Abstract
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is the primary database resource of the Japanese GenomeNet service (http://www.genome.ad.jp/) for understanding higher order functional meanings and utilities of the cell or the organism from its genome information. KEGG consists of the PATHWAY database for the computerized knowledge on molecular interaction networks such as pathways and complexes, the GENES database for the information about genes and proteins generated by genome sequencing projects, and the LIGAND database for the information about chemical compounds and chemical reactions that are relevant to cellular processes. In addition to these three main databases, limited amounts of experimental data for microarray gene expression profiles and yeast two-hybrid systems are stored in the EXPRESSION and BRITE databases, respectively. Furthermore, a new database, named SSDB, is available for exploring the universe of all protein coding genes in the complete genomes and for identifying functional links and ortholog groups. The data objects in the KEGG databases are all represented as graphs and various computational methods are developed to detect graph features that can be related to biological functions. For example, the correlated clusters are graph similarities which can be used to predict a set of genes coding for a pathway or a complex, as summarized in the ortholog group tables, and the cliques in the SSDB graph are used to annotate genes. The KEGG databases are updated daily and made freely available (http://www.genome.ad.jp/kegg/).
INTRODUCTION
The GenomeNet (http://www.genome.ad.jp/) was established in September 1991 under the Human Genome Program of the then Ministry of Education, Science and Culture of Japan as a network of databases and computational services for genome research and related research areas in molecular and cellular biology (1). The GenomeNet is currently operated by the Bioinformatics Center of Kyoto University focusing more on functional genomics and proteomics but still supporting most of the major molecular biology databases. The primary resource of the GenomeNet is the Kyoto Encyclopedia of Genes and Genomes (KEGG) (2). The KEGG project was initiated in May 1995, aimed at understanding the basic principles, as well as practical utilities, of the relations between genomic information and higher order functional information.
While new high-throughput experimental technologies, such as DNA chips, are continuously developed and elaborated to decipher the genome, it is also extremely important to fully make use of the data and knowledge accumulated by traditional experiments in all areas of biomedical sciences. KEGG computerizes such data and knowledge not as text information to be read by humans but as graph information to be manipulated by machines. The KEGG/PATHWAY database contains reference diagrams for molecular pathways and complexes involving various cellular processes, which can readily be integrated with genomic information. A key to this integration is graph representation (3). Mathematically, a graph is a set of nodes and a set of edges. In KEGG the genome is a graph of genes that are one-dimensionally connected and the pathway is a graph of gene products (mostly proteins but including RNAs and complexes) with more complicated patterns of connections. By matching genes in the genome and gene products in the pathway, KEGG can be utilized to predict protein interaction networks and associated cellular functions.
From the perspective of graph representation, the chemical object of protein three-dimensional structure is a graph consisting of atoms (nodes) and atomic interactions (edges). The molecular biological object of protein sequence is a graph of one-dimensionally connected amino acids (nodes). The KEGG data object is a graph consisting of genes or proteins as its nodes and various types of relations or interactions as its edges, as summarized in Table 1. Thus, KEGG is a complementary resource to the existing databases on sequences and three-dimensional structures, focusing on higher level information about interactions and relations of genes or proteins.
Table 1. The KEGG databases.
| Database | Data object (graph) | Node | Edge | Content |
|---|---|---|---|---|
| GENES | Genome | Gene | Adjacency | Gene catalogs of completely sequenced genomes and some partial genomes |
| SSDB | Protein universe | Protein | Sequence similarity | Ortholog/paralog relations of all protein coding genes in complete genomes |
| PATHWAY | Network | Gene product or subnetwork | Generalized protein interaction | Generalized protein interaction networks (pathways and complexes) involving various cellular processes |
| LIGAND | Chemical universe | Compound | Reaction | Chemical compounds and chemical reactions that are relevant to cellular processes |
| EXPRESSION | Transcriptome | Gene | Expression similarity | Microarray gene expression profiles |
| BRITE | Proteome | Protein | Direct interaction | Protein–protein interactions and relations |
GENES DATABASE
Gene annotation
As of 7 September 2001 the GENES database contains 240 943 gene entries derived from the complete genomes of 45 bacteria, 10 archaea and 4 eukaryotes (budding yeast, nematode, fruit fly and thale cress) as well as the genomes of human, mouse and fission yeast. This number is larger than any protein sequence database, SWISS-PROT, PIR or PRF, which contain sequence entries representing over 10 000 organisms, and suggests the difficulty of annotating all known proteins. The complete genome sequence is deposited in the public databases of GenBank, EMBL and DDBJ, with the best annotations of individual genes at the time of publication. However, despite the fact that new gene functions are continuously uncovered because of the availability of complete genome information, the annotations are not updated in the public databases except for a few well-maintained genomes. The KEGG/GENES database is a third-party annotation database attempting to incorporate the most up-to-date information and also to provide standardized annotation across species.
