Abstract
Transcription factors (TFs) modulate gene expression by regulating the accessibility of promoter DNA to RNA polymerases (RNAPs) in bacteria. The MerR family TFs are a large class of bacterial proteins unique in their physiological functions and molecular action: they function as transcription repressors under normal circumstances, but rapidly transform to transcription activators under various cellular triggers, including oxidative stress, imbalance of cellular metal ions, and antibiotic challenge. The promoters regulated by MerR TFs typically contain an abnormal long spacer between the –35 and –10 elements, where MerR TFs bind and regulate transcription activity through unique mechanisms. In this review, we summarize the function, ligand reception, DNA recognition, and molecular mechanism of transcription regulation of MerR-family TFs.
Keywords: RNA polymerase, gene transcription, gene expression, transcription factor, MerR
Introduction
Unlike in eukaryotes, where the labor of gene transcription is split to three DNA-dependent RNA polymerases, i. e., polymerases I, II, and III [1], the gene transcription in bacteria solely replies on the single DNA-dependent RNA polymerase (RNAP) [2]. The bacterial RNAP partners with a set of transcription initiation factors (named σ factors) to form RNAP holoenzymes that are responsible for transcription of distinct gene programs [3]. In addition, a bacterial genome encodes hundreds of transcription factors (TFs) (~ 6% of their total gene count), which respond to environmental and cellular signals through their signal-reception domain (or ligand-binding domain) and modulate transcription of genes by directly binding to promoter DNA through their DNA-binding domain [4].
Bacterial transcription repressors occupy core promoter regions and inhibit transcription through preventing RNAP from engaging with promoter DNA, while the mechanism of bacterial transcription activation is much more complicated than that of transcription repression. The canonical Class I and Class II transcription activation models suggest that transcription activators interact with both RNAP and the upstream of core promoter region to increase local enrichment of RNAP at their regulated gene promoters and/or to facilitate subsequent promoter unwinding process [ 5, 6]. Distinct from the canonical transcription repression/activation models, the MerR-family TFs occupy the core promoter region to either repress or activate transcription of downstream genes depending on the cellular signals [ 7, 8].
In this review, we summarize the physiological function, ligand reception, DNA recognition, and mechanism of transcription regulation of MerR-family TFs. Recent comprehensive reviews are recommended for readers who are interested in mechanism of bacterial transcription and general bacterial transcription regulation [ 5, 6, 9, 15].
Transcription Initiation by Bacterial RNAP
Most bacterial RNAP core enzymes are composed of five subunits, two identical α subunits, one β subunit, one β′ subunit, and one ω subunit [16]. The ω subunit is absent in certain bacterial species and additional small accessory subunits are gained in specific bacterial phyla [17]. The bacterial RNAP holoenzyme is composed of the RNAP core enzyme and one of σ factors which anchor their DNA-recognition domains (σ 2, σ 3.1, and σ 4 of σ 70-type factors; region I and III for σ 54-type factors) on the surface of RNAP core enzyme and thread the linker domain (σ 3.2 of σ 70-type factors; region II for σ 54-type factors) into the active-site cleft [ 18– 23].
During transcription initiation, a RNAP-σ holoenzyme first recognizes the distal end of double-stranded core promoter DNA, i. e., the –35 element and/or extended –10 element, through sequence-specific interaction with σ 4 and/or σ 3.1 [ 24– 26]. The engagement of the distal end of promoter DNA presents its proximal end, i. e., the –10 element, on the surface of σ 2, where the DNA unwinding initiates [ 27, 28]. The σ 4/σ 2 distance and the –35/–10 spacer length (optimally 17 base pairs; bp) are matched to allow the –35 and –10 elements to be recognized by σ 4 and σ 2 in concert [ 24, 29]. At the beginning of DNA unwinding, the base pair at the –11 position of the –10 element is disrupted and the most-conserved adenosine at the –11 position of non-template strand is flipped out and secured by a pocket of σ 2 domain [ 28, 30, 31]. The DNA unwinding subsequently propagates to the downstream of promoter DNA and the unwound region of promoter DNA is stabilized in the main cleft of RNAP through base-specific pocket recognition at specific positions (T –7, G –6, and G +2 of the non-template strand) and electrostatic attraction interactions with the phosphate backbone [ 30, 32]. The resulting RNAP-promoter open complex (RPo) containing a ~13 bp unwound transcription bubble is competent for primer-dependent initiation (using a short RNA primer typically in length of 2–5 nt and one initiating NTP) or de novo initiation (using two initiating NTPs) of RNA synthesis ( Figure 1A) [ 24, 27, 29, 33– 35].
Figure1 .
Models for bacterial transcription initiation and canonical transcription activation(A) The model of RNAP-promoter DNA closed complex (RPc; upper panel) and RNAP-promoter DNA open complex (RPo; lower panel). In RPc, the double-stranded –35 element is recognized by σ4 in a sequence-specific manner, while the double-stranded –10 element is presented onto the surface of σ2 and restrained by sequence nonspecific electrostatic attraction interactions. In RPo, a ~13 bp transcription bubble is unwound and stabilized inside of RNAP. The base pair of –11 position is forced open by the W-dyad (W433 and W434 in E. coli RNAP σ70), and the adenine base of –11A of the non-template strand is recognized and secured in the –11 pocket. The other domains of σ factors are hidden for clarity. (B) The models for Class I (upper panel) and Class II (lower panel) transcription activation. A Class I transcription activator binds to the upstream of core promoter region and makes interactions with the C-terminal domain of the RNAP-α subunit (RNAP-αCTD). A Class II transcription activator binds to the proximal upstream of the core promoter region and makes interactions with both the RNAP-αCTD and σ4.
Class I and Class II Transcription Activation
The E. coli catabolic-activated protein (CAP), also named cAMP-responsive protein (CRP), servers as the prototype of bacterial transcription factor for studying the molecular mechanism of transcription activation [ 5, 36]. The mode of transcription activation by CAP could be divided into two major classes based on the locations of CAP-binding cis element (CAP box) on promoter DNA. In the Class I mode of transcription activation, the CAP box is located at the upstream of core promoter region on the same face of the DNA helix as the UP and –35 elements (for example, the CAP boxes are usually centered at positions –62, –72, –83, and –93 of promoter DNA) [ 37, 38]. The E. coli CAP dimer, which binds to the CAP box at these positions, bends the upstream promoter towards RNAP to establish interaction with RNAP-α C-terminal domain (CTD) in the cryo-EM structure of E. coli CAP Class I transcription activation complex [ 39, 40]. The Class II CAP box is located at the proximal upstream of core promoter DNA, i. e., the CAP box is centered at position –41.5, that partially overlaps with the –35 element [ 38, 41]. The crystal structure of T. thermophilus TAP (a homolog of E. coli CAP) Class II transcription activation complex shows that the DNA-bound TAP makes interactions with both RNAP-α CTD and the σ 4 domain [42]. Despite differences in location of cis elements and contact regions on RNAP, transcription factors bind to the upstream of promoter DNA and make bipartite interactions with promoter DNA and RNAP in both Class I and Class II transcription activation models ( Figure 1B) [ 5, 6, 36, 37].
The MerR-family TFs
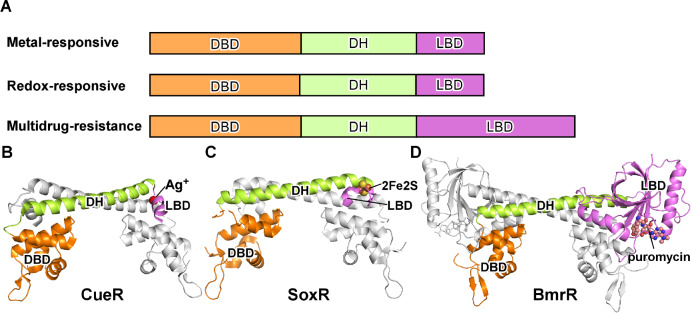
MerR-family TFs are a large family of bacterial TFs that share unique structural and mechanistic features. They typically contain an N-terminal DNA-binding domain (DBD), a central dimerization helix (DH), and a C-terminal ligand-binding domain (LBD) ( Figure 2A) [7]. Like most bacterial TFs, MerR-family TFs function as dimers. The two protomers interact with each other in a ‘head-to-head’ manner. The dimer interface is mainly contributed by the coiled-coil interaction of the central dimerization helices and inter-protomer DBD-LBD interaction ( Figure 2B). The MerR-family TFs could be further categorized into three subfamilies based on their physiological functions: the metal-responsive MerR-family TFs, the redox-responsive MerR-family TFs, and the multidrug-resistance MerR-family TFs.
Figure2 .
