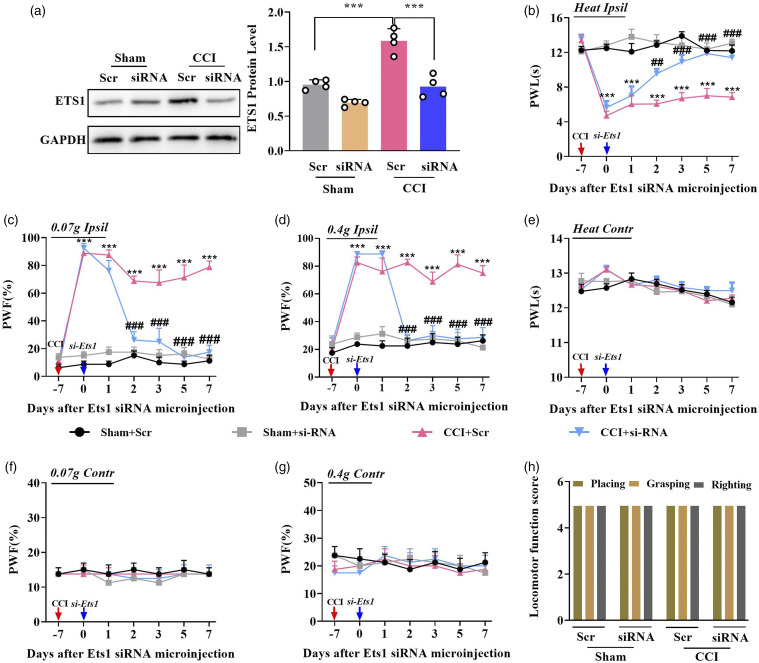
Next, we investigated whether ETS1 regulates neuropathic pain. To this end, we designed an siRNA (targeting −441 of Ets1 mRNA, transcription start site as +1) as described previously31 to knock down ETS1 expression. Compared with the scrambled siRNA, microinjection of siRNA-Ets1 reversed the CCI-induced increase in ETS1 in the ipsilateral L3/4 DRGs on day 3 after siRNA injection in CCI mice with 7 days after surgery. As expected, siRNA-Ets1 microinjection, but not scrambled siRNA, attenuated nerve-injury-induced thermal pain and mechanical pain from day 2 after injection; this antinociception lasted at least 5 days on the ipsilateral side (Figures 3(b)–(d)), but no such behavioral changes were observed on the contralateral side (Figures 3(e)–(g)). These treatments did not cause injury to locomotor function (Figure 3(h)). Collectively, these data suggest that ETS1 may be a critical antinociception regulator.
Figure 3.
Blocking DRG ETS1 upregulation alleviates neuropathic pain development and maintenance in mouse. (a) ETS1 protein levels in the ipsilateral L3/4 DRGs 7 days after CCI surgery in mice with microinjection of Ets1 siRNA (siRNA) and scrambled siRNA (Scr); n = 8 mice per group. **p < 0.01 versus the corresponding sham Scr + CCI Scr group. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (b–d) Effects of microinjection of Ets1 siRNA and scrambled siRNA into the ipsilateral L3/4 DRGs after CCI or sham surgery on ipsilateral paw withdrawal responses to mechanical (c, d) and thermal stimulation (b); n = 8 mice per group. ***p < 0.001 versus the corresponding sham Scr + CCI Scr group. ###p < 0.001 versus the corresponding CCI Scr + CCI siRNA group. Data were analyzed by two-way ANOVA followed by post hoc Tukey test. (e–g) Effects of microinjection of Ets1 siRNA and scrambled siRNA into the ipsilateral L3/4 DRGs after CCI or sham surgery on contralateral paw withdrawal responses to mechanical (f, g) and thermal stimulation (e); n = 8 mice per group. No significance versus the corresponding sham Scr + CCI Scr group. No significance versus the corresponding CCI Scr + CCI siRNA group. Data were analyzed by two-way ANOVA followed by post hoc Tukey test. (h) Locomotor function trial, including placing, grasping, and righting reflexes; n = 8 per group, five trials. No significance: Data were analyzed by one-way ANOVA followed by post hoc Tukey test.
