Abstract
Clear cell renal carcinoma (ccRCC) is histologically defined by its cytoplasmic lipid deposits. Lipid metabolism disorder largely increases the risk of ccRCC. In this study, we aimed to investigate the biological functions and molecular mechanisms of carnitine palmitoyl transferase 1A (CPT1A) in ccRCC. Our results showed that CPT1A is decreased in ccRCC clinical samples and cell lines compared with that in normal samples. Lentivirus overexpressing CPT1A was used to investigate the neoplastic phenotypes of ccRCC, and the results showed that lipid accumulation and tumor growth are attenuated both in vitro and in vivo. In addition, CPT1A prevents cholesterol uptake and lipid accumulation by increasing the peroxisome proliferator-activated receptor α (PPARα) level through regulation of Class B scavenger receptor type 1 (SRB1) and cluster of differentiation 36 (CD36). Furthermore, PI3K/Akt signaling pathway promotes tumor cell proliferation in ccRCC, which is related to the enhanced expression of CD36. Functionally, weakened CPT1A expression is critical for lipid accumulation to promote ccRCC development. Collectively, our research unveiled a novel function of CPT1A in lipid metabolism via PPARα/CD36 axis, which provides a new theoretical explanation for the pathogenesis of ccRCC. Targeting CPT1A may be a potential therapeutic strategy to treat ccRCC.
Keywords: CD36, cholesterol, CPT1A, growth, PPARα, renal cancer
Introduction
According to GLOBOCAN worldwide cancer statistics, approximately 338,000 new kidney cancer cases were diagnosed in 2012, while 143,000 patients succumbed to the disease [1]. Clear cell renal carcinoma (ccRCC) is the most frequent histological subtype, accounting for >75% of all RCC cases [2]. Over the past years, great progress has been made in identifying the genetic alterations driving ccRCC development [ 3, 4]; however, the prognosis of these patients is still poor [5].
In a recent study, a strong association was proposed between obesity and cancers [6], implying that dyslipidemia is a major factor for tumorigenesis. Solid tumor cells require extracellular fatty acid as a nutrient source, especially under metabolic stress conditions [7]. ccRCC cells are characterized by the accumulation of cholesterol, cholesterol esters, and other lipids [ 8, 9], and a decrease in the expression of specific fatty acid oxidation (FAO) enzyme has also been found to be correlated with an increase in tumor stage, size, and grade, with a concomitant decrease in the survival [ 10, 11]. However, the underlying key fat enzymes that affect the tumor cells are yet unclear in ccRCC.
Carnitine palmitoyl acyltransferase 1A (CPT1A) is located on the outer mitochondrial membrane and is a rate-limiting fatty acid oxidation (FAO) enzyme transporting fatty acid into mitochondria for further oxidation by converting acyl-CoA into acyl-carnitines [ 12, 13]. In addition to its function as an FAO enzyme in cancer cells, CPT1A is engaged in various cellular processes owing to the identification of interacting proteins that are associated with tumor development by regulating lipid metabolism through FAO in nasopharyngeal carcinoma [14], breast cancer [15], and lymphocytic leukemia [16]. The expression and activity of CPT1A are decreased in the kidney of ccRCC patients compared with that in normal kidney, and the poor outcome is associated with low expression of CPT1A in tumors in The Cancer Genome Atlas (TCGA) [17]. At present, the mechanisms underlying the dysregulation of lipid homeostasis in ccRCC are not precise, especially with respect to CPT1A loss.
Peroxisome proliferator-activated receptor α (PPARα) is a member of the nuclear receptor superfamily of transcription factors that control nutrient sensing and transcriptional regulation of metabolic pathways, especially fatty acid transport and FAO [18]. The downregulation of PPARα significantly decreases the expression of CPT1A and subsequently the rate of FAO in melanoma cells [19]. Activated PPAR-γ coactivator-1α (PPARGC1A, PGC1α) is translocated into the nucleus, which promotes the expression of PPARα in lipid metabolism [20], and the membrane fatty acid transporter cluster of differentiation 36 (CD36) regulates downstream PGC1α involved in lipid and energy metabolism [21]. The expression of CD36 also activates Nrf2 [22], which has been described as the downstream pathway for PI3K/Akt activation [23]. In hepatocellular carcinoma (HCC), cartilage oligomeric matrix protein (COMP)/CD36 signaling causes phosphorylation of Akt, resulting in the upregulation of tumor-progressive genes [24]. Targeting these pathways has been proven to be an effective strategy for inhibiting the growth in ccRCC because the PI3K/Akt pathway is highly activated in ccRCC, and targeting this pathway, either alone or with other drugs, has great potential for ccRCC treatment [25].
Cancer cells have developed mechanisms for the accumulation of lipid, including excessive intracellular cholesterol, to support cell growth. However, whether the accumulation of lipids is a byproduct of altered metabolism in ccRCC or whether it contributes to disease development is yet to be determined. Herein, we found that overexpression of CPT1A reverses the lipid deposition phenotype and biological phenotype in vivo and in vitro functionally, and demonstrated that CPT1A is a regulator gene of lipid accumulation via PPARα/CD36 axis, which controls the phosphorylation level of Akt in ccRCC development. Together, these findings provide evidence of altered lipid metabolism in ccRCC development, and show that CPT1A is a potential therapeutic target for ccRCC treatment.
Materials and Methods
Bioinformatics analysis
We downloaded the RNA-seq data of ccRCC dataset from TCGA database and analyzed the gene expression of CPT1A through GEPIA ( http://gepia.cancer-pku.cn). The box plot illustrated the association between the expression and clinical features. The relationship between CPT1A expression and patient survival was analyzed by accessing the TCGA database through UALCAN website. Search Tool for the Retrieval of Interacting Genes (STRING; http://string.embl.de) database was used to evaluate the protein-protein interaction (PPI) information. The relationships among the PPARα and CPT1A, PGC1α were analyzed by the Multiple Gene Analysis through Gene expression profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn). Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) functional analysis are common useful methods for annotating gene functions and identifying biological attributes for genome or transcriptome data. The GO and KEGG pathway analysis were performed by KOBAS 3.0 ( http://kobas.cbi.pku.edu.cn) web tool to explore the biological pathways of CPT1A enrichment in ccRCC.
Patients and samples
ccRCC and the corresponding adjacent non-tumorous renal tissue specimens were collected from 20 cases of ccRCC patients who underwent operation at Department of Organ Transplantation, the First Affiliated Hospital of Kunming Medical University from July 2016 to March 2018. Tumors were pathologically confirmed in all patients, and none of them received any adjuvant treatment, such as radiotherapy, chemotherapy, or immunotherapy prior to surgery. After collection, all specimens were snap-frozen in liquid nitrogen and stored at –80°C for protein extraction. Histological and pathological diagnoses of all specimens were confirmed by the Department of Pathology, the First Affiliated Hospital of Kunming Medical University. Written informed consent of all patients involved in the study was obtained before operation and the protocol of the present study was approved by the Ethics Committee of Kunming Medical University (No. KMMU2021MEC019).
Cell culture and reagents
The human ccRCC cell lines, ACHN, Caki-1, and 786-O, and normal kidney epithelial HK2 cell line were obtained from Kunming Institute of Zoology, Chinese Academy of Sciences (Kunming, China). 786O cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (HyClone, Logan, USA); ACHN, HK-2, and Caki-1 were cultured in Dulbecco’s modified Eagle medium (DMEM)/High Glucose (HyClone). Both media were supplemented with 10% fetal bovine serum (FBS; HyClone). All cells were cultured at 37°C in a 5% CO 2 incubator. Etomoxir was obtained from Sigma-Aldrich (Munich, Germany), WY14643 was from Selleck (Houston, USA), and sulfosuccinimidyl oleate (SSO) was purchased from APExBio (Houston, USA).
Lentivirus transfection
Lentivirus overexpressing CPT1A and the corresponding control vector were purchased from GeneChem (Shanghai, China). Lentivirus overexpressing CPT1A was transfected into Caki-1 and 786O cells, according to the manufacturer’s instructions. The experiments were conducted for 48 h, and the overexpression efficiency was verified by western blot analysis. Puromycin (5 μg/mL; Sigma-Aldrich) was used to generate cell lines stably expressing CPT1A.
