Abstract
The nuclear receptors (NRs) are an evolutionarily related family of transcription factors, which share certain common structural characteristics and regulate the expressions of various genes by recognizing different response elements. NRs play important roles in cell differentiation, proliferation, survival and apoptosis, rendering them indispensable in many physiological activities including growth and metabolism. As a result, dysfunctions of NRs are closely related to a variety of diseases, such as diabetes, obesity, infertility, inflammation, the Alzheimer′s disease, cardiovascular diseases, prostate and breast cancers. Meanwhile, small-molecule drugs directly targeting NRs have been widely used in the treatment of above diseases. Here we summarize recent progress in the structural biology studies of NR family proteins. Compared with the dozens of structures of isolated DNA-binding domains (DBDs) and the striking more than a thousand of structures of isolated ligand-binding domains (LBDs) accumulated in the Protein Data Bank (PDB) over thirty years, by now there are only a small number of multi-domain NR complex structures, which reveal the integration of different NR domains capable of the allosteric signal transduction, or the detailed interactions between NR and various coregulator proteins. On the other hand, the structural information about several orphan NRs is still totally unavailable, hindering the further understanding of their functions. The fast development of new technologies in structural biology will certainly help us gain more comprehensive information of NR structures, inspiring the discovery of novel NR-targeting drugs with a new binding site beyond the classic LBD pockets and/or a new mechanism of action.
Keywords: nuclear receptor, structure, drug target, small-molecule drug
Introduction
The study on nuclear receptors (NRs) became a unique research filed in the middle of 1980s, when the molecular cloning of several hormone receptors revealed that they share a common architecture especially in the domain composition [1]. Due to their critical functions in many physiological processes and direct connections to a variety of human diseases, NRs have long been exploited as therapeutic drug targets. By some estimates, NR ligands constitute about 15%–20% of the small-molecule drugs on the pharmaceutical market worldwide [ 2– 4]. At present, drugs directly targeting different NRs have been widely used in the treatment of various diseases, such as tamoxifen and evista targeting the estrogen receptor (ER) used in breast cancer and osteoporosis respectively [5], casodex targeting the androgen receptor (AR) used in prostate cancer [ 6, 7], targretin targeting the retinoid X receptor (RXR) used in skin cancer [ 8, 9], and glitazones targeting the peroxisome proliferator-activated receptor-gamma (PPARγ) used in type II diabetes [5]. In addition, a large number of compounds with stronger binding affinities and better specificities are currently in the research or development stages for new NR-targeting drugs [ 10, 11].
There are totally 48 human NRs, half of which have known endogenous ligands ( Table 1). Based on the protein sequence homology and functional analysis, a typical NR polypeptide can be roughly divided into 5 to 6 regions, which are represented by A-F from the N- to C-terminus respectively ( Figure 1) [ 12, 13]. N-terminal regulatory domain (NTD, i.e. the A/B region) is also called the ligand-independent transcription activation function-1 (AF-1) domain, which shows a low sequence conservation and a high conformational flexibility, with no high-resolution structure
Table1 The number of NR-related structures in PDB database till now
|
Names |
Nomenclature |
Ligand |
DBD |
LBD |
Others |
LBD-LBD |
Multi |
Reference |
|
TRα |
NR1A1 |
Thyroid hormones |
0 |
8 |
1 |
|||
|
TRβ |
NR1A2 |
Thyroid hormones |
1 |
22 |
1 |
|||
|
RAR-α |
NR1B1 |
Retinoic acid |
4 |
6 |
2 |
|||
|
RAR-β |
NR1B2 |
Retinoic acid |
8 |
1 |
1 |
|||
|
RAR-γ |
NR1B3 |
Retinoic acid |
11 |
|||||
|
PPARα |
NR1C1 |
Fatty acids |
55 |
|||||
|
PPARβ |
NR1C2 |
Fatty acids |
44 |
|||||
|
PPARγ |
NR1C3 |
Fatty acids |
238 |
|||||
|
Rev-erbα |
NR1D1 |
Orphan |
4 |
4 |
||||
|
Rev-erbβ |
NR1D2 |
Orphan |
6 |
|||||
|
RORα |
NR1F1 |
Cholesterol |
3 |
|||||
|
RORβ |
NR1F2 |
Retinoic acid |
3 |
|||||
|
RORγ |
NR1F3 |
Orphan |
141 |
|||||
|
LXRα |
NR1H3 |
Oxysterols |
8 |
3 |
||||
|
LXRβ |
NR1H2 |
Oxysterols |
24 |
1 |
||||
|
FXR |
NR1H4 |
Bile acids |
82 |
3 |
||||
|
VDR |
NR1I1 |
Vitamin D |
4 |
100 |
||||
|
PXR |
NR1I2 |
Xenobiotics |
42 |
2 |
||||
|
CAR |
NR1I3 |
Xenobiotics |
1 |
3 |
||||
|
HNF4α |
NR2A1 |
Orphan |
1 |
5 |
1 |
|||
|
HNF4γ |
NR2A2 |
Orphan |
1 |
|||||
|
RXRα |
NR2B1 |
Retinoic acid |
18 |
58 |
20 |
5 |
||
|
RXRβ |
NR2B2 |
Retinoic acid |
6 |
1 |
||||
|
RXRγ |
NR2B3 |
Retinoic acid |
2 |
|||||
|
TR2 |
NR2C1 |
Orphan |
|
|||||
|
TR4 |
NR2C2 |
Orphan |
1 |
|||||
|
TLL |
NR2E2 |
Orphan |
|
|||||
|
PNR |
NR2E3 |
Orphan |
1 |
|||||
|
COUP-TFI |
NR2F1 |
Orphan |
1 |
|
||||
|
COUP-TFII |
NR2F2 |
Orphan |
1 |
|||||
|
EAR2 |
NR2F6 |
Orphan |
|
|||||
|
ERα |
NR3A1 |
Estradiol-17-β |
2 |
298 |
||||
|
ERβ |
NR3A2 |
Estradiol-17-β |
36 |
|||||
|
ERRα |
NR3B1 |
Orphan |
4 |
|||||
|
ERRβ |
NR3B2 |
DES |
1 |
2 |
||||
|
ERRγ |
NR3B3 |
DES |
29 |
|||||
|
GR |
NR3C1 |
Cortisol |
40 |
28 |
||||
|
MR |
NR3C2 |
Aldosterones |
2 |
28 |
||||
|
PR |
NR3C3 |
Progesterone |
1 |
18 |
||||
|
AR |
NR3C4 |
Testosterone |
1 |
95 |
1 |
|||
|
Nur77 |
NR4A1 |
Orphan |
4 |
17 |
||||
|
NURR1 |
NR4A2 |
Orphan |
3 |
5 |
||||
|
NOR1 |
NR4A3 |
Orphan |
|
|||||
|
SF1 |
NR5A1 |
Orphan |
1 |
8 |
||||
|
LRH-1 |
NR5A2 |
Orphan |
2 |
21 |
1 |
1 |
||
|
GCNF |
NR6A1 |
Orphan |
1 |
|||||
|
DAX-1 |
NR0B1 |
Orphan |
2 |
|||||
|
SHP |
NR0B2 |
Orphan |
7 |
DBD, DNA-binding domain; LBD, ligand-binding domain; Others, other family proteins; LBD-LBD, homo- or heterodimer of NR LBD; Multi, Multi-domain complex of NRs.
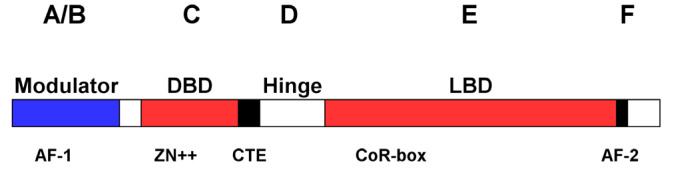
Figure1 .
Structural organization of nuclear receptors
Typical NRs can be roughly divided into five to six functional regions from N-terminus to C-terminus, designated as A–F respectively. A/B domain containing activation function-1 (AF-1) is a regulatory domain; C region is the DNA-binding domain; D is the hinge area; and E/F is the ligand-binding domain containing AF-2.
available yet [119]. DNA-binding domain (DBD), locating at the C region, has been well understood as one of the hallmarks of this NR family of transcription factors. The D region of NRs is a highly variable and flexible hinge region, which connects DBD and the ligand-binding domain (LBD). Nuclear localization signals (NLSs) of some NRs are located in the C and D regions, which affect the intracellular transport and subcellular distribution of NRs. The E/F region, i.e. the LBD, is the largest and most targetable domain of NRs. Transcription activation function-2 (AF-2) domain is also located in this region [119].
Over the past 30 years, the structures of a large number of NRs have been solved, since the cloning of first NR glucocorticoid receptor (GR/NR3C1) by Ronald Evans and colleagues ( Table 1) [120]. As a result, the basic structural information of NRs is relatively clear, especially for the well-studied DBD and LBD regions. A systematic summary of the structural information on NRs has been presented by Fraydoon Rastinejad and colleagues [ 2, 121, 122], with the emphasis that the crystal structures of multi-domain homo- and hetero-dimeric complexes provide important insights, to fully reveal the allosteric regulatory mechanisms of NRs.
To date, the majority of NRs have had their structures (at least for certain domains) published, except for the testicular receptor 2 (TR2/NR2C1), tailless (TLL/NR2E2), V-erbA-related protein 2 (EAR2/NR2F6) and neuron-derived orphan receptor 1 (NOR1/NR4A3) ( Table 1). However, it is worth noting that most of the structural information available has been heavily concentrated on several NRs [i.e. PPARγ, retinoid-related orphan receptor-gamma (RORγ), farnesoid X-activated receptor (FXR), 1,25-dihydroxyvitamin D3 receptor (VDR), RXRα, ERα, GR, and AR], whose functions are very important and well-studied. At present, PPARγ and ERα have contributed the largest number of NR structures (both over 200) in PDB, and most of them are LBDs in complex with synthetic derivatives of endogenous ligands ( Table 1). Meanwhile, there is little information about structures of NRs in complex with other family of proteins, when excluding the classic LXXLL motif-containing peptides derived from the steroid receptor coactivator (SRC) proteins. In recent years, structures of several large complexes containing NRs, SRC and p300 have been visualized using the cryo-electron microscopy (cryo-EM), although at only very low resolutions. With the rapid development of cryo-EM technology, the structural basis of how NRs function in concert with other partner proteins and in response to various ligands, would be fully illustrated soon. In addition, new drug discovery strategies such as proteolysis targeting chimera (PROTAC) have emerged and enriched the approaches to develop novel drugs targeting NRs directly.
