SUMMARY
In North America, the Lyme disease agent, Borrelia burgdorferi, is commonly transmitted by the black-legged tick, Ixodes scapularis. Tick saliva facilitates blood feeding and enhances pathogen survival and transmission. Here, we demonstrate that I. scapularis complement C1q-like protein 3 (IsC1ql3), a tick salivary protein, directly interacts with B. burgdorferi and is important during the initial stage of spirochetal infection of mice. Mice fed upon by B. burgdorferi-infected IsC1ql3-silenced ticks, or IsC1ql3-immunized mice fed upon by B. burgdorferi-infected ticks, have a lower spirochete burden during the early phase of infection compared with control animals. Mechanically, IsC1ql3 interacts with the globular C1q receptor present on the surface of CD4+ and CD8+ T cells, resulting in decreased production of interferon γ. IsC1ql3 is a C1q-domain-containing protein identified in arthropod vectors and has an important role in B. burgdorferi infectivity as the spirochete transitions from the tick to vertebrate host.
Graphical abstract
In brief
Tang et al. report an Ixodes scapularis tick salivary gland protein IsC1ql3, which contributes to the initial stage of the Lyme disease agent Borrelia burgdorferi’s infection of mice. IsC1ql3 inhibits B. burgdorferi-induced IFN-γ production by targeting T cells. These findings demonstrate its immunomodulatory role.
INTRODUCTION
Lyme disease, caused by Borrelia burgdorferi, is one of the most common vector-borne diseases in North America. It results in almost 40,000 cases diagnosed annually in the United States, and the actual number of infections may be 10 times greater according to recent estimates by the Centers for Disease Control and Prevention.1 B. burgdorferi is mainly transmitted by the black-legged tick, Ixodes scapularis. During tick feeding, spirochetes enter the skin at the bite site and then migrate to distal tissues such as the spleen, heart, and joints. I. scapularis attaches to its host for several days and introduces saliva into the dermis. Components in tick saliva facilitate blood feeding and can affect pathogen transmission, either by modulating the host microenvironment at the bite site or by directly interacting with the pathogen.2,3 B. burgdorferi may therefore usurp the immunomodulatory properties of saliva proteins to more efficiently infect the vertebrate host.4
Several salivary proteins dampen immune responses at the tick bite site in vertebrates. A 15-kDa tick salivary gland protein (Salp15) inhibits T cell activation and interleukin (IL)-2 signaling.4,5 The tick saliva lectin pathway inhibitor (TSLPI) binds mannose-binding lectin and inhibits the lectin complement pathway.6 Furthermore, histamine released from basophils and mast cells can influence tick feeding depending on when it is released. Histamine can help increase blood flow during engorgement but can also cause inflammation at the bite site. Ticks not only secrete histamine-binding proteins to reduce the detrimental effects of histamine but also encode a histamine release factor (tHRF) to induce blood flow. Blocking tHRF by the passive transfer of tHRF antiserum to mice can diminish B. burgdorferi infectivity.7 Our recent study also showed that targeting 19 I. scapularis proteins by mRNA vaccination has the potential to disrupt tick feeding and alter B. burgdorferi transmission.3 Therefore, understanding how the Lyme disease spirochete interacts with the vector and host during the initial stage of infection will enhance our knowledge of the pathogenesis of this important tick-borne disease and potentially lead to new treatment and prevention strategies.
The C1q-domain-containing (C1qDC) proteins are characterized by a globular C1q (gC1q) domain in their C terminus. C1qDC proteins contain a collagen region in the N terminus or only have a short N-terminal signal peptide, coiled coil, and other sequence motifs.8 C1qDCs are involved in various physiological processes, including immune and metabolic responses.9,10 One of the best-studied C1qDC proteins, complement component 1q (C1q), is important in the activation of the mammalian classical complement pathway.11 In addition, the C1qDC protein adiponectin is an adipocyte-derived hormone and is involved in influencing a variety of biological processes and biochemical events, such as lipid metabolism, insulin sensitivity, and inflammation.12 In the same adipokine family, closely related C1q/tumor necrosis factor (TNF)-related proteins (CTRPs) have been found to have similar effects.13 C1qDC proteins in invertebrates can also serve as pattern recognition receptors,14-16 and are involved in phagocytosis, inflammation, and the oxidative stress response. C1qDC homologs have not yet been characterized in arthropod vectors of human disease. Recently, we identified a C1qDC protein in the black-legged tick by blasting the tick genome with mammalian adiponectin, which we named the I. scapularis complement C1q-like protein 3 (IsC1ql3). The mammalian adiponectin homolog in ticks, IsC1ql3, regulates tick lipid metabolism by interacting with the tick adiponectin receptor-like protein, and these interactions are critical for B. burgdorferi colonization of I. scapularis.17 In the present study, we define a role for IsC1qI3 in B. burgdorferi infection of mice and demonstrate how a tick-borne pathogen co-opts a vector protein to facilitate infection of the vertebrate host.
RESULTS
Characterization of I. scapularis IsC1ql3
The I. scapularis complement C1q-like protein 3 (IsC1ql3) gene (XP_002415101.2) encodes a protein of 181 amino acids with an N-terminal signal peptide and a globular C1q (gC1q) domain (Figure 1A). The predicted 3D protein structure of the IsC1ql3 gC1q domain has a high degree of similarity to the mammalian complement C1q-like protein 3 with a global model quality estimation (GMQE) value of 0.72 (Figure S1A). Similar to the mammalian C1ql3 structure,9 IsC1ql3 is predicted to form a trimer and has a jelly-roll-like structure (Figure S1B). Phylogenetic tree analysis shows that IsC1ql3 is related to C1q protein, adiponectin, and C1q/TNF-related protein CTRP13 (C1ql3) (Figure 1A). IsC1ql3 is also similar to the C1qDC proteins in shellfish, which function as pattern recognition receptors.16 InterProScan (https://www.ebi.ac.uk/interpro/) and NCBI BLASTp searches show that IsC1ql3 homologs are not present in insects (e.g., Drosophila and mosquitoes) (Figure S2A), but homologs with 62%–96% identity are evident in ticks, including Ixodes persulcatus (GenBank: KAG0410021.1), Dermacentor silvarum (GenBank: XP_037565300.1), Rhipicephalus sanguineus (GenBank: XP_037514814.1), and Rhipicephalus microplus (GenBank: XP_037291008.1). The gC1q domain is highly conserved among these tick species (Figure S2B). Interestingly, IsC1ql3 and the homologs in ticks are only present in Ixodidae (hard) ticks but not Argasidae (soft) ticks, suggesting a specific evolutionary pattern or function of the Ixodidae C1qDC proteins.
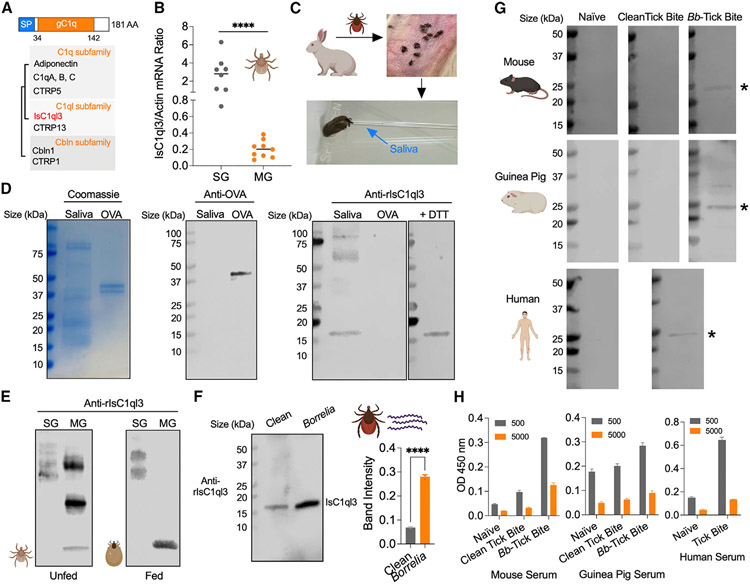
Figure 1. Tick salivary protein IsC1ql3 is induced by B. burgdorferi, and IsC1ql3 antibodies are elicited in animals by natural tick bites.
(A) IsC1ql3 encodes a signal peptide (SP) and a gC1q domain. Simplified tree diagram showing the relationships between C1qDCs and IsC1ql3 (highlighted in red). The tree was adapted from Ye et al.18
(B) RT-qPCR assessment of IsC1ql3 transcript levels in the nymphal tick salivary glands (SGs) and midgut (MG). Each dot represents one biological replicate. Statistical significance was assessed using a non-parametric Mann-Whitney test (****p < 0.0001).
(C) Saliva collection from I. scapularis adults.
(D) SDS-PAGE and western blots of IsC1ql3 in tick saliva. SDS-PAGE gels were run under non-reducing conditions except where specified. Western blots were probed with polyclonal mouse anti-IsC1ql3 or mouse anti-ovalbumin (OVA) sera as a control. The 50 mM reducing agent dithiothreitol (DTT) was applied to reduce IsC1ql3 disulfide-linked multimers or undefined complexes.
(E) Western blots of IsC1ql3 in unfed and fed tick SGs and MG under non-reducing conditions.
(F) Western blots of IsC1ql3 in clean (B. burgdorferi-free) and infected tick saliva. The SDS-PAGE gel was run under reducing conditions. Data are represented as mean ± SD from three replicate experiments. Statistical significance was assessed using an unpaired t test (****p < 0.0001).
(G) IsC1ql3 antibodies are elicited by natural tick bites. Western blots of recombinant IsC1ql3 (rIsC1ql3) by probing with naive serum and serum from mice, guinea pigs, and humans bitten by clean (B. burgdorferi-free) and B. burgdorferi-infected ticks. The asterisks indicate rIsC1ql3.
(H) ELISA analysis of rIsC1ql3 by probing with naive serum and serum from mice and guinea pigs bitten by clean (B. burgdorferi-free) and B. burgdorferi-infected ticks. Humans exposed to tick bites were also examined. Data are represented as mean ± SD from three replicate experiments.
IsC1ql3 is expressed in the I. scapularis gut, salivary gland, and saliva
To investigate the potential function of IsC1ql3, we first evaluated the IsC1ql3 expression profile in the tick gut and salivary glands. IsC1ql3 mRNA was detected in both the gut and salivary glands of ticks (Figure 1B). Notably, salivary glands expressed significantly higher levels of IsC1ql3 compared with the gut (p < 0.0001), suggesting that IsC1qI3 may have a role in processes associated with this organ, such as blood feeding and pathogen transmission. As IsC1ql3 has a signal peptide cleavage site between positions 19 and 20 (Figure 1A), we posited that IsC1ql3 could be secreted out of tick salivary cells into saliva. To examine if IsC1ql3 is present in saliva, we generated recombinant IsC1ql3 (rIsC1ql3) protein with the signal peptide in a Drosophila expression system17 and produced high-titer polyclonal antibodies by immunizing a group of mice (Figure S3). We then utilized the murine anti-IsC1ql3 sera to probe tick saliva, which was harvested from fed ticks using a glass capillary tube (Figure 1C). Immunoblot analysis showed that IsC1ql3 is secreted into saliva (Figure 1D). The predicted molecular weight of endogenous IsC1ql3 without a signal peptide is 17 kDa and is similar to the observed mass seen by western blot (Figure 1D), suggesting IsC1ql3 is a soluble component of saliva. Sera from mice immunized with ovalbumin (OVA) was used as a control (Figures 1D and S3). Under non-reducing conditions, several extra bands were observed with proteins in the range of 50–75 kDa and 75–100 kDa. We explored bands further by first demonstrating that IsC1ql3 has no N-glycoprotein sites as predicted by NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/), indicating that post-translational modifications are not likely the cause of these bands. We then applied the reducing agent dithiothreitol and found that IsC1ql3 in saliva migrated as a monomer with a molecular mass of approximately 17 kDa on immunoblot (Figure 1D). Since IsC1ql3 has a cysteine residue (Figure S1B), it is possible that IsC1ql3 may form disulfide-linked multimers with itself like other C1qDC family proteins (e.g., adiponectin and C1ql3),9,18,19 or IsC1ql3 may bind other undefined protein(s) to form higher order complexes.
We further examined IsC1ql3 protein expression in the tick gut and salivary gland and found that, under non-reducing condi tions, IsC1ql3 appears to be found in an undefined complex in the salivary gland of either unfed or fed nymphal ticks (Figure 1E). Only IsC1ql3 monomers were observed in the gut of fed ticks, suggesting that unknown enzyme(s) in a blood meal may reduce disulfide bonds with IsC1ql3 itself or plasma proteins in the blood meal.
