Abstract
The world’s population is projected to increase by two billion by 2050, resulting in food and energy insecurity. Oilseed crops have been identified as key to address these challenges: they produce and store lipids in the seeds as triacylglycerols that can serve as a source of food/feed, renewable fuels, and other industrially-relevant chemicals. Therefore, improving seed oil content and composition has generated immense interest. Research efforts aiming to unravel the regulatory pathways involved in fatty acid synthesis and to identify targets for metabolic engineering have made tremendous progress. This review provides a summary of the current knowledge of oil metabolism and discusses how photochemical activity and unconventional pathways can contribute to high carbon conversion efficiency in seeds. It also highlights the importance of 13C-metabolic flux analysis as a tool to gain insights on the pathways that regulate oil biosynthesis in seeds. Finally, a list of key genes and regulators that have been recently targeted to enhance seed oil production are reviewed and additional possible targets in the metabolic pathways are proposed to achieve desirable oil content and quality.
Keywords: carbon conversion efficiency, embryo culture, fatty acid synthesis, isolated plastids, lipid storage, metabolic flux analysis, oilseed, triacylglycerol
Introduction
Depending on plant species, seeds accumulate various proportions of biomass components, such as proteins, starch and lipids. In seeds, storage oils are mainly in the form of triacylglycerols (TGs), as an energy reserve utilized during germination and post-germinative growth. These oilseeds have a profound agricultural and industrial significance, utilized predominantly in food processing and preparation, and as a renewable resource for various industrial applications (Jaworski and Cahoon, 2003). Because of their structural similarity with long-chain hydrocarbons, TGs can replace petroleum-based products, such as diesel, lubricants, paints, coatings or inks (Cahoon et al., 2007; Durrett et al., 2008). The renewable biofuels derived from oilseeds produce ~85% less carcinogens during combustion than petroleum-based diesel fuels, presenting advantages in terms of sustainability, environment and health (Atadashi et al., 2012; Hasan and Rahman, 2017). Because of its popularity as a renewable resource, the consumption of seed oil has been increasing simultaneously with the rapidly growing population and upgraded standards of living (Marchive et al., 2014; Samarth and Mahanwar, 2015). To meet these rising demands, there is an urgent need to develop new oilseed cultivars with improved oil content and composition (Singer et al., 2013; Xu et al., 2018). Many research efforts have been focused into the improvement of seed oil over the years using conventional or molecular-assisted breeding approaches, as well as more targeted genetic manipulation.
The selection of candidate genes that can be engineered relies on understanding the biochemical regulations that control carbon partitioning during de novo fatty acid synthesis (FAS) in seeds. Biosynthesis of TGs starts from FAS in plastids which relies on a cycle of condensation, reduction and dehydration reactions that extend an acyl-chain linked to an acyl carrier protein (ACP) by two carbon units per cycle. These fatty acids (FAs) are then assembled into TGs in the endoplasmic reticulum (ER), or used in other metabolic processes, such as chain elongation and acyl editing. The carbon precursor for FAS is acetyl-CoA, which is generated from the oxidative decarboxylation of pyruvate through the pyruvate dehydrogenase complex (Johnston et al., 1997). Determining the efficiency of the developing embryo in converting this carbon source into oil and other biomass components (e.g. proteins, and carbohydrates) known as carbon conversion efficiency (CCE), is important to evaluate the potential for improving seed oil quality (Goffman et al., 2005; Alonso et al., 2007; Allen et al., 2009; Lonien and Schwender, 2009; Chen and Shachar-Hill, 2012; Cocuron et al., 2019; Carey et al., 2020; Tsogtbaatar et al., 2020). CCE results from the sum of all catabolic and anabolic metabolic processes which varies in developing embryos from different oilseeds, classified as “green” or “non-green”, depending on the presence or absence of chlorophyll during seed filling (Goffman et al., 2005; Allen et al., 2009; Lonien and Schwender, 2009; Carey et al., 2020; Tsogtbaatar et al., 2020). However, to decipher the biochemical pathways underlying the CCE in each species, a more quantitative analysis of the carbon fluxes through the central metabolism—which conducts the vast majority of biochemical carbon transformation—is needed. Steady state metabolic flux analysis (MFA) has been useful to gain a quantitative assessment of the carbon flux through central metabolism based on 13C-labeling, which may guide genetic engineering (Libourel & Shachar-Hill, 2008; Lee et al., 2011; Chen & Shachar-Hill, 2012; Kim et al., 2012; Kruger et al., 2012; O’Grady et al., 2012; Shachar-Hill, 2013). In parallel, several strategies have been employed to genetically manipulate FA composition and plant lipid metabolism in order to increase the FA content in oilseeds. To this end, the “push, pull, package, and protect” strategy has been implemented at various degrees; it consists of manipulating the expression of genes to boost the synthesis of FAs (“push”), or increase TG assembly reactions (“pull”), or improve the storage of FAs into lipid droplets (LDs) (“package”), or prevent the degradation of stored lipids (“protect”), or any combination of these.