The function of the gene annotation in KEGG is to assign ortholog identifiers (4), which is done manually by the web-based annotation tool for the GENES relational database. The ortholog identifier is associated with standard definition of gene function. The ortholog identifier also represents a node of the protein interaction network (pathway or complex) in the PATHWAY database. In fact, the ortholog identifier was introduced as an extension of the EC number for the metabolic pathway in order to automatically generate organism-specific pathways by matching genes in the genome against gene products in the reference pathway.
Access methods
The GENES database can be accessed by three methods, although there are numerous links leading to this database in the KEGG system. First, the text information describing GENES, which is stored in the accession number, gene names and definition fields, can be searched by the DBGET/LinkDB system (5). The search can be made against all organisms, individual organisms or groups of organisms as shown in the KEGG table of contents page (http://www.genome.ad.jp/kegg/kegg2.html). Secondly, the pathway information that is matched with GENES can be examined by the hierarchical text browser (get_htext program). The link specified by ‘KEGG’ for each organism in the table of contents displays a functionally categorized gene catalog according to reconstructed organism-specific pathways. Thirdly, the positional information of GENES in the chromosome can be examined by the Java-based genome map browser (http://www.genome.ad.jp/kegg/java/launcher.html). Genes are color coded in both the whole view window and the zoom-up window according to the functional categorization.
Whenever available, the original version of the gene catalog is also maintained in KEGG in order to compare with the original authors’ classification of genes. The gene catalog and the genome map are linked to the original database rather than the GENES database in this case.
SSDB DATABASE
Graph of best–best relations
The SSDB database is a new addition to the KEGG suite of databases. SSDB contains the information about amino acid sequence similarities among all protein-coding genes in the complete genomes, which is computationally generated from the GENES database. All possible pairwise genome comparisons are performed by the SSEARCH program (6), and the gene pairs with the Smith–Waterman similarity score of 100 or more are entered in SSDB. As of 7 September 2001 there are 41 745 353 similarity relations derived from 55 × 55 genome comparisons. In addition, SSDB contains the information about best hits and best–best hits (bidirectional best hits). The relationship between gene x in genome A and gene y in genome B is called best–best hits when x is the best hit of query y against all genes in A and vice versa, and it is often used as an operational definition of ortholog (7). SSDB is a huge graph consisting of protein-coding genes as its nodes and similarity relations as its edges. We call this graph the protein gene universe, or simply the protein universe.
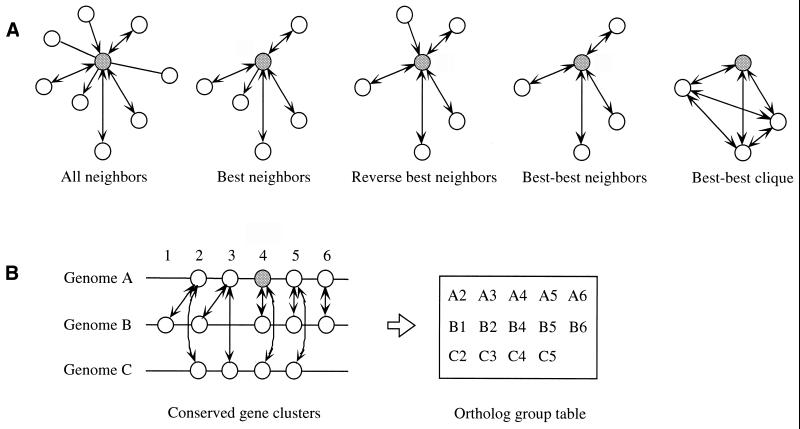
When only the edges of best–best relations are considered, the graph becomes much simpler and can be used effectively to find functional links, especially groups of orthologous genes as partial cliques and possible connections among them (A.Nakaya and M.Kanehisa, manuscript in preparation). In comparison with standard sequence similarity searches by BLAST or FASTA, the search result of SSDB is easier to interpret because of the additional information about best–best hits (Fig. 1A depicts the SSDB graph features).