Immunohistochemical (IHC) staining
Tissue sections (5 μm) were dewaxed and rehydrated before antigen retrieval. After incubation with 3% H 2O 2 at room temperature for 15 min, FBS was used to block the sections. Then, the slides were incubated with primary antibodies overnight at 4°C, followed by incubation with HRP-conjugated secondary antibody for 1 h at room temperature. 3,3′-diaminobenzidine tetrahydrochloride (DAB) was used to visualize the immune complexes, and the sections were counterstained with hematoxylin. The primary polyclonal rabbit antibodies against CPT1A (1:200; Abcam, Cambridge, UK) and CD36 (1:100; Abcam) were used. Images were acquired using a microscope (Nikon, Tokyo, Japan). Percentage of positive cell scores was divided into 0 (0%–5%), 1 (6%–35%), 2 (36%–70%), and 3 (more than 70%). The intensity of protein expression was determined as 0 (no staining), 1 (weakly staining), 2 (moderately staining) and 3 (strongly staining). The final score was calculated using the percentage score × staining intensity score as follows: 0 (−), 2–3 (+), 4–6 (++), and > 6 (+++). Final score < 4 was defined as high expression. The scores were determined by two independent pathologists in a blinded manner.
Oil red O (ORO) staining
ORO staining was conducted to compare lipid accumulation in tumor and paracancerous tissues in ccRCC according to the manufacturer’s instructions. Cells plated in 6-well plates at triplicate were rinsed with PBS twice, and fixed with 10% formaldehyde for 1 h. They were then rinsed with 60% isopropanol for 5 min, stained with 3 mg/mL ORO (Solarbio, Beijing, China) for 4 min, and washed with water three times. After freshly frozen, OCT-embedded tumor sections were stained with a similar protocol after sectioning into 8-μm slices on a cryostat. Briefly, fresh and frozen tissue sections were placed in propylene glycol for 2 minand then incubated in ORO solution for 6 min. A mixture of 85% propylene glycol was prepared in distilled water, the tissue sections were incubated in this mixture for 1 min, and the slides were rinsed in distilled water and examined under an inverted phase contrast microscope (Nikon). For ORO quantification, the cells were dried, and 250 μL of isopropanol was added and incubated for 3 min; the OD value of the eluted solution was measured at 510 nm on a SpectraMax Plus 384 microplate reader (Molecular Devices, San Jose, USA).
Real-time polymerase chain reaction (RT-PCR)
Total RNA was isolated using Trizol reagent (TaKaRa, Dalian, China). RT-PCR was performed using standard procedures. Briefly, RNA samples were reverse transcribed into cDNA using Revertaid TM First Strand cDNA Synthesis kit (Fermentas, Vilnius, Lithuania), and quantitative PCR was performed using SYBR Green qPCR super-mix UDG (Invitrogen, Carlsbad, USA) on a quantitative PCR machine (Bio-Rad, Hercules, USA). The conditions were as follows: initial denaturation at 95°C for 30 s, 40 cycles at 95°C for 30 s and 60°C (annealing temperature) for 30 s and extended at 65°C for 20 s. The expression level was quantified based on the 2 -∆∆Ct method and normalized to that of β-actin. The sequences of primers used were as follows: CPT1A, forward 5′-TTCAGTTCACGGTCACTCCG-3′, reverse 5′-TGACCACGTTCTTCGTCTGG-3′; and β-actin, forward 5′-GGCTGTGCTATCCCTGTACG -3′, reverse 5′-TTGATCTTCATTGTGCTGGGTG-3′.
Western blot analysis
Western blot analysis was performed using standard procedures. Tissues and cells were lysed in Mammalian Protein Extraction Reagent (Thermo Scientific, Waltham, USA) containing a protease inhibitor cocktail (Roche Diagnostic, Indianapolis, USA). The proteins were separated by 12% polyacrylamide/bisacrylamide gel electrophoresis and transferred to PVDF membranes (Millipore, Billerica, USA). The proteins were probed with primary antibodies followed by a HRP-conjugated secondary antibody. Immunodetection was carried out with the Immobilon Western Chemiluminescent HRP substrate (Millipore). Normalization was conducted by blotting the same samples with an antibody against β-actin. Densitometry of the protein bands was quantified with Quantity One software (Bio-Rad), and the values were expressed relative to β-actin (control for loading and transfer). At least three independent experiments were performed for each cell type studied. Primary antibodies against the following proteins were used: CPT1A, pPI3K, Akt, pAkt, SRB1, CD36, β-actin, and PPARα (1:5000; Abcam); Bcl-2, Bax, and Caspase 3 (1:3000; Affbiotech, Cincinnati, USA); and PGC1α (1:1000; Cell Signaling Technology, Beverly, USA).
MTS assay
An equivalent of 5×10 3 cells/well (200 μL/well) was cultured in a 96-well microtiter plate. At each time point, 20 μL of sterile MTS dye (5 mg/mL; Promega, Beijing, China) was added to each well and incubated for another 4-5 h at 37°C. The absorbance was measured at 490 nm on a microplate reader.
Total cholesterol (TC) and total glyceride (TG) quantification tests
TC content determination kit (Solarbio) was utilized to assay the total triglyceride content of the cell samples with or without CPT1A overexpression, according to the manufacturer’s protocol. Briefly, 3×10 5 cells were collected and reacted with the reagents provided. The OD was measured spectrophotometrically at a wavelength of 420 nm on a microplate reader.
TG content determination kit (Solarbio) was used and TG content was detected with a similar protocol.
Colony formation assay
About 500 cells/well were cultured in a 6-well plate for 21 days. The medium was refreshed every 3 days. The culture was terminated after colonies with >50 cells were formed. Subsequently, the cells were fixed in 4% paraformaldehyde, stained with crystal violet (Yeasen, Shanghai, China), and images were acquired under an inverted phase contrast microscope (Nikon), and the number of colonies were counted with ten randomly chosen fields.
Enzyme-linked immunosorbent assay
Mitochondria were isolated from normal and tumor tissue after homogenization in an isotonic cold sucrose solution (30% sucrose; 1× PBS) and centrifugation at 600 g for 15 min at 4°C. The supernatant was subjected to centrifugation again at 12,000 g for 20 min. The resulting pellet was resuspended in 2 mL homogenization buffer and washed by centrifugation at 7000 gfor 10 min, and finally suspended in 0.5 mL homogenization buffer. The concentration of mitochondria was quantified by BCA assay. An equivalent of 10 μg total protein was used for the CPT1A activity assay using a detection kit (Jianglai Biotechnology, Shanghai China).
Immunofluorescence staining
Cells were fixed with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.3% Triton X-100. After being blocked with donkey serum for 1 h at room temperature, the cells were incubated with primary antibody at 4°C overnight and then incubated with fluorescence-conjugated secondary antibodies (1:1000; Invitrogen) at room temperature in the dark for 1 h. Subsequently, the cells were washed three times with PBS and mounted with Prolong Gold anti-fade reagent containing DAPI (Invitrogen). The cell images were captured under a fluorescence microscope (Olympus, Tokyo, Japan).
Cholesterol uptake assay
The cholesterol uptake assay was performed using a Cholesterol Uptake Assay kit (ab236212; Abcam) according to the manufacturer’s instructions. Briefly, cells or vehicle control were treated with 100 μL serum-free culture medium containing 20 μg/mL NBD cholesterol overnight. Then, the cells were collected by centrifugation at 250 g for 5 min, and 100–500 μL Assay Buffer was added before analyzing by flow cytometry using an FL1 (FITC) channel (FloMax 2.82; Partec: CyFlow® Space, Goerlitz, Germany).