IsC1ql3 antibodies are elicited by natural tick bites and enhanced by B. burgdorferi infection
Recently, we demonstrated that IsC1ql3 is significantly induced in replete nymphal tick guts after feeding on B. burgdorferi-infected mice.17 As IsC1qI3 is highly expressed in the salivary glands, and B. burgdorferi moves through the salivary gland during migration from the tick, we examined whether B. burgdorferi infection enhances IsC1qI3 secretion into saliva. We collected saliva from both B. burgdorferi-infected and uninfected ticks. Immunoblot with anti-B. burgdorferi sera confirmed the infection of the collected saliva from infected ticks (Figure 1F). Western blotting analysis using tick saliva (15 μg total protein) and anti-IsC1ql3 sera showed that B. burgdorferi infection significantly increases IsC1ql3 protein secretion in saliva from salivary gland (p < 0.0001) (Figure 1F). We further quantified IsC1ql3 protein in tick saliva by capture ELISA (Figure S4A) with mouse and rabbit anti-IsC1ql3 sera (Figure S3). The amount of IsC1ql3 per tick was estimated to be approximately 0.88 ng/tick and 1.16 ng/tick in B. burgdorferi-uninfected and -infected adult ticks, respectively, assuming I. scapularis produce approximately 1 mL of saliva (Figure S4B).
We then examined the development of IsC1ql3 antibodies following tick bite. We assessed sera from mice and guinea pigs bitten by B. burgdorferi-infected and -uninfected ticks and from humans with a recent tick bite but without any evidence of clinical B. burgdorferi infection. Mouse, guinea pig, and human sera recognized rIsC1ql3 after tick infestation (Figure 1G). The ELISAs suggested that IsC1ql3 antibodies are elicited by natural tick bites and potentially enhanced by B. burgdorferi infection (Figure 1H).
IsC1ql3 directly interacts with a B. burgdorferi protein ligand through the gC1q domain
Since B. burgdorferi can manipulate IsC1ql3 expression and many C1qDC proteins have high affinity for microbes, we examine whether B. burgdorferi can directly interact with IsC1ql3. A pull-down assay demonstrated that rIsC1ql3 bound to B. burgdorferi (Figure 2A). We then examined whether IsC1ql3 can directly interact with B. burgdorferi through its gC1q domain. We generated the recombinant gC1q domain of IsC1ql3 as a His-tagged protein (rIsC1ql3-gC1q) in a prokaryotic expression system and performed flow-cytometry-based binding assays. rIsC1ql3-gC1q bound to B. burgdorferi, while two other tick proteins, I. scapularis protein disulfide isomerase A3 (IsPDIA3)20 and 14-kDa salivary gland protein (SALP14)21 showed only non-specific background activity (Figure 2B). To further validate the interaction between IsC1ql3-gC1q and B. burgdorferi, we performed ELISAs with B. burgdorferi lysates, rIsC1ql3-gC1q, and rSALP14 proteins. rIsC1ql3-gC1q exhibited a dose-dependent interaction with whole-cell B. burgdorferi lysate, while weak non-specific binding was observed with rSALP14, which did not increase with concentration (Figure 2C). The immunofluorescence assay further confirmed the binding of IsC1ql3-gC1q and B. burgdorferi (Figure 2D). To further investigate the potential B. burgdorferi ligand that interacts with IsC1ql3, we performed ELISA-based binding assays with protease-treated B. burgdorferi lysates. After treatment with proteinase K, B. burgdorferi significantly diminished the ability to bind to IsC1ql3 (Figure 2E). Together, these studies reveal that IsC1ql3 can directly interact with a B. burgdorferi protein ligand through the gC1q domain. We further examined whether the interaction between B. burgdorferi and IsC1ql3 can influence spirochete viability, a critical requirement for B. burgdorferi to establish infection. rIsC1ql3 had no effect on B. burgdorferi viability as assessed by the BacTiter-Glo Microbial Cell Viability Assay (Figure 2F).
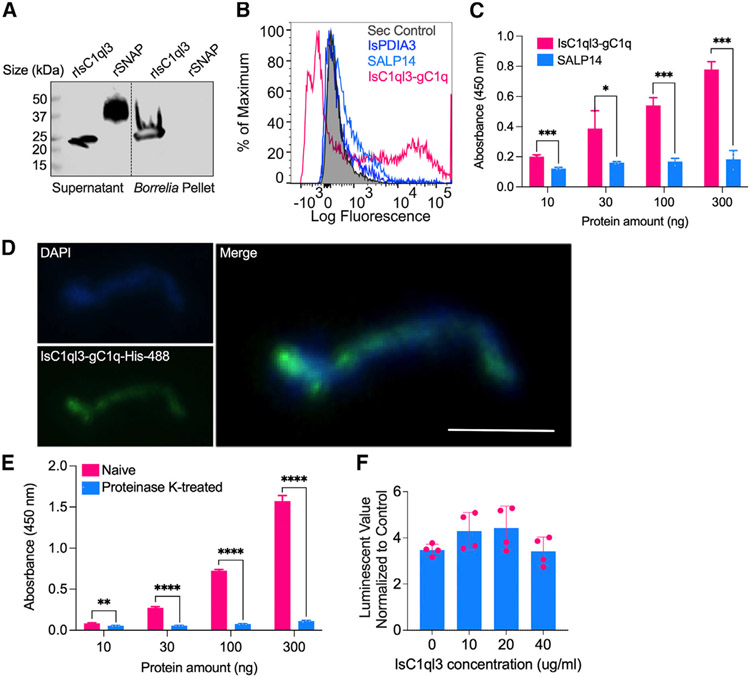
Figure 2. Tick salivary protein IsC1ql3 interacts directly with a B. burgdorferi protein ligand through the gC1q domain.
(A) Binding of rIsC1ql3 to B. burgdorferi was analyzed by a pull-down assay under reducing conditions. Horseradish peroxidase (HRP) V5-tag monoclonal antibody was used to detect protein. rIsC1ql3 was only detected in the B. burgdorferi pellet. The Aedes aegypti synaptosomal-associated protein (rSNAP) was used as a negative control.
(B) Interaction between rIsC1ql3-gC1q and B. burgdorferi as analyzed by flow cytometry. The recombinant I. scapularis protein disulfide isomerase A3 (rIsPDIA3) and 14-kDa salivary gland protein (rSALP14) were used as negative controls. The background of Alexa Fluor 488-His antibody alone with B. burgdorferi is shown in gray shaded region.
(C) ELISA results show the interaction of tick IsC1ql3-gC1q and SALP14 with B. burgdorferi lysate. The lysate was immobilized on microtiter wells and probed with increasing concentrations (10–300 ng) of either rIsC1ql3-gC1q or rSALP14 protein. rIsC1ql3-gC1q showed a dose-dependent interaction with whole-cell B. burgdorferi lysate. Three biological replicates with three technical replicates were included in this assay, and statistical significance was assessed using an unpaired t test (*p < 0.05, ***p < 0.001). Data are represented as mean ± SD.
(D) Binding of rIsC1ql3-gC1q to B. burgdorferi as analyzed by immunofluorescence assay. B. burgdorferi bound to rIsC1ql3-gC1q was measured using an Alexa Fluor 488-conjugated 6X-His monoclonal antibody. Bar: 5 μm.
(E) After treatment with proteinase K, B. burgdorferi binding to IsC1ql3 was markedly reduced. Three biological replicates with three technical replicates were included in this assay, and statistical significance was assessed using an unpaired t test (**p < 0.01, ****p < 0.0001). Data are represented as mean ± SD.
(F) rIsC1ql3 has no effect on B. burgdorferi viability as determined by BacTiter-Glo assay. Each dot represents one replicate. Data are represented as mean ± SD from three replicate experiments.
IsC1ql3-knockdown ticks transmit B. burgdorferi less efficiently to mice
Since IsC1ql3 expression in tick saliva is induced by B. burgdorferi infection and IsC1ql3 can physically interact with B. burgdorferi, we examined whether tick IsC1ql3 has a role in influencing B. burgdorferi transmission from the vector to vertebrate host. We silenced IsC1qI3 gene expression in the salivary gland of infected nymphs by RNA interference (RNAi) and determined whether ticks lacking IsC1ql3 are less capable of transmitting B. burgdorferi to mice. We previously showed that silencing IsC1ql3 had no effect on tick feeding behavior,17 which would suggest that any influence of IsC1ql3 on B. burgdorferi infection is independent of the ability of the tick to take a blood meal. In addition, the B. burgdorferi burden in the infected ticks after engorgement was comparable between ds GFP and ds IsC1ql3 treatments (p > 0.05) (Figure 3A); however, the mice gorged upon by IsC1ql3-knockdown infected nymphal ticks had a significantly lower B. burgdorferi burden in ear tissue at 7 days after infection than the control group (p < 0.05) (Figure 3B). At the later time points, 14 and 21 days, infection levels were unchanged (p > 0.05), suggesting that IsC1ql3 facilitates the establishment of early B. burgdorferi infection in the vertebrate host but does not alter long-term infection.
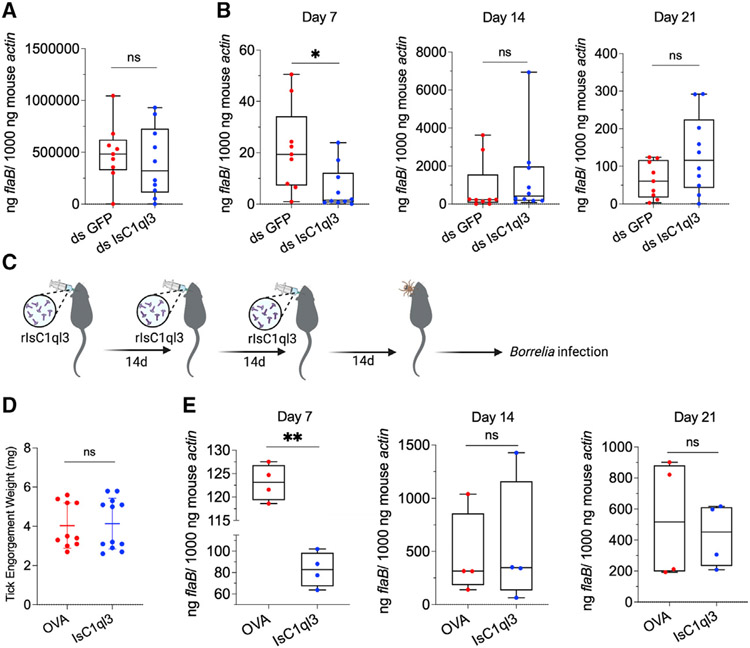
Figure 3. The tick salivary protein IsC1ql3 modulates early infection of mice.
(A) The B. burgdorferi burden in infected ticks after engorgement was comparable between ticks given ds GFP (control) or ds IsC1ql3. Each dot represents one biological replicate. Statistical significance was assessed using a non-parametric Mann-Whitney test (ns, p > 0.05).
(B) B. burgdorferi-infected nymphs microinjected with ds IsC1qI3 or ds GFP were fed on uninfected naive mice to assess transmission of the spirochete. The infection of B. burgdorferi in murine skin 7, 14, and 21 days after infection was determined. Silencing of the tick IsC1qI3 gene significantly decreased the B. burgdorferi burden in murine ear tissue at 7 days following the bite of infected ticks. Each dot represents one biological replicate. Statistical significance was assessed using a non-parametric Mann-Whitney test (*p < 0.05).
(C) Active immunization of mice with IsC1qI3 by injecting 10 μg of rIsC1qI3 protein or OVA (control) three times at 2-week intervals. The immunized mice were then challenged by B. burgdorferi-infected nymphs. The infection of B. burgdorferi in murine skin 7, 14, and 21 days after infection was determined. The antibody titers after the last dose and before tick challenge can be found in Figure S3.
(D) The engorgement weights of nymphs feeding on IsC1ql3-or OVA-immunized mice were comparable. Each dot represents one biological replicate.
(E) Active immunization with rIsC1ql3 protein significantly decreased the B. burgdorferi burden in murine ear tissue at 7 days after the bite of infected ticks. Each dot represents one biological replicate. Statistical significance was assessed using an unpaired t test (**p < 0.01; ns, p > 0.05).
Immunization with IsC1ql3 reduces B. burgdorferi early infection of mice
Next, we examined whether blocking IsC1ql3 in vivo affects tick-borne B. burgdorferi infection. Mice were immunized with 10 μg rIsC1ql3 or with OVA (control). Three weeks after the third immunization, all IsC1ql3-immunized mice exhibited high antibody titers against IsC1ql3 (Figure S3B). Then three B. burgdorferi-infected nymphal ticks were placed on each IsC1ql3-immunized or OVA-immunized mouse and the ticks were allowed to feed to repletion (Figure 3C). The engorgement weights of nymphs feeding on IsC1ql3- or OVA-immunized mice were comparable (Figure 3D), suggesting that IsC1ql3 immunization had no effect on tick feeding behavior, consistent with the data that IsC1qI3-silenced ticks feed normally on mice.17 The B. burgdorferi burden in mice was then examined by qPCR to determine the effect of IsC1ql3 blockade on B. burgdorferi transmission. Compared with the control group, all the mice immunized with IsC1ql3 had a significantly lower B. burgdorferi burden at 7 days (p < 0.01) (Figure 3E). B. burgdorferi levels in mouse skin at 14 days and 21 days post infection were comparable (p > 0.05), further suggesting that IsC1qI3 is only important during the early phase of infection.