Figure 1.
(A) Definition of SSDB graph features. Circles represent protein sequences, and the features around a given sequence (shaded) are shown. Arrows indicates the relationship between the query sequence and the best-hit sequence (see text). (B) By incorporating positional coupling of genes in the genome, SSDB can be used to identify conserved gene clusters, which can then be used to create or modify an ortholog group table. Note that the KEGG ortholog group table represents an additional feature, namely functional coupling of gene products in the pathway or complex.
On top of the SSDB graph, additional edges can be included to further identify various functional links. By incorporating the edges that represent adjacent genes on the chromosome, the gene clusters or operons that are conserved among multiple genomes can be identified (Fig. 1B). Other types of edges include common sequence motifs and common folds in the three-dimensional structures. As part of the SSDB database, sequence motifs in PROSITE (8) and Pfam (9) are precomputed for all proteins in the GENES database.
Access methods
The SSDB database is served by a separate server (http://ssdb.genome.ad.jp/). By specifying a gene of an organism, it is possible to search all neighbors of similar sequences above a given threshold, which is equivalent to usual sequence similarity searches, or to search selected neighbors including best–best neighbors, best neighbors and reverse best neighbors, which tends to produce functionally more meaningful results. SSDB can also be used to find conserved gene clusters (operons) as contiguous sets of best–best neighbors.
The information on sequence motifs is not fully integrated with SSDB yet. Separate searches can be made for sequence motifs in a given sequence or a given set of sequences, or for sequences with given motifs. Because motifs are precomputed, the search for all proteins with a given Pfam motif, for example, can be performed instantaneously.
SSDB for improving gene annotations
The SSDB database is utilized in other parts of the KEGG system, such as the genome map comparison that displays a dot matrix of similar genes. SSDB is also critical to gene annotations in KEGG. When a new genome sequence is publicly released, it is incorporated into the KEGG/GENES database and the DBGET/LinkDB system usually within 1 or 2 days. However, in order to start assigning ortholog identifiers, the SSDB computation must be performed, which may take up to 1 week depending on the genome size. Then, the annotation of ortholog identifiers is performed manually using GFIT (7) and other tools. Thus, the reconstructed pathways and the resulting gene catalog can be made publicly available several weeks afterwards. In order to cope with the rapidly increasing number of complete genomes, the detection of SSDB cliques is being implemented to partly automate ortholog identifier assignments, as well as to identify missed annotations.
PATHWAY DATABASE
Generalized protein interaction network
The data object stored in the PATHWAY database is called the generalized protein interaction network (3,10), or simply the network, which is a network of gene products (nodes) with three types of interactions or relations (edges): enzyme–enzyme relations which are two enzymes catalyzing successive reaction steps in the metabolic pathway, direct protein–protein interactions such as binding and phosphorylation, and gene expression relations involving transcription factors and target gene products. The generalized protein interaction network is drawn manually as a graphical pathway diagram (pathway map), and it is also stored as a set of binary relations. The set of binary relations is a computable form of the network information, but at the moment only the enzyme–enzyme relations are maintained (http://www.genome.ad.jp/brite/ECrel/ecrel.xl) where a relation consists of a pair of nodes (enzymes) and an edge (common compound) in between.