Cholesterol depletion analysis
Cholesterol depletion was verified using a Cell-based Cholesterol Assay kit (ab133116; Abcam), according to the manufacturer’s instructions. A total of 3×10 4 cells were seeded in each well of a 96-well plate. The next day, cells were treated with experimental compounds or vehicle control for 48–72 h. U18666A, a cholesterol transport inhibitor, was used as a positive control (provided at a concentration of 2.5 mM). U18666A was used with a serial of dilutions starting at 1.25 μM. Cells were grown overnight. Most of the culture medium was removed from the wells and the cells were fixed with Cell-Based Assay Fixative Solution for 10 min. The cells were washed with Cholesterol Detection Wash Buffer three times, with 5 min each. The Filipin III Stock Solution (prepared as described in Pre-Assay Preparation) was diluted at 1:100 using Cholesterol Detection Assay Buffer. Filipin III Solution (100 μL) was added to each well and incubated in the dark for 30–60 min. Then the cells were washed with wash buffer two times, with 5 min each. Finally, the staining was examined using a fluorescent microscope (Olympus) with an excitation wavelength of 340–380 nm and emission wavelength of 385–470 nm.
Tumor xenograft study in mice
Four-week-old female BALB/c-nude mice were obtained from the Experimental Animal Center of Kunming Medical University (KMMU), and housed in an environment at a temperature of 22±1°C, relative humidity of 50%±1%, and a 12/12 hlight/dark cycle. All animal experiments (including mice euthanasia) were carried out in compliance with the regulations and guidelines of the institutional animal center of KMMU and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) and the Institutional Animal Care and Use Committee (IACUC) guidelines (No. KMMU2021149).
Caki-1 cells (2×10 6) with or without CPT1A overexpression were subcutaneously injected into the armpit. At 14 days post-transplantation, tumor volume was measured every 3 days. Mice were sacrificed 5 weeks after cells were transplanted. All 12 mice were euthanized with pentobarbital sodium by intraperitoneal injection at a dose of 150 mg/kg, the euthanasia was confirmed by cervical dislocation, and the xenograft tumors were dissected for weighing and photographing. Fresh tumor tissues were stored at –80°C for sectioning and staining.
Statistical analysis
All assays were performed at least three times, and data are expressed as the mean ± standard deviation (SD). Student’s t-test was performed to determine significance. P<0.05 were considered statistically significant. All statistical analyses were carried out using GraphPad Prism 6.0 (La Jolla, USA).
Results
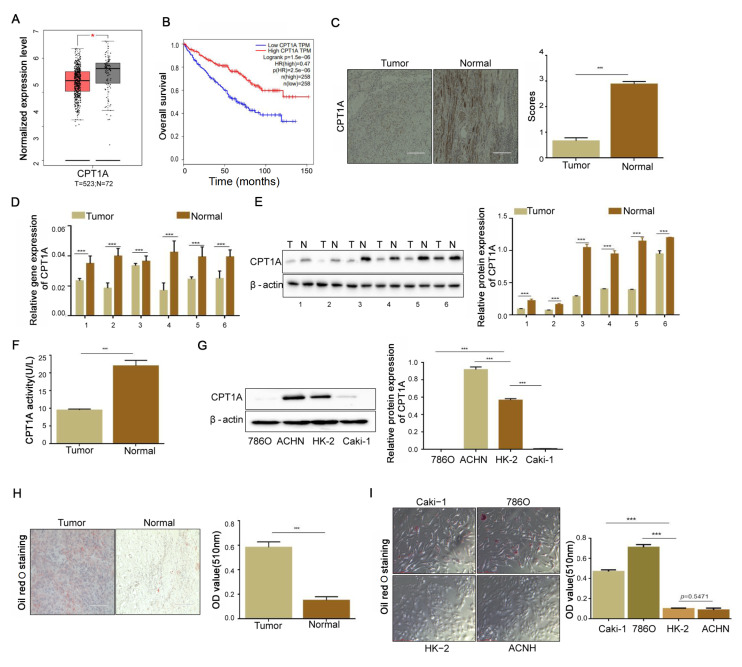
Deletion of CPT1A in ccRCC cells facilitates lipid accumulation
RNA-seq data of patients with ccRCC were downloaded from the TCGA database. Low expression of CPT1A was closely related to the survival and prognosis of patients ( Figure 1A,B). We initially stained the ccRCC tissue by IHC, and the results showed that the percentage of CPT1A-positive cells, which appear as brown-yellow or tan-brown ( Figure 1C), was obviously decreased comparing with that in adjacent normal tissues ( P<0.001). The downregulation of CPT1A in ccRCC clinical samples was further verified by RT-PCR and immunoblotting ( Figure 1D,E). ELISA assay showed a low CPT1A activity in ccRCC ( P<0.001; Figure 1F). We also compared the basal level of CPT1A in different renal cell lines, including normal kidney epithelial cell lines (HK-2), renal adenocarcinoma (ACHN), and ccRCC cell lines (Caki-1 and 786O). The results showed that the expression of CPT1A was lower in ccRCC cell lines than that in HK-2 ( P<0.001) but higher in renal adenocarcinoma cell line ACHN ( P<0.001; Figure 1G). Since the function of CPT1A is to transport long-chain fatty acids into the mitochondrion, we speculated that its downregulation might reduce the lipid droplets in ccRCC cells. Next, we noticed more lipid deposition in ccRCC tissues than in adjacent normal tissues ( P<0.001; Figure 1H), which was further confirmed in cell lines ( Figure 1I). These results demonstrated that CPT1A was downregulated in ccRCC, which might be linked to abnormal lipid accumulation in ccRCC.
Figure1 .
Deficiency of CPT1A increases the lipid formation and cell proliferation in ccRCC(A) Analysis of CPT1A mRNA expression between ccRCC and adjacent tissues through the GEPIA website. (B) The TCGA database accessed through the UALCAN website was used to analyze the relationship between CPT1A expression and the patient survival rate. (C) The expression of CPT1A protein in ccRCC and adjacent tissues revealed by IHC. Scale bar = 50 μm. The percentage of tissue cores displaying low or high CPT1A staining in ccRCC tissues. (D) The RT-qPCR results obtained from 12 cases of ccRCC tissues and adjacent normal tissues showed that CPT1A mRNA expression is downregulated in ccRCC tissues. (E) Western blot analysis revealed a downregulation of CPT1A protein expression in ccRCC tissues. (F) The activity of CPT1A in ccRCC tissues comparing with that in normal tissues detected by ELISA kit. (G) Western blot analysis of CPT1A protein expression in one normal kidney epithelial cell HK-2 and three other RCC cells Caki-1, 786O and ACHN. (H) Lipid deposition in tumor and adjacent ccRCC tissues by Oil red O staining. Scale bar = 50 μm. (I) Lipid deposition in one normal kidney epithelial cell and three RCC cells identified by Oil red O staining. Scale bar = 50 μm. Values are shown as the mean±SD. ***P<0.001 (Student’s t-test).