Mimicking ETS1 upregulation causes neuropathic pain-like behavior
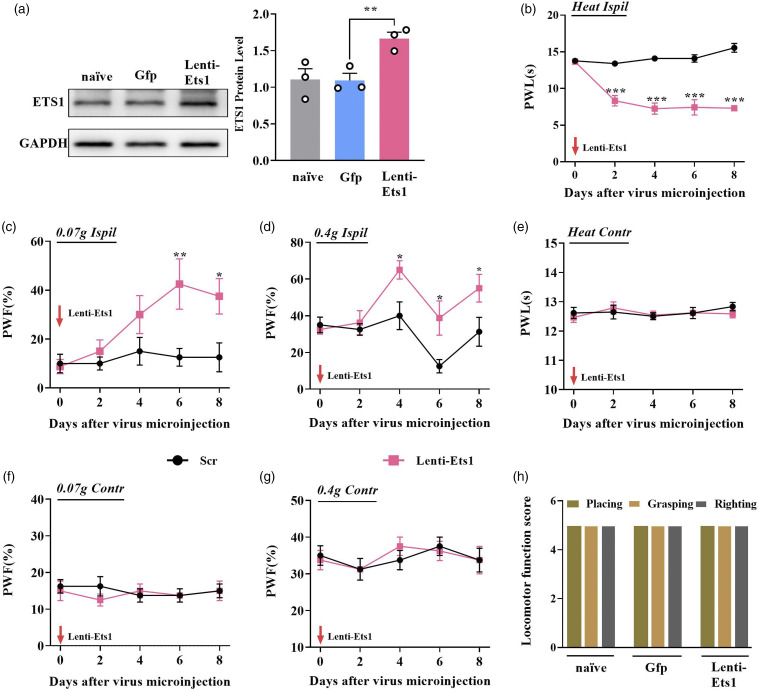
We further examined whether upregulating DRG ETS1 could initiate the genesis of nociceptive hypersensitivity. A full-length Ets1 lentivirus expression vector was constructed. Microinjection of Lenti-Ets1 but not Lenti-Gfp into unilateral L3/4 DRGs of naïve adult mice enhanced the expression of ETS1 by 50.3% on day 6 after injection (Figure 4(a)). Behaviorally, ETS1 overexpression through microinjection of Lenti-Ets1 not only increased the frequency of hind paw withdrawal in response to light and heavy mechanical stimulus (Figures 4(c) and (d)) but also shortened hind paw lifting time after thermal stimulus on days 4, 6, and 8 after injection (Figure 4(b)). However, contralateral side did not observe these responses (Figures 4(e)–(g)). In addition, no locomotor impairments after these treatments were observed in naïve mice (Figure 4(h)). Taken together, these data suggest that DRG ETS1 upregulation is sufficient for the induction of nociceptive hypersensitivity.
Figure 4.
Overexpression of ETS1 in DRG evoked neuropathic pain-like symptoms in naïve mice. (a) Overexpression of ETS1 with Lenti-Ets1 increased the expression of ETS1 in the ipsilateral DRGs of naïve mice as measured by western blotting. Gfp was used as a control for Lenti-Ets1. *p < 0.05 versus Gfp; n = 6. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (b–d) Effects of microinjection of Lenti-Ets1 into the ipsilateral L3/4 DRGs on the ipsilateral PWF in response to 0.07 g (c) and 0.4 g (d) von Frey filament stimuli and PWL in response to heat stimuli (b) at indicated days after microinjection; n = 6 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001 versus Gfp. Data were analyzed by two-way ANOVA followed by post hoc Tukey test. (e–g) Effects of microinjection of Lenti-Ets1 into the ipsilateral L3/4 DRGs on the contralateral PWF in response to 0.07 g (f) and 0.4 g (g) von Frey filament stimuli and PWL in response to heat stimuli (e) at the indicated days after microinjection; n = 6 mouse per group. No significance versus Gfp. Data were analyzed by two-way ANOVA followed by post hoc Tukey test. (h) Locomotor function trial, including placing, grasping, and righting reflexes; n = 8 per group, five trials. No significance versus naïve: Data were analyzed by one-way ANOVA followed by post hoc Tukey test.
ETS1 contributes to neuropathic pain via targeting HDAC1
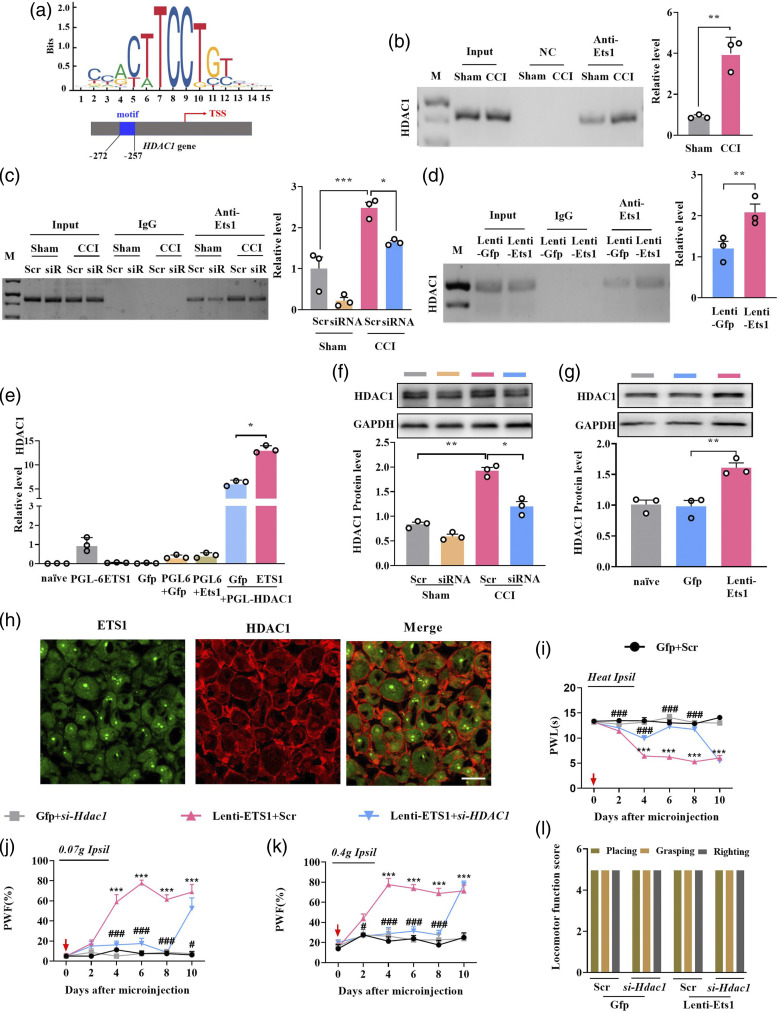
How does ETS1 regulate neuropathic pain? As histone deacetylase HDAC1 is a key initiator in neuropathic pain, we investigated whether ETS1 regulated nociception behavior by targeting HDAC1. Using an online JASPAR program, one ETS1 binding motif (5′-AGATCTTCCTGACTC-3′, −272 to −257, transcription start site designated as +1) was observed in the Hdac1 promotor (Figure 5(a)). The ChIP results showed that Hdac1 promotor fragments containing the binding motifs could be amplified from complexes immunoprecipitated with ETS1 antibody in sham L3/4 DRGs (Figure 5(b)), indicating that ETS1 specifically binds to the Hdac1 promotor. The binding activity was significantly increased on day 7 post CCI-induced nerve injury (Figure 5(b)). ETS1 knockdown through microinjection of Ets1-siRNA markedly blocked the CCI-induced enhanced binding accounts of ETS1 on the Hdac1 promotor on day 2 after siRNA in CCI mice with 7 days after surgery (Figure 5(c)). Whereas ETS1 overexpression by microinjecting Lenti-ETS1 increased binding accounts (Figure 5(d)). The augment in binding activity may have been due to HDAC1 upregulation in ipsilateral L3/L4 DRGs following CCI.