The MerR-family TFs(A) The schematic of three categories of MerR-family TFs. DBD, DNA-binding domain; DH, dimerization helix; LBD, ligand-binding domain. (B) The structure of E. coli CueR dimer, a representative member of the metal-responsive MerR TFs, adapted from the crystal structure of Ag+-bound E. coli CueR-DNA complex (PDB: 4WLW). One protomer is colored in gray, and the second protomer is colored in orange (DBD), green (DH), and pink (LBD). The Ag+ is shows in gray sphere. (C) The structure of E. coli SoxR dimer, a representative member of the redox-responsive MerR TFs, adapted from the crystal structure of oxidated SoxR-DNA complex (PDB: 2ZHG). The [2Fe-2S] cluster is shown as sphere. (D) The structure of B. subtilis BmrR dimer, a representative member of the multidrug-resistance MerR TFs, adapted from the crystal structure of puromycin-bound BmrR-DNA complex (PDB: 1EXI). The puromycin is shown as sphere.
The metal-responsive MerR-family TFs
The metal-responsive MerR-family TFs contain members that specifically recognize various metal cations with +1 charge, for example Cu +, Ag +, and Au +, or +2 charge, for example Cd 2+, Zn 2+, Pb 2+, Ni 2+, and Co 2+ ( Table 1). These patrol metal sensors rapidly respond to the elevated cellular ion concentration and subsequently ignite defense programs by activating expression of their regulated operons. For example, MerR, the founding member of MerR-family TFs, regulates the expression of the mercury-resistance operons that encode an enzyme catalyzing reduction of Hg 2+ to volatile Hg (MerA), transcription factors (MerD and MerR), a Hg 2+ scavenging protein Hg 2+ (MerP), and Hg 2+ transporters [ 43– 47]. The R. metalliduransPbrR regulates expression of the Pb 2+-resistance operon pbrABCDthat encodes a Pb(II) efflux transporter (PbrA), a Pb(II)-binding protein(PbrD), and two function unknown proteins (PbrB and PbrC) [ 48, 49]. The P. putida CadR maintains cellular concentration of Cd 2+ by controlling the expression of a cadmium transporter (CadA) [ 50, 51].
Table1 The summary of representative MerR-family TFs
|
Regulator |
Ligand |
Organism |
Regulated gene |
Reference |
|
Metal-responsive MerR-family TFs | ||||
|
MerR |
Hg + |
Tn 21/Tn 501 |
Mercury-resistance operon merTP (C/ F)AD(E) |
|
|
ZntR |
Zn 2+, Cd 2+, Pb 2+ |
E. coli |
zntA, a metal ion efflux ATPase |
|
|
CueR |
Cu +, Ag +, Au + |
E. coli |
Copper-tolerance genes copA and cueO |
|
|
PmtR |
Zn 2+ |
P. mirabilis |
A zinc-binding protein |
|
|
PbrR |
Pb 2+ |
R. metallidurans |
Pb 2+-resistance operon pbrABCD |
|
|
ZccR |
Zn 2+, Cd 2+, Co 2+ |
B. pertussis |
A metal ion efflux ATPase |
|
|
CadR |
Cd 2+ |
P. putida |
cadA, a cadmium transporter |
|
|
CoaR |
Co 2+ |
SynechocystisPCC 6803 |
coaT, a metal ion efflux ATPase |
|
|
NimR |
Ni 2+ |
H. influenzae |
Ni 2+ uptake transporter (NikKLMQO) |
|
|
Redox-responsive MerR-family TFs | ||||
|
SoxR |
Superoxide |
E. coli |
soxS, a transcription factor regulating antioxidant genes |
|
|
NmlR |
Formaldehyde |
H. influenzae |
adhC and estD, detoxification of formaldehyd e |
|
|
Multidrug-resistance MerR-family TFs | ||||
|
TipA |
Cyclic thiopeptide |
Streptomyces |
Thiostrepton-resistant genes |
|
|
NolA |
Genistein |
B. japonicum |
nodD2, a transcription factor regulating nodulation genes |
|
|
BmrR |
Multidrug |
B. subtilis |
bmr, a multidrug-efflux pump |
|
|
BltR |
Multidrug |
B. subtilis |
blt, a multidrug-efflux pump |
|
|
Mta |
Multidrug |
B. subtilis |
bmr and blt, two multidrug-efflux pumps |
|
|
BrlR |
c-di-GMP |
P. aeruginosa |
mexAB-oprM and mexEF-oprN |
|
|
MerR-family TFs with other functions | ||||
|
Rv1828 |
Fatty acids |
M. tuberculosis |
Unknown |
|
|
Rv3334 |
Unknown |
M. tuberculosis |
kstR, a transcription factor regulating lipid catabolism |
These patrol metal sensors also sense overloaded essential metal ions and activate gene programs to maintain their homeostasis. For example, the E. coli CueR responds to the elevated concentration of cellular free Cu + and activates the expression of copper efflux ATPase (CopA) and multi-copper oxidase (CueO) [ 63, 97, 98]. E. coli ZntR maintains the cellular homeostasis of Zn 2+ by regulating the expression of a zinc transporter (ZntA) [ 59, 99].
The redox-responsive MerR-family TFs
Reactive oxygen species (ROS) that induce cellular and genetic damages are produced as an unavoidable consequence of the aerobic lifestyle [100]. The redox-responsive MerR-family TFs are one of the guardians sensing cellular oxidative stress in bacteria [101]. SoxR [71] and NmlR (also named AdhR) [ 75, 102] are two of the currently reported members of the redox-responsive MerR-family TFs. Although SoxR and NmlR fall into the same group, they employ distinct mechanisms to sense oxidative stress. The C-terminal metal binding loop of SoxR coordinates a [2Fe-2S] metal cluster that is proposed to change its overall charge state upon oxidation [ 103– 105]. Once activated, SoxR increases the expression of soxS gene, encoding an AraC-family transcription factor that controls the expressions of various antioxidant and damage repair proteins [ 71, 72]. NmlR detoxifies oxidative damages induced by formaldehyde as well as other ROS generators [ 75, 76, 106]. In H. influenzae, NmlR controls the expressions of AdhC and EstD that work sequentially to convert formaldehyde into formic acid in a glutathione (GSH)-dependent manner [107]. Although biochemical studies suggested that the activity of NmlR is zinc-dependent [75], the crystal structures of apo or DNA-bound NmlR revealed absence of any coordinated metal ions [76].
The multidrug-resistance MerR-family TFs
The third category of MerR-family TFs is composed of multidrug-resistance receptors that recognize a broad spectrum of exogenous toxic compounds ( Table 1). The representative proteins in this category include TipA [ 77, 78], BmrR [ 84, 85], BltR [ 85, 108], Mta [86], BrlR [ 90– 93, 109], and MrR [110]. TipA has two forms (TipA L and TipA S), both of which can sense the self-encoded natural ribosomal inhibitor, thiostrepton [ 77– 79, 111]. Upon forming a covalent bond with thiostrepton, TipA activates the expressions of its own and other proteins necessary for thiostrepton resistance [ 79, 112]. B. subtilisencodes three members of the MerR subfamily, BltR, BmrR and Mta [ 84– 86]. Mta functions as a master TF that activates the expressions of both multidrug efflux transporters Blt and Bmr; while BltR and BmrR specifically regulate the expressions of the multidrug transporters Blt and Bmr, respectively. BrlR is important for antimicrobial tolerance of P. aeruginosaby regulating the expressions of the multidrug efflux pumps mexAB-oprM and mexEF-oprN [ 90, 92].