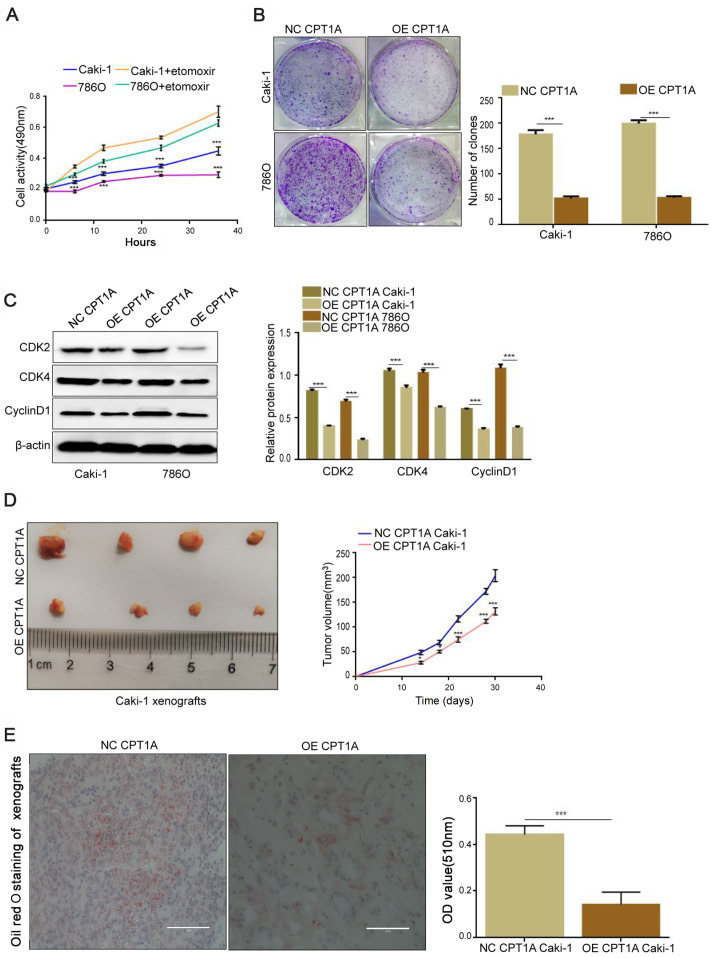
CPT1A inhibits ccRCC growth via inhibiting cholesterol uptake and lipid accumulation
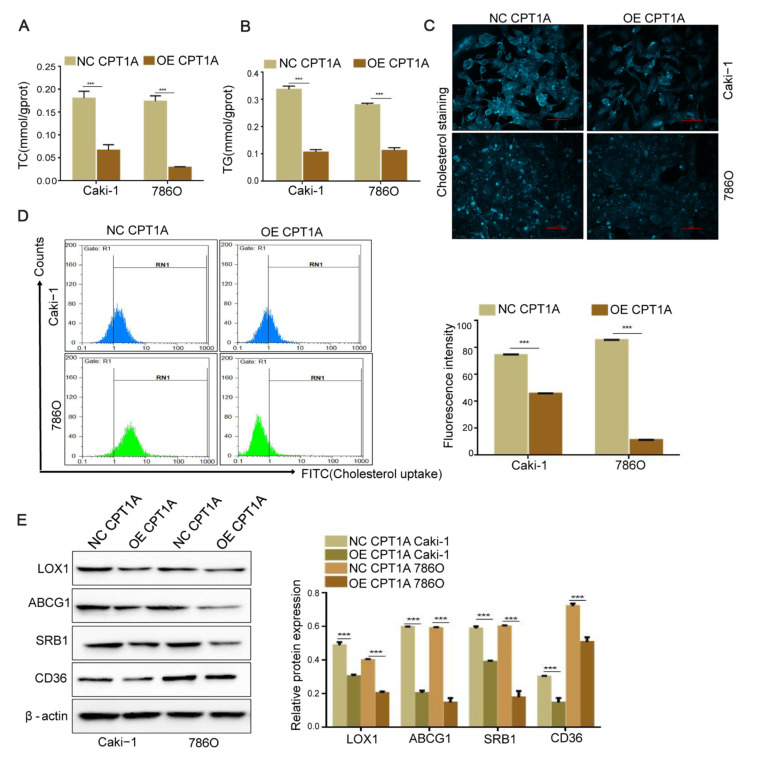
We examined whether these malignant cells could utilize lipid as an alternative energy source due to the low expression of fatty acid transporter CPT1A in ccRCC. The cells were incubated with etomoxir (80 μM), a CPT1A enzymatic inhibitor, for 12 h. The results showed an increasing cell proliferation when CPT1A was inhibited ( Figure 2A). In the colony formation assay, the number of colonies was reduced in CPT1A overexpressing cells compared with that in the corresponding control group ( P<0.001; Figure 2B). Furthermore, immunoblotting was used to detect the expressions of cell proliferation-related proteins, such as Cyclin-dependent kinase 2 (CDK2), CDK4, and Cyclin protein D1 (CyclinD1). The results showed that the expressions of CDK2, CDK4, and CyclinD1 were downregulated ( P<0.001), indicating that cell growth was impeded when CPT1A was overexpressed ( Figure 2C). We also performed a xenograft assay using CPT1A overexpression Caki-1 cells and its normal control cells. Mice that were injected with CPT1A-overexpressing cells showed smaller tumor volume than its normal control group ( Figure 2D). The above results indicated that CPT1A overexpression decreases the proliferation of ccRCC in vitroand in vivo. In addition, the frozen sections of tumor tissues from nude mice were stained with ORO, and a decreased lipid accumulation was observed when CPT1A was overexpressed in vivo( P<0.001; Figure 2E). Meanwhile, the contents of TC and TG were decreased in the CPT1A-overexpressing cells ( P<0.001; Figure 3A,B), and intracellular cholesterol accumulation was weakened when CPT1A was overexpressed ( Figure 3C). Flow cytometry assay revealed that CPT1A overexpression could arrest the cholesterol uptake in ccRCC cells ( P<0.001; Figure 3D), leading to less intracellular cholesterol accumulation. The levels of cholesterol uptake-related proteins determined by western blot analysis indicated that CD36, ATP-binding cassette transporters G1 (ABCG1), SRB1, and lectin-like oxLDL receptor 1 (LOX1) were markedly decreased ( P<0.001; Figure 3E). Collectively, these findings demonstrated that CPT1A suppresses ccRCC growth via inhibiting cholesterol uptake and lipid accumulation.
Figure2 .
CPT1A overexpression prevents lipid accumulation and cell proliferation in vivo and in vitro (A) Cell proliferation of Caki-1 and 786O incubated with etomoxir (80 μM) at 0, 6, 12, 24, and 36 h detected by MTS assay. (B) Colony formation assay of Caki-1 and 786O stably transfected with CPT1A overexpression lentivirus indicated that overexpression of CPT1A could inhibit the proliferation in ccRCC. (C) The expressions of CDK2, CDK4 and CyclinD1 detected by western blot analysis. (D) Orthotopic xenograft transplanted with NC (n = 6) and OE-CPT1A/Caki-1 cells (n = 6). Tumor volume at day 14 was measured in each group. (E) Oil red O staining of frozen sections of tumor tissue from nude mice. Scale bar = 50 μm. Values are shown as the mean±SD. *P<0.05, ***P<0.001 (Student’s t-test).
Figure3 .
CPT1A overexpression decreases the cholesterol absorption and intracellular lipid accumulation(A,B) Contents of TC and TG in CPT1A-overexpressing Caki-1, 786O cell and their corresponding normal control cell lines were detected by ELISA kits. (C) Cholesterol deposition in CPT1A-overexpressing Caki-1 and 786O cells and their corresponding normal control cells. Scale bar = 50 μm. (D) Cholesterol absorption between CPT1A-overexpressing Caki-1 and 786O and their corresponding normal control cells detected by Flow cytometry. (E) The protein expressions of CD36, ABCG1, SRB1, LOX1 in CPT1A-overexpressing Caki-1, and 786O cell and their corresponding normal control cell lines determined by western blot analysis. Values are shown as the mean±SD. ***P<0.001 (Student’s t-test).