Figure 5.
ETS1 positively regulates HDAC1 expression by targeting the promotor. (a) Schematic representation of ETS1 action on HDAC1 promotor region. The ETS1 binding region located in −272 ∼ −257 of mouse HDAC1 promotor (transcription start site designated as +1) is marked in blue. (b–d) ChIP qPCR assay for ETS1 binding with HDAC1 promotor fragments, ipsilateral L3/4 DRGs on day 7 post-CCI or sham surgery (b); ipsilateral L3/4 DRGs on day 7 post-CCI or sham surgery with microinjection of Ets1 siRNA (siRNA) and scrambled siRNA (Scr) (c) and ipsilateral L3/4 DRGs microinjected with Lenti-Ets1 or Lenti-Gfp (d); PCR and DNA gel electrophoresis were performed to verify the binding results. Input, total purified fragments. M, ladder marker. IgG, negative control. n = 6. *p < 0.05, **p < 0.01 versus the sham group. Data were analyzed by Student’s t-test and one-way ANOVA followed by post hoc Tukey test. (e) Luciferase reporter analysis of ETS1 positive regulation of the transcription of HDAC1 with co-transfection of the reporter plasmid. *p < 0.05 versus the corresponding vectors; n = 3 repeats per treatment. The HDAC1 promotor bound by ETS1 was inserted into the luciferase promotor in the pGL6 vector. The constructed or empty pGL6 and ETS1 overexpression plasmid PCD-ETS1 were co-transfected into HEK293 T cells; H2O was used as control. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (f) HDAC1 protein expression in the ipsilateral L3/4 DRGs 7 days after CCI surgery in mice with microinjection of Ets1 siRNA (siRNA) and scrambled siRNA (Scr); n = 6 mice per group. *p < 0.05, **p < 0.01 versus sham group plus scrambled siRNA. Data were analyzed by two-way ANOVA followed by post hoc Tukey test. (g) HDAC1 protein expression in the ipsilateral L3/4 DRG 8 days after viral microinjection into the ipsilateral L3/4 DRGs in mice; n = 6 per group. **p < 0.01 versus the corresponding group, naïve as a control. Data were analyzed with one-way ANOVA followed by post hoc Tukey test. (h) Co-localization of ETS1 (green) with HDAC1 (red) in mice L3/4 DRGs neurons. Scale bar, 30 mm. (i–k) Effects of microinjection of Lenti-Ets1 mixed with siRNA Hdac1 (si-Hdac1) into the ipsilateral L3/4 DRGs on the ipsilateral PWF in response to 0.07 g (j) and 0.4 g (k) von Frey filament stimuli and PWL in response to heat stimuli (i) at the indicated days after microinjection and scrambled siRNA (Scr) as a control; n = 8 per group. ***p < 0.001 versus the corresponding group. Data were analyzed by two-way ANOVA followed by post hoc Tukey test. (l) Locomotor function trial, including placing, grasping, and righting reflexes; n = 8 per group, five trials. No significance versus naïve. Data were analyzed by one-way ANOVA followed by post hoc Tukey test.