IsC1ql3 attenuates the cellular immune response to infection with B. burgdorferi
We next investigated the potential mechanism through which IsC1ql3 facilitates B. burgdorferi infection in mice. Because the mammalian C1q protein, as part of the C1 complex, is central to activation of the antibody-dependent classical complement pathway, we assessed whether tick IsC1ql3 could manipulate the mammalian complement system. Since C1q binds to C1r and C1s to activate the complement cascade, we tested whether IsC1ql3 could bind C1r or C1s by ELISA. The data showed that IsC1ql3 did not interact with C1r or C1s (Figures S5A and S5B). Indeed, IsC1ql3 lacks a collagen-like domain, which is required for binding to C1r and C1s. Furthermore, IsC1ql3 did not interact with the Fc region of monomeric immunoglobulin (Ig) G antibodies (Figure S5C). Taken together, IsC1ql3 may be not involved in the complement pathway cascade.
In addition to the C1q protein, the same gC1q domain has been found in numerous non-complement proteins, which are involved in diverse immunomodulatory activities.9,22-25 Therefore, we hypothesized that IsC1ql3 may alter host immune responses, potentially influencing immune-pathway-related gene expression. We therefore isolated murine splenocytes, which consist of a variety of immune cell populations (e.g., T lymphocytes and macrophages), and incubated the splenocytes with 1 μg/mL rIsC1ql3 or bovine serum albumin (BSA) as control for 6 h (Figure 4A). IsC1ql3 did not affect the viability of splenocytes (Figure S6). Cells were stimulated with lipopolysaccharide (LPS) and the expression of cytokine and chemokine genes, which were listed in our previous study,20 was examined. We found that IsC1ql3 significantly decreased the expression of two cytokines, C-C motif chemokine ligand 3 (CCL3) and interferon γ (IFN-γ) (Figure 4B and S7). Cytokines play an important role in the pathogenesis of Lyme disease by regulating the immune responses against B. burgdorferi,26 and B. burgdorferi induces IFN-γ and CCL3.27,28 We then used B. burgdorferi as the stimulation agent to examine the immunomodulatory role of IsC1ql3. Similar to LPS, IsC1ql3 could inhibit B. burgdorferi-induced IFN-γ expression; however, CCL3 expression was not influenced by IsC1ql3 (Figure 4C). Overall, these results confirm that IsC1ql3 modulates response of immune cells that produce IFN-γ, a signature proinflammatory cytokine.
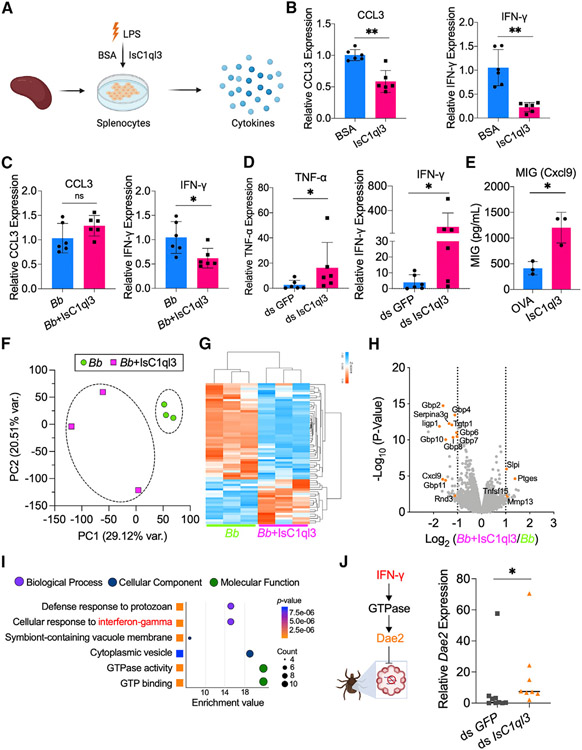
Figure 4. Tick salivary protein IsC1ql3 attenuates the host immune response to B. burgdorferi.
(A) The isolated splenocytes were incubated with 1 μg/mL rIsC1ql3 or BSA and stimulated by lipopolysaccharides (LPS) for 6 h. The cytokine genes expression was then evaluated by qPCR.
(B) IsC1ql3 significantly inhibits CCL3 and IFN-γ gene expression in splenocytes upon LPS stimulation. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (**p < 0.01).
(C) The isolated splenocytes were incubated with B. burgdorferi (1 × 106 cells/mL) alone or mixture with 1 μg/mL rIsC1ql3 for 6 h. IFN-γ gene expression was then evaluated by qPCR. IsC1ql3 significantly inhibits IFN-γ gene expression in splenocytes upon B. burgdorferi infection, but with no effect on CCL3 expression. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (*p < 0.05; ns, p > 0.05).
(D) B. burgdorferi-infected nymphs microinjected with ds IsC1qI3 or ds GFP were fed on clean mice for 72 h to assess cytokine genes expression at the tick bite site. IFN-γ and TNF-α gene expression was significantly upregulated in the absence of tick IsC1ql3. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (*p < 0.05).
(E) Increase in proinflammatory chemokine MIG (CXCL9) was observed in all the IsC1qI3-immunized mice compared with the control mice after B. burgdorferi-infected tick bite. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using an unpaired t test (*p < 0.05).
(F) Principal-component analysis (PCA) of transcriptome data from splenocytes stimulated by B. burgdorferi with or without IsC1ql3.
(G) Cluster dendrogram and heatmap of transcriptome data from splenocytes stimulated by spirochetes with or without IsC1ql3. Each column represents biological replicates. The phylogenetic relationships of differentially expressed genes are shown on the right tree. The top tree indicates the cluster relationship of the sequenced samples.
(H) Volcano plot of differentially expressed genes. The genes related to IFN-γ are highlighted in orange. The gene names can be found in Table S1.
(I) GO enrichment analysis of transcriptome data from splenocytes stimulated by B. burgdorferi spirochetes with or without IsC1ql3. The second-level GO terms are shown in the plot, and enrichment analysis was performed using functional annotation tool DAVID.
(J) Mammalian IFN-γ induces a tick Rho-like GTPase, resulting in expression of antimicrobial peptide Dae2.29 IsC1ql3 silencing caused increased IFN-γ level in blood-meal-induced tick Dae2 expression during tick feeding. Each dot represents one biological replicate. Statistical significance was assessed using a non-parametric Mann-Whitney test (*p < 0.05).
Tick IsC1ql3 inhibits IFN-γ expression at the tick bite site
Since IsC1ql3 influences immune-pathway related gene expression in vitro, we examined whether IsC1ql3 affects host immune responses at the bite site in vivo by silencing IsC1ql3. B. burgdorferi-infected nymphs were injected with GFP or IsC1ql3 dsRNA and fed on mice for 72 h to assess cytokine gene expression at the bite site. RT-qPCR analysis revealed that the transcription of IFN-γ and TNF-α, among the genes analyzed, were significantly upregulated in the absence of IsC1ql3 (Figures 4D and S8). We also examined whether blocking IsC1ql3 would affect cytokine expression. IsC1qI3-immunized mice were challenged with B. burgdorferi-infected nymphs. Upon being bitten by the infected ticks, serum cytokine profiles were assessed using a murine cytokine/chemokine array. IFN-γ was not detectable; however, increases in the IFN-γ-induced chemokine MIG (CXCL9)30 were observed in all the IsC1qI3-immunized mice compared with controls (Figures 4E and S9). This suggests that, when IsC1qI3 was blocked during tick feeding, the mice had a stronger proinflammatory response to B. burgdorferi.
IsC1ql3-associated host transcriptional responses to B. burgdorferi
To further understand the immunomodulation roles of IsC1ql3, we utilized RNA sequencing (RNA-seq) to investigate the effect of IsC1ql3 on the murine transcriptome. We compared the transcriptome of splenocytes stimulated by B. burgdorferi, with or without IsC1ql3. Principal-component analysis (PCA) and cluster dendrograms revealed that IsC1ql3-incubated cells formed a separate cluster from the cells without IsC1ql3 after B. burgdorferi stimulation (Figures 4F and 4G). We found that 63 genes were differentially expressed (p < 0.05 and fold change ≥2), 43 genes were downregulated, and 20 genes were upregulated when incubated with IsC1ql3 (Table S1). Interestingly, the top genes of the 43 downregulated genes were all IFN-γ-induced genes, including 10 IFN-γ inducible GTPases (IFN-γ inducible 47-kDa GTPase [Iigp1], guanylate binding protein [Gbp2], Gbp11, Gbp10, Gbp8, Gbp4, Gbp6, Gbp7, T cell specific GTPase 1 [Tgtp1], and Rho family GTPase 3 [Rnd3]), Cxcl9, and serine and cysteine protease inhibitor (Serpina3g) (Figure 4H). In contrast, four of the 20 upregulated genes were IFN-γ-repressed genes, including secretory leukocyte protease inhibitor (Slpi), TNF superfamily member 15 (Tnfsf15), matrix metallopeptidase 13 (Mmp13), and prostaglandin E synthase (Ptges) (Figure 4H). Based on Gene Ontology (GO) functional classification, the most differentially expressed genes in the biological process category were involved in defense responses to protozoa (GO: 0042832) and cellular responses to interferon-γ (GO: 0071346) (Figure 3I). Therefore, IsC1ql3 mainly inhibits IFN-γ production, including multiple IFN-γ-inducible GTPases. Smith et al.29 identified a signaling cascade whereby IFN-γ in the incoming blood meal induces a tick Rho-like GTPase (IGTPase), which enhances expression of the antimicrobial peptide Dae2, limiting B. burgdorferi levels (Figure 4J). Since IsC1ql3 inhibits IFN-γ production in mammals, we investigated whether IsC1ql3 silencing caused increased IFN-γ levels in the incoming blood meal, which would then induce tick Dae2 expression. The result showed that Dae2 expression was significantly induced in the fed tick gut after silencing IsC1ql3 (p < 0.05) (Figure 4J), which may limit B. burgdorferi survival. This is consistent with our previous observation that IsC1ql3-silenced nymphs had a marked reduction of the B. burgdorferi burden in the gut.17 Taken together, IsC1ql3 attenuates the host immune response by inhibiting IFN-γ production, which has been shown to limit B. burgdorferi survival in both mammals and ticks.2,29,31
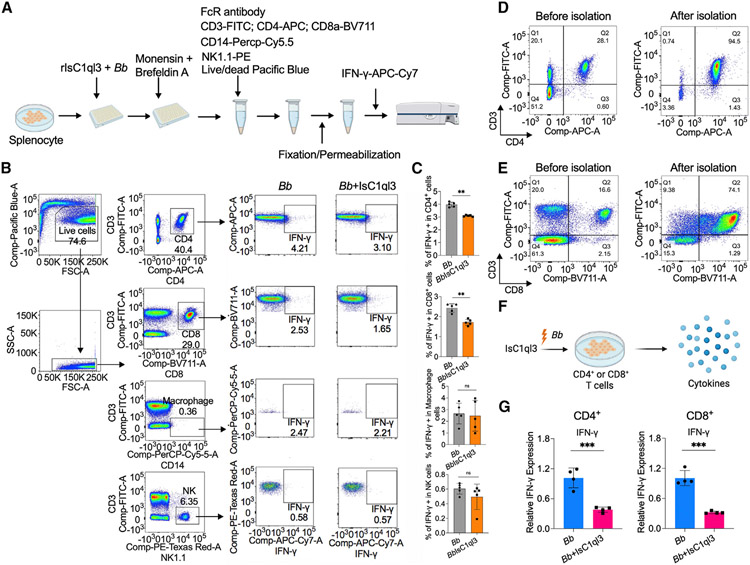
IsC1ql3 inhibits B. burgdorferi-induced IFN-γ production by targeting T cells
IFN-γ is primarily secreted by T cells, macrophages, and natural killer (NK) cells.32,33 To investigate which immune cell(s) were targeted by IsC1ql3 to regulate IFN-γ production, CD4+, CD8+ T cells, macrophages, and NK cells were isolated from murine splenocytes by intracellular cytokine staining of IFN-γ using flow cytometry (Figure 5A). We stimulated the cells with B. burgdorferi since B. burgdorferi induces the production of IFN-γ by NK or T cells.27 We found that IsC1ql3 significantly inhibited IFN-γ production in CD4+ and CD8+ T cells (p < 0.01) but not in macrophages or NK cells (Figures 5B and 5C). We further utilized IL-12 as an IFN-γ activator, since it evokes IFN-γ responses in B. burgdorferi-exposed cells.34 In this case, IsC1ql3 still significantly inhibited IFN-γ production in CD4+ and CD8+ T cells, and macrophages and NK cells also produced less IFN-γ when incubated with IsC1ql3 (Figure S10). Based on the intracellular cytokine staining data, it is likely that IsC1ql3 mainly targets CD4+ and CD8+ T cells to repress B. burgdorferi-activated IFN-γ production. To further confirm the influence of IsC1ql3 on CD4+ and CD8+ T cells, we isolated CD4+ and CD8+ T cells from splenocytes (Figure 5D and 5E) and then incubated the cells with B. burgdorferi and IsC1ql3 (Figure 5F). The results showed that IsC1ql3 inhibits B. burgdorferi-induced IFN-γ production in CD4+ or CD8+ T cells (Figure 5G).