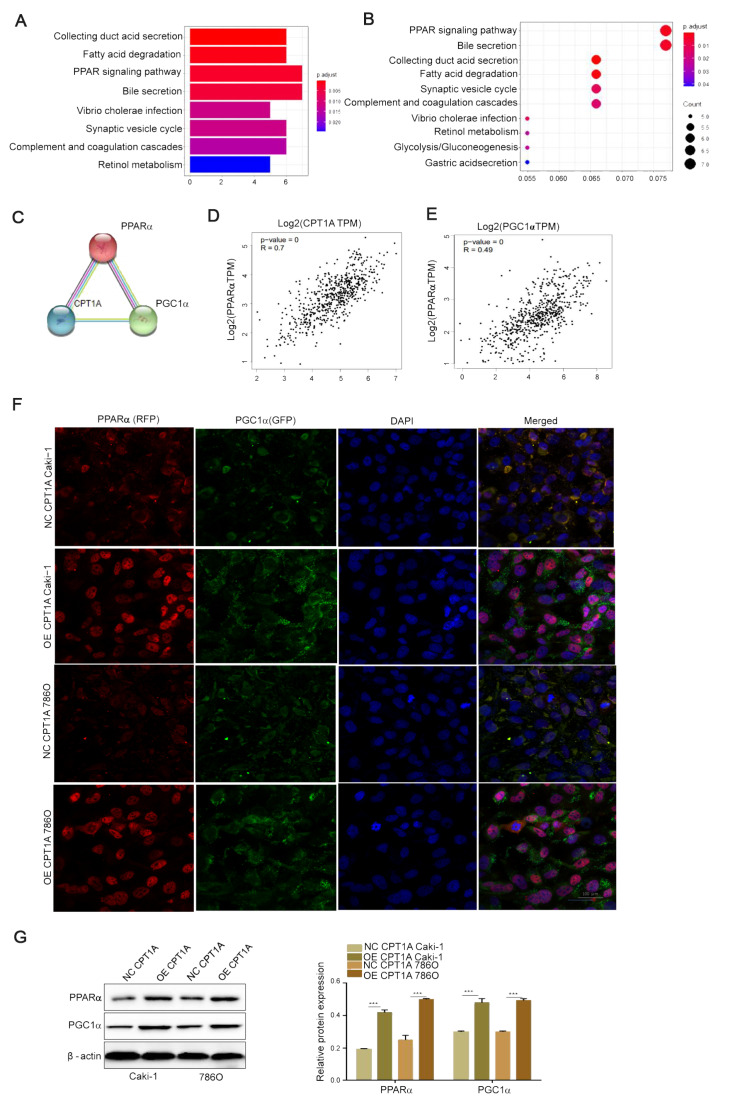
The potential relationships among CPT1A, PPARα and PGC1α
In order to explore the biological function of CPT1A in lipid metabolism in ccRCC, the KEGG pathway and GO functional analyses were performed using the Cluster Profiler package. Under the cutoff criteria gene size ≥ 20 and false discovery rate (FDR) < 0.01, high expression samples were found to be enriched in the PPAR signaling pathways ( Figure 4A,B). PPRAα is a key biomarker gene of ccRCC [26]. PPI network and miRNA-gene network were constructed and analyzed by https://string-db.org and gene expression profiling interactive analysis (GEPIA). The STRING tool revealed that CPT1A, PPARα, and PGC1α proteins interact with each other in ccRCC ( Figure 4C), and multiple gene analysis through GEPIA showed that the related coefficient of CPT1A and PPARα in ccRCC was R=0.7 ( P=0), and that of PPARα and PGC1α was R=0.49 ( P=0) ( Figure 4D,E). Furthermore, immunofluorescence staining confirmed that PPARα and PGC1α were co-expressed in ccRCC cells, and the expressions of PPARα and PGC1α were increased in CPT1A-overexpressing cells ( Figure 4F). In order to confirm this, we detected the protein levels of PPARα and PGC1α in CPT1A overexpressed cells, and found that the expression of PPARα was increased ( P<0.001), and the expression of PGC1α was also increased ( P<0.001; Figure 4G). Our results suggested that there is a potential relationship among CPT1A, PPARα and PGC1α.
Figure4 .
The potential relationship among CPT1A, PPARα and PGC1α(A,B) KEGG and GO were used to map pathway database. KEGG pathways analysis of the genes in the top four modules. (C) The STRING website was used to analyze the interaction of proteins with CPT1A-related proteins. CPT1A was input into STRING, and the main cluster was formed. (D,E) Multiple genes correlation analysis between CPT1A and PPARα (R = 0.7) and between PGC1α and PPARα (R = 0.49) using GEPIA website. (F) The expressions of PPARα and PGC1α in CPT1A overexpression Caki-1 and 786O cells and their corresponding normal control cells detected by immunofluorescence assay. Scale bar = 100 μm. (G) The protein expressions of PPARα and PGC1α in CPT1A-overexpressing Caki-1, 786O cells and their corresponding normal control cell lines determined by western blot analysis. Values are shown as the mean±SD. ***P<0.001 (Student’s t-test).
CPT1A suppresses the activity of PI3K/Akt signaling via inhibiting CD36
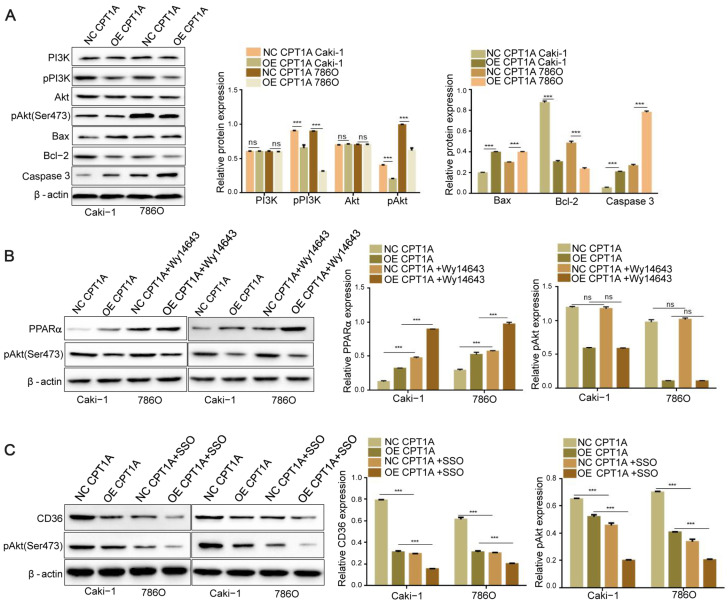
To elucidate the regulatory pathways implicated in the effect of CPT1A overexpression on ccRCC cell metabolism, we examined the expressions of related proteins in the PI3K/Akt signaling pathway by western blot analysis. As a result, we found that CPT1A overexpression significantly decreased the levels of pPI3K and pAkt without affecting their corresponding total protein levels ( P<0.001), and the expressions of Bax and Caspase 3 were increased but the expression of Bcl-2 was decreased ( P<0.001; Figure 5A). PPARα can regulate liver cell energy metabolism remodeling via Akt signaling [27]. To evaluate weather phosphorylated Akt level in ccRCC is regulated by PPARα or CD36 in ccRCC, we promoted the expression of PPARα by treatment with WY14643 (an activator of PPARα) for 24 h and detected the level of phosphorylated Akt by western blot analysis. The results showed that the expression of PPARα was increased in ccRCC cells when CPT1A was overexpressed, and treatment with WY14643 could further induced the expression PPARα ( P<0.001). The level of phosphorylated Akt was decreased when CPT1A was overexpressed ( P<0.001), but the level phosphorylated Akt was not further decreased by treatment with WY14643 ( P>0.05; Figure 5B). Then, we treated the CPT1A-overexpressing cells with SSO (an inhibitor of CD36) for 24 h to investigate whether the impeded activity of CD36 could aggravate the decreased level of phosphorylated Akt induced by CPT1A overexpression. The results showed that the expression of CD36 was decreased with CPT1A overexpression ( P<0.001). Treatment with SSO could reduce the expression of CD36 and further decreased the level of phosphorylated Akt ( P<0.001; Figure 5C). These results indicated that CPT1A inhibits the phosphorylation of Akt via inhibiting CD36.
Figure5 .
CPT1A suppresses the activity of PI3K/Akt signaling via downregulating CD36(A) Bax, Bcl-2, Caspase 3, p-Akt, Akt, p-PI3K, and PI3K protein levels in different groups (OE/NC CPT1A Caki-1, OE/NC CPT1A 786O). (B) Caki-1 and 786O cells were treated with 50 μM of Wy14643 for 24 h. PPARα and p-Akt levels in different groups. (C) After cells were treated with the inhibitor of CD36 (SSO, 25 μM)for 24 h, CD36 and p-Akt protein levels were determined in different groups. Values are shown as the mean±SD. ns: no significance; ***P<0.001 (Student’s t-test).
Discussion
ccRCC, charactered by a reprogramming of energetic metabolism, involves different processes, such as aerobic glycolysis [ 28– 30]. Complex I is inhibited by NADH dehydrogenase 1 alpha subcomplex 4-like 2 (NDUFA4L2), an HIF-1 target gene encoding a regulatory protein that attenuates mitochondrial oxygen consumption [31], and Rpn10 directly promotes inhibitor of nuclear factor-kappa B alpha (IκBα) degradation through the ubiquitin-proteasome system (UPS) to promote cell proliferation, migration, and invasion in ccRCC [32]. Previous studies have demonstrated that elevated lipogenesis and accumulation of intracellular lipid droplets in tumors contribute to cell membrane synthesis, cell growth, and transformation [ 33– 35]. The metabolic adaptation in ccRCC resulting in lipid storage is necessary for ccRCC development [36]. The 36-kDa phospholipid-binding protein annexin A3 (AnxA3) in the endocytic compartment negatively modulates lipid storage in ccRCC cells by interfering with the vesicular trafficking involved in lipid uptake and accumulation [37].