This review focuses on FAS in seeds, highlighting the limitation and challenges involved in performing earlier experiments with isolated plastids and the relevance of 13C-MFA to decipher the pathways that contribute to oil biosynthesis, discussing how photochemical activity and unconventional pathways may contribute to higher efficiency of carbon conversion, assessing key genes and regulators that have been recently targeted to enhance seed oil content, and proposing alternative targets/strategies to achieve desirable oil content and quality.
Fatty acid synthesis and elongation in seeds
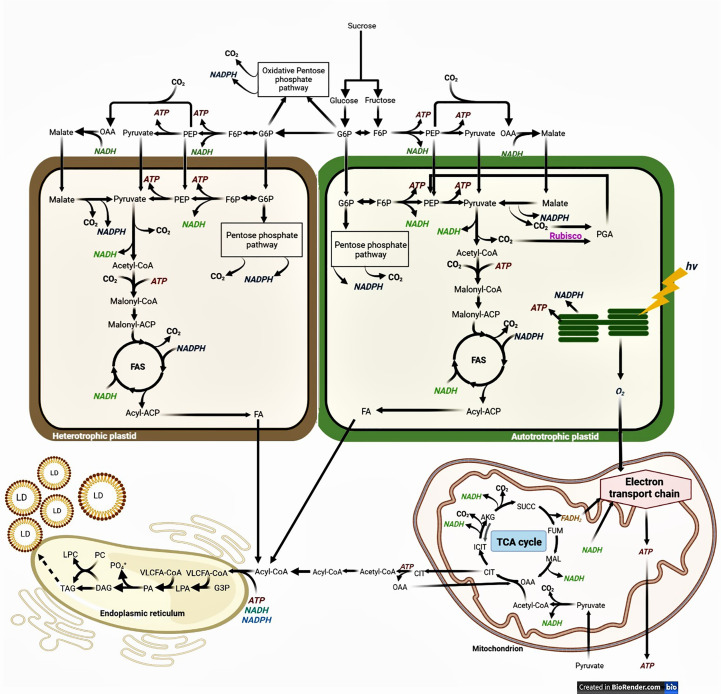
The schematic mechanism of FAS and FA elongation for green and non-green embryos, and the possible sources of carbon precursors, energy, and reductants, are depicted in Figure 1 . The disaccharide sucrose represents the major form in which photosynthetically assimilated carbon is transported into oil seeds. The hexose phosphates generated by the cleavage of sucrose can be metabolized through the glycolysis and the oxidative pentose phosphate pathway (OPPP) which can be found both in the cytosol and in the plastids. A major route for carbon going into FAS may involve the cytosolic glycolytic pathway until phosphoenolpyruvate (PEP) and pyruvate, which may be imported into the plastid and undergo decarboxylation to form acetyl-coenzyme A (acetyl-CoA) via the plastidic pyruvate dehydrogenase complex. Acetyl-CoA carboxylase (ACCase) is the first committed step for the FAS: it uses energy to carboxylate acetyl-CoA into malonyl-CoA which is transferred to the Acyl Carrier Protein (ACP) by the malonyl-CoA-ACP transacylase. Then, malonyl-ACP is condensed with acetyl-CoA by the 3-ketoacyl-ACP synthase III (KAS III), generating 3-ketobutyryl-ACP. The 3-ketoacyl-ACP reductase utilizes NADPH to reduce 3-ketobutyryl-ACP into 3-hydroxybutyryl-ACP which is dehydrated by 3-hydroxyacyl-ACP dehydratase to form trans-Δ2-butenoyl-ACP. The reduction of the double bond uses NAD(P)H to convert trans-Δ2-butenoyl-ACP into butyryl-ACP which in then condensed with malonyl-CoA by the 3-ketoacyl-ACP synthase I (KAS I) to generate 3-ketoacyl-ACP. This cycle is repeated to elongate saturated FA chains till 16:0-ACP, and then, the 3-ketoacyl-ACP synthase II (KAS II) performs the last elongation step to synthesize 18:0-ACP. The stearoyl-ACP Δ9-desaturase desaturates 18:0-ACP into 18:1-ACP. Plastidic de novo FAS ends when the FA thioesterase removes the ACP group from acyl backbones.
Figure 1.