Next, we determined whether ETS1 acts directly on the translation of HDAC1; to this end, we carried out in vitro and in vivo verification experiments. First, a 799-bp-length Hdac1 promotor fragment containing the binding motifs was cloned upstream of pGL6, a luciferase reporter vector, and co-transfected into HEK293T cells with the full-length ETS1 expression plasmid. As expected, co-transfection of the ETS1 expression plasmid, but not the control Gfp plasmid, increased the luciferase activity of the reporter 2.11-fold (Figure 5(e)). These in vitro data indicate that ETS1 initiates the expression of HDAC1. As expected, knockdown of ETS1 with siRNA reversed the upregulation of HDAC1 protein expression caused by nerve injury in the injured DRGs on day 3 after ETS1 siRNA injection in CCI mice, whereas the scrambled siRNA did not produce these effects (Figure 5(f)). On the contrary, upregulation of ETS1 by lentivirus injection increased the HDAC1 protein expression in L3/4 DRGs on day 5 after injection (Figure 5(g)). Given that approximately 73% of ETS1-labelled DRG neurons were positive for HDAC1 (Figure 5(h)), it is very likely that ETS1 directly regulates HDAC1 expression in DRGs.
Finally, we examined behaviorally whether DRG Ets1 upregulated nociception hypersensitivity via the mediation of Hdac1. As expected, pain hypersensitivity to mechanical and heat stimulus developed in the group microinjected with Lenti-Ets1 alone, whereas hyperalgesia disappeared in the groups injected with the Lenti-ETS1 together with Hdac1 siRNA (Figures 5(i)–(k)). No locomotor changes were observed in these treated mice (Figure 5(l)). The above results indicate that ETS1 may be involved in the neuropathic pain development and maintenance in the injured DRG at least partly via targeting HDAC1.
ETS1 participates in central sensitization in the dorsal spinal horn
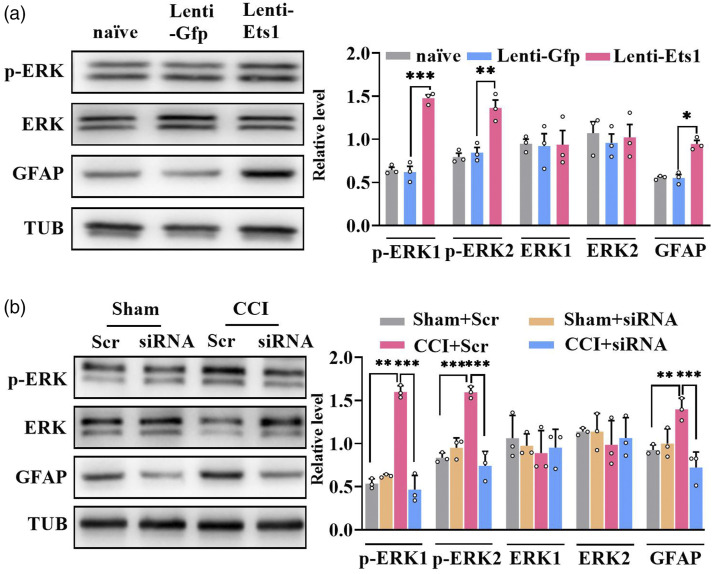
We next sought to establish whether spinal sensitization was caused by DRG ETS1 under neuropathic pain conditions. We found that CCI nerve injury caused significant increases in phosphorylated extracellular signal-regulated kinase 1/2 (p-ERK1/2, a marker of neuronal hyperactivation) and glial fibrillary acidic protein (GFAP, a marker of astrocyte hyperactivation) levels on the injured side of the spinal dorsal horn; these increases were reversed by microinjection of Ets1 siRNA but not its scrambled siRNA in the ipsilateral L3/4 DRGs on day 2 after microinjection (Figure 6(b)). However, in contrast to the results for p-ERK1/2 and GFAP, the level of total ERK1/2 did not change in the dorsal horn after these treatments (Figure 6(b)). Conversely, DRG microinjection of Lenti-Ets1 but not Lenti-Gfp induced increases in p-ERK1/2 and GFAP levels in the ipsilateral side dorsal horn on day 5 after injection (Figure 6(a)), indicating hyperactivity of neurons and astrocytes. As anticipated, no change in total ERK1/2 level was observed in the dorsal horn after microinjection (Figure 6(a)). This evidence strongly suggests that ETS1 participates in neuropathic pain via spinal sensitization.
Figure 6.
ETS1 participates in central sensitization in the spinal cord. (a) Expression levels of spinal cord proteins p-ERK1/2, ERK1/2, and GFAP in the ipsilateral L3/4 DRG 8 days after viral microinjection in mice; n = 6 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001 versus the corresponding Lenti-Gfp group. Data were analyzed by one-way ANOVA followed by post hoc Tukey test. (b) Expression levels of spinal cord p-ERK1/2, ERK1/2, and GFAP in the ipsilateral L3/4 DRGs 7days after CCI surgery in mice with microinjection of Ets1 siRNA (siRNA) and scrambled siRNA (Scr); n = 6 mice per group. **p < 0.05, ***p < 0.001 versus sham group plus scrambled siRNA. Data were analyzed by two-way ANOVA followed by post hoc Tukey test.