Figure 5. Tick salivary protein IsC1ql3 inhibits B. burgdorferi-induced IFN-γ production by mainly targeting CD4+ and CD8+ T cells.
(A) Work flow to detect IFN-γ production in CD4+, CD8+ T cells, macrophages, and NK cells after incubating with B. burgdorferi (1 × 106 cells/mL) alone or mixture with 1 μg/mL rIsC1ql3 for 6 h by flow cytometry.
(B) Flow cytometry and gating strategy to quantify IFN-γ production in CD4+, CD8+ T cells, macrophages, and NK cells.
(C) Percentage of CD4+, CD8+ T cells, macrophages, and NK cells expressing IFN-γ. IsC1ql3 significantly inhibits IFN-γ production in CD4+ and CD8+ cells upon B. burgdorferi infection. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (**p < 0.01).
(D) Isolation of CD4+ T cells from splenocytes. The purity of the isolated CD4+ T cells was determined by flow cytometry.
(E) Isolation of CD8+ T cells from splenocytes. The purity of the isolated CD8+ T cells was determined by flow cytometry.
(F) The purified CD4+ or CD8+ T cells were incubated with spirochetes (1 × 106 cells/mL) alone or as a mixture with 1 μg/mL rIsC1ql3 for 6 h. The IFN-γ gene expression was then evaluated by qPCR.
(G) IsC1ql3 significantly inhibits IFN-γ gene expression in CD4+ or CD8+ T cells upon B. burgdorferi infection. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (*p < 0.05).
IsC1ql3 interacts with gC1qR on immune cells to modulate the immune response
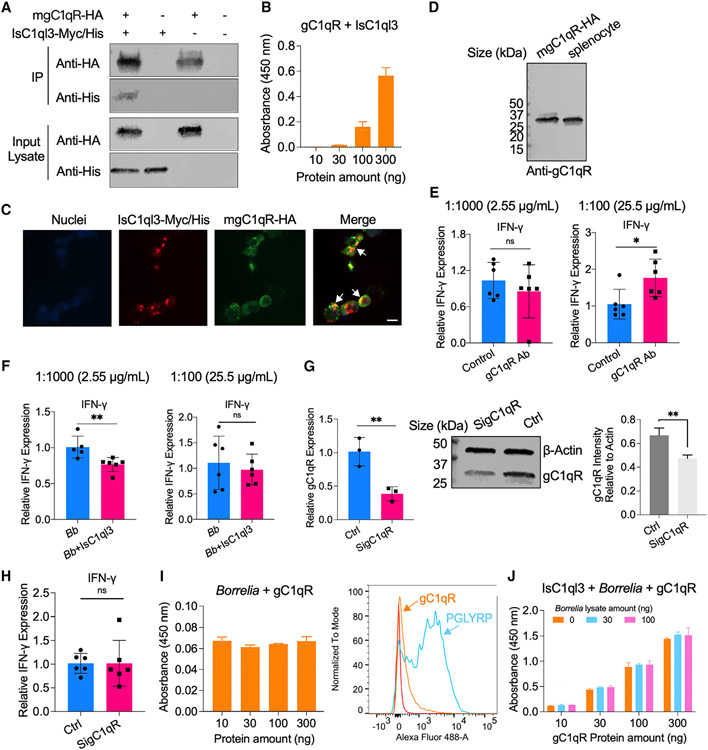
We further explored how IsC1ql3 regulates cytokine production in immune cells. A candidate receptor for the gC1q domain-containing proteins is a multi-functional pattern recognition protein, gC1qR.35,36 We investigated this target because gC1qR regulates IFN-γ production by T cells.37 gC1qR also plays an important role in microbial pathogenesis and immune regulation. For instance, hepatitis C virus core protein binds to gC1qR on the surface of T cells, resulting in reduced production of cytokines, including IFN-γ and IL-12.38-39 We therefore examined whether IsC1ql3-mediated IFN-γ inhibition in T cells is mediated through gC1qR. Co-immunoprecipitation (coIP) assay indicated that IsC1ql3 interacted with gC1qR as demonstrated by the detection of IsC1ql3 protein with HA-Tag immunoprecipitation of mgC1qR-HA (Figure 6A). To confirm the coIP results, we performed an ELISA with gC1qR, and we found that IsC1ql3 bound gC1qR in a dose-dependent manner (Figure 6B). We further utilized microscopy to determine whether IsC1ql3 can colocalize with gC1qR, and the co-immunolocalization assay demonstrated that IsC1ql3 bound on the surface of cells with partial colocalization to gC1qR (Figure 6C). The western blot assay further confirmed that gC1qR is expressed in splenocytes (Figure 6D). Therefore, gC1qR on immune cells may serve as a receptor for IsC1ql3.
Figure 6. IsC1ql3 interacts with gC1qR on the surface of immune cells and modulates host immune responses.
(A) gC1qR-HA interacted with IsC1ql3-Myc/His in coIP assays with co-transfecting plasmids encoding mouse mgC1qR-HA and/or IsC1ql3-Myc/His into HEK293T cells.
(B) ELISA results show the interaction of IsC1ql3 and gC1qR. The rgC1qR protein was immobilized on microtiter wells and probed with increasing concentrations (10–300 ng) of rIsC1ql3 protein. rIsC1ql3 showed dose-dependent interaction with rgC1qR protein. Three biological replicates with three technical replicates were included in this assay. Data are represented as mean ± SD.
(C) Co-immunolocalization of gC1qR-HA and IsC1ql3-Myc/His. gC1qR-HA was detected using a rabbit anti-HA polyclonal antibody and goat anti-rabbit IgG labeled with Alexa Fluor 488 (green) secondary antibody. IsC1ql3-Myc/His was detected using a mouse anti-His polyclonal antibody and goat anti-mouse IgG labeled with Alexa Fluor 549 (red) secondary antibody. The white arrows indicate examples of colocalization. Bar: 10 μm.
(D) gC1qR is expressed in the splenocytes as determined by western blot.
(E) The splenocytes were incubated with gC1qR antibody (Ab) at a concentration of 1:1,000 (2.55 μg/mL) and 1:100 (25.5 μg/mL). The IFN-γ gene expression was then evaluated by qPCR. The gC1qR polyclonal antibody stimulates IFN-γ gene expression at the concentration of 1:100. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (*p < 0.05; ns, p > 0.05).
(F) The splenocytes were incubated with 25.5 and 2.55 μg/mL gC1qR antibody for 20 min and then the cells were stimulated with a mixture of IsC1ql3 and B. burgdorferi. IFN-γ gene expression was then evaluated by qPCR. The polyclonal antibody against gC1qR blocked interactions with IsC1ql3 in splenocytes at the concentration of 1:100. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (**p < 0.01; ns, p > 0.05).
(G) gC1qR expression was silenced by delivering siRNA into splenocytes. qPCR and immunoblotting confirmed silencing of gC1qR. Control (Ctrl) group was transfected with Allstars negative control (Qiagen). Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using an unpaired t test (**p < 0.01).
(H) Silencing of gC1qR affected IsC1ql3-mediated IFN-γ inhibition. Each dot represents one biological replicate. Data are represented as mean ± SD. Statistical significance was assessed using a non-parametric Mann-Whitney test (ns, p > 0.05).
(I) B. burgdorferi did not interact with gC1qR as determined by ELISA and flow cytometry. The peptidoglycan recognition protein 1 (PGLYRP1) was used as the positive control.38 Three biological replicates with three technical replicates were included in this assay. Data are represented as mean ± SD.
(J) Competitive ELISA showed that the interaction between gC1qR and IsC1ql3 was not altered in the absence or presence of B. burgdorferi. The rIsC1ql3 protein with or without B. burgdorferi lysate was probed with increasing concentrations (10–300 ng) of gC1qR protein. Three biological replicates with three technical replicates were included in this assay. Data are represented as mean ± SD.
We then investigated whether IsC1ql3 interacts with gC1qR in a manner that contributes to IFN-γ production. In particular, we examined whether antibody against gC1qR could block IsC1ql3-mediated IFN-γ production in immune cells. We first determined the effect of gC1qR antibody on gC1qR-mediated IFN-γ production (Figure 6E). The results showed that incubation of gC1qR antibody (against gC1qR fusion protein, amino acids 74–282) for 6 h exhibited a dose-dependent induction of IFN-γ in splenocytes. Specifically, gC1qR antibody significantly induced IFN-γ expression at a concentration of 25.5 μg/mL (1:100); however, changes in IFN-γ expression were not observed at the concentration of 2.55 μg/mL (1:1,000) (Figure 6E). We then incubated the splenocytes with 25.5 μg/mL or 2.55 μg/mL gC1qR antibody for 20 min and stimulated the cells with a mixture of IsC1ql3 and B. burgdorferi. We still found that IsC1ql3 significantly inhibited IFN-γ production in splenocytes after blocking with 2.55 μg/mL gC1qR antibody; however, gC1qR antibody blocked IsC1ql3-mediated IFN-γ inhibition at the concentration of 25.5 μg/mL (Figure 6F). We further silenced gC1qR expression by delivering small interfering RNA (siRNA) into splenocytes (Figure 6G) and also found that silencing of gC1qR affected IsC1ql3-mediated IFN-γ inhibition (Figure 6H). These data suggest that IsC1ql3 may interfere with gC1qR in a manner that contributes to IFN-γ production.
Since IsC1ql3 has affinity for both B. burgdorferi and gC1qR, and many types of microbial pathogens possess strong affinity for gC1qR to regulate cytokine expression during infection,40 it is possible that IsC1ql3 prevents B. burgdorferi from binding gC1qR to escape host immune responses. Therefore, we further examined whether B. burgdorferi could bind to gC1qR. B. burgdorferi showed no direct interaction with gC1qR based on ELISA and flow cytometry assays (Figure 6I). We further investigated whether B. burgdorferi and gC1qR competitively bind IsC1ql3 or form a tripartite complex. The competitive ELISA showed that the interaction between gC1qR and IsC1ql3 was not altered in the absence or presence of B. burgdorferi (Figure 6J).
DISCUSSION
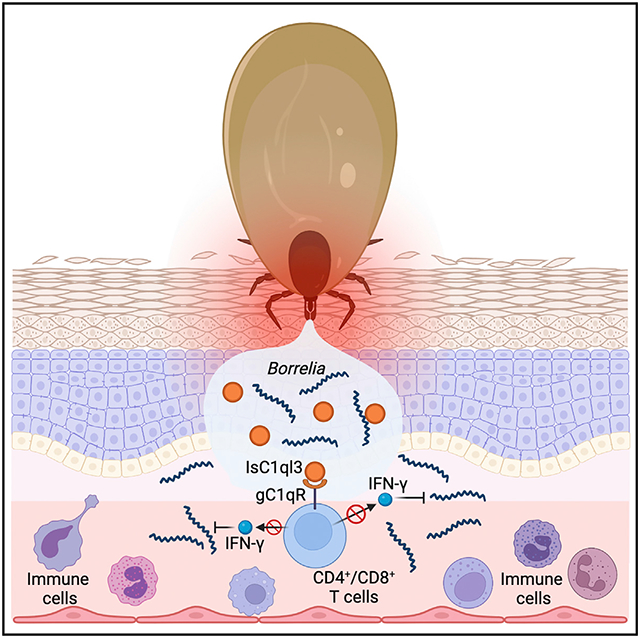
Understanding the interface between the mammalian host and I. scapularis may lead to alternative strategies for counteracting tick-borne diseases. Tick salivary proteins contain immunomodulatory molecules that can alter the local host environment, thereby influencing tick feeding and/or the transmission of pathogens.41 In this study, we characterized an I. scapularis salivary protein, IsC1ql3, which is a C1qDC protein identified in arthropod vectors. IsC1ql3 is important during the initial stage of spirochetal infection of mice as silencing of the IsC1ql3 gene in ticks or blocking IsC1ql3 via immunization significantly reduces B. burgdorferi levels during the initial phase of tick-transmitted infection of mice.