DNA Recognition of MerR-family TFs
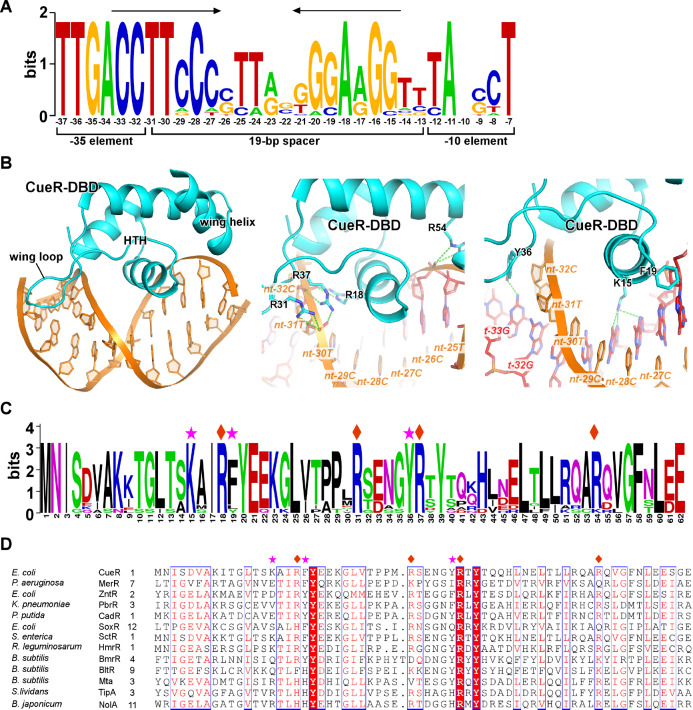
The N-terminal winged helix-turn-helix (wHTH) domain of the MerR-family TFs recognizes their cognate long palindromic cis elements that are located at the spacer region between the –35 and –10 elements of their regulated promoters [ 51, 64, 73, 76, 87] ( Figure 3A). Both the central helix-turn-helix and two wings (a wing loop and a wing HTH) make direct interactions with DNA. Residues in the central helix and wing loop insert into the major and minor grooves of dsDNA, respectively, and make sequence-specific H-bond and Van der Waal interactions ( Figure 3B). These residues are conserved in homologs of a certain MerR-family TF that recognize the same DNA sequence motifs ( Figure 3C) but vary in MerR-family members that recognize different DNA sequence motifs ( Figure 3D), suggesting that they are responsible for sequence-specific recognition. Meanwhile, several positively charged residues make extensive interactions with the phosphate backbones of DNA and function as clamps to stabilize the protein-DNA interactions during DNA conformational transition. These clamp residues are conserved in the majority of MerR-family TFs, highlighting the importance of these residues, and suggesting that most MerR-family TFs employ the same set of residues to anchor and distort dsDNA ( Figure 3D).
Figure3 .
DNA recognition of MerR-family TFs(A) The consensus DNA sequence logo of E. coli CueR regulated promoters. The palindromic repeats are highlighted by arrows. The positions are numbered respective to the transcription start site (+1). (B) The interaction between the CueR-DBD and dsDNA. The sequence nonspecific interactions between backbone phosphates of DNA and residues of CueR-DBD are shown in the middle panel. The base-specific interactions made by CueR-DBD are shown in the right panel. (C) The consensus protein sequence logo of CueR from various bacterial species. (D) The multiple-sequence alignment of DBD from multiple MerR TFs. The residues contacting backbone phosphates are labeled by diamonds and the residues making base-specific interactions are labeled by asterisks.
Signal Reception of MerR-family TFs
In contrast to the conserved wHTH fold of the N-terminal DBD, the C-terminal LBD of the MerR-family TFs varies radically in both length and sequence, conferring the diversity of ligand recognition to this family. In general, the C-terminal LBD of MerR-family TFs is able to coordinate metal cations, sense cellular oxidative condition, and recognizes a broad spectrum of exogenic toxic organic chemicals ( Table 1).
The metal- and redox-responsive MerR-family TFs contain a short C-terminal metal binding loop for coordinating metal ions or metal clusters. The metal-responsive MerR-family TFs exhibit ultra-sensitivity towards their cognate metal ions with reported dissociation constants from micromolar level to nanomolar levels [ 50, 69, 97, 113]. O’Halloran′s group has reported zeptomolar (10 –21 M) and femtomolar (10 –15 M) sensitivity for ZntR/Zn 2+ and CueR/Cu +, respectively, under the metal buffering experimental conditions [ 114– 116]. The metal sensors of the MerR family exhibit stringent selectivity towards metal ions of different charge states but are tolerant to ions with the same charge state and the same valence shell configuration.
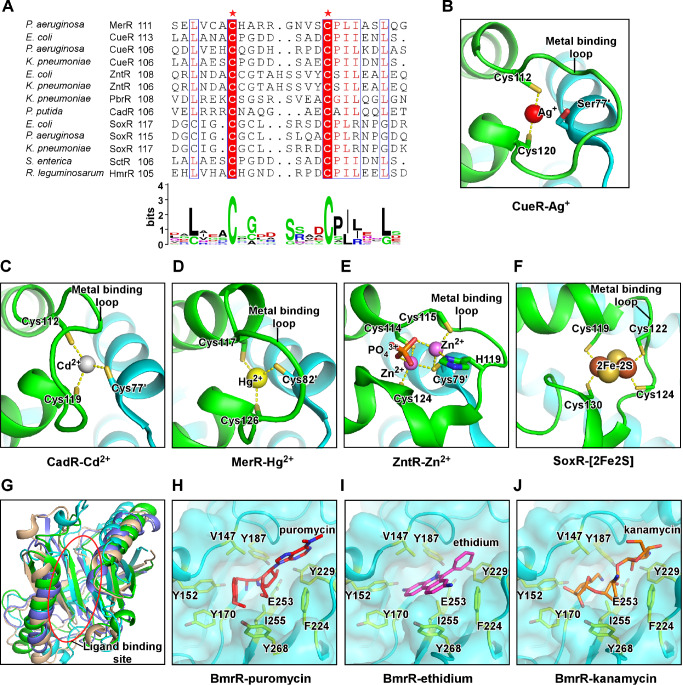
The C-terminal metal binding loop along with nearby residues creates the metal coordination site that determinates the ion selectivity. Almost all members of the MerR subfamily contain two conserved cysteine residues at both ends of the metal binding loop ( Figure 4A) [114]. The two cysteine residues (Cys112 and Cys120) of E. coli CueR make two coordinate covalent bonds to Cu + with an essentially linear S-Cu-S bond angel, while a serine (Ser77) residue from the other protomer inserts into the ion pocket to stabilize Cys112 conformation ( Figure 4B) [114]. The two conserved cysteine residues (Cys112 and Cys119) in P. putida CadR also participate into the tetrahedral coordination network by making two coordinate covalent bonds with Cd 2+, while another cysteine (Cys77′) forms the third tetrahedral coordinate covalent bond ( Figure 4C) [51]. As for MerR, the two conserved cysteines (Cys117 and Cys126 in Tn501 MerR) and the cysteine from the other protomer (Cys82′ in Tn501 MerR) form three coordinate covalent bonds with Hg 2+ in a planar trigonal coordination geometry ( Figure 4D) [ 117, 118]. Structural superimposition and sequence alignment suggest that the +1/+2 ion selectivity of metal-responsive MerR-family TFs is mainly determined by the identity of the residue extended from the other protomer (Ser77′ in E. coli CueR, Cys77′ in P. putidaCadR, or Cys79′ in E. coli ZntR) that inserts into the ion coordination site [ 51, 114]. It is notable that certain MerR proteins harbor residues that allow coordinating the second ion either at the same site ( E. coli ZntR) ( Figure 4E) or at a new remote site ( P. putidaCadR) [ 51, 114]. Besides the conserved cysteines residues (Cys119 and Cys130), the C-terminal metal binding loop of E. coliSoxR contains two more cysteines residues (Cys122 and Cys124) to accommodate a [2Fe-2S] cluster, of which each of the Fe atoms is stabilized by two coordinate valent bonds made by two cysteine residues ( Figure 4F). In summary, MerR proteins respond to metal or oxidative stresses by either directly coordinating cognat metal ions or an oxygen-sensitive metal cluster [2Fe-2S]. The specificity is encoded in the ligand-coordination site, where the number and position of cysteine residues pre-define coordination geometry of their corresponding ligands.
Figure4 .
The signal reception of MerR-family TFs(A) The multiple sequence alignment and consensus protein sequence logo for the ligand-binding domain of MerR-family TFs. The two mostly conserved cysteines are highlighted and labeled by asterisks. (B) E. coli CueR coordinates one molecule of Ag+ through Cys112 and Cys120 of its metal-binding loop. The Ser77’ of the other protomer restrains the conformation of Cys112 (PDB: 4WLW). (C) P. putida CadR coordinates one molecule of Cd+ through Cys112 and Cys119 from one protomer and Cys77’ from the other protomer (PDB: 6JGX). (D) B. megaterium MerR coordinates one molecule of Hg2+ through Cys117 and Cys126 from one protomer and Cys82’ from the other protomer (PDB: 5CRL). (E) E. coli ZntR coordinates two molecules of Zn+ through Cys114, Cys115, H119, and Cys124 from one protomer, Cys79’ from the other protomer, and a phosphate group (PDB: 1Q08). (F) E. coli SoxR coordinates the [2Fe-2S] cluster through residues Cys119, Cys 122, Cys 124, and Cys 130 of the same protomer. (G) Structure superimposition of the ligand-binding domains of four multidrug-resistance MerR TFs, B. subtilis BmrR (cyan; PDB: 3IAO), E. coli EcmrR (green; PDB: 6XLA), P. aeruginosa BrlR (blue; PDB: 5XBT), and E. coli SbmC (light brown; PDB: 1JYH; DNA Gyrase inhibitory protein, Gyrl). (H-J) Detailed presentation of the ligand-binding pocket of B. subtilis BmrR occupied by puromycin (PDB: 3Q3D), ethidium (PDB: 3Q2Y), and kanamycin (PDB: 3Q5R). Residues V147 and I255 server as a hydrophobic pincer. Residues Y152, Y170, Y187, F224, Y229, and Y268 server as an aromatic ring to accommodate drugs with distinct chemical structures. Residue E253 makes auxiliary polar interactions with the drugs.