Discussion
DRGs contain the cell bodies of primary sensory neurons such as nociceptive neurons. Peripheral nerve injury causes maladaptive molecular changes in DRG cell bodies and axons; these changes result in hypersensitivity and hyperexcitability of central nervous system neurons (central sensitization) and are crucial for the development and maintenance of neuropathic pain.32 Therefore, understanding the molecular mechanisms in DRGs underlying nerve injury is essential for the treatment of neuropathic pain. Here, we show for the first time that peripheral nerve injury induced an increase in expression of TF ETS1 in DRG neurons. Blockade of ETS1 attenuates neuropathic pain through reversing the nerve-injury-induced increase in expression of HDAC1 (a key initiator of neuropathic pain). It appears that ETS1 may serve as a management target in neuropathic pain.
TFs recognize specific DNA sequences to control gene transcription. DRG TFs have emerged as essential participants in the development and maintenance of chronic pain. Pou4f3 regulates neuropathic pain by promoting DRG-specific DS-lncRNA (long non-coding RNA) transcription in DRG; reversing the decrease in Pou3f4 alleviates CCI-induced pain hypersensitivity.31 Nerve-injury-specific lncRNAs cooperate with TF ELF1 to increase CCL2 expression, leading to the production of neuropathic pain symptoms.23 Although a strong association of TFs with neuropathic pain has been established, it remains challenging to determine which and how TFs control gene expression.33 In this work, we demonstrate the involvement of DRG E26 avian leukemia oncogene 1, 5′ domain (ETS1), a TF, in CCI-induced neuropathic pain.
ETS1 consists of a conserved ETS DNA-binding region that recognizes the core consensus DNA sequence GGAA/T. ETS1 can act either as a transcriptional activator or a repressor of many genes and participates in stem cell development, cellular senescence and death, and tumorigenesis.34 Accumulating evidence shows that ETS1 is associated with a variety of diseases such as Jacobson syndrome and systemic lupus erythematosus.35,36 Yet, it is unknown whether ETS1 regulates pain. In the present study, peripheral nerve injury robustly increased levels of ETS1 protein expression in DRGs injured by both CCI and SNI surgery, but CFA inflammatory pain did not change the expression level of DRG ETS1, indicating that DRG ETS1 may be a specific regulator of nerve-injury-induced neuropathic pain. In addition, nerve injury did affect the expression of ETS1 mRNA in DRGs. Therefore, we speculate that the self-regulation of ETS1 is likely to involve a post-transcriptional mechanism; this needs to be further explored in future. Moreover, ETS1 was predominantly expressed in small- and medium-diameter neurons, which are closely associated with sensory neurons in the spinal dorsal horn. Furthermore, microinjection of Ets1 siRNA caused the degradation of Ets1 mRNA and the suppression of ETS1 protein, as a result, the blockade of the increased ETS1 in the injured DRGs attenuated CCI-induced mechanical allodynia and heat hyperalgesia and mimicking the upregulation of DRG ETS1 induced neuropathic-pain-like behavior. However, the motor function of mice was not affected by the relevant manipulations. Importantly, we observed corresponding alterations in the hyperexcitement of spinal neurons and astrocytes in response to ETS1 changes in DRGs. Thus, our data support the hypothesis that DRG ETS1 contributes to the development and maintenance of neuropathic pain.
Histone deacetylase HDAC1 is a regulator of histone acylation modification and plays an important part in nervous-system-related disorders including retinoblastoma, Rett syndrome, and myeloma37–39 via governing the structural modification of chromosomes and gene expression.40 HDAC1 can be widely expressed in nervous tissues including spinal cord,18 DRG,19 hippocampus, and cortex.41 A growing number of studies show that HDAC1 is associated with the processing of different types of pain, including inflammation pain,20 cancer pain,19 and nerve-injury-sourced neuropathic pain;42 thus, it seems to be a critical initiator of pain.18,43,44 Suppression of HDAC1 activity in the dorsal horn or DRGs can alleviate partial nerve ligation18 or cancer-related neuropathic pain behavior.19 Although HDAC1 has been shown to be involved in pain,43 how it is regulated has remained elusive. Here, we found a conserved binding site of ETS1, spanning 799 bp from the −272 to −257 bp region in the HDAC1 promotor region, and further in vitro and in vivo molecular data showed that ETS1 bound to the motif site of HDAC1 promotor and regulated the expression of HDAC1. Behaviorally, ETS1-upregulation-induced nociception behavior was blocked by HDAC1 knockdown with siRNA. Briefly, upregulation of ETS1 leads to the increase of HADC1 expression, while previous studies have shown that the increased HDAC1 leads to the genesis of neuropathic pain,43 and then the knockdown of HDAC1 expression with Hdac1-siRNA alleviated pain sensitivity. Our evidence establishes Ets1 as a controller in HDAC1 transcription under the conditions of neuropathic pain. A previous study showed that TF Sp1 also participates in the regulation of neuropathic pain via targeting the HDAC1/c-JUN complex.43 Therefore, whether there are other TFs that could independently or in combination participate in the regulation of DRG HDAC1 in neuropathic pain should be investigated in the future.