The complement system is an important component of innate immunity42 and an attractive target for microbial manipulation of host defenses.39 Several tick salivary proteins or B. burgdorferi surface proteins interfere with the mammalian complement cascade.6,43,44 C1q is the recognition component of the C1 complex and initiates the classical complement cascade upon activation. IsC1ql3 has a gC1q domain; however, it is not involved in manipulating the host complement system. Instead, our study demonstrated that IsC1ql3 facilitates the transmission of B. burgdorferi by counteracting lymphocyte-mediated host immune responses, thereby providing a survival advantage to B. burgdorferi in the early phase of murine infection. Specifically, IsC1ql3 attenuates the host responses to B. burgdorferi by inhibiting IFN-γ production. Indeed, IFN-γ has been associated with host defense against B. burgdorferi,2,31 and tick saliva and salivary gland extract downregulates IFN-γ production in T cells stimulated with concanavalin A.45
Our study further demonstrates that IsC1ql3 inhibits IFN-γ production by interacting with gC1qR on CD4+ or CD8+ T cells. Many microbes exploit gC1qR-dependent regulatory pathways to manipulate host immunity. gC1qR has been reported to interact with some viral and bacterial proteins, including hepatitis C virus and Staphylococcus aureus protein A, suggesting that gC1qR may facilitatea shared mechanism of pathogen-mediated immune suppression.22,46 Importantly, gC1qR downregulates mammalian cytokine production, including IL-12 and IFN-γ, in T lymphocytes.47-51 We show that B. burgdorferi does not interact directly with gC1qR. Instead, B. burgdorferi interacts with IsC1ql3 and co-opts IsC1ql3 to manipulate gC1qR-mediated IFN-γ signaling in host T cells, therefore attenuating host immune responses. The immunosuppressive effect is common in C1qDC proteins. For instance, C1q/TNF-Related Protein-3 (CTRP3), as an LPS antagonist, specifically and effectively inhibits the binding of LPS to its receptor, TLR4/MD-2, leading to the inhibition of proinflammatory pathways.52 Moreover, the adiponectin paralog CORS-26 exerts anti-inflammatory effects in LPS-treated monocytic cells, and CORS-26 also reduces IL-6 and TNF-α secretion.53 In addition to proinflammatory inhibition, C1qDC proteins in invertebrates can serve as pattern recognition receptors.14-16 For instance, the C1qDCs in pacific oysters such as Crassostrea gigas have high affinity for bacterial LPS or peptidoglycan and may exert functions in the immune responses against invading microbes.14,16 Although IsC1ql3 can recognize and directly interact with B. burgdorferi, it does not serve as a pattern recognition receptor in ticks to activate the immune system against B. burgdorferi.
IsC1ql3 homologs are not present in insects, including disease-transmitted vectors such as mosquitoes and tsetse flies, suggesting that IsC1ql3 may be specifically exploited by tick-borne pathogens to enhance infectivity. In addition, IsC1ql3 and its homologs are only present in Ixodidae ticks, and not Argasidae ticks, which suggests that blood-feeding adaptations occurred independently in these distinct tick families. The divergence of Ixodidae ticks and Argasidae ticks was predicted to be in the late Cretaceous period (120–92 million years ago), and the major tick families evolved to have different hematophagy processes.54 Therefore, the origin and evolution of IsC1ql3 may be related to blood-feeding adaptations in Ixodidae ticks. We speculate that IsC1ql3 may have been acquired from other species (e.g., vertebrate host) by horizontal gene transfer a long time ago, since this phenomenon has been demonstrated with tick-adrenomedullin (TAM). TAMs are conserved in all vertebrates and Ornithodoros ticks but not in any other invertebrates, including Argas ticks.55 Ornithodoros ticks horizontally acquired vertebrate adrenomedullin and employed it to facilitate blood feeding.
In summary, this study elucidates how a specific tripartite relationship between B. burgdorferi, a component of tick saliva, and the vertebrate host response influences infectivity of the Lyme spirochete during the initial phase of infection. IsC1ql3 is unusual for several reasons. First, it has a role in altering the IFN-γ response, which is special among C1q-related proteins. Second, it is important in two stages of the spirochete life cycle: colonization of I. scapularis17 and infection of the vertebrate host, whereas all I. scapularis proteins identified so far function at one specific interval of the Borrelia life cycle. This distinctive interaction between B. burgdorferi and IsC1qI3 demonstrates the diverse mechanisms that the Lyme disease agent uses to survive in the arthropod and vertebrate hosts.
Limitations of the study
Our study demonstrates that IsC1ql3 may form disulfide-linked multimers with itself or bind other undefined protein(s) to form higher-order complexes. The exact components of the complexes and the potential structure of the IsC1ql3 multimers still require further investigation. In addition, IsC1ql3 can directly interact with a B. burgdorferi protein ligand on the surface of the spirochete, and this ligand(s) is likely present in both the tick and mammal. Elucidating the protein ligand (s) and the avidity of the binding would enhance our understanding of this important vector-pathogen interaction. Our studies also suggest that IsC1ql3 does not interact with the Fc region of monomeric IgG antibodies. However, C1q has a higher affinity for hexameric IgG, and complement can be activated by IgG hexamers.56 Therefore, additional studies should be conducted with antibody-antigen immune complex.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiaotian Tang (xiaotian.tang@yale.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
The RNA-seq data are available in the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information under the accession number: GSE207307.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics statement
All the experiments in this study were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal protocols were approved by the Institutional Animal Care and Use Committee at the Yale University School of Medicine.
Animals, ticks, spirochetes, and cells
Six-week-old female C3H/HeN and C57BL/6J mice were purchased from the Charles River Laboratories and Jackson Laboratory. All mice were maintained in a pathogen-free facility at Yale University. To obtain spirochetes-infected mice, The mice were injected subcutaneously with 100 μL of 1 × 105 cells/mL B. burgdorferi (strain N40).
Pathogen-free I. scapularis larvae and adults were obtained from the Oklahoma State University (Stillwater, OK). The larval ticks were fed on pathogen-free C3H/HeN mice and allowed to molt to nymphs. To generate B. burgdorferi-infected nymphs, the larvae were placed on spirochetes-infected mice. All the ticks were maintained at 85% relative humidity with a 14h light and 10h dark period at 23°C.
The spirochetes B. burgdorferi (strain N40) were grown in Barbour-Stoenner-Kelly H (BSK-H) complete medium (Sigma-Aldrich, #B8291) in a 33°C setting incubator. The live cell density was determined by dark field microscopy and hemocytometer (INCYTO, #DHC-N01).
Human embryonic kidney HEK293T cells (ATCC, #CRL-3216) were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, ThermoFisher, #11965-118) media supplemented with 10% Fetal Bovine Serum (FBS, Sigma, #12306C-500). Drosophila S2 cells (ATCC, #CRL-1963) were grown in Schneider’s Drosophila media (Gibco, #21720-024) with 10% FBS at 28°C. Murine splenocytes were isolated from mice as described in Uraki et al.59 Briefly, the spleens were minced in RPMI 1640 medium (Sigma-Aldrich, #R4130) and forced through a 70 μm cell-strainer nylon mesh (Fisherbrand, #22-363-548). After a single wash with PBS and red blood cells lysis with NH4Cl, the cells were resuspended in RPMI 1640 medium. Murine CD4+ and CD8+ T cells were isolated from splenocytes with MojoSort™ mouse CD4 and CD8 T cell isolation kits (BioLegend, #480005 and #480007). The purity of the isolated cells was evaluated by flow cytometry with the antibodies of FITC anti-mouse CD3 antibody (BioLegend, #100204), APC anti-mouse CD4 antibody (BioLegend, #100412), and Brilliant Violet 711™ anti-mouse CD8a antibody (BioLegend, #100747).
METHOD DETAILS
Bioinformatic analysis
The homologs of IsC1ql3 (XP_002415101.2) were identified based on InterProScan (https://www.ebi.ac.uk/interpro/) and BLASTp search. Multiple alignment of protein sequences was performed using the Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The IsC1ql3 protein structure homology-modelling was performed in SWISS-MODEL (https://swissmodel.expasy.org/).
Gene expression evaluation by quantitative real-time PCR (qPCR)
To evaluate gene expression of target genes in ticks, pathogen-free, B. burgdorferi-infected, unfed, or fed I. scapularis nymphs were dissected under the dissecting microscope to get guts and salivary glands. The RNA from dissected guts and salivary glands were purified by Trizol (Invitrogen, #15596-018) following the manufacturer’s protocol, and cDNA was synthesized using the iScript cDNA Synthesis Kits (Bio-Rad, #1708891). qPCR was performed using iQ SYBR Green Supermix (Bio-Rad, #1725124) with an initial denaturing step of 2 min at 95°C and 45 amplification cycles consisting of 20 s at 95°C followed by 15 s at 60°C, and 30 s at 72°C. The target genes and corresponding primer sequences are shown in Table S2.
Tick saliva collection
Tick saliva was collected from adult ticks as previously described.4 Briefly, pathogen-free or B. burgdorferi-infected adult ticks were placed on rabbits and collected after feeding to repletion. Then the ticks were immobilized on microscope slides with double-sided tape. Tick mouth parts were placed inside capillary tubes, and 2 μL of 50 mg/mL pilocarpine was applied. Saliva was then collected for 2–3 h, and the concentration was quantified using the BCA Protein Estimation kit (ThermoFisher Scientific, #23225).
Purification of recombinant proteins
The IsC1ql3 gene full length with signal peptide was PCR amplified from tick nymph cDNA using the primer pair listed in Table S2, then cloned into the BglII and XhoI sites of the pMT/BiP/V5-His vector (Invitrogen, #V413020). The recombinant protein was expressed and purified using the Drosophila Expression System as described previously.60 The protein was purified from the supernatant by TALON metal affinity resin (Clontech, #635606) and eluted with 150 mM imidazole. The eluted samples were filtered through a 0.22-μm filter and concentrated with a 10-kDa concentrator (MilliporeSigma, #Z740203) by centrifugation at 4°C. Recombinant protein purities were assessed by SDS-PAGE using 4–20% Mini-Protean TGX gels (Bio-Rad, #4561094) and quantified using the BCA Protein Estimation kit (ThermoFisher Scientific, #23225).
The gC1q domain of IsC1ql3 gene was PCR-amplified from tick nymph cDNA using the primer pair listed in Table S2, then cloned into the NdeI and HindIII sites of the pET-28a (+) vector (Addgene, #69864-3). Recombinant protein was expressed in Escherichia coli (E. coli) and was further purified by Ni-NTA agarose (QIAGEN, #30230) as described by the manufacturer. The eluted samples were filtered through a 0.22-μm filter and concentrated with a 3-kDa concentrator (MilliporeSigma, #C7719) by centrifugation at 4°C. Recombinant protein purities were assessed and quantified as described above.
Mouse immunization and antiserum preparation
C3H/HeN mice were immunized once with either 10 μg of rIsC1ql3 or ovalbumin emulsified in complete Freud’s adjuvant (Thermo Fisher Scientific, #77140) followed by two boosts of the respective antigen with Incomplete Freud’s adjuvant (Thermo Fisher Scientific, #77145) every two weeks. Blood was collected from each mouse and the serum was obtained by centrifugation. The antibody titers in serum were tested by ELISA and Western blot to confirm antigen-specific antibodies for each mouse.
Rabbit immunization and antiserum preparation
Similar to the antibody preparation of mice, rabbits were immunized subcutaneously with 150 μg rIsC1ql3 proteins in complete Freund’s adjuvant and boosted three times at two-week intervals with 150 μg rIsC1ql3 proteins in incomplete Freund’s adjuvant. The rabbits were euthanized, and the sera were obtained by cardiac puncture two weeks after the final boost. The reactivity to rIsC1ql3 protein was examined by ELISA and Western blot.
Western blot to detect IsC1ql3 protein in tick saliva and tissues
To examine if IsC1ql3 exists in tick saliva, the collected tick saliva and ovalbumin (control) were first separated by SDS-PAGE, and then transferred onto a 0.45-m-pore-size polyvinylidene difluoride (PVDF) membrane (Bio-Rad, #1620177) and processed for immunoblotting. The blots were blocked in 1% non-fat milk in PBS for 60 min. Mouse polyclonal anti-IsC1ql3 antibody or mouse polyclonal anti-OVA antibody was diluted in 0.05% PBST at 1:500 and incubated with the blots for 1 h at room temperature or 4°C overnight. HRP-conjugated secondary antibody (1:2500, Invitrogen, #62-6520) was diluted in PBST and incubated for 1 h at room temperature. After washing with PBST, the immunoblots were imaged and quantified with an LI-COR Odyssey imaging system. To detect IsC1ql3 protein in the tick gut and salivary gland, unfed or fed ticks were dissected and the tissues were lysed by RIPA lysis and extraction buffer (Thermo Fisher Scientific, #89900). The western blots were conducted as described above. For all the western blots, the SDS-PAGE gels were run under non-reducing conditions except where specified.
Capture ELISA to quantify IsC1ql3 in tick saliva
The IsC1ql3 concentration in tick saliva was quantified by capture ELISA as previously described.4 Briefly, the murine IsC1qI3 antiserum was first coated as the primary or capture antibody. Then the collected tick saliva was added and specific amounts of recombinant IsC1qI3 protein were used as standards. After washing, rabbit IsC1qI3 antiserum was used as the detection antibody. The concentration of IsC1qI3 in tick saliva was quantified based on the standard curve of rIsC1ql3.
