Abstract
PlantCARE is a database of plant cis-acting regulatory elements, enhancers and repressors. Regulatory elements are represented by positional matrices, consensus sequences and individual sites on particular promoter sequences. Links to the EMBL, TRANSFAC and MEDLINE databases are provided when available. Data about the transcription sites are extracted mainly from the literature, supplemented with an increasing number of in silico predicted data. Apart from a general description for specific transcription factor sites, levels of confidence for the experimental evidence, functional information and the position on the promoter are given as well. New features have been implemented to search for plant cis-acting regulatory elements in a query sequence. Furthermore, links are now provided to a new clustering and motif search method to investigate clusters of co-expressed genes. New regulatory elements can be sent automatically and will be added to the database after curation. The PlantCARE relational database is available via the World Wide Web at http://sphinx.rug.ac.be:8080/PlantCARE/.
INTRODUCTION
The complete genome of the dicotyledonous model plant Arabidopsis thaliana being available since the end of 2000 is the first rough blueprint of a plant. The initial step in unraveling this genome was finding the genes and their structure, leading to an estimated number of more than 25 500 genes (1). The next step is now the study of the function of individual genes and their interaction with other genes. Expression of a gene is an essential part of its function and its expression profile the key element towards achieving a full functional description of each gene.
Large-scale transcriptome expression analyses, such as microarrays, produce sets of co-expressed genes. The working hypothesis is then to assume that among the co-expressed genes some genes will also be co-regulated. By looking for over-represented oligonucleotide sequences, regulatory elements can be found, which are shared by some of the promoter sequences of genes from a given gene cluster (2). The knowledge on plant promoters is of major interest in biotechnology and will offer the possibility to control gene expression in many areas. Here, we describe the present status of the PlantCARE database (3), its content and analysis tools that are currently available.
DATABASE STATUS AND AVAILABILITY
At present, we have collected 417 cis-acting regulatory elements, of which 150 are from monocotyledonous species, 263 from dicotyledonous species and four from conifers, describing approximately 160 individual promoters from higher plant genes. The database can be queried on names of transcription factor (TF) sites, motif sequence, function, species, cell type, gene, TF and literature references. These queries result in a listing of entries with links to other information within the database or beyond through accession numbers from other databases, such as EMBL, GenBank, TRANSFAC (4) and MEDLINE. The World Wide Web interface has been improved in several ways to facilitate usage and querying by the user.
NEW IMPLEMENTATIONS
New programs designed to identify new regulatory elements in silico from transcriptome data are now made available through the PlantCARE web site. These are a new quality-based clustering method (5) and a motif search algorithm called Motif Sampler (6), and a probabilistic approach based on Gibbs Sampling (7,8) which looks for over-represented motifs in upstream regions. We also provide the possibility to send new data to our database. In this way we would like to encourage direct online submission of data concerning plant promoters, which would enable a faster growth of the data put at the disposal of the community. However, for different reasons, we have implemented a submission form that does not append the data directly to the database. The data will only be made available for searches on promoter sequences after curation.
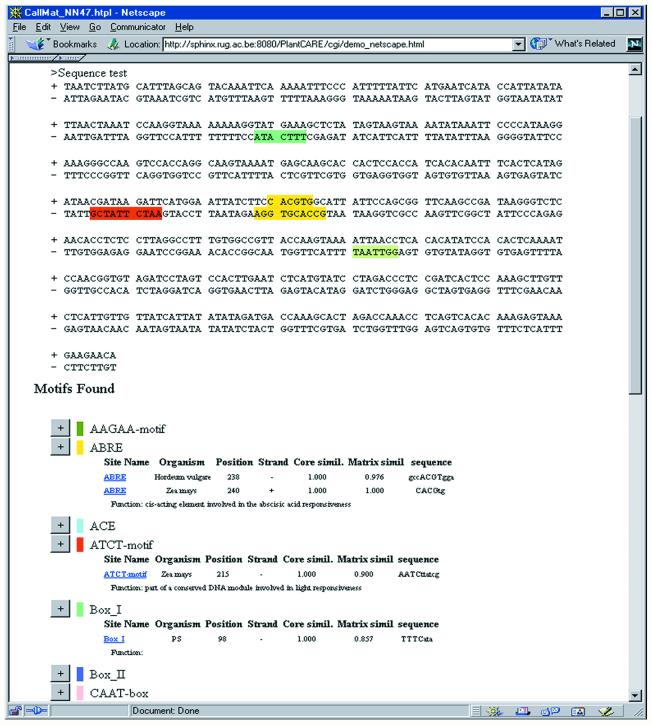
For each site of a particular species, a positional matrix has been generated that can be used with the MatInspector program (9). The data from PlantCARE is accessible through queries. Upon submission of a promoter sequence by a user, the new ‘Search for CARE’ implementation presents a dynamic HTML page with the PlantCARE TF sites highlighted on the sequence. The resulting report also shows a filtered list of sites found (see Fig. 1 and complementary demo at http://sphinx.rug.ac.be:8080/PlantCARE/cgi/demo.html). The filtering is basically intended to remove redundancy but will be extended to look for combinations of sites, as this will lower the high amount of hits obtained when looking for single TF sites. Information regarding TF site, organism, motif position, strand, core similarity, matrix similarity, motif sequence and function are listed whereas the potential sites are mapped on the query sequence. Links allow the characteristics of each site to be displayed and point to sequences in which the TF site was described. The database has been adapted to allow the storage of combinations of TF sites.
Figure 1.
Output in dynamic HTML of ‘Search for CARE’. See also the demo at http://sphinx.rug.ac.be:8080/PlantCARE/cgi/demo.html.
FUTURE PROSPECTS
The PlantCARE database is updated on a regular basis. Considering the large number of biological articles of interest, we are investigating a way to automate this task. We also aim at storing into the database the information that has been retrieved from our in silico motif predictions by using microarray data and will try to develop a strategy through collaborations to check in vitro the potential functionality in order to validate motifs.
CITATION OF THE PlantCARE DATABASE
Users are asked to cite this article when publishing results that have been obtained using the PlantCARE database.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by a grant from IWT: project STWW-980396. P.R. is Research director of INRA (Institut National de la Recherche Agronomique, France). Y.M. and Y.V.P. are post-doctoral researchers of the FWO.
REFERENCES
- 1.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana.Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 2.Zhang M.Q. (1999) Promoter analysis of co-regulated genes in the yeast genome. Comput Chem., 23, 233–250. [DOI] [PubMed] [Google Scholar]
- 3.Rombauts S., Déhais,P., Van Montagu,M. and Rouzé,P. (1999) PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res., 27, 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer.F. (2000) TRANSFAC: an integrated system for gene expression regulation Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thijs G., Moreau,Y., De Smet,F., Mathys,J., Lescot,M., Rombaults,S., Rouze,P., De Moor,B and Marchal,K. (2001) INCLUSive: INtegrated CLustering, Upstream sequence retrieval and motif Sampling. Bioinformatics, in press [DOI] [PubMed] [Google Scholar]
- 6.Thijs G., Marchal,K., Lescot,M., Rombauts,S., De Moor,B., Rouzé,P. and Moreau,Y. (2001) A Gibbs Sampling method to detect over-represented motifs in upstream regions of co-expressed genes. Proceedings of the Fifth Annual International Conference on Computational Molecular Biology (RECOMB), ACM Press, New York, Montréal, Canada, pp. 296–302.
- 7.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F., and Wootton,J.C. (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 8.Neuwald A.F., Liu,J.S. and Lawrence,C.E. (1995) Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci., 4, 1618–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]