Despite we demonstrated that ETS1 regulates neuropathic pain via targeting HDAC1, there are several limitations under the experiential conditions. Firstly, exception for the HDAC1, what other downstream targets ETS1 have are still identified in future. Secondly, here we used siRNA to knockdown ETS1 expression, given that siRNA may frequently cause the off-target effect, the other strategies such as genetic mice with conditionally knocking out Est1 gene in DRG needs to be carried out to further confirm the current results. Finally, DRGs microinjection may cause cell damage and affect pain behavior in mice, and although it has been verified in previous articles that injected DRGs can still maintain structural integrity and cell number unchanged,23,29 in future work, a method without DRG microinjection is required to be developed to deliver manipulation tools.
In summary, our results demonstrate for the first time that ETS1 contributes to neuropathic pain via controlling HDAC1 in DRG. Blockage of upregulated ETS1 in DRGs relieves nerve-injury-induced neuropathic pain. Our study provides a novel mechanism underlying neuropathic pain. ETS1 may become a promising target for the treatment of neuropathic pain in a periphery-dependent manner.
Footnotes
Author contributions: F-Q L. conceived the project. F-Q L. and Z-Q P. supervised all experiments. H-L Z., S-Y S., T J, M Z. performed the experiments; H-L Z., S-Y S., T J, M Z., Q L, K Y., R W, Y Z, Z-Q P., and F-Q L. analyzed data; H-L Z., and F-Q L. wrote the draft of manuscript. Z-Q P. discussed the results and edited the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The work was supported by grants from Key project of Shanghai Chongming District (CKY2019-1), Jiang Su-Specially Appointed Professor Project, Natural Science Foundation of Jiangsu Province (BK20201460), and Key project of the Natural Science Foundation of Jiangsu Education Department (22KJA320008).
ORCID iD
Fuqing Lin https://orcid.org/0000-0002-6478-0013
References
- 1.Bouhassira D, Lantéri-Minet M, Attal N, Laurent B, Touboul C. Prevalence of chronic pain with neuropathic characteristics in the general population. Pain 2008; 136: 380–387. DOI: 10.1016/j.pain.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 2.Jensen MP, Chodroff MJ, Dworkin RH. The impact of neuropathic pain on health-related quality of life: review and implications. Neurology 2007; 68: 1178–1182. [DOI] [PubMed] [Google Scholar]
- 3.van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: a systematic review of epidemiological studies. Pain 2014; 155: 654–662. DOI: 10.1016/j.pain.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Ye J, Ding H, Ren J, Xia Z. The publication trend of neuropathic pain in the world and China: a 20 years bibliometric analysis. J Headache Pain 2018; 19: 110. DOI: 10.1186/s10194-018-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alsaloum M, Waxman SG. iPSCs and DRGs: stepping stones to new pain therapies. Trends Mol Med 2022; 28: 110–122. DOI: 10.1016/j.molmed.2021.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2010; 2: 657–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang F, Ma SB, Tian ZC, Cui YT, Cong XY, Wu WB, Wang FD, Li ZZ, Han WJ, Wang TZ, Sun ZC, Zhang FL, Xie RG, Wu SX, Luo C. Nociceptor-localized cGMP-dependent protein kinase I is a critical generator for central sensitization and neuropathic pain. Pain 2021; 162: 135–151. DOI: 10.1097/j.pain.0000000000002013. [DOI] [PubMed] [Google Scholar]
- 8.Zhang ZJ, Guo JS, Li SS, Wu XB, Cao DL, Jiang BC, Jing PB, Bai XQ, Li CH, Wu ZH, Lu Y, Gao YJ. TLR8 and its endogenous ligand miR-21 contribute to neuropathic pain in murine DRG. J Exp Med 2018; 215: 3019–3037. DOI: 10.1084/jem.20180800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett-Sinha LA. Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci 2013; 70: 3375–3390. DOI: 10.1007/s00018-012-1243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seidel JJ, Graves BJ. An ERK2 docking site in the pointed domain distinguishes a subset of ETS transcription factors. Genes Dev 2002; 16: 127–137. DOI: 10.1101/gad.950902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackereth CD, Scharpf M, Gentile LN, MacIntosh SE, Slupsky CM, McIntosh LP. Diversity in structure and function of the Ets family PNT domains. J Mol Biol 2004; 342: 1249–1264. DOI: 10.1016/j.jmb.2004.07.094. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, Xue H, Zhu J, Yang L, Zhang F, Qian R, Lin W, Wang Y. ESE1 is associated with neuronal apoptosis in lipopolysaccharide induced neuroinflammation. Neurochem Res 2016; 41: 2752–2762. [DOI] [PubMed] [Google Scholar]