Western blot and ELISA to detect IsC1ql3 antibody in mammalian serum
To examine if IsC1ql3 antibodies are elicited by natural tick bites in mammalian serum, western blots of rIsC1ql3 were probed with naive serum and serum from mice, guinea pig and human bitten by clean (B. burgdorferi-free) and B. burgdorferi-infected ticks. The SDS-PAGE gels were run under reducing conditions. ELISA assays were also conducted. Briefly, 200 ng rIsC1ql3 were coated in a 96-well plate overnight. Samples were blocked with 1% BSA followed by incubation with naive serum and serum from mice, guinea pig and human bitten by clean (B. burgdorferi-free) and B. burgdorferi-infected ticks for 1 h at 37°C. After washing and incubating with Goat anti-mouse (1:2000, Invitrogen, #62-6520), anti-Guinea pig (1:2000, Abcam, #ab6908), and anti-human IgG HRP conjugated secondary antibody (1:2000, Sigma-Aldrich, #AP309P), KPL Sureblue TMB Microwell Peroxidase substrate, 1-component (Seracare, #5120-0077) was added. The reaction was stopped with 2 M sulfuric acid, and absorbance was read at 450 nm.
Pull-down assay
The pull-down assay was conducted to examine the interaction between B. burgdorferi and tick IsC1ql3 as previously described.6,17 Briefly, B. burgdorferi (~106–107 cells/mL) were incubated with recombinant IsC1ql3 or the control Aedes aegypti synaptosomal-associated protein (SNAP) for 2 h at 4°C. B. burgdorferi was then pelleted and the pellet and supernatant were separated. The pellet was washed 5–8 times in 1.5 mL PBS/0.1% BSA and was resuspended in the same volume as the supernatant. Equal volumes of supernatant and pellet were used to run Western blot as described above. HRP V5-tag monoclonal antibody (1:1000, Invitrogen, #R961-25) was used to detect protein. The SDS-PAGE gels were run under reducing conditions.
Flow cytometry to validate B. burgdorferi-IsC1ql3 interaction
B. burgdorferi was cultured to a density of ~106–107 cells/mL and harvested at 5000 × g for 15 min. Cells were washed twice with PBS and then blocked in 1% BSA for 1 h at 4°C. After spin, the pellet was suspended and incubated with I. scapularis protein disulfide isomerase A3 (rIsPDIA3), 14 kDa salivary gland protein (rSALP14), and rIsC1ql3 at 4°C for 2 h. After a co-incubation, spirochetes were washed three times with PBS and fixed in 2% PFA. After washing, the spirochetes were probed anti 6X-His monoclonal antibody-conjugated to Alexa Fluor 488 (Invitrogen, #MA1-21315-488) and run through BD LSRII (BD bioscience). The data was then analyzed by FlowJo.
ELISA to validate B. burgdorferi-IsC1ql3 interactions
B. burgdorferi was cultured to a density of ~106–107 cells/mL and harvested at 5000 × g for 15 min. Cells were washed twice with PBS, pelleted and lysed using Bug-buster Protein Extraction Reagent (Novagen, #70921-3). Protein concentration in the lysate was measured by absorbance at 280 nm. For the protease assay, the B. burgdorferi lysate was incubated in the presence or absence of proteinase K (QIAGEN, #19131) for 30 min. In a 96-well plate, wells were coated with 200 ng of B. burgdorferi lysate. Samples were blocked with 1% BSA followed by incubation with either rIsC1ql3 or rSALP14 protein at varying concentrations (10–300 ng) for 1 h at 37°C. After washing and incubating with HRP Anti-6X His tag antibody (Abcam, #ab3553), KPL Sureblue TMB Microwell Peroxidase substrate, 1-component (Seracare, #5120-0077) was added. The reaction was stopped with 2 M sulfuric acid, and absorbance was read at 450 nm.
Immunofluorescence assay
B. burgdorferi was cultured to a density of ~106–107 cells/mL, washed two times with PBS and blocked in 1% BSA for 1 h at 4°C. B. burgdorferi was then incubated with rIsC1ql3 at 4°C for 2 h. After fixation in 2% PFA, the spirochetes were probed anti 6X-His monoclonal antibody-conjugated to Alexa Fluor 488 (ThermoFisher, #MA1-21315-488) for 1 h. B. burgdorferi were then stained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (Invitrogen, #P36935). After staining, the fluorescence signals were examined with an EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific).
Effects of RNAi silencing the tick IsC1ql3 gene on B. burgdorferi transmission
Fed-nymph gut cDNA was prepared as described above and used as template to amplify segment of IsC1ql3 gene. The PCR primers with T7 promoter sequences are shown in Table S2. Double-stranded RNA (dsRNA) were synthesized using the MEGAscript RNAi kit (Invitrogen, #AM1626M) using PCR-generated DNA template that contained the T7 promoter sequence at both ends. The dsRNA quality was examined by agarose gel electrophoresis. DsRNA of the Aequorea victoria green fluorescent protein (GFP) was used as a control. B. burgdorferi-infected nymphs were injected in tick body with dsRNA using glass capillary needles as described by Narasimhan and colleagues.21 A group of four GFP or IsC1ql3 dsRNA injected B. burgdorferi-infected nymphs were placed on each C3H/HeN mouse and allowed to feed to repletion. Ticks were placed on the mouse head/back between the ears. At 7,14, and 21 days-post tick detachment, the mice were anesthetized, and skin was aseptically punch biopsied and the spirochete burden was assessed by qPCR. Ticks fed near the head area and skin punch biopsies were collected from the pinnae/ears.
Effects of active immunization on B. burgdorferi transmission
For active immunization, C3H/HeN mice were immunized subcutaneously with 10 μg IsC1ql3 or ovalbumin in complete Freund’s adjuvant and boosted twice every two weeks with the same amount of IsC1ql3 or ovalbumin in incomplete Freund’s adjuvant. The generation of antibodies was determined by ELISA. Two weeks after the final immunization, the mice were fed on by B. burgdorferi-infected ticks (four tick each). The B. burgdorferi burden in murine skin at 7, 14, and 21 days were determined by qPCR as described above.
ELISA to examine IsC1ql3 and complement proteins interactions
The interactions between IsC1ql3 and mammal complement proteins were examined by ELISA as described above, with recombinant human complement C1r subcomponent protein (Abclonal, #RP00142) and recombinant human complement C1s protein (R&D Systems, # 2060-SE-010).
Cell viability assay
To test if IsC1ql3 has effect on B. burgdorferi viability, 106 cells/mL B. burgdorferi were incubated with 1 X PBS, 10, 20, and 40 μg/mL rIsC1ql3 at 33°C for 48 h in a final volume of 250 μL. The viability of B. burgdorferi was determined as described in Gupta et al.,38 using BacTiter-Glo Microbial Cell Viability Assay kit (Promega, #G8230), which is based on quantitation of the ATP present by measuring luminescence.
Similarly, to test whether IsC1ql3 has an effect on splenocytes viability, 106 cells/mL splenocytes were incubated with 1 X PBS, 5, 10, 20 μg/mL rIsC1ql3 or BSA for 6 h. The viability of splenocytes were then evaluated using the CellTiter-Glo luminescent cell viability assay kit (Promega, #G7570).
Cytokine quantification by qPCR and mouse cytokine/chemokine arrays
We determined the effects of IsC1ql3 on host cytokine production in vitro and in vivo. For in vitro assays, the murine splenocytes, CD4+ and CD8+ T cells were isolated as described above. The isolated cells were incubated with 1 μg/mL rIsC1ql3 or BSA (control) and stimulated by 10 ng/mL LPS or 106 B. burgdorferi for 6 h. The cells and supernatant were harvested, and total RNA was extracted using a RNeasy Mini kit (Qiagen). For in vivo studies, tick IsC1ql3 gene was silenced as described above. Then the silenced nymphal ticks were put on mice ears. After 72 h post tick bite, mice were be sacrificed, and punch biopsy specimens were taken from the tick bite site. Total RNA was extracted, and cDNA was synthesized as described above. The qPCR primers sequences of cytokine genes are shown in Table S2. In addition, we also quantify cytokines production by the Mouse Cytokine/Chemokine Array 32-plex (MD-32) performed by Eve Technologies. Serum collected from each group of mice was sent for cytokine analyses. The cytokines represented by this array are Eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IP-10, KC, LIF, LIX, MCP-1, M-CSF, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, TNFα, and VEGF.
RNA-seq analysis
Total RNA was extracted from murine splenocytes, which were incubated with rIsC1ql3 and stimulated by B. burgdorferi as described above. RNA was isolated using RNeasy Mini kit according to the manufacturer’s instructions (Qiagen, #74104). RNA was submitted for library preparation using TruSeq (Illumina, San Diego, CA, USA) and sequenced using Illumina HiSeq 2500 by paired-end sequencing at the Yale Center for Genome Analysis (YCGA). All the RNA-seq analyses were performed using Partek Genomics Flow software (St. Louis, MO, USA). Specifically, RNA-seq data were trimmed and aligned to the mouse genome (mm10) using STAR (v2.7.3a).57 The aligned reads were quantified to Ensembl Transcripts release 100 using the Partek E/M algorithm58 and the subsequent steps were performed on gene-level annotation followed by total count normalization. The gene-level data were normalized by dividing the gene counts by the total number of reads followed by the addition of a small offset (0.0001). Principal components analysis (PCA) and gene expression heatmap were performed using default parameters for the determination of the component number, with all components contributing equally in Partek Flow. Volcano plot was performed on the genes that were differentially expressed across the conditions (p < 0.05 and fold change ≥ 2 for each comparison). GO enrichment analysis were conducted using the functional annotation tool DAVID (https://david.ncifcrf.gov/tools.jsp).
Flow cytometry to quantify IFN-γ in immune cells
The isolated murine splenocytes were incubated with B. burgdorferi (1 × 106 cells/mL) alone or mixture with 1 μg/mL rIsC1ql3 for 6 h. The inhibitors of intracellular protein transport, monensin (Invitrogen, #00-4505-51) and brefeldin A (Invitrogen, #00-4506-51) were added during the last hours of in vitro activation. The cells were then incubated with Fc receptor antibody (TruStain FcX™ anti-mouse CD16/32) (BioLegend, #101320), and the antibodies of FITC anti-mouse CD3 antibody (BioLegend, #100204), APC anti-mouse CD4 antibody (BioLegend, #100412), and Brilliant Violet 711™ anti-mouse CD8a antibody (BioLegend, #100747), PerCP/Cyanine5.5 anti-mouse CD14 antibody (BioLegend, #123313), PE anti-mouse NK-1.1 antibody (BioLegend, #108708), and LIVE/DEAD™ fixable violet stain kit (Invitrogen, #L34955) on ice for 20 min. Then the cells were fixed and permeabilized using BD Cytofix/Cytoperm fixation/permeabilization solution (BD Biosciences, #555028). Cells were washed and stained by intracellular staining for 30 min at 4°C using APC/Cyanine7 anti-mouse IFN-γ antibody (BioLegend, #505849). The samples were run through BD LSRII (BD bioscience). The data was then analyzed by FlowJo.
Co-immunoprecipitation (coIP) assay
HEK293T cells were transfected with 1 μg plasmids encoding mouse mgC1qR-HA (sinobiological, #MG50199-CY) and/or IsC1ql3-Myc/His. At 48 h, the cells were lysed by RIPA lysis and extraction buffer (Thermo Fisher Scientific, #89900), and precipitated using Pierce™ HA-Tag IP/Co-IP Kit (Thermo Scientific, #26180) as described in the manual. Input (the cell lysate) and the the immunoprecipitated eluates were loaded to SDS-PAGE gel, and then run the western blow as described above. The SDS-PAGE gels were run under reducing conditions. The antibodies of mouse anti HA-Tag mAb (ABclonal, #AE008), HRP Anti-6X His tag antibody (Abcam, #ab3553), or rabbit anti-Isc1ql3 antiserum were used to detect the proteins.
Co-immunofluorescence assay
HEK293T cells were transfected with 1 μg plasmid encoding mouse mgC1qR-HA (sinobiological, #MG50199-CY) and/or IsC1ql3-Myc/His. At 48 h, the cells were washed with PBS and fixed in 4% PFA for 15 min at room temperature. Then, the cells were blocked in 1% BSA in PBS for 1 h and subsequently immunolabeled with mouse anti HA-Tag mAb (ABclonal, #AE008) and rabbit anti-Isc1ql3 antiserum. Cells were washed with PBS three times and then immunolabeled with secondary antibodies of goat anti-rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 (1:100, Invitrogen, #A-21428) and goat anti-mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:100, Invitrogen, #A-11001) for 1 h at room temperature. Nuclei were stained with DAPI (Invitrogen, #D9542). After staining, the fluorescence signals were examined with an EVOS FL Auto Cell Imaging System (Thermo Fisher Scientific).
Antibody blocking of gC1qR to test IFN-γ expression
The isolated splenocytes were incubated with the polyclonal antibody of gC1qR (ABclonal, #A1883) (1:100, 25.5 μg/mL and 1:1000, 2.55 μg/mL) and then incubated with 1 μg/mL rIsC1ql3 and stimulated by 106 B. burgdorferi spirochetes for 6 h. The cells and supernatant were harvested, and the cytokine IFN-γ expression was evaluated as described above.