Simplified schematic biochemical pathway of fatty acid and TG synthesis in higher plants. Fatty acid synthesis (FAS) and lipid droplet (LD) formation in heterotrophic and in autotrophic embryos. ACP, acyl carrier protein; AKG, α-ketoglutarate; CIT, citrate; CoA, coenzyme A; DAG, diacylglycerol; FA, fatty acid; F6P, fructose 6-phosphate; FUM, fumarate; G6P, glucose 6-phosphate; hv, light; ICIT, isocitrate; LD, lipid droplet; LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; MAL, malate; OAA, oxaloacetate; PA, phosphatidic acid; PC, phosphatidylcholine; PGA, phosphoglycerate; PEP, phosphoenolpyruvate; PGA, phosphoglycerate; SUCC, succinate; TCA, tricarboxylic acid; TG, triacylglycerol; VLCFA, very-long-chain fatty acid.
Beyond the complex network defined by interconnected OPPP and glycolytic pathways, additional metabolic routes may supply precursors for de novo FAS in the plastid ( Figure 1 ). During the conversion of pyruvate into acetyl-CoA, there is a concomitant loss of the carboxylic group of the pyruvate (released as CO2) which has a significant impact on the CCE of developing embryos. In autotrophic plastids of green embryos, ribulose-1,5-bisphophate carboxylase/oxygenase (Rubisco) is able to re-fix the released CO2 apart from the Calvin cycle. This “Rubisco shunt” involves the conversion of hexose-phosphates and triose-phosphates to ribulose-1,5-bisphosphate (RuBP) by the non-oxidative reactions of the OPPP, and the subsequent fixation of CO2 onto RuBP by Rubisco and cleavage in two 3-phosphoglycerate (PGA) (Schwender et al., 2004). PGA can be further metabolized to pyruvate and then to FAs via acetyl-CoA. This metabolic route offsets the loss of CO2 at reactions, such as the OPPP and pyruvate dehydrogenase, thus increasing the CCE in developing embryos. Another route for the synthesis of plastidial pyruvate (and then acetyl-CoA) is through decarboxylation of imported malate by the plastidial NADP-dependent malic enzyme (pNADP-ME) (Smith et al., 1992; Shearer et al., 2004). Supply of malate relies on either the translocation of mitochondrial malate or the carboxylation of PEP into oxaloacetate (OAA) via the cytosolic phosphoenolpyruvate carboxylase, followed by the conversion of OAA into malate catalyzed by NAD-dependent malate dehydrogenase (King et al., 1998) which happens in both green and non-green seeds. The production of acetyl-CoA from imported malate requires the successive action of the pNADP-ME and the plastidic pyruvate dehydrogenase, which presents the disadvantage of generating 2 molecules of CO2 for each 4-carbon malate molecule ( Figure 1 ).