The multidrug-resistance MerR TFs have a Gyrl-like ligand-binding domain (LBD) at their C-terminal domain that is much larger than the metal-binding loop of the metal- and redox-responsive members of the family ( Figure 4G) [119]. Structure superimposition of high-resolution crystal structures of ligand-bound multidrug-resistance MerR proteins reveals that ligands are recognized by a small and rigid pocket of LBD nearby the DNA-binding domain ( Figure 4H–J) [ 87, 93, 110, 120]. The pocket is surrounded by two aliphatic residues functioning as a hydrophobic pincer pair to anchor drugs, a set of aromatic residues allowing docking of drugs with distinct chemical structures, and a trio of acidic residues making auxiliary H-bonds with polar moiety of drugs ( Figure 4H–J) [120]. Besides the common drug-binding pocket in all multidrug-resistance subfamily of MerR proteins, additional pockets were identified in P. aeruginosa BlrR and E. coli EcmrR [ 93, 110].
The Signal-induced Conformational Change
Most MerR-family TFs interact with DNA both in the absence and in the presence of ligands, while the state of ligand occupancy defines the shape of dsDNA and determines the outcome of transcription [ 60, 72, 121, 122]. Recently reported crystal structures of MerR-TF/DNA complex in the activated state all revealed a highly distorted DNA, suggesting a unified mechanism of transcription activation [ 51, 64, 73, 76, 87]. The crystal structures of apo-CueR/DNA (repressive complex) and Ag +-CueR/DNA (activated complex) determined by O’ Halloran’s lab provided excellent opportunity to understand the conformational change induced by ligand binding [64].
The structures show that the C-terminal metal binding loop of apo CueR is disordered but becomes folded upon Ag + binding. The folded metal binding loop establishes new interaction with nearby structure units and transmits the ligand-biding signal to the distance change of two DBDs. The refolded metal binding loop wedges into the interface of the nearby dimerization helix and DBD. Such event on one hand causes a slight inward rotation of the nearby DBD, and on the other hand causes a small ‘scissors’ movement of dimerization helix resulting in further inward rotation of DBD at the other end. Because both the half palindromic repeats of DNA are tightly anchored by DBD, the conformational change of DBD forces kinking and under twisting of their associated DNA. Other metal-responsive MerR-family TFs likely use the same strategy to trigger the allosteric movement upon metal binding. However, it is still to be determined how the redox-responsive MerR-family TFs, such as SoxR that coordinates the [2Fe-2S] metal cluster and adopts ordered conformation in both the oxidative and reduced conditions, interchanging their conformations [ 73, 101].
The signal-induced conformational change is less determined for multidrug-resistance MerR TFs, as structural information for direct comparison of TF/DNA complexes in the absence and presence of ligand is unavailable. The crystal structures of drug-bound BmrR-DNA complexes and cryo-EM structures of E. coli EcmrR-RPo revealed direct interaction between DBD and LBD, suggesting that drug binding might affect the LBD-DBD interaction leading to DNA distortion through an unknown signal transmission manner [ 87, 110].
Transcription Repression by MerR-family TFs
The gene promoters regulated by MerR-family TFs typically possess abnormally long spacers (19–20 bp) between the –35 and –10 elements compared with the optimal length (17±1 bp) ( Figure 3A) [7]. The unique long spacer is essential for regulation, as shortening the spacer renders promoters irresponsible to activation by their cognate MerR TFs [ 123– 125]. The strict requirement of the –35/–10 spacer length (17±1 bp) is defined by the distance of σ 4 and σ 2, which are anchored near the RNA exit channel and the top of RNAP main cleft, respectively [ 18, 19]. The unique structure architecture of RNAP-σ 70 holoenzyme allows sequential recognition of the –35 and –10 elements of bacterial gene promoters containing optimal spacer length [30]. Increase of spacer length in 2–3 bp results in increase of the –35/–10 element distance in 6.8–10.2 Å and rotation in 72°–108° around the helix axis [64]. Consequently, the –10 element of promoter DNA, when its –35 element is bound by σ 4, is extended and rotated away from the σ 2 domain, preventing further DNA unwinding and resulting in a very low basal transcription activity.
Binding of apo MerR-family TFs further inhibits the weak basal transcription activity of their regulated promoter DNA [ 64, 65]. The crystal structure of apo CueR-DNA shows that the engagement of apo CueR shifts the trajectory of promoter dsDNA further away from the σ 2 domain and restrains the dsDNA in the inactive shape [64]. Although the apo CueR-dsDNA is the only reported crystal structure of apo MerR-TF/DNA, footprinting data support that other metal-responsive MerR TFs probably also interact with a straight dsDNA in the absence of ligand binding [ 121, 126].
Transcription Activation by MerR-family TFs
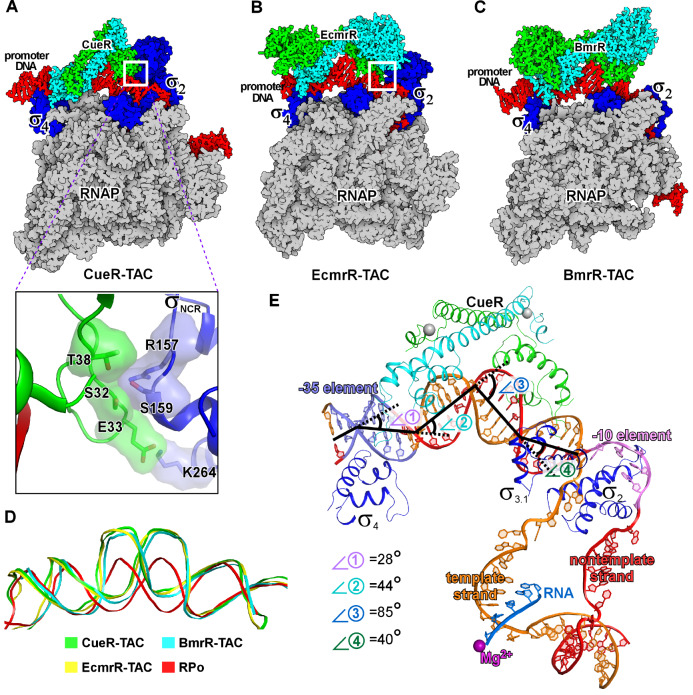
The long-term debate regarding the transcription activation mechanism of MerR-family TFs is whether and what extent the MerR TF-RNAP interaction contributes to transcription activation [ 7, 121, 127]. The canonical Class I and Class II transcription activators bridge RNAP and promoter DNA by making bipartite interaction; the interaction with RNAP is necessary as mutating the activator-RNAP interface substantially reduces transcription activation activity [37]. However, the concept of requirement of TF-RNAP interaction for transcription activation is challenged by recent cryo-EM structures of transcription activation complexes comprising MerR-family TFs, E. coli CueR-TAC, B. subtilis BmrR-TAC, and E. coli MrR-TAC ( Figure 5A–C) [ 65, 88, 110, 128].
Figure5 .
The cryo-EM structures of transcription activation complexes comprising MerR-family TFsThe cryo-EM structures of (A) E. coli CueR transcription activation complex (PDB: 6XH7), (B) E. coli EcmrR transcription activation complex (PDB: 6XL5), and (C) B. subtilis BmrR transcription activation complex (PDB: 7CKQ). The insertion box shows the small surface patch of CueR-DBD that interacts with the σNCR. (D) Structure superimposition of the upstream dsDNAs of the above three transcription activation complexes and that of a bacterial RPo (PDB: 6OUL). (E) The kinks of the upstream promoter DNA at positions −35 (⦟1, 28°), −30 (⦟2, 44°), −24 (⦟3, 85°) and −18 (⦟4, 40°) in the cryo-EM structure of E. coli CueR-TAC (PDB: 6LDI). Kink 1 at −35 is induced by CueR and σ704, and kinks 2, 3 and 4 are induced by the CueR dimer.