Silencing of gC1qR expression by siRNA
The isolated splenocytes were transiently transfected with 50 nM gC1qR siRNA mixture of 5′-UAAUUUAGCCUCCGUGCCGTT-3′ (ThermoFisher, #159915) and 5′-UCACGGUCACUUUCAACAT-3′ (W.N. Keck, Yale), using TransIT 2020 (Mirus, #MIR5404). After 40 h post transfection, the cells were harvested for evaluating gC1qR expression and immunoblotting. After confirming silencing of gC1qR, the cells were incubated with 1 μg/mL rIsC1ql3 and stimulated by 106 B. burgdorferi spirochetes for 6 h. The cells and supernatant were harvested, and the cytokine IFN-γ expression was evaluated as described above.
Competitive ELISA
In a 96-well plate, each well was coated with 200 ng rIsC1ql3. Samples were then blocked with 1% BSA followed by incubation with 30 ng, 100 ng or without B. burgdorferi lysate. The rgC1qR protein was added at varying concentrations (10–300 ng) for 1 h at 37°C. After washing, the protein was incubated with a polyclonal gC1qR antibody (ABclonal, #A1883), followed by Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP (Invitrogen, #31460). Then KPL Sureblue TMB Microwell Peroxidase substrate, 1-component (Seracare, #5120-0077) was added. The reaction was stopped with 2 M sulfuric acid, and absorbance was read at 450 nm.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance of differences observed in experimental and control groups was analyzed using GraphPad Prism version 8.0 (GraphPad Software, Inc., San Diego, CA). Non-parametric Mann-Whitney test or unpaired t test were utilized to compare the mean values of control and tested groups, and p < 0.05 was considered significant.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD3 antibody | BioLegend | Cat# 100204; RRID:AB_312661 |
| APC anti-mouse CD4 antibody | BioLegend | Cat# 100412; RRID:AB_312697 |
| Brilliant Violet 711™ anti-mouse CD8a antibody | BioLegend | Cat# 100747; RRID:AB_11219594 |
| Goat anti-Mouse IgG (H + L) Secondary Antibody, HRP | Invitrogen | Cat# 62-6520; RRID:AB_2533947 |
| Goat Anti-Guinea pig IgG H&L (HRP) | Abcam | Cat# ab6908; RRID:AB_955425 |
| Goat Anti-Human IgG Antibody, HRP conjugate | Sigma-Aldrich | Cat# AP309P; RRID:AB_11212373 |
| V5 Tag Monoclonal Antibody, HRP | Invitrogen | Cat# R961-25; RRID:AB_2556565 |
| 6x-His Tag Monoclonal Antibody (HIS.H8), Alexa Fluor™ 488 | Invitrogen | Cat# MA1-21315-488; RRID:AB_2610645 |
| HRP Anti-6X His tag® antibody | Abcam | Cat# ab3553; RRID:AB_303900 |
| TruStain FcX™ (anti-mouse CD16/32) Antibody | BioLegend | Cat# 101320; RRID:AB_1574975 |
| PerCP/Cyanine5.5 anti-mouse CD14 antibody | BioLegend | Cat# 123313; RRID:AB_2074185 |
| PE anti-mouse NK-1.1 antibody | BioLegend | Cat# 108708; RRID:AB_313395 |
| APC/Cyanine7 anti-mouse IFN-γ antibody | BioLegend | Cat# 505849; RRID:AB_2616697 |
| Mouse anti HA-Tag mAb | ABclonal | Cat# AE008; RRID:AB_2770404 |
| Goat anti-rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Invitrogen | Cat# A-21428; RRID:AB_2535849 |
| Goat anti-mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A-11001; RRID:AB_2534069 |
| GC1qR/C1QBP Rabbit pAb | ABclonal | Cat# A1883; RRID:AB_2763916 |
| Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP | Invitrogen | Cat# 31460; RRID:AB_228341 |
| Bacterial and virus strains | ||
| Borrelia burgdorferi (N40) | Dr. Erol Fikrig Laboratory | N/A |
| E. coli BL21(DE3) Chemically Competent Cell | Thermo Scientific | Cat# EC0114 |
| Biological samples | ||
| Mouse spleen tissue | This manuscript | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Barbour-Stoenner-Kelly H (BSK-H) complete medium | Sigma-Aldrich | Cat# B8291 |
| Dulbecco’s Modified Eagle’s Medium | ThermoFisher | Cat# 11965-118 |
| Schneider’s Drosophila media | Gibco | Cat# 21720-024 |
| RPMI 1640 medium | Sigma-Aldrich | Cat# R4130 |
| Trizol | Invitrogen | Cat# 15596-018 |
| iQ SYBR Green Supermix | Bio-Rad | Cat# 1725124 |
| TALON metal affinity resin | Clontech | Cat# 635606 |
| Ni-NTA agarose | QIAGEN | Cat# 30230 |
| Complete Freud’s adjuvant | Thermo Fisher Scientific | Cat# 77140 |
| Incomplete Freud’s adjuvant | Thermo Fisher Scientific | Cat# 77145 |
| RIPA lysis and extraction buffer | Thermo Fisher Scientific | Cat# 89900 |
| SureBlue™ TMB 1-Component Microwell Peroxidase Substrate | Seracare | Cat# 5120-0077 |
| Bugbuster Protein Extraction Reagent | Novagen | Cat# 70921-3 |
| Proteinase K | QIAGEN | Cat# 19131 |
| 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) | Invitrogen | Cat# P36935 |
| Monensin | Invitrogen | Cat# 00-4505-51 |
| Brefeldin A | Invitrogen | Cat# 00-4506-51 |
| BD Cytofix/Cytoperm fixation/ permeabilization solution | BD Biosciences | Cat# 555028 |
| TransIT®-2020 Transfection Reagent | Mirus | Cat# MIR5404 |
| Recombinant IsC1ql3 protein | This manuscript | N/A |
| Recombinant Ixodes scapularis protein disulfide isomerase A3 | Dr. Erol Fikrig Laboratory | N/A |
| Recombinant 14 kDa salivary gland protein | Dr. Erol Fikrig Laboratory | N/A |
| Recombinant Aedes aegypti synaptosomal-associated protein | Dr. Erol Fikrig Laboratory | N/A |
| C1QBP Protein, Mouse, Recombinant | Sinobiological | Cat# 50199-M07E |
| Critical commercial assays | ||
| MojoSort™ mouse CD4 T cell isolation kits | BioLegend | Cat# 480005 |
| MojoSort™ mouse CD8 T cell isolation kits | BioLegend | Cat# 480007 |
| iScript cDNA Synthesis Kits | Bio-Rad | Cat# 1708891 |
| BCA Protein Estimation kit | ThermoFisher Scientific | Cat# 23225 |
| MEGAscript RNAi kit | Invitrogen | Cat# AM1626M |
| BacTiter-Glo Microbial Cell Viability Assay kit | Promega | Cat# G8230 |
| CellTiter-Glo luminescent cell viability assay kit | Promega | Cat# G7570 |
| RNeasy Mini kit | Qiagen | Cat# 74104 |
| LIVE/DEAD™ fixable violet stain kit | Invitrogen | Cat# L34955 |
| Pierce™ HA-Tag IP/Co-IP Kit | Thermo Scientific | Cat# 26180 |
| Deposited data | ||
| RNA-seq data | This manuscript | GSE207307 |
| Experimental models: Cell lines | ||
| Human: HEK293T cells | ATCC | Cat# CRL-3216 |
| D. melanogaster S2 cells | ATCC | Cat# CRL-1963 |
| Experimental models: Organisms/strains | ||
| C3H/HeN mice | Charles River Laboratories | N/A |
| C57BL/6J mice | Jackson Laboratory | Stock #: 000664; RRID: IMSR_JAX:000664 |
| Black-legged tick (Ixodes scapularis) | Oklahoma State University | N/A |
| Oligonucleotides | ||
| See Table S2 for primers | This manuscript | N/A |
| gC1qR siRNA: 5′-UAAUUUAGCCUCCGUGCCGTT-3′ | ThermoFisher | Cat# 159915 |
| gC1qR siRNA: 5′-UCACGGUCACUUUCAACAT-3′ | This manuscript | N/A |
| Recombinant DNA | ||
| Plasmid: pMT/BiP/V5-His vector | Invitrogen | Cat# V413020 |
| Plasmid: pET-28a (+) vector | Addgene | Cat# 69864-3 |
| Plasmid: C1QBP cDNA ORF Clone, Mouse, C-HA tag | Sinobiological | Cat# MG50199-CY |
| Plasmid: IsC1ql3-Myc/His | This manuscript | N/A |
| Software and algorithms | ||
| Prism | GraphPad | RRID:SCR_002798 |
| FlowJo | BD Biosciences | https://www.flowjo.com/ |
| Partek Genomics Flow software | Partek | https://www.partek.com/ |
| STAR | Dobin et al. 201357 | https://github.com/alexdobin/STAR |
| Partek E/M algorithm | Xing et al. 200658 | N/A |
| BioRender software | BioRender | https://biorender.com/ |
| Other | ||
| Hemocytometer | INCYTO | Cat# DHC-N01 |
| 10% Fetal Bovine Serum | Sigma | Cat# 12306C-500 |
| 70 μm cell-strainer nylon mesh | Fisherbrand | Cat# 22-363-548 |
| 10-kDa concentrator | MilliporeSigma | Cat# Z740203 |
| Mouse anti-IsC1ql3 serum | This manuscript | N/A |
| Rabbit anti-IsC1ql3 serum | This manuscript | N/A |
| 4-20% Mini-Protean TGX gels | Bio-Rad | Cat# 4561094 |
| 3-kDa concentrator | MilliporeSigma | Cat# C7719 |
| Polyvinylidene difluoride (PVDF) membrane | Bio-Rad | Cat# 1620177 |
Highlights.
Tick salivary protein IsC1ql3 directly interacts with B. burgdorferi
IsC1ql3 contributes to the initial stage of B. burgdorferi infection of mice
IsC1ql3 interacts with the globular C1q receptor present on the surface of T cells
IsC1ql3 inhibits B. burgdorferi-induced IFN-γ production
ACKNOWLEDGMENTS
This work was supported by grants from the NIH (AI126033, AI138949) and the Steven and Alexandra Cohen Foundation. This research was also supported in part by the Howard Hughes Medical Institute Emerging Pathogens Initiative. We sincerely thank Ms. Kathleen DePonte and Mr. Ming-Jie Wu for their excellent technical assistance. We would like to acknowledge that figures were created using BioRender.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111673.