As of 7 September 2001 the PATHWAY database contains 5761 entries including 201 reference pathway diagrams and 83 ortholog group tables, as well as 14 960 enzyme–enzyme relations. From the manually drawn reference pathways, many organism-specific pathways are automatically generated according to the ortholog identifier assignments in the GENES database. The total number of gene product nodes that appear on the KEGG pathways is approximately 6000, and roughly one-quarter to one-third of the genes in a bacterial or archaeal genome can be mapped to one or more pathway diagrams. The ortholog group tables contain the information about correlated clusters, which are common subgraphs among multiple graphs (11). In this case a correlated cluster represents a relationship between the positional correlation of genes in the genome and the functional correlation of gene products in the network, such as a set of genes in a conserved gene cluster (operon) forming a subpathway or a complex (Fig. 1B). The total number of genes in the KEGG ortholog group tables is approximately 26 000, which is ∼10% of the total number of genes in the GENES database.
Access methods
The network information of the KEGG/PATHWAY database is hierarchically categorized into four levels. According to our view on the hierarchy and modularity of cellular functions, the top level is categorized into metabolism, genetic information processing, environmental information processing, and the rest named cellular processes. In addition, a new top category of human diseases is being introduced (see Table 2 for the top two levels). The third level corresponds to a pathway diagram and/or an ortholog group table, which is a collection of genes and proteins. The PATHWAY database can best be viewed by following this hierarchy top-down in the KEGG table of contents page (http://www.genome.ad.jp/kegg/kegg2.html) where the top level item of metabolism is designated by ‘Metabolic pathways’ and the rest of the top level items are designated by ‘Regulatory pathways’. Alternatively, the hierarchy may be used bottom-up starting from the KEGG gene catalogs for individual organisms. In addition, the text information describing PATHWAY entries can be searched by the DBGET/LinkDB system.
Table 2. Hierarchical organization of network information in KEGG/PATHWAY.
| Metabolism | Genetic information processing |
| Carbohydrate metabolism | Transcription |
| Energy metabolism | Translation |
| Lipid metabolism | Sorting and degradation |
| Nucleotide metabolism | Replication and repair |
| Amino acid metabolism | Environmental information processing |
| Metabolism of other amino acids | Membrane transport |
| Metabolism of complex carbohydrates | Signal transduction |
| Metabolism of complex lipids | Ligand–receptor interaction |
| Metabolism of cofactors and vitamins | Cellular processes |
| Metabolism of other substances | Cell motility |
| Cell cycle and cell division | |
| Cell death | |
| Development | |
| Human diseases | |
| Neurodegenerative disorder |
OTHER DATABASES
LIGAND
Originally, the LIGAND database (http://www.genome.ad.jp/ligand/) was developed as a value-added database (12) for the enzyme nomenclature of the International Union of Biochemistry and Molecular Biology (IUBMB). This portion is maintained as the ENZYME section of LIGAND, which is linked to and from the KEGG metabolic pathway. Currently, efforts are being made to add more data in the COMPOUND section and the REACTION section of LIGAND. The COMPOUND section contains chemical structures of metabolites and other chemical compounds, including drugs and xenobiotic chemicals, and the REACTION section contains chemical reactions, mostly enzymatic reactions, represented as conversions of chemical structures. As described elsewhere in this issue (13), a web-based chemical structure search is now available for the COMPOUND and REACTION sections, which are stored in the ISIS system.
EXPRESSION
Despite the efforts to establish data repositories for gene expression data, most useful data are dispersed on the World Wide Web, i.e. located at authors’ FTP sites, possibly because of the lack of mandatory data submission requirement as for sequence data. The KEGG/EXPRESSION database (http://www.genome.ad.jp/kegg/expression/) is not a data repository, but it collects gene expression data from many laboratories in Japan as part of our collaborative research projects. Currently, microarray gene expression data for Synechocystis and Bacillus subtilis are publicly made available in this database.
The EXPRESSION database is handled with the Java-based graphical viewers. Each experiment can be examined with the array image viewer and the Scatter plot viewer, and a series of experiments can be examined by the cluster viewer once hierarchical cluster analysis is performed. All these viewers are tightly integrated with the PATHWAY and GENES databases, so that expression patterns and clusters can be mapped to the KEGG pathways or chromosomal positions, in order to make sense of the expression data.