In this review, we present a comprehensive overview of structures related to NRs in the Protein Data Bank (PDB) database, as well as some perspectives in the future development of small-molecule drugs directly targeting NRs.
DBD Structures of NRs
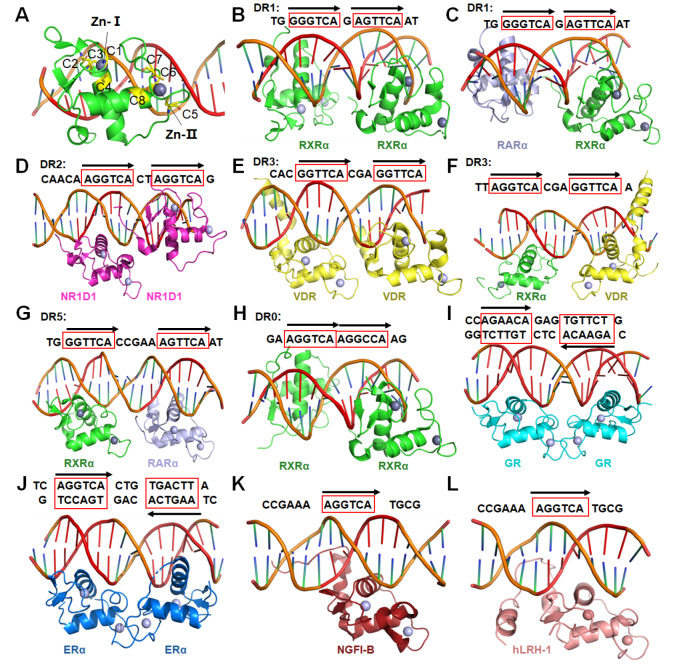
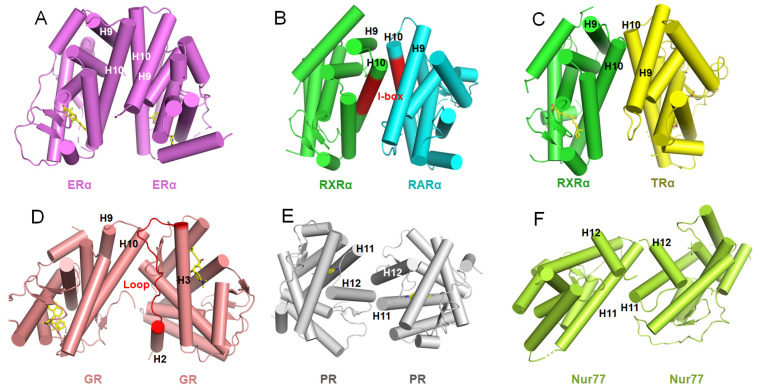
The DBDs of NRs possess highly conserved protein sequences, and their structures and functions have been extensively studied. The typical NR DBD structure conforms to a Zn-finger subtype of transcription factors, in which eight highly conserved cysteine residues coordinate two zinc ions, setting the NRs apart from other DNA-binding proteins ( Figure 2A). In the PDB database, RXR and GR have contributed the most DBD-DNA complex structures of NRs ( Table 1). RXR can act as the dimer partner for several NRs including itself. Then these NR dimers bind to different response elements on the genomic DNA, and regulate transcription in a cell and gene-specific manner. The number of spacer nucleotides between two NR-binding half-sites, was initially defined for various RXR heterodimers in a simplified manner by the ‘DR (direct repeat) 1–5 rule’: i.e. RXR-RXR, and retinoic acid receptor (RAR)-RXR (DR1), RXR-RAR (DR2), RXR-VDR (DR3), RXR-TR (DR4) and RXR–RAR (DR5) ( Figure 2B–G) [ 21, 23, 59, 123– 126]. Interestingly, a recent study reported the structure of RXR DBD homodimer in complex with Hoxb13 DR0 DNA ( Figure 2H) [23]. On the contrary, receptors for androgens (AR), corticosteroids (GR/MR), progesterone (PR) or estrogens (ER) typically bind DNA sequences configured as inverted repeats of the half-site sequences AGAACA or AG/AGTCA, respectively ( Figure 2I–J); while the remaining members of the family bind half-sites of the sequence AGGTCA/AGGCTA, arranged singly ( Figure 2K,L) or as direct repeats [ 2, 127]. The advent of next-generation sequencing, in combination with chromatin immunoprecipitation (ChIP-seq), has allowed for the identification of NR-binding sites in a genome-wide manner and in various cell types under different pathophysiological conditions [128]. Furthermore, these studies have revealed that NR response elements can vary from monomeric half-sites to nearly perfect inverted or direct repeats, and have demonstrated the cooperation between NRs and other transcription factors, such as the forkhead box A1 (FOXA1) (for AR and ER), activating protein-1 (AP-1) [for GR and liver X receptor (LXR)], and CCAAT/enhancer-binding protein (C/EBP) (for PPAR-γ and LXR) [128].
Figure2 .
Interactions of DBDs with different response elements
(A) The typical structure of NR DBD (PDB code: 4CN5). (B–H) Homo- or hetero-dimers of NR DBD complexes with DR0 to DR5 response elements (PDB codes: 4CN5, 1DSZ, 1GA5, 1KB2, 1YNW, 6XWG, and 6XWH, respectively). (I,J) Homodimers of GR and ERα bound to inverted repeats of the half-site sequences (PDB codes: 5CBX, 4AA6). (K,L) NGFI-B and hLRH-1 bound to single half-sites of DNA sequence (PDB codes: 1CIT, 5L0M).
LBD Structures of NRs
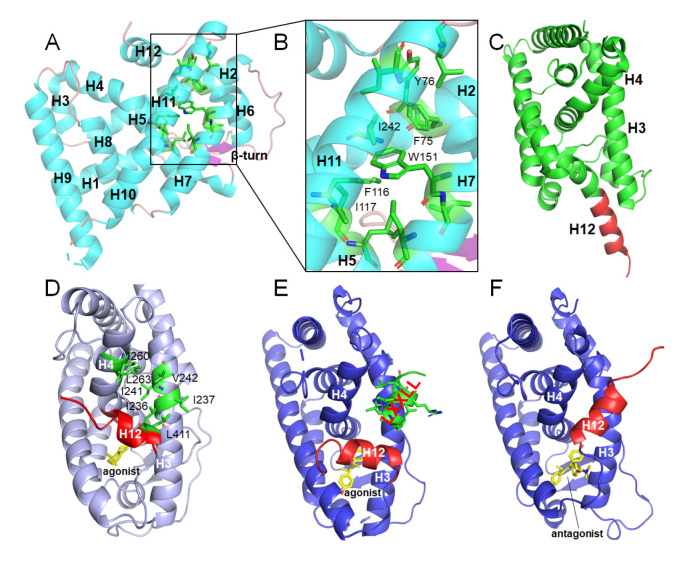
The NR LBDs usually show a similar overall conformation, consisting of 11–13 α helices and 1 β-turn that together form a three-layered helical sandwich ( Figure 3A). The typical ligand binding pocket is embedded in the interior of LBD, with the core position composed of amino acids derived from the helices H2, H5, H7, H11, and H12. The volume of pockets can vary from zero to more than 1500 Å 3 ( Figure 3B) [129]. LBD plays an important role in the transcriptional regulation of classical NRs, and its mechanism of action has been clearly revealed. Structural analysis shows that LBD works like a “mousetrap” and ligand binding can induce the conformational change of helix H12 [27]. When ligands bind into pockets, H12 as the “gate” of the “mousetrap”, will move towards LBD and fold back to close the entrance of the ligand binding pocket ( Figure 3C,D). Moreover, the H12 adopts different conformations when the pocket binds agonists or antagonists. For agonists, H12, H3 and H4 form a hydrophobic coactivator binding surface, where the LXXLL motif can be well integrated ( Figure 3D,E). While for antagonists, H12 moves away from the excited position, masks or destroys the binding sites of coactivators, and forms the binding interface for corepressors ( Figure 3F) [130]. Under different signal stimulations, NRs interact with different coregulators and conduct different functions, activating or repressing the expression of downstream genes. However, there are also several NRs not conforming to the above mechanism. For example, RORγ, immediate-early response protein NOT (Nurr1) and nuclear hormone receptor Nur77 are in the transcriptionally activated conformation independent of ligands ( Figure 3A). Especially, the pockets of three members of the NR4A subfamily ( Table 1) are completely occupied by hydrophobic amino acids, and the hydrophobic binding regions of co-regulators are also occupied by some polar amino acids ( Figure 3B). As a result, the transcriptional regulation mechanism of this type of NRs is still unclear.
Figure3 .
Structural characteristics and classical transcriptional regulation mechanisms of NR LBDs
(A) Overall conformation of Nur77 LBD shown as cartoon in cyan (PDB code: 3V3E). (B) Ligand binding pocket of Nur77 is occupied by several hydrophobic amino acids. (C) Inactive state of the apo-LBD, H12 color in red (PDB code: 6HN6). (D) Active state of RARγ holo-LBD, agonist shown as sticks in yellow, hydrophobic amino acids (shown as sticks in green) in H3, H4, H5 forming a hydrophobic interaction surface (PDB code: 2LBD). (E) Active state of ERα holo-LBD, agonist shown as sticks in yellow, coactivator GRIP1 with the LXXLL motif color in green (PDB code: 3ERD). (F) ERα LBD bound with antagonist and H12 color in red occupying the co-activator binding site (PDB code: 3ERT).
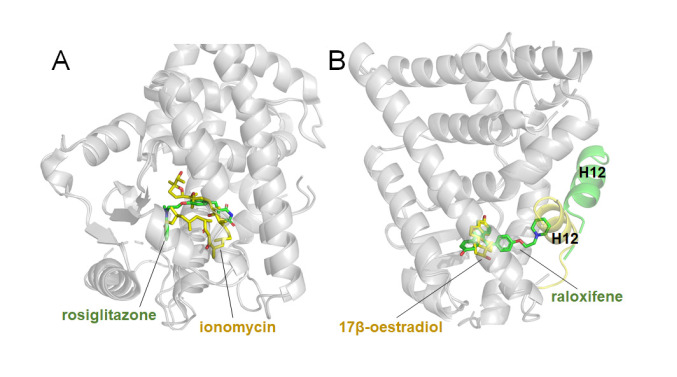
As mentioned earlier, most of NR structures reported in PDB are the complexes of LBDs with endogenous ligands and their derivatives. By now, the numbers of LBD structures for PPARγ and ERα have reached 238 and 298, respectively ( Table 1). Rosiglitazone is one of the thiazolidinedione (TZD) drugs for diabetes treatment, and it is an insulin sensitizer with a high affinity and specificity to activate PPARγ. However, TZDs also display severe adverse effects, giving rise to fluid retention, weight gain, liver toxicity and cardiovascular diseases, which are prevalent among diabetic patients [ 131– 133]. Based on the structure of PPARγ LBD in complex with rosiglitazone, researchers designed and discovered more effective agonists. Ionomycin interacts with the PPARγ LBD in a unique binding mode ( Figure 4A) and effectively improves hyperglycaemia and insulin resistance, with reduced side effects compared with TZDs in the mouse model of diabetes [133]. Another example among NR drugs is raloxifene, which is a selective ERα modulator used in osteoporosis. The crystal structures of ERα LBD in complex with the endogenous estrogen, 17β-oestradiol, and the selective antagonist raloxifene, indicate that agonist and antagonist bind at the same site within the core pocket of LBD, but demonstrate different binding modes ( Figure 4B) [134]. These small-molecule bound complex structures not only help reveal their mechanisms of action, but also provide valuable information for the structure-based drug design targeting NRs.