REFERENCES
- 1.Rosenberg R, Lindsey NP, Fischer M, Gregory CJ, Hinckley AF, Mead PS, Paz-Bailey G, Waterman SH, Drexler NA, Kersh GJ, et al. (2018). Vital signs: trends in reported vectorborne disease cases—United States and Territories, 2004–2016. MMWR Morb. Mortal. Wkly. Rep 67, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurokawa C, Lynn GE, Pedra JHF, Pal U, Narasimhan S, and Fikrig E (2020). Interactions between Borrelia burgdorferi and ticks. Nat. Rev. Microbiol 18, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sajid A, Matias J, Arora G, Kurokawa C, DePonte K, Tang X, Lynn G, Wu M-J, Pal U, Strank NO, et al. (2021). mRNA vaccination induces tick resistance and prevents transmission of the Lyme disease agent. Sci. Transl. Med 13, eabj9827. [DOI] [PubMed] [Google Scholar]
- 4.Anguita J, Ramamoorthi N, Hovius JWR, Das S, Thomas V, Persinski R, Conze D, Askenase PW, Rincón M, Kantor FS, and Fikrig E (2002). Salp15, an Ixodes scapularis salivary protein, inhibits CD4+ T cell activation. Immunity 16, 849–859. [DOI] [PubMed] [Google Scholar]
- 5.Ramamoorthi N, Narasimhan S, Pal U, Bao F,Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, et al. (2005). The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature 436, 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuijt TJ, Coumou J, Narasimhan S, Dai J, DePonte K, Wouters D, Brouwer M, Oei A, Roelofs JJTH, van Dam AP, et al. (2011). A tick mannose-binding lectin inhibitor interferes with the vertebrate complement cascade to enhance transmission of the lyme disease agent. Cell Host Microbe 10, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai J, Narasimhan S, Zhang L, Liu L, Wang P, and Fikrig E (2010). Tick histamine release factor is critical for Ixodes scapularis engorgement and transmission of the lyme disease agent. PLoS Pathog. 6, e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carland TM, and Gerwick L (2010). The C1q domain containing proteins: where do they come from and what do they do? Dev. Comp. Immunol 34, 785–790. [DOI] [PubMed] [Google Scholar]
- 9.Ressl S, Vu BK, Vivona S, Martinelli DC, Südhof TC, and Brunger AT (2015). Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure 23, 688–699. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi T, Kamon J, Minokoshi Y, Ito Y, Waki H, Uchida S, Yamashita S, Noda M, Kita S, Ueki K, et al. (2002). Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med 8, 1288–1295. [DOI] [PubMed] [Google Scholar]
- 11.Kishore U, and Reid KB (1999). Modular organization of proteins containing C1q-like globular domain. Immunopharmacology 42, 15–21. [DOI] [PubMed] [Google Scholar]
- 12.Nigro E, Perrotta F, Monaco ML, Polito R, Pafundi PC, Matera MG, Daniele A, and Bianco A (2020). Implications of the adiponectin system in non-small cell lung cancer patients: a case-control study. Biomolecules 10, 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, and Lodish HF (2008). Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-γ agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem. J 416, 161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv Z, Qiu L, Wang M, Jia Z, Wang W, Xin L, Liu Z, Wang L, and Song L (2018). Comparative study of three C1q domain containing proteins from pacific oyster Crassostrea gigas. Dev. Comp. Immunol 78, 42–51. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Wang L, Zhang H, Zhou Z, Siva VS, and Song L (2012). A C1q domain containing protein from scallop Chlamys farreri serving as pattern recognition receptor with heat-aggregated IgG binding activity. PLoS One 7, e43289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong Y, Liu Z, Wu Z, Han Z, Wang L, and Song L (2019). A novel globular C1q domain containing protein (C1qDC-7) from Crassostrea gigas acts as pattern recognition receptor with broad recognition spectrum. Fish Shellfish Immunol. 84, 920–926. [DOI] [PubMed] [Google Scholar]
- 17.Tang X, Cao Y, Arora G, Hwang J, Sajid A, Brown CL, Mehta S, Marín-López A, Chuang Y-M, Wu M-J, et al. (2021). The Lyme Disease agent co-opts adiponectin receptor-mediated signaling in its arthropod vector. Elife 10, e72568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye JJ, Bian X, Lim J, and Medzhitov R (2020). Adiponectin and related C1q/TNF-related proteins bind selectively to anionic phospholipids and sphingolipids. Proc. Natl. Acad. Sci. USA 117, 17381–17388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei Z, Peterson JM, and Wong GW (2011). Metabolic regulation by C1q/TNF-related protein-13(CTRP13): activation OF AMP-activated protein kinase and suppression of fatty acid-induced JNK signaling. J. Biol. Chem 286, 15652–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, Rosen C, Arora G, Gupta A, Booth CJ, Murfin KE, Cerny J, Marin Lopez A, Chuang Y-M, Tang X, et al. (2020). An Ixodes scapularis protein disulfide isomerase contributes to Borrelia burgdorferi colonization of the vector. Infect. Immun 88, e00426–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narasimhan S, Montgomery RR, DePonte K, Tschudi C, Marcantonio N, Anderson JF, Sauer JR, Cappello M, Kantor FS, and Fikrig E (2004). Disruption of Ixodes scapularis anticoagulation by using RNA interference. Proc. Natl. Acad. Sci. USA 101, 1141–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agarwal V, Ahl J, Riesbeck K, and Blom AM (2013). An alternative role of C1q in bacterial infections: facilitating Streptococcus pneumoniae adherence and invasion of host cells. J. Immunol 191, 4235–4245. [DOI] [PubMed] [Google Scholar]
- 23.Fox S, Ryan KA, Berger AH, Petro K, Das S, Crowe SE, and Ernst PB (2015). The role of C1q in recognition of apoptotic epithelial cells and inflammatory cytokine production by phagocytes during Helicobacter pylori infection. J. Inflamm 12. 51–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao L, Chen S, Sherchan P, Ding Y, Zhao W, Guo Z, Yu J, Tang J, and Zhang JH (2018). Recombinant CTRP9 administration attenuates neuroinflammation via activating adiponectin receptor 1 after intracerebral hemorrhage in mice. J. Neuroinflammation 15, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koltes JE, Arora I, Gupta R, Nguyen DC, Schaid M, Kim J.-a., Kimple ME, and Bhatnagar S (2019). A gene expression network analysis of the pancreatic islets from lean and obese mice identifies complement 1q like-3 secreted protein as a regulator of β-cell function. Sci. Rep 9. 10119–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Petzke MM, Iyer R, Wu H, and Schwartz I (2008). Pattern of proinflammatory cytokine induction in RAW264.7 mouse macrophages is identical for virulent and attenuated Borrelia burgdorferi. J. Immunol 180, 8306–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma Y, Seiler KP, Tai K-F, Yang L, Woods M, and Weis JJ (1994). Outer surface lipoproteins of Borrelia burgdorferi stimulate nitric oxide production by the cytokine-inducible pathway. Infect. Immun 62, 3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strle K, Drouin EE, Shen S, El Khoury J, McHugh G, Ruzic-Sabljic E, Strle F, and Steere AC (2009). Borrelia burgdorferi stimulates macrophages to secrete higher levels of cytokines and chemokines than Borrelia afzelii or Borrelia garinii. J. Infect. Dis 200, 1936–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith AA, Navasa N, Yang X, Wilder CN, Buyuktanir O, Marques A, Anguita J, and Pal U (2016). Cross-species interferon signaling boosts microbicidal activity within the tick vector. Cell Host Microbe 20, 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter SL, Müller M, Manders PM, and Campbell IL (2007). Induction of the genes for Cxcl9 and Cxcl10 is dependent on IFN-γ but shows differential cellular expression in experimental autoimmune encephalomyelitis and by astrocytes and microglia in vitro. Glia 55, 1728–1739. [DOI] [PubMed] [Google Scholar]
- 31.Moore MW, Cruz AR, LaVake CJ, Marzo AL, Eggers CH, Salazar JC, and Radolf JD (2007). Phagocytosis of Borrelia burgdorferi and Treponema pallidum potentiates innate immune activation and induces gamma interferon production. Infect. Immun 75, 2046–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boehm U, Klamp T, Groot M, and Howard JC (1997). Cellular responses to interferon-γ. Annu. Rev. Immunol 15, 749–795. [DOI] [PubMed] [Google Scholar]
- 33.Darwich L, Coma G, Peña R, Bellido R, Blanco EJJ, Este JA, Borras FE, Clotet B, Ruiz L, Rosell A, et al. (2009). Secretion of interferon-γ by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology 126, 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van de Schoor FR, Vrijmoeth HD, Brouwer MA, Ter Hofstede HJ, Lemmers HL, Dijkstra H, Boahen CK, Oosting M, Kullberg B-J, and Hovius JW (2022). Borrelia burgdorferi is a poor inducer of interferon-gamma: amplification induced by interleukin-12. Infect. Immun 90, e0055821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghebrehiwet B, Lim B-L, Peerschke EI, Willis AC, and Reid KB (1994). Isolation, cDNA cloning, and overexpression of a 33-kD cell surface glycoprotein that binds to the globular" heads" of C1q. J. Exp. Med 179, 1809–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pednekar L, Pathan AA, Paudyal B, Tsolaki AG, Kaur A, Abozaid SM, Kouser L, Khan HA, Peerschke EI, Shamji MH, et al. (2016). Analysis of the interaction between globular head modules of human C1q and its candidate receptor gC1qR. Front. Immunol 7, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai X, Liu K, Fang H, Zhang Q, Gao X, Liu F, Zhou S, Wang X, Niu Y, Hong Y, et al. (2021). Mitochondrial C1qbp promotes differentiation of effector CD8+T cells via metabolic-epigenetic reprogramming. Sci. Adv 7, eabk0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta A, Arora G, Rosen CE, Kloos Z, Cao Y, Cerny J, Sajid A, Hoornstra D, Golovchenko M, Rudenko N, et al. (2020). A human secretome library screen reveals a role for Peptidoglycan Recognition Protein 1 in Lyme borreliosis. PLoS Pathog. 16, e1009030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hovingh ES, Van den Broek B, and Jongerius I (2016). Hijacking complement regulatory proteins for bacterial immune evasion. Front. Microbiol 7, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hosszu KK, Valentino A, Peerschke EI, and Ghebrehiwet B (2020). SLE: novel postulates for therapeutic options. Front. Immunol 11, 583853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pham M, Underwood J, and Oliva Chávez AS (2021). Changing the recipe: pathogen directed changes in tick saliva components. Int. J. Environ. Res. Public Health 18, 1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarma JV, and Ward PA (2011). The complement system. Cell Tissue Res. 343, 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hart T, Nguyen NTT, Nowak NA, Zhang F, Linhardt RJ, Diuk-Wasser M, Ram S, Kraiczy P, and Lin Y-P (2018). Polymorphic factor H-binding activity of CspA protects Lyme borreliae from the host complement in feeding ticks to facilitate tick-to-host transmission. PLoS Pathog. 14, e1007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie J, Zhi H, Garrigues RJ, Keightley A, Garcia BL, and Skare JT (2019). Structural determination of the complement inhibitory domain of Borrelia burgdorferi BBK32 provides insight into classical pathway complement evasion by Lyme disease spirochetes. PLoS Pathog. 15, e1007659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramachandra RN, and Wikel SK (1995). Effects of Dermacentor andersoni (Acari: Ixodidae) salivary gland extracts on Bos indicus and B. taurus lymphocytes and macrophages: in vitro cytokine elaboration and lymphocyte blastogenesis. J. Med. Entomol 32, 338–345. [DOI] [PubMed] [Google Scholar]
- 46.Lukácsi S, Mácsik-Valent B, Nagy-Baló Z, Kovács KG, Kliment K, Bajtay Z, and Erdei A (2020). Utilization of complement receptors in immune cell-microbe interaction. FEBS Lett. 594, 2695–2713. [DOI] [PubMed] [Google Scholar]
- 47.Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, and Hahn YS (2000). Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J. Clin. Invest 106, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao ZQ, Nguyen DT, Hiotellis AI, and Hahn YS (2001). Hepatitis C virus core protein inhibits human T lymphocyte responses by a complement-dependent regulatory pathway. J. Immunol 167, 5264–5272. [DOI] [PubMed] [Google Scholar]
- 49.Yao ZQ, Eisen-Vandervelde A, Ray S, and Hahn YS (2003). HCV core/gC1qR interaction arrests T cell cycle progression through stabilization of the cell cycle inhibitor p27Kip1. Virology 314, 271–282. [DOI] [PubMed] [Google Scholar]
- 50.Waggoner SN, Cruise MW, Kassel R, and Hahn YS (2005). gC1q receptor ligation selectively down-regulates human IL-12 production through activation of the phosphoinositide 3-kinase pathway. J. Immunol 175, 4706–4714. [DOI] [PubMed] [Google Scholar]
- 51.Waggoner SN, Hall CHT, and Hahn YS (2007). HCV core protein interaction with gC1q receptor inhibits Th1 differentiation of CD4+ T cells via suppression of dendritic cell IL-12 production. J. Leukoc. Biol 82, 1407–1419. [DOI] [PubMed] [Google Scholar]
- 52.Kopp A, Bala M, Buechler C, Falk W, Gross P, Neumeier M, Schölmerich J, and Schäffler A (2010). C1q/TNF-related protein-3 represents a novel and endogenous lipopolysaccharide antagonist of the adipose tissue. Endocrinology 151, 5267–5278. [DOI] [PubMed] [Google Scholar]
- 53.Weigert J, Neumeier M, Schäffler A, Fleck M, Schölmerich J, Schütz C, and Buechler C (2005). The adiponectin paralog CORS-26 has anti-inflammatory properties and is produced by human monocytic cells. FEBS Lett. 579, 5565–5570. [DOI] [PubMed] [Google Scholar]
- 54.Mans BJ, Louw AI, and Neitz AWH (2002). Evolution of hematophagy in ticks: common origins for blood coagulation and platelet aggregation inhibitors from soft ticks of the genus Ornithodoros. Mol. Biol. Evol 19, 1695–1705. [DOI] [PubMed] [Google Scholar]
- 55.Iwanaga S, Isawa H, and Yuda M (2014). Horizontal gene transfer of a vertebrate vasodilatory hormone into ticks. Nat. Commun 5, 3373. [DOI] [PubMed] [Google Scholar]
- 56.Diebolder CA, Beurskens FJ, De Jong RN, Koning RI, Strumane K, Lindorfer MA, Voorhorst M, Ugurlar D, Rosati S, Heck AJR, et al. (2014). Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing Y, Yu T, Wu YN, Roy M, Kim J, and Lee C (2006). An expectation-maximization algorithm for probabilistic reconstructions of full-length isoforms from splice graphs. Nucleic Acids Res. 34, 3150–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uraki R, Hastings AK, Marin-Lopez A, Sumida T, Takahashi T, Grover JR, Iwasaki A, Hafler DA, Montgomery RR, and Fikrig E (2019). Aedes aegypti AgBR1 antibodies modulate early Zika virus infection of mice. Nat. Microbiol 4, 948–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schuijt TJ, Narasimhan S, Daffre S, DePonte K, Hovius JWR, Van’t Veer C, van der Poll T, Bakhtiari K, Meijers JCM, Boder ET, et al. (2011). Identification and characterization of Ixodes scapularis antigens that elicit tick immunity using yeast surface display. PLoS One 6, e15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are available in the Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information under the accession number: GSE207307.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.