Integrated lipid-omics-transcriptomics approach revealed that ccRCC tissues exhibit a reprogramming of fatty acid metabolism in association with altered expressions of lipid metabolism-associated genes such as CPT1A, and delineated a lipidomic profile of human ccRCC, which is inhibited by hypoxia inducible factors (HIFs), reducing FA transport into mitochondria, and rerouting FA to LDs for storage [9]. Subsequently, we identified the most dysregulated FAO-enzyme, CPT1A, and investigated its role in ccRCC. In the present study, we found that the expression of CPT1A is negatively correlated with the malignant clinicopathological features of ccRCC. The loss of CPT1A expression in ccRCC clinical samples reduced the CPT1A activity. Moreover, large-scale survival analysis showed that loss of CPT1A expression in ccRCC associated with poor prognosis could serve as an independent prognostic indicator.
Typically, fatty acid synthesis, cholesterol uptake, and β-oxidation are tightly balanced to avoid intracellular lipid accumulation. Reprogrammed cellular metabolic pathways of fatty acid and cholesterol synthesis have been recognized as common metabolic hallmarks in several tumor types [ 38, 39]. CPT1A is underregulated by the lipid metabolism pathway related to PPARα [26], which is confirmed in this study. PPARα is a key transcription factor that regulates the expression of proteins involved in fatty acid uptake and β-oxidation [40]. The deficiency of PPARα contributes to overloading lipid-associated tubular injury, while PPARα agonist exerts an opposite effect [41]. PGC1α is a transcriptional coactivator within the metabolic reprogramming family [42]. PPARα and PGC1α stimulate the transcription of the CPT1A gene [43]. In the present study, PPARα and PGC1α were co-expressed in ccRCC cell lines, as detected by immunofluorescence staining, and the expressions of both molecules were further enhanced by CPT1A overexpression. Consistent with this finding, the protein expression of PPARα was increased in CPT1A-overexpressed whole cell lysate. The STRING platform at the protein level and multiple gene analyses through GEPIA also revealed an interaction among CPT1A, PPARα, and PGC1α in ccRCC.
CD36 is counter-regulated by PPARα [44]and is upregulated in ccRCC ( Supplementary Figure S1). CD36, SRB1 and lysosomal integral membrane protein type 2 (LIMP2) are three members of Class B scavenger receptors [45]. In addition to scavenger receptors, SR-A, LOX and CD36 promote lipid uptake, and cholesterol efflux is regulated by SR-B1 in foam cell formation. Interestingly, SRB1 is a transmembrane protein that has been well-characterized as a unique multifunctional receptor for cholesterol influx and efflux mediating cholesterol movement into and out of cells [46]. SRB1 and ABCG1 cholesterol transfer to high-density lipoprotein (HDL), resulting in an insufficient activity of the lipid efflux pathway [47]. Herein, two members of Class B scavenger receptors, CD36 and SRB1, were found to be highly expressed in ccRCC cells. CPT1A overexpression reduces the levels of CD36, ABCG1 and SRB1 proteins. The absence of CPT1A inhibits cholesterol metabolism and causes cholesterol deposition in macrophages [48]. The above phenomena might explain the phenotype of the enhanced cholesterol level and increased intracellular lipid deposition in ccRCC. In the present study, we found that CPT1A deficiency increases the intracellular cholesterol absorption and lipid accumulation via the PPARα/CD36 axis, while overexpression of CPT1A reduces the cholesterol absorption and intracellular lipid accumulation.
ccRCC is characterized by large intracellular lipid droplets (LDs) containing free and esterified cholesterol, and elevated dietary cholesterol promotes tumor growth. Blocking exogenous sources of cholesterol is sufficient to cause ccRCC cell apoptosis and decreased PI3K/Akt signaling [49], which is consistent with our results. The phosphorylation of Akt promotes cancer development [50], and Akt is frequently activated in RCC, which is linked to the survival and progression of cancers [51]. In the present study, CPT1A overexpression reduces the intracellular cholesterol accumulation and maintains the phosphorylation Akt at a low level in ccRCC cells, which could be further reduced by SSO, an inhibitor of CD36. Akt is a master regulator of cell survival under stress conditions [52]. Cloning test and xenograft tumor studies in nude mice showed that CPT1A overexpression inhibited cell proliferation and decreased the protein expressions of CDK2, CDK4 and CyclinD1 in ccRCC. Meanwhile, mitochondrial apoptosis-related proteins, including Bax and Caspase 3 were increased, and Bcl-2 was decreasing in CPT1A-overexpressing cells with increased cell apoptosis. Together, these findings indicated that the deficiency of CPT1A increases the cholesterol influx, leading to cholesterol accumulation, and the increased cholesterol level related to PPARα/CD36 axis promotes the phosphorylated Akt to accelerate cell proliferation in ccRCC. However, the potential molecular mechanisms about mutual regulation mechanism between CPT1A and PPARα need to be further explored in ccRCC. In further studies, whether increased cholesterol accumulation induced by CD36 overexpression is responsible for the activity of phosphorylated Akt to promote cell proliferation in ccRCC needs to be further verified.
Due to ccRCC chemotherapy resistance, anti-angiogenesis is one of the most innovative targeted therapies for ccRCC. In recent years, a better understanding of the molecular basis of RCC has led to the introduction of anti-angiogenic therapies for this tumor. Cytotoxic drugs or TKIs, such as sunitinib, have been used for ccRCC therapy in clinic, although these drugs yield partial responses in a minority of patients, with no evidence of complete responses [53]. Nevertheless, the combinations of inhibitors of the PD1/PD-L1 axis with VEGF inhibitors or cytotoxic T-lymphocyte antigen (CTLA)-4 inhibitors have shown promising efficacy in metastatic renal cell carcinorma (mRCC) [54]. Considering this, we will explore the effect of CPT1A on cytotoxic drugs or TKIs such as sunitinib in the future, which may reveal new function of CPT1A in ccRCC clinical therapy.
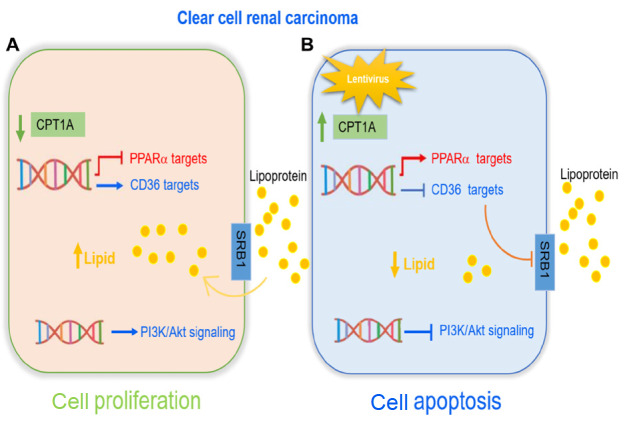
In conclusion, long-chain fatty acid transferase CPT1A may play a unique role in the biological behavior of ccRCC and our study provides new insights into the molecular mechanisms of lipid disorder-promoted cell proliferation in ccRCC. Dysregulated cholesterol absorption and lipid accumulation in ccRCC are induced by the loss of CPT1A, indicating its potential role as both prognostic marker and therapeutic target in ccRCC. Our data extend the understanding of the important role of CPT1A in ccRCC metabolism and development, and provide a potential target for the exploration of therapeutic strategies in ccRCC. This is the first report demonstrating that CPT1A overexpression inhibits the tumor growth in vivo and in vitroby decreasing the cholesterol absorption and lipid accumulation via PPARα/CD36 axis and suppressing Akt phosphorylation in ccRCC. These interactions are illustrated in Figure 6. Thus, the restoration of the activity of CPT1A via PPARα/CD36 axis may be a promising ccRCC therapeutic strategy, which provides new hope for patient treatment.