Figure4 .
NR LBD structures in complex with different small molecules
(A) Superimposition of PPARγ LBD in complex with ionomycin (stick in yellow) and rosiglitazone (stick in green) (PDB codes: 4FGY, 2PRG). (B) Superimposition of ERα LBD in complex with endogenous agonist, 17β-oestradiol (stick in yellow) and antagonist raloxifene (stick in green), related H12 shown as cartoon color in yellow and green respectively (PDB codes: 1ERE, 1ERR).
For the majority of NRs, their activations and functions depend on the dimerization. Generally, NRs possess two dimerization sites, one in DBD and the other in LBD. LBD is considered as the major dimerization site for NRs [22]. Ligand binding not only changes the conformation of LBDs to coordinate with the coregulator proteins, but also promotes the dimerization of NRs [22]. According to the analysis of LBD structures in the PDB database, most of the NRs utilize the classical dimerization sites (H10 and H9 or even Loops 8-9 and H7) to form dimers ( Figure 5A–C). The size of dimerization interface of ERα homodimer is about 1700 Å 2, while those for the RXRα homodimer and its heterodimer with RARα LBD are only 950 Å 2 and 970 Å 2, respectively ( Figure 5A,B) [135]. The smaller dimerization interface of RXRα is related to its biological function, making RXRα relatively easy to depolymerize and form new homo- or heterodimers with other NRs [136]. The heterodimerization of RXR with RAR, TR and VDR via LBDs mainly depends on the I-box located in H9-H10 region, as shown by their dimeric crystal structures ( Figure 5B,C) [ 22, 137– 139]. However, not all NRs dimerize in this way. GR homodimerizes through the hydrophobic interactions of some hydrophobic amino acids in the H2-H3 loop region, with a dimerization area of 623 Å 2 ( Figure 5D) [140]. PR homodimerizes through hydrogen bonds and salt bridges formed in the H11-H12 region, and the dimerization surface area is about 600 Å 2, which is much smaller than those of classical NRs ( Figure 5E) [97]. Another NR with a small dimer interface is Nur77, whose asymmetry interface is mediated by hydrogen bonds and salt bonds in the H11-H12 region with a surface area of only about 590 Å 2 ( Figure 5F) [ 105, 106].
Figure5 .
Different dimerization interfaces of NR LBDs
(A–C) Homo- or hetero-dimers through H9 and H10. The I-box of RXRα and RARα colored in red (PDB codes: 1ERR, 1DKF, 3UVV, respectively). (D) Homodimer interface of GR (PDB code: 6DXK). (E,F) Homodimers of PR and Nur77 through H11 and H12 (PDB codes: 1ZUC, 3V3E).
Structures of Full-length and Multi-domain Receptor Complexes
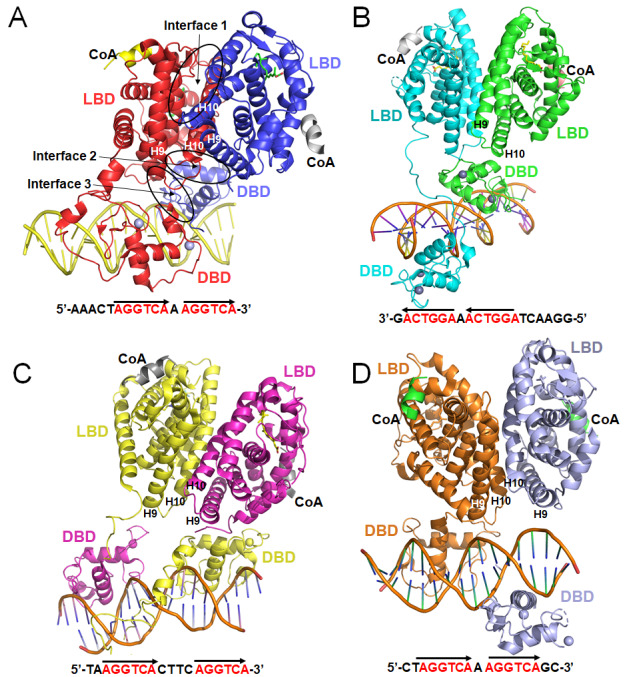
NRs are multi-domain transcription factors that bind to specific DNA elements and regulate the expressions of downstream genes. How DNA binding influences the NR conformation, and how the conformations of DBD and LBD change collaboratively in the process of transcriptional regulation, are still poorly understood. Previous structural studies have mainly focused on the isolated DNA or ligand-binding segments, i.e. DBDs and LBDs. To date, only 4 crystal structures of full-length or multi-domain NR complexes bound to DNA have been reported, with 3 of them by Fraydoon Rastinejad and colleagues.
The structure of PPARγ and RXRα, published in 2008 as a heterodimer bound to DNA, ligands and coactivator peptides, is the first structure of multi-domain NR complex, in which PPARγ-RXRα polypeptides reside in a polar arrangement set by the 5′ extension of the DR1 and the lone spacer base pair [70]. PPARγ occupies most of the DR1 including its specific 5′ element. Three interfaces link PPARγ and RXRα, including those that are DNA-dependent. The PPARγ LBD cooperates with both DBDs to enhance the response-element binding ( Figure 6A).
Figure6 .
Four multi-domain complex structures of NRs
(A) The PPARγ-RXRα (colored in red and blue, respectively) heterodimer on DNA response element with DR1 (PDB code: 3E00). (B) The HNF-4α homodimer bound to DR1 (PDB code: 4IQR). (C) The LXRβ-RXRα (colored in yellow and magenta, respectively) heterodimer on DR4 (PDB code: 4NQA). (D) The RARβ-RXRα (colored in orange and purple, respectively) heterodimer bond to DR1 (PDB code: 5UAN). The key DBD-interacting region at the H9 and H10 of LBD is marked with capital letters.
HNF-4α is the most abundant DNA-binding protein in liver, where about 40% of the actively transcribed genes have at least one HNF-4α response element [ 141, 142]. To understand the extent of domain integration of HNF-4α, Rastinejad group solved and analyzed the crystal structure of HNF-4α, an obligate homodimer, bound to its DNA element and coactivator-derived peptides in 2013 ( Figure 6B) [65]. The LBD and DBD portions match their previously determined isolated structures. Both DBDs are in register with their half-sites, interacting with the major grooves. The DBD of the upstream subunit and the hinge region of the downstream subunit, form an important domain-domain interface of the complex, which is similar to the one in the PPARγ-RXRα complex. The manner in which the two LBDs cooperate to interact with the upstream DBD suggests that the physical integration of all three domains may be required for a high-affinity DNA binding, as further proved by the physiological and biochemical experiments [143].
The LXRs are physiologically important oxysterol-dependent NRs. LXRs are master regulators of lipid and cholesterol metabolism [ 144, 145], inflammation [146], neural development [147], cancer [ 148, 149], and other physiological processes. Furthermore, LXRβ has protective effects upon dopamine neurons [ 150, 151] and modulates the cytotoxic functions of microglia [152]. Thus, LXRs are key pharmaceutical targets in a variety of diseases. Gustafsson group reported the crystal structure of the RXRα-LXRβ heterodimer on its cognate element, an AGGTCA direct repeat spaced by 4 nt (DR4) in 2014 ( Figure 6C) [52]. This complex shows an extended X-shaped arrangement, with DBD and LBD crossed, which is different from the other 3 multi-domain complexes ( Figure 6C). Compared with previous NR structures, it reveals the flexibility in NR organization and suggests a role for RXRα in the adaptation of heterodimeric complexes to DNA.
The RAR and RXR proteins are among the most intensively studied NRs for their structural properties, but most structural characterizations to date have focused on the isolated LBD domains ( Table 1). In 2017, Rastinejad group reported the crystal structure of the multi-domain RARβ-RXRα heterodimer bound to DR1 DNA, ligands and coactivator peptides [26]. The DBD and LBD of RARβ are physically connected. However, the corresponding two domains of RXRα are spatially displaced from each other without any physical contacts, and each of them locates on the opposite side of the double-strand DNA. Both RXRα and RARβ adopt the active conformation at their LBDs. This conformation is defined by both receptors having their H12 appropriately positioned by ligands to facilitate the recruitment of coactivator LXXLL motifs ( Figure 6D).
By analyzing the structures of 4 multi-domain complexes and comparing with the structures of their individual LBDs and DBDs, we found that they are essentially identical at the single domain level. These observations suggest that none of these domains undergoes major internal distortions in adopting the quaternary state of the multi-domain heterodimers. Through structural comparison and analysis, we can also learn the following lessons. (1) The flexible hinge region not only physically connects DBD and LBD, but also plays important regulatory roles in the interactions between LBD and DBD, as well as those between DBD and DNA. In addition, the hinge region can interact with DNA directly and play special roles in stabilizing the formation of the whole complex. (2) Specific response elements, proper ligands and coactivators are needed to obtain a stable complex crystal structure. Therefore, the success of future crystallization efforts on new multi-domain NR complexes would require a lot of trial and error in the combination of these three NR partners above. (3) LBD plays key roles in the formation of homodimers or heterodimers, including the formation of its various interfaces with DBD ( Figure 6). These unique interfaces may offer new opportunities for the discovery of NR-targeting drugs with a novel mechanism of action. (4) At present, there is still no clear structural information for the NTD region, which is very flexible and not conserved among NRs.