In these structures, the MerR-family transcription factors reside on one face of the upstream promoter DNA, while the RNAP-σ 70 (or RNAP-σ ) holoenzyme recognizes the -35 and -10 elements from the opposite face. The architecture explains the paradox that MerR-TFs activate transcription while occupying the core promoter region, an interaction typically represses transcription due to steric hinderance between TFs and RNAP. Intriguingly, although RNAP and MerR-TF occupy the same core promoter regions, BmrR makes interaction with neither RNAP core enzyme nor σ A factor in the cryo-EM structure of B. subtilis BmrR-TAC [88], while E. coli CueR and EcmrR only contact a small surface patch of the non-conserved region of σ 70 factor (σ NCR) in the cryo-EM structures of E. coli CueR-TAC and E. coli EcmrR-TAC ( Figure 5A) [110]. The TF-σ NCR interaction is not essential for their transcription activation activity, because MerR TFs (CueR and ZntR) in E. coli are able to activate transcription of their regulated promoters using evolutionarily distant RNAPs ( i. e. RNAPs from M. tuberculosis, T. thermophilus, and S. coelicolor) that have completely different σ NCR domains [65]. The TF-σ NCR interactions observed in E. coli CueR-TAC and EcmrR-TAC, however, contribute to the transcription activation by providing auxiliary bridge interaction between RNAP and DNA [ 110, 128].
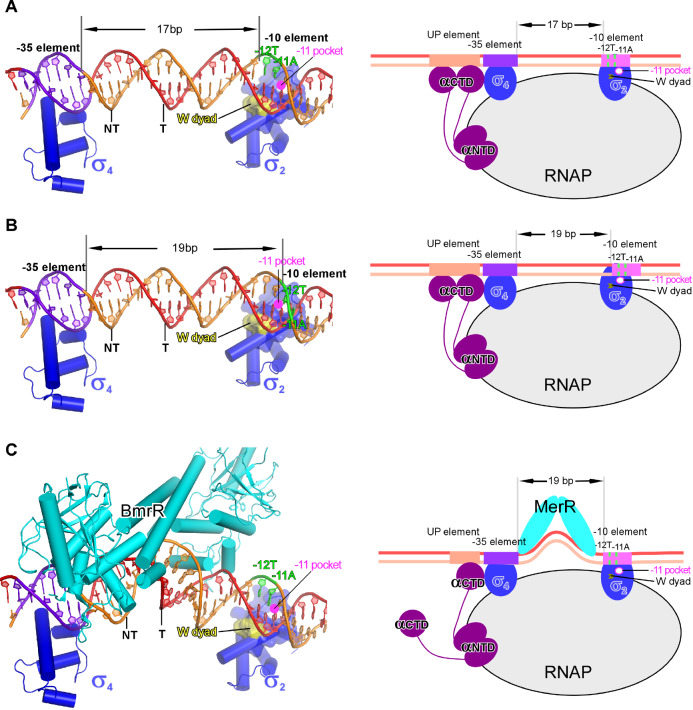
Several lines of evidence point to a ‘DNA distortion’ mechanism of transcription activation by the MerR-family TFs. The DNA distortion induced by ligand-bound MerR was initially proposed based on results of DNA mobility and footprinting assays [126], and subsequently directly visualized in crystal structures of ligand-bound MerR-family TFs complexed with DNA, including crystal structures of CueR-Ag +/DNA, CadR-Cd 2+/DNA, SoxR-[2Fe-2S]/DNA, and NmlR-Ni 2+/DNA [ 51, 64, 73, 76, 87]. The most striking feature of the distorted DNA is the 90° kink at the center of the palindromic dyad, where the two central base pair steps are often broken and the minor groove becomes significantly wider even than the canonical major groove, resulting in a A-DNA-like structure ( Figure 5D). The distal ends of their cognate dsDNA are still gripped by the wHTH domains, resulting in a ‘Ω’-like shape of the dsDNA ( Figure 5D,E). Such conformational switch of dsDNA results in pronounced changes both in the distance and in the phase angle between the -35 and -10 elements. Detailed comparison between the canonical B-formed dsDNA and activator-bound dsDNA revealed that the DNA distortion shortens the –35/–10 distance by ~7Å and reduces the phase angles of dsDNA by 72°, close to those of a 17-bp spacer promoter ( Figure 6) [64].
Figure6 .
The DNA-distortion mechanism of transcription activation by MerR-family TFs(A) The –35 and –10 elements are properly aligned on the protein surface of σ4 and σ2. The –11A of the nontemplate strand DNA is close to the –11 pocket. The panel figure is prepared from a B. subtilis RPc model comprising a promoter DNA with 17-bp –35/–10 spacer. (B) The –11A of the nontemplate strand DNA is rotated away from the –11 pocket in the B. subtilis RPc model comprising a promoter DNA with 19-bp –35/–10 spacer. (C) The BmrR-induced central kink realigns the −35 and −10 elements to a proper space and phase for simultaneous engagement by σ4 and σ2 according to the structure model of a BmrR-RPc complex. Yellow surfaces (left) and arrows (right) show the tryptophan dyad; pink circles show the –11 pocket.
In summary, the structural and the biochemical evidence provides strong support for the DNA distortion paradigm of allosteric transcriptional control by the MerR-family TFs. Such mode of transcription activation doesn’t necessitate the RNAP-TF interaction, and thus is distinct from the transcription activation mechanism of canonical Class I and Class II transcription activation modes.
Conclusion
This review discussed the studies of the past years, regarding the unique mechanism of transcription activation of the MerR-family TFs with focus on the structural and biochemical data. Intriguingly, a conceptional similar DNA-distortion mechanism has also been employed by the eukaryotic TATA-box binding protein (TBP) during eukaryotic polymerase II transcription initiation. A recent study in Seok’s lab identified FruR, a GalR-LacI family transcription factor in V. cholerae, which binds to its cis element located between the -35 and -10 element with a 20-bp spacer and likely uses a similar DNA-distortion mechanism to regulate the expressions of its target genes [129]. These discoveries suggest that such DNA-distortion mechanism might be more widely employed by eukaryotic and prokaryotic transcription factors than we expected. The unique RNAP-contact-independent action, the ultra-sensitivity and selectivity towards cognate ligands, and the stringent transcription regulation make the MerR-family TFs ideal transcription modules in synthetic biology uses.
Acknowledgments
The authors apologize to colleagues whose work could not be cited due to the scope and space limits.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
The work was supported by the grants from the National Key Research and Development Program of China (No. 2018YFA0900701), the Strategic Priority Research Program of CAS (No. XDB29020000), the National Natural Science Foundation of China (No. 31822001), and the Shanghai Science and Technology Innovation Program (No. 19JC1415900).
References
- 1.Roeder RG, Rutter WJ. Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature. . 1969;224:234–237. doi: 10.1038/224234a0. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlin M, Berg P. Deoxyribo ucleic acid-directed synthesis of ribonucleic acid by an enzyme from Escherichia coli . Proc Natl Acad Sci U S A. . 1962;48:81–94. doi: 10.1073/pnas.48.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feklistov A, Sharon BD, Darst SA, Gross CA. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol. . 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- 4.Seshasayee AS, Sivaraman K, Luscombe NM. An overview of prokaryotic transcription factors: a summary of function and occurrence in bacterial genomes. Subcell Biochem. 2011,52: 7-23. [DOI] [PubMed]
- 5.Browning DF, Busby SJW. Local and global regulation of transcription initiation in bacteria. Nat Rev Microbiol. . 2016;14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- 6.Browning DF, Butala M, Busby SJW. Bacterial transcription factors: regulation by pick "N" mix. J Mol Biol. . 2019;431:4067–4077. doi: 10.1016/j.jmb.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The merr family of transcriptional regulators. FEMS Microbiol Rev. . 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 8.Baksh KA, Zamble DB. Allosteric control of metal-responsive transcriptional regulators in bacteria. J Biol Chem. . 2020;295:1673–1684. doi: 10.1074/jbc.REV119.011444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Boyaci H, Campbell EA. Diverse and unified mechanisms of transcription initiation in bacteria. Nat Rev Microbiol. . 2021;19:95–109. doi: 10.1038/s41579-020-00450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts JW. Mechanisms of bacterial transcription termination. J Mol Biol. . 2019;431:4030–4039. doi: 10.1016/j.jmb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Busby SJW. Transcription activation in bacteria: ancient and modern. Microbiology. . 2019;165:386–395. doi: 10.1099/mic.0.000783. [DOI] [PubMed] [Google Scholar]