Figure6 .
Diagram of the function of CPT1A in ccRCC progression(A) ccRCC cells have an excessive lipid accumulation caused by CPT1A deficiency. CPT1A deficiency inhibits PPARα level and promotes CD36 level, leading to an increased lipid accumulation which in turn promotes an activity of PI3K/Akt signaling. (B) Overexpression of CPT1A promotes ccRCC cell apoptosis by suppressing the activity of PI3K/Akt signaling and decreasing intracellular lipid accumulation. Mechanistically, CPT1A overexpression downregulates the expressions of CD36 and SRB1, and upregulates the expression of PPARα.
Supporting information
Supplementary Data
Supplementary data is available at Acta Biochimica et Biophysica Sinicaonline.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (Nos. 81960462, 81760455, 31960200, 31660246 and 31960145) and Yunnan Province Applied Research Funds (Nos. 2018FE468-001, 202001AY070001-179, 2018FB120 and 2019FB091).
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. . 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Inamura K. Renal cell tumors: understanding their molecular pathological epidemiology and the 2016 WHO classification. Int J Mol Sci. . 2017;18:2195. doi: 10.3390/ijms18102195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diaz de Leon A, Pirasteh A, Costa DN, Kapur P, Hammers H, Brugarolas J, Pedrosa I. Current challenges in diagnosis and assessment of the response of locally advanced and metastatic renal cell carcinoma. RadioGraphics. . 2019;39:998–1016. doi: 10.1148/rg.2019180178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motzer RJ, Escudier B, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, et al. Nivolumab versus everolimus in patients with advanced renal cell carcinoma: updated results with long‐term follow‐up of the randomized, open‐label, phase 3 CheckMate 025 trial. Cancer. . 2020;126:4156–4167. doi: 10.1002/cncr.33033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. . 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 6.Herrington CS, Poulsom R, Coates PJ. Recent advances in pathology: the 2020 annual review issue of the journal of pathology. J Pathol. . 2020;250:475–479. doi: 10.1002/path.5425. [DOI] [PubMed] [Google Scholar]
- 7.VandeKopple MJ, Wu J, Auer EN, Giaccia AJ, Denko NC, Papandreou I. HILPDA regulates lipid metabolism, lipid droplet abundance, and response to microenvironmental stress in solid tumors. Mol Cancer Res. . 2019;17:2089–2101. doi: 10.1158/1541-7786.MCR-18-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avgerinos KI, Spyrou N, Mantzoros CS, Dalamaga M. Obesity and cancer risk: emerging biological mechanisms and perspectives. Metabolism. . 2019;92:121–135. doi: 10.1016/j.metabol.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Lucarelli G, Ferro M, Loizzo D, Bianchi C, Terracciano D, Cantiello F, Bell LN, et al. Integration of lipidomics and transcriptomics reveals reprogramming of the lipid metabolism and composition in clear cell renal cell carcinoma. Metabolites. . 2020;10:509. doi: 10.3390/metabo10120509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, Lu J, Han L, Wang X, Man Q, Liu S. Prognostic significance of two lipid metabolism enzymes, HADHA and ACAT2, in clear cell renal cell carcinoma. Tumor Biol. . 2016;37:8121–8130. doi: 10.1007/s13277-015-4720-4. [DOI] [PubMed] [Google Scholar]
- 11.Wettersten HI, Hakimi AA, Morin D, Bianchi C, Johnstone ME, Donohoe DR, Trott JF, et al. Grade-dependent metabolic reprogramming in kidney cancer revealed by combined proteomics and metabolomics analysis. Cancer Res. . 2015;75:2541–2552. doi: 10.1158/0008-5472.CAN-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system-from concept to molecular analysis. Eur J Biochem. . 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- 13.Mørkholt AS, Wiborg O, Nieland JGK, Nielsen S, Nieland JD. Blocking of carnitine palmitoyl transferase 1 potently reduces stress-induced depression in rat highlighting a pivotal role of lipid metabolism. Sci Rep. . 2017;7:2158. doi: 10.1038/s41598-017-02343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Q, Tan Z, Shi F, Tang M, Xie L, Zhao L, Li Y, et al. PGC1α/CEBPB/CPT1A axis promotes radiation resistance of nasopharyngeal carcinoma through activating fatty acid oxidation. Cancer Sci. . 2019;110:2050–2062. doi: 10.1111/cas.14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balaban S, Shearer RF, Lee LS, van Geldermalsen M, Schreuder M, Shtein HC, Cairns R, et al. Adipocyte lipolysis links obesity to breast cancer growth: adipocyte-derived fatty acids drive breast cancer cell proliferation and migration. Cancer Metab. . 2017;5:1. doi: 10.1186/s40170-016-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricciardi MR, Mirabilii S, Allegretti M, Licchetta R, Calarco A, Torrisi MR, Foà R, et al. Targeting the leukemia cell metabolism by the CPT1a inhibition: functional preclinical effects in leukemias. Blood. . 2015;126:1925–1929. doi: 10.1182/blood-2014-12-617498. [DOI] [PubMed] [Google Scholar]
- 17.Du W, Zhang L, Brett-Morris A, Aguila B, Kerner J, Hoppel CL, Puchowicz M, et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat Commun. . 2017;8:1769. doi: 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray AJ, Montgomery HE, Feelisch M, Grocott MPW, Martin DS. Metabolic adjustment to high-altitude hypoxia: from genetic signals to physiological implications. Biochem Soc Trans. . 2018;46:599–607. doi: 10.1042/BST20170502. [DOI] [PubMed] [Google Scholar]
- 19.Aloia A, Müllhaupt D, Chabbert CD, Eberhart T, Flückiger-Mangual S, Vukolic A, Eichhoff O, et al. A fatty acid oxidation-dependent metabolic shift regulates the adaptation of BRAF-mutated melanoma to MAPK inhibitors . Clin Cancer Res. . 2019;25:6852–6867. doi: 10.1158/1078-0432.CCR-19-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen YP, Tsai CW, Shen CY, Day CH, Yeh YL, Chen RJ, Ho TJ, et al. Palmitic acid interferes with energy metabolism balance by adversely switching the SIRT1-CD36-fatty acid pathway to the PKC zeta-GLUT4-glucose pathway in cardiomyoblasts. J Nutral Biochem. . 2016;31:137–149. doi: 10.1016/j.jnutbio.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Manio MCC, Matsumura S, Masuda D, Inoue K. CD36 is essential for endurance improvement, changes in whole-body metabolism, and efficient PPAR-related transcriptional responses in the muscle with exercise training. Physiol Rep. . 2017;5:e13282. doi: 10.14814/phy2.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grajchen E, Wouters E, van de Haterd B, Haidar M, Hardonnière K, Dierckx T, Van Broeckhoven J, et al. CD36-mediated uptake of myelin debris by macrophages and microglia reduces neuroinflammation. J Neuroinflammation. . 2020;17:224. doi: 10.1186/s12974-020-01899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prieto P, Cuenca J, Través PG, Fernández-Velasco M, Martín-Sanz P, Boscá L. Lipoxin A4 impairment of apoptotic signaling in macrophages: implication of the PI3K/Akt and the ERK/Nrf-2 defense pathways. Cell Death Differ. . 2010;17:1179–1188. doi: 10.1038/cdd.2009.220. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Wang C, Wang Y, Sun L, Liu Z, Wang L, Song T, et al. HSCs-derived COMP drives hepatocellular carcinoma progression by activating MEK/ERK and PI3K/AKT signaling pathways. J Exp Clin Cancer Res. . 2018;37:231. doi: 10.1186/s13046-018-0908-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, German P, Bai S, Barnes S, Guo W, Qi X, Lou H, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. . 2015;42:343–353. doi: 10.1016/j.jgg.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu F, Wu S, Gou X. Identification of biomarkers and potential molecular mechanisms of clear cell renal cell carcinoma. Neoplasma. . 2018;65:242–252. doi: 10.4149/neo_2018_170511N342. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Xu L, Lu B, Zhao M, Li L, Sun W, Qiu Z, et al. LncRNA H19/microRNA-675/PPARα axis regulates liver cell injury and energy metabolism remodelling induced by hepatitis B X protein via Akt/mTOR signalling. Mol Immunol. . 2019;116:18–28. doi: 10.1016/j.molimm.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Lucarelli G, Loizzo D, Franzin R, Battaglia S, Ferro M, Cantiello F, Castellano G, et al. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev Mol Diagnostics. . 2019;19:397–407. doi: 10.1080/14737159.2019.1607729. [DOI] [PubMed] [Google Scholar]