- 13.Yu HL, Wang LZ, Zhang LL, Chen BL, Zhang HJ, Li YP, Xiao GD, Chen YZ. ESE1 expression correlates with neuronal apoptosis in the hippocampus after cerebral ischemia/reperfusion injury. Neural Regen Res 2019; 14: 841–849. DOI: 10.4103/1673-5374.249232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo H, Liu Z, Liu B, Li H, Yang Y, Xu ZD. Virus-mediated overexpression of ETS-1 in the ventral hippocampus counteracts depression-like behaviors in rats. Neurosci Bull 2019; 35: 1035–1044. DOI: 10.1007/s12264-019-00412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H-L, Wang L-Z, Zhang L-L, Chen B-L, Zhang H-J, Li Y-P, Xiao G-D, Chen Y-Z. ESE1 expression correlates with neuronal apoptosis in the hippocampus after cerebral ischemia/reperfusion injury. Neural Regen Res 2019; 14: 841–849. DOI: 10.4103/1673-5374.249232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo H, Liu Z, Liu B, Li H, Yang Y, Xu Z-QD. Virus-mediated overexpression of ETS-1 in the ventral hippocampus counteracts depression-like behaviors in rats. Neurosci Bull 2019; 35: 1035–1044. DOI: 10.1007/s12264-019-00412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanna MD, Guandalini L, Romanelli MN, Galeotti N. The new HDAC1 inhibitor LG325 ameliorates neuropathic pain in a mouse model. Pharmacol Biochem Behav 2017; 160: 70–75. DOI: 10.1016/j.pbb.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Denk F, Huang W, Sidders B, Bithell A, Crow M, Grist J, Sharma S, Ziemek D, Rice ASC, Buckley NJ, McMahon SB. HDAC inhibitors attenuate the development of hypersensitivity in models of neuropathic pain. Pain 2013; 154: 1668–1679. DOI: 10.1016/j.pain.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X-T, Hu X-F, Zhu C, Zhou K-X, Zhao W-J, Zhang C, Han X, Wu C-L, Wei Y-Y, Wang W, Deng J-P, Chen F-M, Gu Z-X, Dong Y-L. Suppression of histone deacetylases by SAHA relieves bone cancer pain in rats via inhibiting activation of glial cells in spinal dorsal horn and dorsal root ganglia. J Neuroinflammation 2020; 17: 125. DOI: 10.1186/s12974-020-01740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Yang Y, Wang Y, Yang F, Li C-L, Wang X-L, Li Z, Chen J. Selective class I histone deacetylase inhibitors suppress persistent spontaneous nociception and thermal hypersensitivity in a rat model of bee venom-induced inflammatory pain. Sheng Li Xue Bao 2015; 67: 447–454. [PubMed] [Google Scholar]
- 21.Xie Y, Li Z, Xu H, Ma J, Li T, Shi C, Jin J. Downregulation of Sp1 inhibits the expression of HDAC1/SOX10 to alleviate neuropathic pain-like behaviors after spinal nerve ligation in mice. ACS Chem Neurosci 2022; 13: 1446–1455. DOI: 10.1021/acschemneuro.2c00091. [DOI] [PubMed] [Google Scholar]
- 22.Bennett G, Al-Rashed S, Hoult SJR, Brain DS. Nerve growth factor induced hyperalgesia in the rat hind paw is dependent on circulating neutrophils. Pain 1998; 77: 315–322. DOI: 10.1016/S0304-3959(98)00114-6. [DOI] [PubMed] [Google Scholar]
- 23.Du S, Wu S, Feng X, Wang B, Xia S, Liang L, Zhang L, Govindarajalu G, Bunk A, Kadakia F, Mao Q, Guo X, Zhao H, Berkman T, Liu T, Li H, Stillman J, Bekker A, Davidson S, Tao Y-X. A nerve injury-specific long noncoding RNA promotes neuropathic pain by increasing Ccl2 expression. J Clin Invest 2022; 132: 132. DOI: 10.1172/JCI153563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao L-Y, Zhang M, Tao Y, Xu H, Liu Q, Yang K, Wei R, Zhou H, Jin T, Liu X-D, Xue Z, Shen W, Cao J-L, Pan Z. miRNA-22 upregulates in dorsal horn neurons and is essential for inflammatory pain. Oxid Med Cell Longev 2022; 2022: 8622388–8622423. DOI: 10.1155/2022/8622388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Zheng B, Du S, Han G, Zhao H, Wu S, Jia S, Bachmann T, Bekker A, Tao Y-X. Eukaryotic initiation factor 4 gamma 2 contributes to neuropathic pain through down-regulation of Kv1.2 and the mu opioid receptor in mouse primary sensory neurones. Br J Anaesth 2021; 126: 706–719. DOI: 10.1016/j.bja.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia S, Wei G, Bono J, Pan Z, Zheng B, Wang B, Adaralegbe A, Tenorio C, Bekker A, Tao Y-X. TET1 overexpression attenuates paclitaxel-induced neuropathic pain through rescuing K1.1 expression in primary sensory neurons of male rats. Life Sci 2022; 297: 120486. DOI: 10.1016/j.lfs.2022.120486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Z, Zhang M, Ma T, Xue Z-Y, Li G-F, Hao L-Y, Zhu L-J, Li Y-Q, Ding H-L, Cao J-L. Hydroxymethylation of microRNA-365-3p regulates nociceptive behaviors via Kcnh2. J Neurosci 2016; 36: 2769–2781. DOI: 10.1523/JNEUROSCI.3474-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y-K, Lu Y-G, Zhao X, Zhang J-B, Zhang F-M, Chen Y, Bi L-B, Gu J-H, Jiang Z-J, Wu X-M, Li Q-Y, Liu Y, Shen J-X, Liu X-J. Cytokine activin C ameliorates chronic neuropathic pain in peripheral nerve injury rodents by modulating the TRPV1 channel. Br J Pharmacol 2020; 177: 5642–5657. DOI: 10.1111/bph.