Structure of NRs in Complex with Other Family Proteins
The homo- or hetero-dimerization of certain NRs is one of the key steps in their transcriptional regulation of downstream genes. Meanwhile, cofactor recruitment also plays important roles in the regulatory function of NRs. For example, the N-terminal AF-1 region of Nur77 can enhance the ability of transcriptional activation by recruiting coactivators such as SRCs, P300, P300/CBP-associated factor (PCAF) [153]. Protein arginine methyltransferases 1 (PRMT1) can enhance the transcriptional activation of Nur77 by cooperating with SRC2 [154]. Nur77 and Nurr1 can also inhibit the transcription of downstream genes by recruiting the repressor FHL2 (four and a half LIM domains protein-2) [155] or repressor cofactor complexes [ 156, 157]. However, the detailed recognition mechanism during the recruitment process is still missing, due to the lack of structures of protein-protein complex between cofactors and NRs.
In addition to the transcriptional regulation, in specific cell lines, NRs can also play other roles through protein-protein interactions. Nur77 directly associates with p65 to block its binding to the κB element [105]. However, this function of Nur77 is countered by the LPS-activated p38a phosphorylation of Nur77. In this process, Nur77 regulates the inflammatory response through direct interactions with p65 and p38a [105]. The direct interaction between 1-(3,4,5-trihydroxyphenyl)nonan-1-one (THPN) and Nur77-LBD helps form a suitable surface that can bind to mitochondria outer protein Nix, and then triggers autophagy in the human melanoma Mel-11, ME4405 and MM200 cells [104]. Therefore, analyzing the complex structures of NRs and other family proteins (especially those identified outside of the classical NR pathways), may provide a new angle for the drug design targeting NRs.
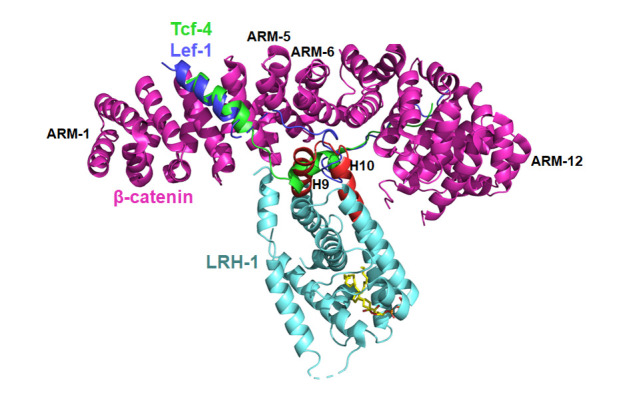
At present, the β-catenin armadillo repeat in complex with the liver receptor homolog-1 (LRH-1) LBD is the only complex structure between NRs and other family proteins that has been solved [112]. As the principal agent of Wnt-dependent effects on cell adhesion, differentiation and cancer, β-catenin engages in multiple protein–protein contacts [ 158, 159], some of which are understood with atomic details [160]. β-Catenin is comprised of a central association region, the armadillo-repeat region (ARM), and N- and C-terminal transactivation domains [161], whereas the N- and C-terminal regions are both intrinsically unstructured [162]. The interaction modes between NRs and β-catenin are complex, but there is evidence that NR LBDs are indispensable for the contact in some cases. Biochemical and genetic evidence reveals that both LRH-1 and AR LBDs bind to the ARM, with AR exhibiting a strong dependence on the ARM-5 and ARM-6 segments ( Figure 7) [ 163– 165]. In this complex structure, the LRH-1 LBD utilizes a novel interaction surface to dock into the positively charged groove at a site that partially overlaps the binding surface for T-cell factor 4 (Tcf-4) and lymphoid enhancer-binding factor 1 (Lef-1) ( Figure 7) [ 166, 167]. This structure suggests an interesting mechanism for the assembly of multifactor transcription complexes. NR LBDs are known to engage in multiple protein–protein contacts with co-regulators [ 168, 169], and β-catenin is shown to bind many proteins including a wide variety of transcription factors [164]. The structural information of this complex indicates the possibility to develop new compounds that inhibit NR functions, by binding directly or impacting allosterically towards key protein interaction surfaces. Thus, the β-catenin binding site of LRH-1 (the H9–H10 of LBD) and other NRs could be a new targeting spot for the future drug discovery. Moreover, it is very interesting that NR LBDs use exactly the same “patch” region (H9–H10) to interact with DBDs, as shown in the four multi-domain NR structures ( Figure 5) [26]. These findings suggest a potential common mechanism for NR LBDs to mediate the protein-protein interactions.
Figure7 .
Complex structure of β-catenin and LRH-1
The overall structure of β-catenin and LRH-1 shown as cartoon in magentas and cyan colors, respectively. Peptides of Tcf-4 and Lef-1 colored in green and blue respectively. H9 and H10 of LRH-1 LBD colored in red (PDB code: 3TX7).
Application of Cro-EM in NR Structure Analysis
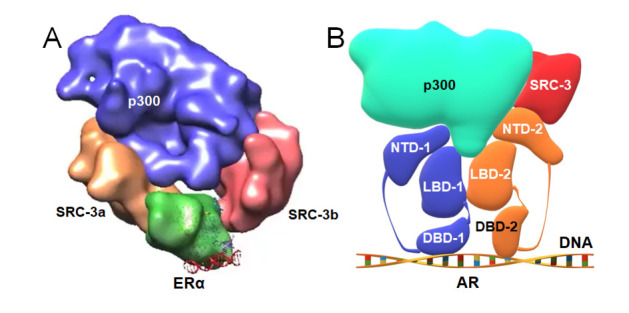
While full-length NRs are very difficult to crystallize, the fast developing cryo-EM technology has also been adopted to unravel the structural features of the NR signaling scaffold, as well as the critical roles of inter-domain communications [170]. Orlov et al. [171] presented the first cryo-EM structure of a 100-kDa complex of VDR-RXR heterodimer and their cognate DNA response element at the 10–15 Å resolution. Following that, other researchers used cryo-EM to determine the quaternary structure of an active complex of ERα/SRC-3/p300 bound to an ERE-DNA fragment at resolution ~25 Å in 2015 ( Figure 8A); the structure of ARE DNA-bound full-length AR at resolution ~12.6 Å, and the structure of ARE DNA-bound AR/SRC-3/p300 complex at resolution ~20 Å in 2020 ( Figure 8B) [ 172, 173]. Both cryo-EM structures of ERα and AR in complex with SRC-3 and p300 indicate that the AF-1 in NTD plays a key role in the SRC-3 and p300 recruitment ( Figure 8). Especially in the AR complex ( Figure 8B), AF-1 in NTD participates in the interactions with SRC-3 and p300 much more than the AF-2 in LBD, which is consistent with previous reports [ 174, 175].
Figure8 .
Cryo-EM models of ERα and AR complexes with cofactors and DNA
(A) The components of ERα homodimer in complex with SRC-3 and p300. (B) Model of homodimer of AR in complex with p300 and SRC-3.
Unfortunately, these current cryo-EM NR structures could not provide more details at the atom level due to their poor resolutions. To push the resolution limit, a number of factors have to be considered and optimized, especially the intrinsic dynamics between NRs and their co-regulators. For example, the application of specific antibodies targeting certain components of the large NR complex may help increase the overall stability. As the crystal structures of three key components of the NR complex (i.e. LBD, DBD and DNA) have been well defined with their unique sizes and shapes, the model building step in solving NR cryo-EM structures would be relatively straight-forward [170]. With more large NR complex structures available, the discovery of new mechanism drugs targeting NRs will be further accelerated.
Conclusion and Future Direction
Until recently, all previous structural research efforts on NR proteins were more focused on and successful with individual DBDs and LBDs, especially for some classic NRs ( Table 1). However, the functions and structures of a considerable number of NRs are still not very clear, especially for TR2, TLL, EAR2 and NOR1. At present, the multi-domain crystal structures of PPARγ-RXRα, HNF4α-HNF4α, LXRβ-RXRα and RARβ-RXRα have been solved. But the structural information about NRs is still not complete, as the N-terminal region with a high flexibility and a low homology still has no high-resolution structure reported yet. Furthermore, no multi-domain structure of orphan NRs has been solved. NRs can be conceptualized as highly dynamic scaffold proteins, where binding of ligand, DNA or transcriptional coregulator proteins can allosterically change the scaffold structure and direct changes in subsequent binding events [170]. Therefore, more full-length NR complex structures are needed to obtain the conformational change information during the transcriptional regulation, which can surely provide a new structural basis for designing better small-molecule drugs. Especially in recent years, a series of major breakthroughs have been made in the development of cryo-EM. It bypasses mysteries in the protein crystallization process, and provides an alternative yet powerful technical approach for the structural analysis of full-length NRs, transcriptional regulatory complexes, as well as the complexes of NRs with other family proteins.
In the past a few years, the induced and targeted protein degradation, based on the PROTAC concept, has gained momentum as a new small-molecule therapeutic strategy [176]. Till now, a large number of PROTAC molecules related to AR and ER have been reported, some of which for the treatment of prostate and breast cancers have entered the phase II clinical stage [ 177, 178]. This technology will bring new opportunities for the research of certain undruggable NRs, especially for some orphan receptors. With the progress of these technologies, new breakthroughs will be achieved in the discovery of novel drugs targeting NRs.
COMPETING INTERESTS
The authors declare that they have no conflict of interest.
Funding Statement
This work was supported by the grants from the National Natural Science Foundation of China (No. 31800664), the National Key R&D Program of China (No. 2018YFE0113000), the Taishan Scholars Program of Shandong (No. tsqn201909004), the University Innovation Group Program of Jinan (No. 2020GXRC006), and the Interdisciplinary Innovative Research Fund of SDU (No. 2020QNQT009).