- 29.Bianchi C, Meregalli C, Bombelli S, Di Stefano V, Salerno F, Torsello B, De Marco S, et al. The glucose and lipid metabolism reprogramming is grade-dependent in clear cell renal cell carcinoma primary cultures and is targetable to modulate cell viability and proliferation. Oncotarget. . 2017;8:113502–113515. doi: 10.18632/oncotarget.23056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ragone R, Sallustio F, Piccinonna S, Rutigliano M, Vanessa G, Palazzo S, Lucarelli G, et al. Renal cell carcinoma: a study through NMR-based metabolomics combined with transcriptomics. Diseases. . 2016;4:7. doi: 10.3390/diseases4010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucarelli G, Rutigliano M, Sallustio F, Ribatti D, Giglio A, Lepore Signorile M, Grossi V, et al. Integrated multi-omics characterization reveals a distinctive metabolic signature and the role of NDUFA4L2 in promoting angiogenesis, chemoresistance, and mitochondrial dysfunction in clear cell renal cell carcinoma. Aging. . 2018;10:3957–3985. doi: 10.18632/aging.101685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang T, Tian W, Zhou Q, Li J, Jiang Z, Chen J, Ge C, et al. Upregulation of Rpn10 promotes tumor progression via activation of the NF-κB pathway in clear cell renal cell carcinoma. Acta Biochim Biophys Sin. . 2021;53:988–996. doi: 10.1093/abbs/gmab078. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Liu X, Wu F, Zhang X, Zhang H, Gao D, Bi D, et al. HADHA overexpression disrupts lipid metabolism and inhibits tumor growth in clear cell renal cell carcinoma. Exp Cell Res. . 2019;384:111558. doi: 10.1016/j.yexcr.2019.111558. [DOI] [PubMed] [Google Scholar]
- 34.Ackerman D, Tumanov S, Qiu B, Michalopoulou E, Spata M, Azzam A, Xie H, et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. . 2018;24:2596–2605.e5. doi: 10.1016/j.celrep.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. . 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou Y, Lu Q, Yao Q, Dong D, Chen B. Identification of novel prognostic biomarkers in renal cell carcinoma. Aging. . 2020;12:25304–25318. doi: 10.18632/aging.104131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bombelli S, Torsello B, De Marco S, Lucarelli G, Cifola I, Grasselli C, Strada G, et al. 36-kDa Annexin A3 isoform negatively modulates lipid storage in clear cell renal cell carcinoma cells. Am J Pathol. . 2020;190:2317–2326. doi: 10.1016/j.ajpath.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 38.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer Lett. . 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Zhai W, Lu H, Dong S, Fang J, Yu Z. Identification of potential key genes and key pathways related to clear cell renal cell carcinoma through bioinformatics analysis. Acta Biochim Biophys Sin. . 2020;52:853–863. doi: 10.1093/abbs/gmaa068. [DOI] [PubMed] [Google Scholar]
- 40.Liu GZ, Hou TT, Yuan Y, Hang PZ, Zhao JJ, Sun L, Zhao GQ, et al. Fenofibrate inhibits atrial metabolic remodelling in atrial fibrillation through PPAR-α/sirtuin 1/PGC-1α pathway. Br J Pharmacol. . 2016;173:1095–1109. doi: 10.1111/bph.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams DK, Stokes C, Horenstein NA, Papke RL. Differential regulation of receptor activation and agonist selectivity by highly conserved tryptophans in the nicotinic acetylcholine receptor binding site. J Pharmacol Exp Ther. . 2009;330:40–53. doi: 10.1124/jpet.109.151225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goto T. A review of the studies on food-derived factors which regulate energy metabolism via the modulation of lipid-sensing nuclear receptors. Biosci Biotechnol Biochem. . 2019;83:579–588. doi: 10.1080/09168451.2018.1559025. [DOI] [PubMed] [Google Scholar]
- 43.Song S, Attia RR, Connaughton S, Niesen MI, Ness GC, Elam MB, Hori RT, et al. Peroxisome proliferator activated receptor α (PPARα) and PPAR gamma coactivator (PGC-1α) induce carnitine palmitoyltransferase IA (CPT-1A) via independent gene elements. Mol Cell Endocrinol. . 2010;325:54–63. doi: 10.1016/j.mce.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Q, Shu F, Gong J, Ding P, Cheng R, Li J, Tong R, et al. Sweroside ameliorates NAFLD in high-fat diet induced obese mice through the regulation of lipid metabolism and inflammatory response. J EthnoPharmacol. . 2020;255:112556. doi: 10.1016/j.jep.2020.112556. [DOI] [PubMed] [Google Scholar]
- 45.Zani IA, Stephen SL, Mughal NA, Russell D, Homer-Vanniasinkam S, Wheatcroft SB, Ponnambalam S. Scavenger receptor structure and function in health and disease. Cells. . 2015;4:178–201. doi: 10.3390/cells4020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ou M, Huang R, Luo Q, Xiong L, Chen K, Wang Y. Characterisation of scavenger receptor class B type 1 in rare minnow ( Gobiocypris rarus) . Fish Shellfish Immunol. . 2019;89:614–622. doi: 10.1016/j.fsi.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 47.Annema W, Nijstad N, Tölle M, de Boer JF, Buijs RVC, Heeringa P, van der Giet M, et al. Myeloperoxidase and serum amyloid A contribute to impaired in vivo reverse cholesterol transport during the acute phase response but not group IIA secretory phospholipase A2. J Lipid Res. . 2010;51:743–754. doi: 10.1194/jlr.M000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura M, Liu J, Yu ZX, Yamazaki T, Yan Y, Kawagishi H, Rovira II, et al. Macrophage fatty acid oxidation inhibits atherosclerosis progression. J Mol Cell Cardiol. . 2019;127:270–276. doi: 10.1016/j.yjmcc.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riscal R, Bull CJ, Mesaros C, Finan JM, Carens M, Ho ES, Xu JP, et al. Cholesterol auxotrophy as a targetable vulnerability in clear cell renal cell carcinoma. Cancer Discov. 2021, doi: 10.1158/2159-8290 . [DOI] [PMC free article] [PubMed]
- 50.Gill JG, Piskounova E, Morrison SJ. Cancer, oxidative stress, and metastasis. Cold Spring Harb Symp Quant Biol. . 2016;81:163–175. doi: 10.1101/sqb.2016.81.030791. [DOI] [PubMed] [Google Scholar]
- 51.He D, Sun X, Yang H, Li X, Yang D. TOFA induces cell cycle arrest and apoptosis in ACHN and 786-O cells through inhibiting PI3K/Akt/mTOR pathway. J Cancer. . 2018;9:2734–2742. doi: 10.7150/jca.26374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Link W. Introduction to foxo biology. Methods Mol Biol. 2019, 1890: 1–9. . [DOI] [PubMed]
- 53.Tamma R, Rutigliano M, Lucarelli G, Annese T, Ruggieri S, Cascardi E, Napoli A, et al. Microvascular density, macrophages, and mast cells in human clear cell renal carcinoma with and without bevacizumab treatment. Urologic Oncol-Semin Original Investigations. . 2019;37:355.e11–355.e19. doi: 10.1016/j.urolonc.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 54.Di Lorenzo G, De Placido S, Pagliuca M, Ferro M, Lucarelli G, Rossetti S, Bosso D, et al. The evolving role of monoclonal antibodies in the treatment of patients with advanced renal cell carcinoma: a systematic review. Expert Opin Biol Ther. . 2016;16:1387–1401. doi: 10.1080/14712598.2016.1216964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.