- 12.Mazumder A, Kapanidis AN. Recent advances in understanding sigma70-dependent transcription initiation mechanisms. J Mol Biol. . 2019;431:3947–3959. doi: 10.1016/j.jmb.2019.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Danson AE, Jovanovic M, Buck M, Zhang X. Mechanisms of sigma(54)-dependent transcription initiation and regulation. J Mol Biol. . 2019;431:3960–3974. doi: 10.1016/j.jmb.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang JY, Mishanina TV, Landick R, Darst SA. Mechanisms of transcriptional pausing in bacteria. J Mol Biol. . 2019;431:4007–4029. doi: 10.1016/j.jmb.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belogurov GA, Artsimovitch I. The mechanisms of substrate selection, catalysis, and translocation by the elongating RNA polymerase. J Mol Biol. . 2019;431:3975–4006. doi: 10.1016/j.jmb.2019.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebright RH. Rna polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. . 2000;304:687–698. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 17.Weiss A, Shaw LN. Small things considered: the small accessory subunits of rna polymerase in gram-positive bacteria. FEMS MicroBiol Rev. . 2015;39:541–554. doi: 10.1093/femsre/fuv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. . 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 19.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. . 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 20.Lin W, Mandal S, Degen D, Cho MS, Feng Y, Das K, Ebright RH. Structural basis of ECF-σ-factor-dependent transcription initiation. Nat Commun. . 2019;10:710. doi: 10.1038/s41467-019-08443-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Fang C, Zhuang N, Wang T, Zhang Y. Structural basis for transcription initiation by bacterial ECF σ factors. Nat Commun. . 2019;10:1153. doi: 10.1038/s41467-019-09096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang C, Li L, Shen L, Shi J, Wang S, Feng Y, Zhang Y. Structures and mechanism of transcription initiation by bacterial ecf factors. Nucleic Acids Res. . 2019;47:7094–7104. doi: 10.1093/nar/gkz470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Darbari VC, Zhang N, Lu D, Glyde R, Wang YP, Winkelman JT, et al. Structures of the RNA polymerase-σ 54 reveal new and conserved regulatory strategies . Science. . 2015;349:882–885. doi: 10.1126/science.aab1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife. . 2015;4:e08504. doi: 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, et al. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol Cell. . 2002;9:527–539. doi: 10.1016/S1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 26.Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. . 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. . 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. . 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble . Mol Cell. . 2015;58:534–540. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Chiu C, Gopalkrishnan S, Chen AY, Olinares PDB, Saecker RM, Winkelman JT, et al. Stepwise promoter melting by bacterial rna polymerase. Mol Cell. . 2020;78:275–288.. doi: 10.1016/j.molcel.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feklistov A, Bae B, Hauver J, Lass-Napiorkowska A, Kalesse M, Glaus F, Altmann KH, et al. RNA polymerase motions during promoter melting. Science. . 2017;356:863–866. doi: 10.1126/science.aam7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin Y, Qayyum MZ, Pupov D, Esyunina D, Kulbachinskiy A, Murakami KS. Structural basis of ribosomal RNA transcription regulation. Nat Commun. . 2021;12:528. doi: 10.1038/s41467-020-20776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skalenko KS, Li L, Zhang Y, Vvedenskaya IO, Winkelman JT, Cope AL, Taylor DM, et al. Promoter-sequence determinants and structural basis of primer-dependent transcription initiation in Escherichia coli . Proc Natl Acad Sci U S A. . 2021;118:e2106388118. doi: 10.1073/pnas.2106388118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernández-Tornero C, Kulbachinskiy A, et al. Structural basis of transcription initiation by bacterial rna polymerase holoenzyme. J Biol Chem. . 2014;289:24549–24559. doi: 10.1074/jbc.M114.584037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Degen D, Ho MX, Sineva E, Ebright KY, Ebright YW, Mekler V, et al. GE23077 binds to the RNA polymerase ‘i’ and ‘i+1’ sites and prevents the binding of initiating nucleotides. eLife. . 2014;3:e02450. doi: 10.7554/eLife.02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DJ, Minchin SD, Busby SJ. Activating transcription in bacteria. Annu Rev Microbiol. . 2012;66:125–152. doi: 10.1146/annurev-micro-092611-150012. [DOI] [PubMed] [Google Scholar]
- 37.Busby S, Ebright RH. Transcription activation by catabolite activator protein (cap) J Mol Biol. . 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 38.Gaston K, Bell A, Kolb A, Buc H, Busby S. Stringent spacing requirements for transcription activation by crp. Cell. . 1990;62:733–743. doi: 10.1016/0092-8674(90)90118-X. [DOI] [PubMed] [Google Scholar]
- 39.Hudson BP, Quispe J, Lara-González S, Kim Y, Berman HM, Arnold E, Ebright RH, et al. Three-dimensional em structure of an intact activator-dependent transcription initiation complex. Proc Natl Acad Sci U S A. . 2009;106:19830–19835. doi: 10.1073/pnas.0908782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu B, Hong C, Huang RK, Yu Z, Steitz TA. Structural basis of bacterial transcription activation. Science. . 2017;358:947–951. doi: 10.1126/science.aao1923. [DOI] [PubMed] [Google Scholar]
- 41.Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class ii cap-dependent promoters: two interactions between cap and rna polymerase. Cell. . 1996;87:1123–1134. doi: 10.1016/S0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y, Zhang Y, Ebright RH. Structural basis of transcription activation. Science. . 2016;352:1330–1333. doi: 10.1126/science.aaf4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown NL, Ford SJ, Pridmore RD, Fritzinger DC. Deoxyribonucleic acid sequence of a gene from the Pseudomonas transposon TN501 encoding mercuric reductase. Biochemistry. . 1983;22:4089–4095. doi: 10.1021/bi00286a015. [DOI] [PubMed] [Google Scholar]
- 44.Barrineau P, Gilbert P, Jackson WJ, Jones CS, Summers AO, Wisdom S. The DNA sequence of the mercury resistance operon of the incfii plasmid nr1. J Mol Appl Genet 1984, 2: 601-619 . [PubMed]
- 45.Barrineau P, Gilbert P, Jackson WJ, Jones CS, Summers AO, Wisdom S. The structure of the mer operon. Basic Life Sci. 1985, 30: 707-718 . [DOI] [PubMed]
- 46.Brown NL, Misra TK, Winnie JN, Schmidt A, Seiff M, Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid r100 and transposon tn501: Further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. . 1986;202:143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- 47.Boyd ES, Barkay T. The mercury resistance operon: from an origin in a geothermal environment to an efficient detoxification machine. Front Microbio. . 2012;3:1–13. doi: 10.3389/fmicb.2012.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borremans B, Hobman JL, Provoost A, Brown NL, van Der Lelie D. Cloning and Functional Analysis of the pbr Lead Resistance Determinant of Ralstonia metallidurans CH34 . J Bacteriol. . 2001;183:5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, Liu X, Wang D, Chen W, Hu Q, Wei T, Zhou W, et al. Structural basis for the selective pb(ii) recognition of metalloregulatory protein pbrr691. Inorg Chem. . 2016;55:12516–12519. doi: 10.1021/acs.inorgchem.6b02397. [DOI] [PubMed] [Google Scholar]
- 50.Lee SW, Glickmann E, Cooksey DA. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator . Appl Environ Microbiol. . 2001;67:1437–1444. doi: 10.1128/AEM.67.4.1437-1444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu X, Hu Q, Yang J, Huang S, Wei T, Chen W, He Y, et al. Selective cadmium regulation mediated by a cooperative binding mechanism in cadr. Proc Natl Acad Sci U S A. . 2019;116:20398–20403. doi: 10.1073/pnas.1908610116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mercury(ii)—thiolate chemistry and the mechanism of the heavy metal biosensor merr. Progress in inorganic chemistry1990. p. 323-412
- 53.Wright JG, Tsang HT, Penner-Hahn JE, O′Halloran TV. Coordination chemistry of the hg-merr metalloregulatory protein: evidence for a novel tridentate mercury-cysteine receptor site. J Am Chem Soc. . 1990;112:2434–2435. doi: 10.1021/ja00162a062. [DOI] [Google Scholar]
- 54.Heltzel A, Gambill D, Jackson WJ, Totis PA, Summers AO. Overexpression and DNA-binding properties of the mer-encoded regulatory protein from plasmid nr1 (tn21) J Bacteriol. . 1987;169:3379–3384. doi: 10.1128/jb.169.7.3379-3384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee IW, Livrelli V, Park SJ, Totis PA, Summers AO. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. Ii. Repressor/activator (merr)-rna polymerase interaction with merop mutants. J Biol Chem. . 1993;268:2632–2639. doi: 10.1016/S0021-9258(18)53821-7. [DOI] [PubMed] [Google Scholar]
- 56.Helmann JD, Wang Y, Mahler I, Walsh CT. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. . 1989;171:222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lund PA, Brown NL. Regulation of transcription in Escherichia coli from the mer and merr promoters in the transposon tn501 . J Mol Biol. . 1989;205:343–353. doi: 10.1016/0022-2836(89)90345-8. [DOI] [PubMed] [Google Scholar]