References
- 1.Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. . 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rastinejad F, Huang P, Chandra V, Khorasanizadeh S. Understanding nuclear receptor form and function using structural biology. J Mol Endocrinol. . 2013;51:T1–T21. doi: 10.1530/JME-13-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, Karlsson A, Al-Lazikani B, Hersey A, Oprea TI, Overington JP. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. . 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porter BA, Ortiz MA, Bratslavsky G, Kotula L. Structure and function of the nuclear receptor superfamily and current targeted therapies of prostate cancer. Cancers (Basel) 2019, 11: 1852 . [DOI] [PMC free article] [PubMed]
- 5.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. . 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 6.Schellhammer PF. An evaluation of bicalutamide in the treatment of prostate cancer. Expert Opin Pharmacother. 2002, 3: 1313–1328 . [DOI] [PubMed]
- 7.See WA, Tyrrell CJ; CASODEX Early Prostate Cancer Trialists’ Group. The addition of bicalutamide 150 mg to radiotherapy significantly improves overall survival in men with locally advanced prostate cancer. J Cancer Res Clin Oncol. 2006, 132: S7–S16 . [DOI] [PMC free article] [PubMed]
- 8.Gniadecki R, Assaf C, Bagot M, Dummer R, Duvic M, Knobler R, Ranki A, Schwandt P, Whittaker S. The optimal use of bexarotene in cutaneous T-cell lymphoma. Br J Dermatol. . 2007;157:433–440. doi: 10.1111/j.1365-2133.2007.07975.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Zhou H, Su Y. Targeting truncated RXRα for cancer therapy. Acta Biochim Biophys Sin. 2016, 48: 49–59 . [DOI] [PMC free article] [PubMed]
- 10.Elix C, Pal SK, Jones JO. The role of peroxisome proliferator-activated receptor gamma in prostate cancer. Asian J Androl. 2018, 20: 238–243 . [DOI] [PMC free article] [PubMed]
- 11.Ishigami-Yuasa M, Kagechika H. Chemical screening of nuclear receptor modulators. Int J Mol Sci. 2020, 21: 5512 . [DOI] [PMC free article] [PubMed]
- 12.Kumar R, Thompson EB. The structure of the nuclear hormone receptors. Steroids. . 1999;64:310–319. doi: 10.1016/S0039-128X(99)00014-8. [DOI] [PubMed] [Google Scholar]
- 13.Thomson SA, Baldwin WS, Wang YH, Kwon G, Leblanc GA. Annotation, phylogenetics, and expression of the nuclear receptors in Daphnia pulex. BMC Genomics. 2009, 10: 500 . [DOI] [PMC free article] [PubMed]
- 14.Ye L, Li YL, Mellström K, Mellin C, Bladh LG, Koehler K, Garg N, Garcia Collazo AM, Litten C, Husman B, Persson K, Ljunggren J, Grover G, Sleph PG, George R, Malm J. Thyroid receptor ligands. 1. agonist ligands selective for the thyroid receptor β 1 . J Med Chem. . 2003;46:1580–1588. doi: 10.1021/jm021080f. [DOI] [PubMed] [Google Scholar]
- 15.Souza PCT, Puhl AC, Martínez L, Aparício R, Nascimento AS, Figueira ACM, Nguyen P, Webb P, Skaf MS, Polikarpov I. Identification of a new hormone-binding site on the surface of thyroid hormone receptor. Mol Endocrinol. . 2014;28:534–545. doi: 10.1210/me.2013-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez L, Nascimento AS, Nunes FM, Phillips K, Aparicio R, Dias SM, Figueira AC, et al. Gaining ligand selectivity in thyroid hormone receptors via entropy. Proc Natl Acad Sci U S A. 2009, 106: 20717-20722 . [DOI] [PMC free article] [PubMed]
- 17.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995, 375: 203–211 . [DOI] [PubMed]
- 18.Borngraeber S, Budny MJ, Chiellini G, Cunha-Lima ST, Togashi M, Webb P, Baxter JD, et al. Ligand selectivity by seeking hydrophobicity in thyroid hormone receptor. Proc Natl Acad Sci U S A. 2003, 100: 15358-15363 . [DOI] [PMC free article] [PubMed]
- 19.Yao B, Wei Y, Zhang S, Tian S, Xu S, Wang R, Zheng W, et al. Revealing a mutant-Induced receptor allosteric mechanism for the thyroid hormone resistance. iScience. 2019, 20: 489–496 . [DOI] [PMC free article] [PubMed]
- 20.Kojetin DJ, Matta-Camacho E, Hughes TS, Srinivasan S, Nwachukwu JC, Cavett V, Nowak J, Chalmers MJ, Marciano DP, Kamenecka TM, Shulman AI, Rance M, Griffin PR, Bruning JB, Nettles KW. Structural mechanism for signal transduction in RXR nuclear receptor heterodimers. Nat Commun. . 2015;6:8013. doi: 10.1038/ncomms9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR–RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. . 2000;19:1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell. . 2000;5:289–298. doi: 10.1016/S1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 23.Osz J, McEwen AG, Bourguet M, Przybilla F, Peluso-Iltis C, Poussin-Courmontagne P, Mély Y, Cianférani S, Jeffries CM, Svergun DI, Rochel N. Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR–RXR. Nucleic Acids Res. . 2020;48:9969–9985. doi: 10.1093/nar/gkaa697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knegtel RM, Katahira M, Schilthuis JG, Bonvin AM, Boelens R, Eib D, van der Saag PT, Kaptein R. The solution structure of the human retinoic acid receptor-β DNA-binding domain. J Biomol NMR. . 1993;3:1–17. doi: 10.1007/BF00242472. [DOI] [PubMed] [Google Scholar]
- 25.Germain P, Kammerer S, Pérez E, Peluso-Iltis C, Tortolani D, Zusi FC, Starrett J, Lapointe P, Daris JP, Marinier A, de Lera AR, Rochel N, Gronemeyer H. Rational design of RAR‐selective ligands revealed by RARβ crystal stucture. EMBO Rep. . 2004;5:877–882. doi: 10.1038/sj.embor.7400235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chandra V, Wu D, Li S, Potluri N, Kim Y, Rastinejad F. The quaternary architecture of RARβ–RXRα heterodimer facilitates domain–domain signal transmission. Nat Commun. . 2017;8:868. doi: 10.1038/s41467-017-00981-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. . 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 28.Thoreau E, Arlabosse JM, Bouix-Peter C, Chambon S, Chantalat L, Daver S, Dumais L, et al. Structure-based design of Trifarotene (CD5789): a potent and selective RARγ agonist for the treatment of acne. Bioorganic Bioorg Med Chem Lett. 2018, 28: 1736–1741 . [DOI] [PubMed]
- 29.Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, Oplinger JA, Kliewer SA, Gampe Robert T. J, McKee DD, Moore JT, Willson TM. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. . 2001;98:13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, Parks DJ, Moore JT, Kliewer SA, Willson TM, Stimmel JB. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARα. Nature. . 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 31.Kamata S, Oyama T, Saito K, Honda A, Yamamoto Y, Suda K, Ishikawa R, Itoh T, Watanabe Y, Shibata T, Uchida K, Suematsu M, Ishii I. Pparα ligand-binding domain structures with endogenous fatty acids and fibrates. iScience. . 2020;23:101727. doi: 10.1016/j.isci.2020.101727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, Kliewer SA, Milburn MV. Molecular recognition of fatty acids by peroxisome proliferator–activated receptors. Mol Cell. . 1999;3:397–403. doi: 10.1016/S1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 33.Lagu B, Kluge AF, Tozzo E, Fredenburg R, Bell EL, Goddeeris MM, Dwyer P, Basinski A, Senaiar RS, Jaleel M, Tiwari NK, Panigrahi SK, Krishnamurthy NR, Takahashi T, Patane MA. Selective PPARδ modulators improve mitochondrial function: potential treatment for duchenne muscular dystrophy (DMD) ACS Med Chem Lett. . 2018;9:935–940. doi: 10.1021/acsmedchemlett.8b00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oberfield JL, Collins JL, Holmes CP, Goreham DM, Cooper JP, Cobb JE, Lenhard JM, Hull-Ryde EA, Mohr CP, Blanchard SG, Parks DJ, Moore LB, Lehmann JM, Plunket K, Miller AB, Milburn MV, Kliewer SA, Willson TM. A peroxisome proliferator-activated receptor ligand inhibits adipocyte differentiation. Proc Natl Acad Sci U S A. . 1999;96:6102–6106. doi: 10.1073/pnas.96.11.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. . 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 36.Shang J, Mosure SA, Zheng J, Brust R, Bass J, Nichols A, Solt LA, Griffin PR, Kojetin DJ. A molecular switch regulating transcriptional repression and activation of PPARγ. Nat Commun. . 2020;11:956. doi: 10.1038/s41467-020-14750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007, 14: 1207–1213 . [DOI] [PMC free article] [PubMed]
- 38.Zhao Q, Khorasanizadeh S, Miyoshi Y, Lazar MA, Rastinejad F. Structural elements of an orphan nuclear receptor–DNA complex. Mol Cell. . 1998;1:849–861. doi: 10.1016/S1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- 39.Phelan CA, Gampe RT, Lambert MH, Parks DJ, Montana V, Bynum J, Broderick TM, et al. Structure of Rev-erbalpha bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol. 2010, 17: 808–814 . [DOI] [PMC free article] [PubMed]
- 40.Mosure SA, Strutzenberg TS, Shang J, Munoz-Tello P, Solt LA, Griffin PR, Kojetin DJ. Structural basis for heme-dependent NCoR binding to the transcriptional repressor REV-ERBβ. Sci Adv. 2021, 7: eabc6479 . [DOI] [PMC free article] [PubMed]
- 41.Woo EJ, Jeong DG, Lim MY, Jun Kim S, Kim KJ, Yoon SM, Park BC, Eon Ryu S. Structural insight into the constitutive repression function of the nuclear receptor REV-ERBβ. J Mol Biol. . 2007;373:735–744. doi: 10.1016/j.jmb.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 42.Santori FR, Huang P, van de Pavert SA, Douglass Jr. EF, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, Rozman D, Horvat S, Rahier A, Mebius RE, Rastinejad F, Nes WD, Littman DR. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. . 2015;21:286–298. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hRORalpha LBD at 1.63 Å: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002, 10: 1697–1707 . [DOI] [PubMed]
- 44.Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORα ligand binding domain in complex with cholesterol sulfate at 2.2 Å. J Biol Chem. . 2004;279:14033–14038. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- 45.Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schüle R, Moras D, Renaud JP. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. EMBO J. . 2001;20:5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stehlin-Gaon C, Willmann D, Zeyer D, Sanglier S, Van Dorsselaer A, Renaud JP, Moras D, Schüle R. All-trans retinoic acid is a ligand for the orphan nuclear receptor RORβ. Nat Struct Mol Biol. . 2003;10:820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 47.de Vries RM, Meijer FA, Doveston RG, Leijten-van de Gevel IA, Brunsveld L. Cooperativity between the orthosteric and allosteric ligand binding sites of RORγt. Proc Natl Acad Sci U S A. 2021, 118: e2021287118 . [DOI] [PMC free article] [PubMed]