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J-Y, Liang L, Gu X, Li Z, Wu S, Sun L, Atianjoh FE, Feng J, Mo K, Jia S, Lutz BM, Bekker A, Nestler EJ, Tao Y-X. DNA methyltransferase DNMT3a contributes to neuropathic pain by repressing Kcna2 in primary afferent neurons. Nat Commun 2017; 8: 14712. DOI: 10.1038/ncomms14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burek DJ, Massaly N, Yoon HJ, Doering M, Morón JA. Behavioral outcomes of complete Freund adjuvant-induced inflammatory pain in the rodent hind paw: a systematic review and meta-analysis. Pain 2022; 163: 809–819. DOI: 10.1097/j.pain.0000000000002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan Z, Du S, Wang K, Guo X, Mao Q, Feng X, Huang L, Wu S, Hou B, Chang Y-J, Liu T, Chen T, Li H, Bachmann T, Bekker A, Hu H, Tao Y-X. Downregulation of a dorsal root ganglion-specifically enriched long noncoding RNA is required for neuropathic pain by negatively regulating RALY-triggered Ehmt2 expression. Adv Sci (Weinh) 2021; 8: e2004515. DOI: 10.1002/advs.202004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang J-H, Song E-M, Do Y-H, Ahn S, Oh J-Y, Hwang T-Y, Ryu Y, Jeon S, Song M-Y, Park H-J. Acupuncture alleviates chronic pain and comorbid conditions in a mouse model of neuropathic pain: the involvement of DNA methylation in the prefrontal cortex. Pain 2021; 162: 514–530. DOI: 10.1097/j.pain.0000000000002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. The human transcription factors. Cell 2018; 172: 650–665. DOI: 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol 2015; 35: 20–38. DOI: 10.1016/j.semcancer.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Conrad S, Demurger F, Moradkhani K, Pichon O, Le Caignec C, Pascal C, Thomas C, Bayart S, Perlat A, Dubourg C, Jaillard S, Nizon M. 11q24.2q24.3 microdeletion in two families presenting features of Jacobsen syndrome, without intellectual disability: role of FLI1, ETS1, and SENCR long noncoding RNA. Am J Med Genet A 2019; 179: 179–1000. DOI: 10.1002/ajmg.a.61113. [DOI] [PubMed] [Google Scholar]
- 36.Leng R-X, Pan H-F, Chen G-M, Feng C-C, Fan Y-G, Ye D-Q, Li X-P. The dual nature of Ets-1: focus to the pathogenesis of systemic lupus erythematosus. Autoimmun Rev 2011; 10: 439–443. DOI: 10.1016/j.autrev.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, Ramos KS. Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res 2009; 665: 20–28. DOI: 10.1016/j.mrfmmm.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahgoub M, Adachi M, Suzuki K, Liu X, Kavalali ET, Chahrour MH, Monteggia LM. MeCP2 and histone deacetylases 1 and 2 in dorsal striatum collectively suppress repetitive behaviors. Nat Neurosci 2016; 19: 1506–1512. DOI: 10.1038/nn.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng L, Zhang A, Liu J, Liu M, Zhang Y. HDAC1 promotes the migration of human myeloma cells via regulation of the lncRNA/Slug axis. Int J Mol Med 2022; 49: 3. DOI: 10.3892/ijmm.2021.5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Riccio A. New endogenous regulators of class I histone deacetylases. Sci Signal 2010; 3: pe1. DOI: 10.1126/scisignal.3103pe1. [DOI] [PubMed] [Google Scholar]
- 41.Kumar V, Kundu S, Singh A, Singh S. Understanding the role of histone deacetylase and their inhibitors in neurodegenerative disorders: current targets and future perspective. Curr Neuropharmacol 2022; 20: 158–178. DOI: 10.2174/1570159X19666210609160017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borgonetti V, Galeotti N. Combined inhibition of histone deacetylases and BET family proteins as epigenetic therapy for nerve injury-induced neuropathic pain. Pharmacol Res 2021; 165: 105431. DOI: 10.1016/j.phrs.2021.105431. [DOI] [PubMed] [Google Scholar]
- 43.Sanna MD, Galeotti N. The HDAC1/c-JUN complex is essential in the promotion of nerve injury-induced neuropathic pain through JNK signaling. Eur J Pharmacol 2018; 825: 99–106. DOI: 10.1016/j.ejphar.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 44.Sanna MD, Guandalini L, Romanelli MN, Galeotti N. The new HDAC1 inhibitor LG325 ameliorates neuropathic pain in a mouse model. Pharmacol Biochem Behav 2017; 160: 70–75. DOI: 10.1016/j.pbb.2017.08.006. [DOI] [PubMed] [Google Scholar]