- 58.Ni′Bhriain NN, Silver S, Foster TJ. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid r100. J Bacteriol. . 1983;155:690–703. doi: 10.1128/JB.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. Zntr is a Zn(ii)-responsive merr-like transcriptional regulator of znta in Escherichia coli . Mol Microbiol. . 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 60.Outten CE, Outten FW, O′Halloran TV. DNA distortion mechanism for transcriptional activation by Zntr, a Zn(ii)-responsive merr homologue in Escherichia coli . J Biol Chem. . 1999;274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- 61.Khan S, Brocklehurst KR, Jones GW, Morby AP. The functional analysis of directed amino-acid alterations in zntr from Escherichia coli . Biochem Biophysl Res Commun. . 2002;299:438–445. doi: 10.1016/S0006-291X(02)02660-8. [DOI] [PubMed] [Google Scholar]
- 62.Pruteanu M, Neher SB, Baker TA. Ligand-controlled proteolysis of the Escherichia coli transcriptional regulator ZntR . J Bacteriol. . 2007;189:3017–3025. doi: 10.1128/JB.01531-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoyanov JV, Hobman JL, Brown NL. Cuer (ybbi) of Escherichia coli is a merr family regulator controlling expression of the copper exporter copa . Mol Microbiol. . 2001;39:502–512. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- 64.Philips SJ, Canalizo-Hernandez M, Yildirim I, Schatz GC, Mondragón A, O′Halloran TV. Allosteric transcriptional regulation via changes in the overall topology of the core promoter. Science. . 2015;349:877–881. doi: 10.1126/science.aaa9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fang C, Philips SJ, Wu X, Chen K, Shi J, Shen L, Xu J, et al. Cuer activates transcription through a DNA distortion mechanism. Nat Chem Biol. . 2021;17:57–64. doi: 10.1038/s41589-020-00653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noll M, Petrukhin K, Lutsenko S. Identification of a novel transcription regulator from proteus mirabilis, pmtr, revealed a possible role of yjai protein in balancing zinc in Escherichia coli . J Biol Chem. . 1998;273:21393–21401. doi: 10.1074/jbc.273.33.21393. [DOI] [PubMed] [Google Scholar]
- 67.Julian DJ, Kershaw CJ, Brown NL, Hobman JL. Transcriptional activation of merr family promoters in cupriavidus metallidurans ch34. Antonie van Leeuwenhoek. . 2009;96:149–159. doi: 10.1007/s10482-008-9293-4. [DOI] [PubMed] [Google Scholar]
- 68.Kidd SP, Brown NL. Zccr—a merr-like regulator from bordetella pertussis which responds to zinc, cadmium, and cobalt. Biochem Biophys Res Commun. . 2003;302:697–702. doi: 10.1016/S0006-291X(03)00249-3. [DOI] [PubMed] [Google Scholar]
- 69.Rutherford JC, Cavet JS, Robinson NJ. Cobalt-dependent transcriptional switching by a dual-effector merr-like protein regulates a cobalt-exporting variant cpx-type atpase. J Biol Chem. . 1999;274:25827–25832. doi: 10.1074/jbc.274.36.25827. [DOI] [PubMed] [Google Scholar]
- 70.Kidd SP, Djoko KY, Ng JQ, Argente MP, Jennings MP, McEwan AG. A novel nickel responsive MerR-like regulator, NIMR, from haemophilus influenzae. Metallomics. . 2011;3:1009–1018. doi: 10.1039/c1mt00127b. [DOI] [PubMed] [Google Scholar]
- 71.Amábile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli : two genes control a superoxide stress regulon . Nucleic Acids Res. . 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hidalgo E, Leautaud V, Demple B. The redox-regulated soxr protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. . 1998;17:2629–2636. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe S, Kita A, Kobayashi K, Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor soxr bound to DNA. Proc Natl Acad Sci U S A. . 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bradley TM, Hidalgo E, Leautaud V, Ding H, Demple B. Cysteine-to-alanine replacements in the Escherichia coli soxr protein and the role of the [2Fe-2S] centers in transcriptional activation . Nucleic Acids Res. . 1997;25:1469–1475. doi: 10.1093/nar/25.8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kidd SP, Potter AJ, Apicella MA, Jennings MP, McEwan AG. NmlR of Neisseria gonorrhoeae : a novel redox responsive transcription factor from the MerR family . Mol MicroBiol. . 2005;57:1676–1689. doi: 10.1111/j.1365-2958.2005.04773.x. [DOI] [PubMed] [Google Scholar]
- 76.Couñago RM, Chen NH, Chang CW, Djoko KY, McEwan AG, Kobe B. Structural basis of thiol-based regulation of formaldehyde detoxification in H. influenzae by a MerR regulator with no sensor region . Nucleic Acids Res. . 2016;44:6981–6993. doi: 10.1093/nar/gkw543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Holmes DJ, Caso JL, Thompson CJ. Autogenous transcriptional activation of a thiostrepton-induced gene in streptomyces lividans. EMBO J. . 1993;12:3183–3191. doi: 10.1002/j.1460-2075.1993.tb05987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiu ML, Folcher M, Griffin P, Holt T, Klatt T, Thompson CJ. Characterization of the covalent binding of thiostrepton to a thiostrepton-induced protein from Streptomyces lividans . Biochemistry. . 1996;35:2332–2341. doi: 10.1021/bi952073e. [DOI] [PubMed] [Google Scholar]
- 79.Chiu ML, Folcher M, Katoh T, Puglia AM, Vohradsky J, Yun BS, Seto H, et al. Broad spectrum thiopeptide recognition specificity of the streptomyces lividans tipal protein and its role in regulating gene expression. J Biol Chem. . 1999;274:20578–20586. doi: 10.1074/jbc.274.29.20578. [DOI] [PubMed] [Google Scholar]
- 80.Myers CL, Harris J, Yeung JCK, Honek JF. Molecular interactions between thiostrepton and the TipAS protein from Streptomyces lividans . ChemBioChem. . 2014;15:681–687. doi: 10.1002/cbic.201300724. [DOI] [PubMed] [Google Scholar]
- 81.Garcia M, Dunlap J, Loh J, Stacey G. Phenotypic characterization and regulation of the nolA gene of Bradyrhizobium japonicum . Mol Plant Microbe Interact. . 1996;9:625–636. doi: 10.1094/MPMI-9-0625. [DOI] [PubMed] [Google Scholar]
- 82.Sadowsky MJ, Cregan PB, Gottfert M, Sharma A, Gerhold D, Rodriguez-Quinones F, Keyser HH, et al. The Bradyrhizobium japonicum nola gene and its involvement in the genotype-specific nodulation of soybeans . Proc Natl Acad Sci U S A. . 1991;88:637–641. doi: 10.1073/pnas.88.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loh J, Stacey G. Nodulation gene regulation in Bradyrhizobium japonicum : a unique integration of global regulatory circuits . Appl Environ Microbiol. . 2003;69:10–17. doi: 10.1128/AEM.69.1.10-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ahmed M, Borsch CM, Taylor SS, Vázquez-Laslop N, Neyfakh AA. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. . 1994;269:28506–28513. doi: 10.1016/S0021-9258(18)46956-6. [DOI] [PubMed] [Google Scholar]
- 85.Ahmed M, Lyass L, Markham PN, Taylor SS, Vázquez-Laslop N, Neyfakh AA. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated . J Bacteriol. . 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baranova NN, Danchin A, Neyfakh AA. Mta, a global merr-type regulator of the bacillus subtilis multidrug-efflux transporters. Mol Microbiol. . 1999;31:1549–1559. doi: 10.1046/j.1365-2958.1999.01301.x. [DOI] [PubMed] [Google Scholar]
- 87.Heldwein EEZ, Brennan RG. Crystal structure of the transcription activator bmrr bound to DNA and a drug. Nature. . 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- 88.Fang C, Li L, Zhao Y, Wu X, Philips SJ, You L, Zhong M, et al. The bacterial multidrug resistance regulator bmrr distorts promoter DNA to activate transcription. Nat Commun. . 2020;11:6284. doi: 10.1038/s41467-020-20134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Newberry KJ, Brennan RG. The structural mechanism for transcription activation by merr family member multidrug transporter activation, n terminus. J Biol Chem. . 2004;279:20356–20362. doi: 10.1074/jbc.M400960200. [DOI] [PubMed] [Google Scholar]
- 90.Chambers JR, Liao J, Schurr MJ, Sauer K. BrlR from P seudomonas aeruginosa is a c-di-GMP-responsive transcription factor . Mol MicroBiol. . 2014;92:471–487. doi: 10.1111/mmi.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liao J, Sauer K. The merr-like transcriptional regulator BRLR contributes to pseudomonas aeruginosa biofilm tolerance. J Bacteriol. . 2012;194:4823–4836. doi: 10.1128/JB.00765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao J, Schurr MJ, Sauer K. The merr-like regulator brlr confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms . J Bacteriol. . 2013;195:3352–3363. doi: 10.1128/JB.00318-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang F, He Q, Yin J, Xu S, Hu W, Gu L. Brlr from Pseudomonas aeruginosa is a receptor for both cyclic DI-gmp and pyocyanin . Nat Commun. . 2018;9:2563. doi: 10.1038/s41467-018-05004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gupta K, Marques CNH, Petrova OE, Sauer K. Antimicrobial tolerance of Pseudomonas aeruginosa biofilms is activated during an early developmental stage and requires the two-component hybrid sags . J Bacteriol. . 2013;195:4975–4987. doi: 10.1128/JB.00732-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Singh S, Sevalkar RR, Sarkar D, Karthikeyan S. Characteristics of the essential pathogenicity factor Rv1828, a MerR family transcription regulator from Mycobacterium tuberculosis . FEBS J. . 2018;285:4424–4444. doi: 10.1111/febs.14676. [DOI] [PubMed] [Google Scholar]