- 48.Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol. 2010, 24: 923–929 . [DOI] [PMC free article] [PubMed]
- 49.Svensson S, Ostberg T, Jacobsson M, Norström C, Stefansson K, Hallén D, Johansson IC, Zachrisson K, Ogg D, Jendeberg L. Crystal structure of the heterodimeric complex of LXR and RXR ligand-binding domains in a fully agonistic conformation. EMBO J. . 2003;22:4625–4633. doi: 10.1093/emboj/cdg456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stachel SJ, Zerbinatti C, Rudd MT, Cosden M, Suon S, Nanda KK, Wessner K, DiMuzio J, Maxwell J, Wu Z, Uslaner JM, Michener MS, Szczerba P, Brnardic E, Rada V, Kim Y, Meissner R, Wuelfing P, Yuan Y, Ballard J, Holahan M, Klein DJ, Lu J, Fradera X, Parthasarathy G, Uebele VN, Chen Z, Li Y, Li J, Cooke AJ, Bennett DJ, Bilodeau MT, Renger J. Identification and in vivo evaluation of liver X receptor β-selective agonists for the potential treatment of alzheimer’s disease . J Med Chem. . 2016;59:3489–3498. doi: 10.1021/acs.jmedchem.6b00176. [DOI] [PubMed] [Google Scholar]
- 51.Williams S, Bledsoe RK, Collins JL, Boggs S, Lambert MH, Miller AB, Moore J, McKee DD, Moore L, Nichols J, Parks D, Watson M, Wisely B, Willson TM. X-ray crystal structure of the liver x receptor β ligand binding domain. J Biol Chem. . 2003;278:27138–27143. doi: 10.1074/jbc.M302260200. [DOI] [PubMed] [Google Scholar]
- 52.Lou X, Toresson G, Benod C, Suh JH, Philips KJ, Webb P, Gustafsson JA. Structure of the retinoid X receptor α-liver X receptor β (RXRα-LXRβ) heterodimer on DNA. Nat Struct Mol Biol. 2014, 21: 277–281 . [DOI] [PubMed]
- 53.Chen H, Chen Z, Zhang Z, Li Y, Zhang S, Jiang F, Wei J, et al. Discovery of new LXRβ agonists as glioblastoma inhibitors. Eur J Med Chem. 2020, 194: 112240 . [DOI] [PubMed]
- 54.Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, Bowman ME, Ferrer JL, Anisfeld AM, Edwards PA, Rosenfeld JM, Alvarez JGA, Noel JP, Nicolaou KC, Evans RM. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell. . 2003;11:1079–1092. doi: 10.1016/S1097-2765(03)00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang N, Zou Q, Xu J, Zhang J, Liu J. Ligand binding and heterodimerization with retinoid X receptor α (RXRα) induce farnesoid X receptor (FXR) conformational changes affecting coactivator binding. J Biol Chem. . 2018;293:18180–18191. doi: 10.1074/jbc.RA118.004652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sasaki H, Masuno H, Kawasaki H, Yoshihara A, Numoto N, Ito N, Ishida H, Yamamoto K, Hirata N, Kanda Y, Kawachi E, Kagechika H, Tanatani A. Lithocholic acid derivatives as potent vitamin d receptor agonists. J Med Chem. . 2021;64:516–526. doi: 10.1021/acs.jmedchem.0c01420. [DOI] [PubMed] [Google Scholar]
- 57.Shaffer PL, Gewirth DT. Structural basis of VDR-DNA interactions on direct repeat response elements. EMBO J. . 2002;21:2242–2252. doi: 10.1093/emboj/21.9.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vanhooke JL, Benning MM, Bauer CB, Pike JW, DeLuca HF. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. . 2004;43:4101–4110. doi: 10.1021/bi036056y. [DOI] [PubMed] [Google Scholar]
- 59.Shaffer PL, Gewirth DT. Structural analysis of RXR–VDR interactions on DR 3 DNA . J Steroid Biochem Mol Biol. . 2004;89-90:215–219. doi: 10.1016/j.jsbmb.2004.03.084. [DOI] [PubMed] [Google Scholar]
- 60.Watkins RE, Wisely GB, Moore LB, Collins JL, Lambert MH, Williams SP, Willson TM, et al. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 2001, 292: 2329–2333 . [DOI] [PubMed]
- 61.Delfosse V, Huet T, Harrus D, Granell M, Bourguet M, Gardia-Parège C, Chiavarina B, et al. Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures. Proc Natl Acad Sci U S A. 2021, 118: e2020551118 . [DOI] [PMC free article] [PubMed]
- 62.Lin W, Wang YM, Chai SC, Lv L, Zheng J, Wu J, Zhang Q, Wang YD, Griffin PR, Chen T. SPA70 is a potent antagonist of human pregnane X receptor. Nat Commun. . 2017;8:741. doi: 10.1038/s41467-017-00780-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suino K, Peng L, Reynolds R, Li Y, Cha JY, Repa JJ, Kliewer SA, Xu HE. The nuclear xenobiotic receptor CAR. Mol Cell. . 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 64.Xu RX, Lambert MH, Wisely BB, Warren EN, Weinert EE, Waitt GM, Williams JD, Collins JL, Moore LB, Willson TM, Moore JT. A structural basis for constitutive activity in the human CAR/RXRα heterodimer. Mol Cell. . 2004;16:919–928. doi: 10.1016/j.molcel.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 65.Chandra V, Huang P, Potluri N, Wu D, Kim Y, Rastinejad F. Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature. . 2013;495:394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dhe-Paganon S, Duda K, Iwamoto M, Chi YI, Shoelson SE. Crystal structure of the HNF4α ligand binding domain in complex with endogenous fatty acid ligand. J Biol Chem. . 2002;277:37973–37976. doi: 10.1074/jbc.C200420200. [DOI] [PubMed] [Google Scholar]
- 67.Han EH, Singh P, Lee IK, Urrutia R, Chi YI. ErbB3-binding protein 1 (EBP1) represses HNF4α-mediated transcription and insulin secretion in pancreatic β-cells. J Biol Chem. . 2019;294:13983–13994. doi: 10.1074/jbc.RA119.009558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wisely GB, Miller AB, Davis RG, Thornquest AD Jr, Johnson R, Spitzer T, Sefler A, et al. Hepatocyte nuclear factor 4 is a transcription factor that constitutively binds fatty acids. Structure. 2002, 10: 1225–1234 . [DOI] [PubMed]
- 69.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. . 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 70.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-γ–RXR-α nuclear receptor complex on DNA. Nature. . 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe M, Fujihara M, Motoyama T, Kawasaki M, Yamada S, Takamura Y, Ito S, Makishima M, Nakano S, Kakuta H. Discovery of a “gatekeeper” antagonist that blocks entry pathway to retinoid X receptors (RXRs) without allosteric ligand inhibition in permissive RXR heterodimers. J Med Chem. . 2021;64:430–439. doi: 10.1021/acs.jmedchem.0c01354. [DOI] [PubMed] [Google Scholar]
- 72.Love JD, Gooch JT, Benko S, Li C, Nagy L, Chatterjee VKK, Evans RM, Schwabe JWR. The structural basis for the specificity of retinoid-X receptor-selective agonists: new insights into the role of helix H12. J Biol Chem. . 2002;277:11385–11391. doi: 10.1074/jbc.M110869200. [DOI] [PubMed] [Google Scholar]
- 73.Chaikuad A, Pollinger J, Rühl M, Ni X, Kilu W, Heering J, Merk D. Comprehensive set of tertiary complex structures and palmitic acid binding provide molecular insights into ligand design for RXR isoforms. Int J Mol Sci. 2020, 21: 8457 . [DOI] [PMC free article] [PubMed]
- 74.Zhou XE, Suino-Powell KM, Xu Y, Chan CW, Tanabe O, Kruse SW, Reynolds R, Engel JD, Xu HE. The orphan nuclear receptor TR4 is a vitamin A-activated nuclear receptor. J Biol Chem. . 2011;286:2877–2885. doi: 10.1074/jbc.M110.168740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan MHE, Zhou XE, Soon FF, Li X, Li J, Yong EL, Melcher K, Xu HE. The crystal structure of the orphan nuclear receptor NR2E3/PNR ligand binding domain reveals a dimeric auto-repressed conformation. PLoS ONE. . 2013;8:e74359. doi: 10.1371/journal.pone.0074359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, Xu HE. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid–activated receptor. PLoS Biol. . 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bosica F, Andrei SA, Neves JF, Brandt P, Gunnarsson A, Landrieu I, Ottmann C, et al. Design of drug-like protein-protein interaction stabilizers guided by chelation-controlled bioactive conformation stabilization. Chemistry. 2020, 26: 7131-7139 . [DOI] [PubMed]
- 78.Sijbesma E, Hallenbeck KK, Leysen S, de Vink PJ, Skóra L, Jahnke W, Brunsveld L, Arkin MR, Ottmann C. Site-directed fragment-based screening for the discovery of protein–protein interaction stabilizers. J Am Chem Soc. . 2019;141:3524–3531. doi: 10.1021/jacs.8b11658. [DOI] [PubMed] [Google Scholar]
- 79.Nettles KW, Bruning JB, Gil G, Nowak J, Sharma SK, Hahm JB, Kulp K, Hochberg RB, Zhou H, Katzenellenbogen JA, Katzenellenbogen BS, Kim Y, Joachimiak A, Greene GL. NFκB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses. Nat Chem Biol. . 2008;4:241–247. doi: 10.1038/nchembio.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schwabe JWR, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. . 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-C. [DOI] [PubMed] [Google Scholar]
- 81.Pike AC, Brzozowski AM, Hubbard RE, Bonn T, Thorsell AG, Engström O, Ljunggren J, Gustafsson JA, Carlquist M. Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. . 1999;18:4608–4618. doi: 10.1093/emboj/18.17.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Phillips C, Roberts LR, Schade M, Bazin R, Bent A, Davies NL, Moore R, Pannifer AD, Pickford AR, Prior SH, Read CM, Scott A, Brown DG, Xu B, Irving SL. Design and structure of stapled peptides binding to estrogen receptors. J Am Chem Soc. . 2011;133:9696–9699. doi: 10.1021/ja202946k. [DOI] [PubMed] [Google Scholar]
- 83.Souza PCT, Textor LC, Melo DC, Nascimento AS, Skaf MS, Polikarpov I. An alternative conformation of ERβ bound to estradiol reveals H12 in a stable antagonist position. Sci Rep. . 2017;7:3509. doi: 10.1038/s41598-017-03774-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kallen J, Schlaeppi JM, Bitsch F, Filipuzzi I, Schilb A, Riou V, Graham A, Strauss A, Geiser M, Fournier B. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor α (ERRα) J Biol Chem. . 2004;279:49330–49337. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 85.Gearhart MD, Holmbeck SMA, Evans RM, Dyson HJ, Wright PE. Monomeric complex of human orphan estrogen related receptor-2 with DNA: a pseudo-dimer interface mediates extended half-site recognition. J Mol Biol. . 2003;327:819–832. doi: 10.1016/S0022-2836(03)00183-9. [DOI] [PubMed] [Google Scholar]