BRITE
Biomolecular Relations in Information Transmission and Expression (BRITE; http://www.genome.ad.jp/brite/) is a database of binary relations for computation and comparison of graphs involving genes and proteins. It is not a fully developed database yet, but its purpose in KEGG is to expand the collection of the generalized protein interactions that underlie the KEGG pathway diagrams, especially direct protein–protein interactions obtained by systematic experiments such as yeast two-hybrid systems, and gene expression relations of transcription factors and transcribed gene products. BRITE will integrate the generalized protein interactions with other diverse sets of binary relations, including sequence similarity relations stored in the SSDB database, expression similarity relations obtained by cluster analysis of the EXPRESSION data, positional correlations in the GENES genome maps and cross-reference links between database entries in the LinkDB database, towards automating logical reasoning steps to understand functions.
OTHER RESOURCES IN GenomeNet
DBGET/LinkDB
DBGET/LinkDB (http://www.genome.ad.jp/dbget/dbget.links.html) is the backbone retrieval system for all GenomeNet databases including a number of molecular biology databases that are mirrored at the GenomeNet. DBGET is based on a flat-file view of molecular biology databases, where the database is considered as a collection of entries. Because cross-reference information is often provided pointing to related entries in other databases, the web of molecular biology databases is a graph consisting of entries (nodes) and cross-references (edges), which is like the World Wide Web consisting of pages (nodes) and hyperlinks (edges). LinkDB is capable of searching this graph and identify entries that are both directly and indirectly related.
Computational tools
GenomeNet provides various computational services (http://www.genome.ad.jp/SIT/), including sequence similarity searches by BLAST and FASTA against all major sequence databases that are updated daily, and sequence motif search by MOTIF, which is an in-house-developed search system, against major motif libraries.
FTP site
All the KEGG data are freely available to academic users by anonymous FTP (http://www.genome.ad.jp/anonftp/).
Acknowledgments
ACKNOWLEDGEMENTS
The computational resource was provided by the Bioinformatics Center, Institute for Chemical Research, Kyoto University. This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Japan Society for the Promotion of Science, and the Japan Science and Technology Corporation.
REFERENCES
- 1.Kanehisa M. (1997) Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem. Sci., 22, 442–444. [DOI] [PubMed] [Google Scholar]
- 2.Kanehisa M. (1997) A database for post-genome analysis. Trends Genet., 13, 375–376. [DOI] [PubMed] [Google Scholar]
- 3.Kanehisa M. (2000) Post-genome Informatics. Oxford University Press, Oxford, UK.
- 4.Kanehisa M. and Goto,S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujibuchi W., Goto,S., Migimatsu,H., Uchiyama,I., Ogiwara,A., Akiyama,Y. and Kanehisa,M. (1998) DBGET/LinkDB: an integrated database retrieval system. Pac. Symp. Biocomput., 683–694. [PubMed] [Google Scholar]
- 6.Pearson W.R. (1996) Effective protein sequence comparison. Methods Enzymol., 266, 227–258. [DOI] [PubMed] [Google Scholar]
- 7.Bono H., Ogata,H., Goto,S. and Kanehisa,M. (1998) Genome Res., 8, 203–210. [DOI] [PubMed] [Google Scholar]
- 8.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. Updated article in this issue: Nucleic Acids Res. (2002), 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. Updated article in this issue: Nucleic Acids Res. (2002), 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanehisa M. (2000) Pathway databases and higher order function. Adv. Protein Chem., 54, 381–408. [DOI] [PubMed] [Google Scholar]
- 11.Ogata H., Fujibuchi,W., Goto,S. and Kanehisa,M. (2000) A heuristic graph comparison algorithm and its application to detect functionally related enzyme clusters. Nucleic Acids Res., 28, 4021–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suyama M., Ogiwara,A., Nishioka,T. and Oda,J. (1993) Searching for amino acid sequence motifs among enzymes: the Enzyme–Reaction Database. Comput. Appl. Biosci., 9, 9–15. [DOI] [PubMed] [Google Scholar]
- 13.Goto S., Okuno,Y., Hattori,M., Nishioka,T. and Kanehisa,M. (2001) LIGAND: database of chemical compounds and reactions in biological pathways. Nucleic Acids Res., 30, 402–404. [DOI] [PMC free article] [PubMed] [Google Scholar]