- 96.Gomez RL, Jose L, Ramachandran R, Raghunandanan S, Muralikrishnan B, Johnson JB, Sivakumar KC, et al. The multiple stress responsive transcriptional regulator Rv3334 of Mycobacterium tuberculosis is an autorepressor and a positive regulator of kstR . FEBS J. . 2016;283:3056–3071. doi: 10.1111/febs.13791. [DOI] [PubMed] [Google Scholar]
- 97.Outten FW, Outten CE, Hale J, O′Halloran TV. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR . J Biol Chem. . 2000;275:31024–31029. doi: 10.1074/jbc.M006508200. [DOI] [PubMed] [Google Scholar]
- 98.Rademacher C, Masepohl B. Copper-responsive gene regulation in bacteria. Microbiology. . 2012;158:2451–2464. doi: 10.1099/mic.0.058487-0. [DOI] [PubMed] [Google Scholar]
- 99.Singh VK, Xiong A, Usgaard TR, Chakrabarti S, Deora R, Misra TK, Jayaswal RK. Zntr is an autoregulatory protein and negatively regulates the chromosomal zinc resistance operon znt of staphylococcus aureus. Mol Microbiol. . 1999;33:200–207. doi: 10.1046/j.1365-2958.1999.01466.x. [DOI] [PubMed] [Google Scholar]
- 100.Storz G, Imlayt JA. Oxidative stress. Curr Opin MicroBiol. . 1999;2:188–194. doi: 10.1016/S1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 101.Pomposiello PJ, Demple B. Redox-operated genetic switches: the soxr and oxyr transcription factors. Trends Biotechnol. . 2001;19:109–114. doi: 10.1016/S0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen TT, Eiamphungporn W, Mäder U, Liebeke M, Lalk M, Hecker M, Helmann JD, et al. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR) . Mol MicroBiol. . 2009;71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hidalgo E, Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing soxr protein. EMBO J. . 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fujikawa M, Kobayashi K, Kozawa T. Direct oxidation of the [2Fe-2S] cluster in soxr protein by superoxide. J Biol Chem. . 2012;287:35702–35708. doi: 10.1074/jbc.M112.395079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kobayashi K, Fujikawa M, Kozawa T. Oxidative stress sensing by the iron–sulfur cluster in the transcription factor, SoxR. J InOrg Biochem. . 2014;133:87–91. doi: 10.1016/j.jinorgbio.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 106.Stroeher UH, Kidd SP, Stafford SL, Jennings MP, Paton JC, McEwan AG. A pneumococcal merr-like regulator and s-nitrosoglutathione reductase are required for systemic virulence. J Infect Dis. . 2007;196:1820–1826. doi: 10.1086/523107. [DOI] [PubMed] [Google Scholar]
- 107.Chen NH, Couñago RM, Djoko KY, Jennings MP, Apicella MA, Kobe B, McEwan AG. A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of neisseria meningitidis in biofilms. Antioxid Redox Signal. . 2013;18:743–755. doi: 10.1089/ars.2012.4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woolridge DP, Vazquez-Laslop N, Markham PN, Chevalier MS, Gerner EW, Neyfakh AA. Efflux of the natural polyamine spermidine facilitated by the Bacillus subtilis multidrug transporter blt . J Biol Chem. . 1997;272:8864–8866. doi: 10.1074/jbc.272.14.8864. [DOI] [PubMed] [Google Scholar]
- 109.Poudyal B, Sauer K. The ABC of biofilm drug tolerance: the merr-like regulator brlr is an activator of ABC transport systems, with pa1874-77 contributing to the tolerance of pseudomonas aeruginosa biofilms to tobramycin. Antimicrob Agents Chemother. . 2018;62:17–17. doi: 10.1128/AAC.01981-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang Y, Liu C, Zhou W, Shi W, Chen M, Zhang B, Schatz DG, et al. Structural visualization of transcription activated by a multidrug-sensing merr family regulator. Nat Commun. . 2021;12:2702. doi: 10.1038/s41467-021-22990-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murakami T, Holt TG, Thompson CJ. Thiostrepton-induced gene expression in streptomyces lividans. J Bacteriol. . 1989;171:1459–1466. doi: 10.1128/jb.171.3.1459-1466.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kahmann JD, Sass HJ, Allan MG, Seto H, Thompson CJ, Grzesiek S. Structural basis for antibiotic recognition by the tipa class of multidrug-resistance transcriptional regulators. EMBO J. . 2003;22:1824–1834. doi: 10.1093/emboj/cdg181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ralston DM, O′Halloran TV. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated merr transcription complex. Proc Natl Acad Sci U S A. . 1990;87:3846–3850. doi: 10.1073/pnas.87.10.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O′Halloran TV, Mondragón A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by cuer. Science. . 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 115.Outten CE, O′Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. . 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 116.Hitomi Y, Outten CE, O′Halloran TV. Extreme zinc-binding thermodynamics of the metal sensor/regulator protein, zntr. J Am Chem Soc. . 2001;123:8614–8615. doi: 10.1021/ja016146v. [DOI] [PubMed] [Google Scholar]
- 117.Wang D, Huang S, Liu P, Liu X, He Y, Chen W, Hu Q, et al. Structural analysis of the Hg(ii)-regulatory protein tn501 merr from Pseudomonas aeruginosa . Sci Rep. . 2016;6:33391. doi: 10.1038/srep33391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang CC, Lin LY, Zou XW, Huang CC, Chan NL. Structural basis of the mercury(ii)-mediated conformational switching of the dual-function transcriptional regulator merr. Nucleic Acids Res. . 2015;43:7612–7623. doi: 10.1093/nar/gkv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moreno A, Froehlig JR, Bachas S, Gunio D, Alexander T, Vanya A, Wade H. Solution binding and structural analyses reveal potential multidrug resistance functions for sav2435 and ctr107 and other gyri-like proteins. Biochemistry. . 2016;55:4850–4863. doi: 10.1021/acs.biochem.6b00651. [DOI] [PubMed] [Google Scholar]
- 120.Bachas S, Eginton C, Gunio D, Wade H. Structural contributions to multidrug recognition in the multidrug resistance (mdr) gene regulator, bmrr. Proc Natl Acad Sci U S A. . 2011;108:11046–11051. doi: 10.1073/pnas.1104850108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.O′Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG. The merr heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. . 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- 122.Sameach H, Narunsky A, Azoulay-Ginsburg S, Gevorkyan-Aiapetov L, Zehavi Y, Moskovitz Y, Juven-Gershon T, et al. Structural and dynamics characterization of the merr family metalloregulator cuer in its repression and activation states. Structure. . 2017;25:988–996.e3. doi: 10.1016/j.str.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 123.Parkhill J, Brown NL. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn 501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. . 1990;18:5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hidalgo E, Demple B. Spacing of promoter elements regulates the basal expression of the soxs gene and converts soxr from a transcriptional activator into a repressor. EMBO J. . 1997;16:1056–1065. doi: 10.1093/emboj/16.5.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lund P, Brown N. Up-promoter mutations in the positively-regulated mer promoter of TnSOl . Nucleic Acids Res. . 1989;17:5517–5528. doi: 10.1093/nar/17.14.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ansari AZ, Bradner JE, O′Halloran TV. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. . 1995;374:370–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 127.Kulkarni RD, Summers AO. MerR cross-links to the α, β, and σ 70 subunits of RNA polymerase in the preinitiation complex at the merTPCAD promoter . Biochemistry. . 1999;38:3362–3368. doi: 10.1021/bi982814m. [DOI] [PubMed] [Google Scholar]
- 128.Shi W, Zhang B, Jiang Y, Liu C, Zhou W, Chen M, Yang Y, et al. Structural basis of copper-efflux-regulator-dependent transcription activation. iScience. . 2021;24:102449. doi: 10.1016/j.isci.2021.102449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoon CK, Kang D, Kim MK, Seok YJ. Vibrio cholerae FruR facilitates binding of RNA polymerase to the fru promoter in the presence of fructose 1-phosphate . Nucleic Acids Res. . 2021;49:1397–1410. doi: 10.1093/nar/gkab013. [DOI] [PMC free article] [PubMed] [Google Scholar]