- 86.Yao B, Zhang S, Wei Y, Tian S, Lu Z, Jin L, He Y, Xie W, Li Y. Structural insights into the specificity of ligand binding and coactivator assembly by estrogen-related receptor β. J Mol Biol. . 2020;432:5460–5472. doi: 10.1016/j.jmb.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 87.Greschik H, Wurtz JM, Sanglier S, Bourguet W, van Dorsselaer A, Moras D, Renaud JP. Structural and functional evidence for ligand-independent transcriptional activation by the estrogen-related receptor 3. Mol Cell. . 2002;9:303–313. doi: 10.1016/S1097-2765(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 88.Suyama K, Kaneko S, Kesamaru H, Liu X, Matsushima A, Kakuta Y, Okubo T, Kasatani K, Nose T. Evaluation of the influence of halogenation on the binding of bisphenol A to the estrogen-related receptor γ. Chem Res Toxicol. . 2020;33:889–902. doi: 10.1021/acs.chemrestox.9b00379. [DOI] [PubMed] [Google Scholar]
- 89.Thouennon E, Delfosse V, Bailly R, Blanc P, Boulahtouf A, Grimaldi M, Barducci A, Bourguet W, Balaguer P. Insights into the activation mechanism of human estrogen-related receptor γ by environmental endocrine disruptors. Cell Mol Life Sci. . 2019;76:4769–4781. doi: 10.1007/s00018-019-03129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frank F, Okafor CD, Ortlund EA. The first crystal structure of a DNA-free nuclear receptor DNA binding domain sheds light on DNA-driven allostery in the glucocorticoid receptor. Sci Rep. . 2018;8:13497. doi: 10.1038/s41598-018-31812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. . 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 92.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009, 324: 407–410 . [DOI] [PMC free article] [PubMed]
- 93.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013, 20: 53–58 . [DOI] [PMC free article] [PubMed]
- 94.Fagart J, Huyet J, Pinon GM, Rochel M, Mayer C, Rafestin-Oblin ME. Crystal structure of a mutant mineralocorticoid receptor responsible for hypertension. Nat Struct Mol Biol. 2005, 12: 554–555 . [DOI] [PubMed]
- 95.Takahashi M, Ubukata O, Homma T, Asoh Y, Honzumi M, Hayashi N, Saito K, Tsuruoka H, Aoki K, Hanzawa H. Crystal structure of the mineralocorticoid receptor ligand‐binding domain in complex with a potent and selective nonsteroidal blocker, esaxerenone (CS‐3150) FEBS Lett. . 2020;594:1615–1623. doi: 10.1002/1873-3468.13746. [DOI] [PubMed] [Google Scholar]
- 96.Matias PM, Donner P, Coelho R, Thomaz M, Peixoto C, Macedo S, Otto N, Joschko S, Scholz P, Wegg A, Bäsler S, Schäfer M, Egner U, Carrondo MA. Structural evidence for ligand specificity in the binding domain of the human androgen receptor. J Biol Chem. . 2000;275:26164–26171. doi: 10.1074/jbc.M004571200. [DOI] [PubMed] [Google Scholar]
- 97.Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. . 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 98.Lusher SJ, Raaijmakers HCA, Vu-Pham D, Kazemier B, Bosch R, McGuire R, Azevedo R, Hamersma H, Dechering K, Oubrie A, van Duin M, de Vlieg J. X-ray structures of progesterone receptor ligand binding domain in its agonist state reveal differing mechanisms for mixed profiles of 11β-substituted steroids. J Biol Chem. . 2012;287:20333–20343. doi: 10.1074/jbc.M111.308403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sack JS, Kish KF, Wang C, Attar RM, Kiefer SE, An Y, Wu GY, Scheffler JE, Salvati ME, Krystek Stanley R. J, Weinmann R, Einspahr HM. Crystallographic structures of the ligand-binding domains of the androgen receptor and its T877A mutant complexed with the natural agonist dihydrotestosterone. Proc Natl Acad Sci U S A. . 2001;98:4904–4909. doi: 10.1073/pnas.081565498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Estébanez-Perpiñá E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, et al. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A. 2007, 104: 16074-16079 . [DOI] [PMC free article] [PubMed]
- 101.Unwalla R, Mousseau JJ, Fadeyi OO, Choi C, Parris K, Hu B, Kenney T, Chippari S, McNally C, Vishwanathan K, Kilbourne E, Thompson C, Nagpal S, Wrobel J, Yudt M, Morris CA, Powell D, Gilbert AM, Chekler ELP. Structure-based approach to identify 5-[4-hydroxyphenyl]pyrrole-2-carbonitrile derivatives as potent and tissue selective androgen receptor modulators. J Med Chem. . 2017;60:6451–6457. doi: 10.1021/acs.jmedchem.7b00373. [DOI] [PubMed] [Google Scholar]
- 102.Meinke G, Sigler PB. DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nat Struct Biol. . 1999;6:471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- 103.Zhan Y, Chen Y, Zhang Q, Zhuang J, Tian M, Chen H, Zhang L, Zhang H, He J, Wang W, Wu R, Wang Y, Shi C, Yang K, Li A, Xin Y, Li TY, Yang JY, Zheng Z, Yu C, Lin SC, Chang C, Huang P, Lin T, Wu Q. The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat Chem Biol. . 2012;8:897–904. doi: 10.1038/nchembio.1069. [DOI] [PubMed] [Google Scholar]
- 104.Wang W, Wang Y, Chen H, Xing Y, Li F, Zhang Q, Zhou B, Zhang H, Zhang J, Bian X, Li L, Liu Y, Zhao B, Chen Y, Wu R, Li A, Yao L, Chen P, Zhang Y, Tian X, Beermann F, Wu M, Han J, Huang P, Lin T, Wu Q. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat Chem Biol. . 2014;10:133–140. doi: 10.1038/nchembio.1406. [DOI] [PubMed] [Google Scholar]
- 105.Li L, Liu Y, Chen H, Li F, Wu J, Zhang H, He J, Xing Y, Chen Y, Wang W, Tian X, Li A, Zhang Q, Huang P, Han J, Lin T, Wu Q. Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation. Nat Chem Biol. . 2015;11:339–346. doi: 10.1038/nchembio.1788. [DOI] [PubMed] [Google Scholar]
- 106.Yang PB, Hou PP, Liu FY, Hong WB, Chen HZ, Sun XY, Li P, Zhang Y, Ju CY, Luo LJ, Wu SF, Zhou JX, Wang ZJ, He JP, Li L, Zhao TJ, Deng X, Lin T, Wu Q. Blocking PPARγ interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression. Proc Natl Acad Sci U S A. . 2020;117:27412–27422. doi: 10.1073/pnas.2002997117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, Walker NPC, Perlmann T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. . 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 108.Rajan S, Jang Y, Kim CH, Kim W, Toh HT, Jeon J, Song B, Serra A, Lescar J, Yoo JY, Beldar S, Ye H, Kang C, Liu XW, Feitosa M, Kim Y, Hwang D, Goh G, Lim KL, Park HM, Lee CH, Oh SF, Petsko GA, Yoon HS, Kim KS. PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function. Nat Chem Biol. . 2020;16:876–886. doi: 10.1038/s41589-020-0553-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. . 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 110.Blind RD, Sablin EP, Kuchenbecker KM, Chiu HJ, Deacon AM, Das D, Fletterick RJ, Ingraham HA. The signaling phospholipid PIP 3 creates a new interaction surface on the nuclear receptor SF-1 . Proc Natl Acad Sci U S A. . 2014;111:15054–15059. doi: 10.1073/pnas.1416740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sablin EP, Krylova IN, Fletterick RJ, Ingraham HA. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol Cell. . 2003;11:1575–1585. doi: 10.1016/S1097-2765(03)00236-3. [DOI] [PubMed] [Google Scholar]
- 112.Yumoto F, Nguyen P, Sablin EP, Baxter JD, Webb P, Fletterick RJ. Structural basis of coactivation of liver receptor homolog-1 by -catenin. Proc Natl Acad Sci U S A. . 2012;109:143–148. doi: 10.1073/pnas.1117036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proc Natl Acad Sci U S A. . 2008;105:18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weikum ER, Tuntland ML, Murphy MN, Ortlund EA. A structural investigation into oct4 regulation by orphan nuclear receptors, germ cell nuclear factor (GCNF), and liver receptor homolog-1 (LRH-1) J Mol Biol. . 2016;428:4981–4992. doi: 10.1016/j.jmb.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu X, Wang Y, Gutierrez JS, Damsker JM, Nagaraju K, Hoffman EP, Ortlund EA. Disruption of a key ligand-H-bond network drives dissociative properties in vamorolone for Duchenne muscular dystrophy treatment. Proc Natl Acad Sci U S A. . 2020;117:24285–24293. doi: 10.1073/pnas.2006890117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci U S A. . 2005;102:9505–9510. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhi X, Zhou XE, He Y, Zechner C, Suino-Powell KM, Kliewer SA, Melcher K, Mangelsdorf DJ, Xu HE. Structural insights into gene repression by the orphan nuclear receptor SHP. Proc Natl Acad Sci U S A. . 2014;111:839–844. doi: 10.1073/pnas.1322827111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Musille PM, Pathak M, Lauer JL, Hudson WJ, Griffin PR, Ortlund EA. Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nat Struct Mol Biol. 2012, 19: 532–S2 . [DOI] [PMC free article] [PubMed]
- 119.Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem J. . 2005;391:449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Brad Thompson E, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. . 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010, 72: 247–272 . [DOI] [PMC free article] [PubMed]
- 122.Khorasanizadeh S, Rastinejad F. Visualizing the architectures and interactions of nuclear receptors. Endocrinology. . 2016;157:4212–4221. doi: 10.1210/en.2016-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao Q, Chasse SA, Devarakonda S, Sierk ML, Ahvazi B, Rastinejad F. Structural basis of RXR-DNA interactions 1 1Edited by P. E. Wright. J Mol Biol. . 2000;296:509–520. doi: 10.1006/jmbi.1999.3457. [DOI] [PubMed] [Google Scholar]
- 124.Osz J, McEwen AG, Poussin-Courmontagne P, Moutier E, Birck C, Davidson I, Moras D, et al. Structural basis of natural promoter recognition by the retinoid X nuclear receptor. Sci Rep. 2015, 5: 8216 . [DOI] [PMC free article] [PubMed]
- 125.Sierk ML, Zhao Q, Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. . 2001;40:12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 126.Li Y, Meng Q, Yang M, Liu D, Hou X, Tang L, Wang X, et al. Current trends in drug metabolism and pharmacokinetics. Acta Pharm Sin B. 2019, 9: 1113–1144 . [DOI] [PMC free article] [PubMed]
- 127.Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Mol Cell Endocrinol. . 2014;382:97–106. doi: 10.1016/j.mce.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 128.Meyer CA, Tang Q, Liu XS. Minireview: applications of next-generation sequencing on studies of nuclear receptor regulation and function. Mol Endocrinol. 2012, 26: 1651–1659 . [DOI] [PMC free article] [PubMed]
- 129.Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: a perspective from structural genomics. Structure. 2003, 11: 741–746 . [DOI] [PubMed]
- 130.Weatherman RV, Fletterick RJ, Scanlan TS. Nuclear-receptor ligands and ligand-binding domains. Annu Rev Biochem. 199, 68: 559–581 . [DOI] [PubMed]
- 131.Yki-Järvinen H. Thiazolidinediones. N Engl J Med. . 2004;351:1106–1118. doi: 10.1056/NEJMra041001. [DOI] [PubMed] [Google Scholar]
- 132.Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med. . 2007;356:2457–2471. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- 133.Zheng W, Feng X, Qiu L, Pan Z, Wang R, Lin S, Hou D, Jin L, Li Y. Identification of the antibiotic ionomycin as an unexpected peroxisome proliferator-activated receptor γ (PPARγ) ligand with a unique binding mode and effective glucose-lowering activity in a mouse model of diabetes. Diabetologia. . 2013;56:401–411. doi: 10.1007/s00125-012-2777-9. [DOI] [PubMed] [Google Scholar]
- 134.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engström O, Öhman L, Greene GL, Gustafsson JÅ, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. . 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 135.Renaud JP, Moras D. Structural studies on nuclear receptors. CMLS Cell Mol Life Sci. . 2000;57:1748–1769. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. . 1998;10:384–391. doi: 10.1016/S0955-0674(98)80015-X. [DOI] [PubMed] [Google Scholar]
- 137.Lee SK, Na SY, Kim HJ, Soh J, Choi HS, Lee JW. Identification of critical residues for heterodimerization within the ligand-binding domain of retinoid X receptor. Mol Endocrinol. 1998, 12: 325–332 . [DOI] [PubMed]
- 138.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature. . 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 139.Chen S, Costa CH, Nakamura K, Ribeiro RCJ, Gardner DG. Vitamin D-dependent suppression of human atrial natriuretic peptide gene promoter activity requires heterodimer assembly. J Biol Chem. . 1999;274:11260–11266. doi: 10.1074/jbc.274.16.11260. [DOI] [PubMed] [Google Scholar]
- 140.Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. . 2002;110:93–105. doi: 10.1016/S0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 141.Sladek FM, Zhong WM, Lai E, Darnell Jr JE. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990, 4: 2353–2365 . [DOI] [PubMed]
- 142.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, et al. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010, 51: 642–653 . [DOI] [PMC free article] [PubMed]
- 143.Jiang G, Lee U, Sladek FM. Proposed mechanism for the stabilization of nuclear receptor DNA binding via protein dimerization. Mol Cell Biol. . 1997;17:6546–6554. doi: 10.1128/MCB.17.11.6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000, 14: 2819–2830 . [DOI] [PMC free article] [PubMed]
- 145.Kalaany NY, Gauthier KC, Zavacki AM, Mammen PPA, Kitazume T, Peterson JA, Horton JD, Garry DJ, Bianco AC, Mangelsdorf DJ. LXRs regulate the balance between fat storage and oxidation. Cell Metab. . 2005;1:231–244. doi: 10.1016/j.cmet.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 146.Hindinger C, Hinton DR, Kirwin SJ, Atkinson RD, Burnett ME, Bergmann CC, Stohlman SA. Liver X receptor activation decreases the severity of experimental autoimmune encephalomyelitis. J Neurosci Res. . 2006;84:1225–1234. doi: 10.1002/jnr.21038. [DOI] [PubMed] [Google Scholar]
- 147.Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, Gustafsson JA. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. . 2002;99:13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nguyen-Vu T, Vedin LL, Liu K, Jonsson P, Lin JZ, Candelaria NR, Candelaria LP, Addanki S, Williams C, Gustafsson JÅ, Steffensen KR, Lin CY. Liver X receptor ligands disrupt breast cancer cell proliferation through an E2F-mediated mechanism. Breast Cancer Res. . 2013;15:R51. doi: 10.1186/bcr3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lo Sasso G, Bovenga F, Murzilli S, Salvatore L, Di Tullio G, Martelli N, D′Orazio A, Rainaldi S, Vacca M, Mangia A, Palasciano G, Moschetta A. Liver X receptors inhibit proliferation of human colorectal cancer cells and growth of intestinal tumors in mice. Gastroenterology. . 2013;144:1497–1507. doi: 10.1053/j.gastro.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 150.Fan X, Kim HJ, Bouton D, Warner M, Gustafsson JA. Expression of liver X receptor is essential for formation of superficial cortical layers and migration of later-born neurons. Proc Natl Acad Sci U S A. . 2008;105:13445–13450. doi: 10.1073/pnas.0806974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim HJ, Fan X, Gabbi C, Yakimchuk K, Parini P, Warner M, Gustafsson JA. Liver X receptor (LXR ): a link between-sitosterol and amyotrophic lateral sclerosis-Parkinson′s dementia. Proc Natl Acad Sci U S A. . 2008;105:2094–2099. doi: 10.1073/pnas.0711599105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dai Y, Tan X, Wu W, Warner M, Gustafsson JÅ. Liver X receptor protects dopaminergic neurons in a mouse model of Parkinson disease. Proc Natl Acad Sci U S A. . 2012;109:13112–13117. doi: 10.1073/pnas.1210833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wansa KD, Harris JM, Muscat GE. The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem. . 2002;277:33001–33011. doi: 10.1074/jbc.M203572200. [DOI] [PubMed] [Google Scholar]
- 154.Lei N, Zhang X, Chen H, Wang Y, Zhan Y, Zheng Z, Shen Y, Wu Q. A feedback regulatory loop between methyltransferase PRMT1 and orphan receptor TR3. Nucleic Acids Res. . 2009;37:832–848. doi: 10.1093/nar/gkn941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kurakula K, van der Wal E, Geerts D, van Tiel CM, de Vries CJ. FHL2 protein is a novel co-repressor of nuclear receptor Nur77. J Biol Chem. . 2011;286:44336–44343. doi: 10.1074/jbc.M111.308999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yao L, He J, Chen H, Wang Y, Wang W, Wu R, Yu C, Wu Q. Orphan receptor TR3 participates in cisplatin-induced apoptosis via Chk2 phosphorylation to repress intestinal tumorigenesis. Curr Alzheimer Rescinogenesis. . 2012;33:301–311. doi: 10.1093/carcin/bgr287. [DOI] [PubMed] [Google Scholar]
- 157.Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. . 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009, 136: 3205-3214 . [DOI] [PubMed]
- 159.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. . 2009;10:468–477. doi: 10.1038/nrm2717. [DOI] [PubMed] [Google Scholar]
- 160.Xu W, Kimelman D. Mechanistic insights from structural studies of β-catenin and its binding partners. J Cell Sci. . 2007;120:3337–3344. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 161.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell. . 1997;90:871–882. doi: 10.1016/S0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 162.Xing Y, Takemaru KI, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W. Crystal structure of a full-length β-catenin. Structure. . 2008;16:478–487. doi: 10.1016/j.str.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Botrugno OA, Fayard E, Annicotte JS, Haby C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J, Schoonjans K. Synergy between LRH-1 and β-catenin induces G1 cyclin-mediated cell proliferation. Mol Cell. . 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 164.Mulholland DJ, Dedhar S, Coetzee GA, Nelson CC. Interaction of nuclear receptors with the Wnt/β-catenin/TCF signaling axis: Wnt you like to know? Endocrine Rev. . 2005;26:898–915. doi: 10.1210/er.2003-0034. [DOI] [PubMed] [Google Scholar]
- 165.Yang F, Li X, Sharma M, Sasaki CY, Longo DL, Lim B, Sun Z. Linking β-catenin to androgen-signaling pathway. J Biol Chem. . 2002;277:11336–11344. doi: 10.1074/jbc.M111962200. [DOI] [PubMed] [Google Scholar]
- 166.Sun J, Weis WI. Biochemical and structural characterization of β-catenin interactions with nonphosphorylated and CK2-phosphorylated Lef-1. J Mol Biol. . 2011;405:519–530. doi: 10.1016/j.jmb.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. TCF4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. . 2001;8:1048–1052. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 168.Lonard DM, Lanz RB, O′Malley BW. Nuclear receptor coregulators and human disease. Endocrine Rev. . 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 169.Xu PL, Shan SF, Kong YY, Xie YH, Wang Y. Characterization of a strong repression domain in the hinge region of orphan nuclear receptor hB1F/hLRH-1. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao. 2003, 35: 909–916 . [PubMed]
- 170.Nwachukwu JC, Nettles KW. The nuclear receptor signalling scaffold: insights from full-length structures. EMBO J. . 2012;31:251–253. doi: 10.1038/emboj.2011.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Orlov I, Rochel N, Moras D, Klaholz BP. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. . 2012;31:291–300. doi: 10.1038/emboj.2011.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yu X, Yi P, Hamilton RA, Shen H, Chen M, Foulds CE, Mancini MA, Ludtke SJ, Wang Z, O′Malley BW. Structural insights of transcriptionally active, full-length androgen receptor coactivator complexes. Mol Cell. . 2020;79:812–823.e4. doi: 10.1016/j.molcel.2020.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yi P, Wang Z, Feng Q, Pintilie GD, Foulds CE, Lanz RB, Ludtke SJ, Schmid MF, Chiu W, O′Malley BW. Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol Cell. . 2015;57:1047–1058. doi: 10.1016/j.molcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, Kung HJ, Brodie AMH, Edwards J, Qiu Y. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion–resistant growth. Cancer Res. . 2009;69:2305–2313. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jenster G, van der Korput HA, Trapman J, Brinkmann AO. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. . 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 176.Scudellari M. Protein-slaying drugs could be the next blockbuster therapies. Nature. . 2019;567:298–300. doi: 10.1038/d41586-019-00879-3. [DOI] [PubMed] [Google Scholar]
- 177.Jan M, Sperling AS, Ebert BL. Cancer therapies based on targeted protein degradation - lessons learned with lenalidomide. Nat Rev Clin Oncol. 2021, 18: 401–417 . [DOI] [PMC free article] [PubMed]
- 178.Mullard A. Targeted protein degraders crowd into the clinic. Nat Rev Drug Discov. . 2021;20:247–250. doi: 10.1038/d41573-021-00052-4. [DOI] [PubMed] [Google Scholar]