FAS also requires stoichiometric amounts of ATP, NADPH, and NADH for each sequential addition of an acetyl unit to the growing chain of the fatty acid ( Figure 1 ). ATP is required for the carboxylation of acetyl-CoA to malonyl-CoA by ACCase, whereas the two reductases of the FAS complex require NADPH and NADH, respectively. Plastids of oilseeds must either take up ATP produced by oxidative phosphorylation in the mitochondria or generate it internally. ATP can be produced in the cytosol via the glycolysis and through the mitochondrial oxidative phosphorylation, and then be imported into the plastids via a nucleotide transporter. A potential source for ATP and reductant within the plastids is generated by the oxidation of sugar phosphates via glycolytic enzymes: glyceraldehyde 3-phosphate dehydrogenase generates NADH whereas phosphoglycerate kinase and pyruvate kinase produce ATP. Likewise, the subsequent conversion of pyruvate into acetyl-CoA by the pyruvate dehydrogenase is accompanied by the production of NADH. The intraplastidial conversion of malate to pyruvate by pNADP-ME constitutes another potential source of NADPH. Finally, the oxidation of sugar phosphates by the plastidial OPPP may also contribute to the production of NADPH. In the case of green oilseeds, light reactions in the thylakoids produce ATP and NADPH that can be used for de novo FAS ( Figure 1 ). The balance between cyclic and non-cyclic photophosphorylation may also adjust respective ATP and NADPH productions to metabolic requirements in autotrophic plastids (Allen et al., 2009).
Following FAS, acyl groups in FA are hydrolyzed by acyl-ACP thioesterases, releasing free acyl chains. These free acyl chains are then activated to CoA esters on the outer membrane of the plastid by long-chain acyl-CoA synthetases prior to their export toward the ER for their elongation into very-long chain fatty acids (VLCFA). Carbon for FA elongation comes from the cleavage of cytosolic citrate into OAA and acetyl-CoA by citrate lyase, reaction that requires ATP ( Figure 1 ). This cytosolic acetyl-CoA is then used by acetyl-CoA carboxylase to synthesize malonyl-CoA, the carbon donor for FA elongation. The elongation of acyl-CoA in the ER involves four sequential enzymatic steps catalyzed by the fatty acid elongase complex: i) the ketoacyl-CoA synthase (KCS) requires energy under the form of ATP to condenses malonyl-CoA with the elongating acyl-CoA; ii) the 3-ketoacyl-CoA reductase (KCR) uses (NAD/PH) to reduce 3-ketoacyl-CoA; iii) the 3-hydroxy acyl-CoA dehydrate (HCD) catalyzes the dehydration of 3-hydroxyacyl-CoA; and iv) the enoyl-CoA reductase (ECR) uses NAD/PH to reduce enoyl-CoA. Similar to FAS, FA elongation requires energy and reductant. ATP may be produced by oxidative phosphorylation in the mitochondria and cytosolic glycolysis which may also provide NADH. The cytosolic pentose-phosphate pathway may contribute to NADPH production in developing embryos with the glucose-6-phosphate and 6-phosphogluconate dehydrogenases ( Figure 1 ).
Finally, the assembly of TGs involves the sequentially esterification of acyl-CoAs to glycerol 3-phosphate (G3P) backbone by membrane-bound acyltransferases ( Figure 1 ). The first acylation yields lysophosphatidic acid (LPA), which in turn is acylated to produce the central metabolite phosphatidic acid (PA). PA is then converted to diacylglycerol (DAG) by the action of PA phosphatase (PAP). In the Kennedy pathway, a third FA is transferred to the vacant position of DAG by diacylglycerol acyltransferase (DGAT), the only enzymatic reaction of the pathway exclusively committed to TG biosynthesis (Cao and Huang, 1986; Cao and Huang, 1987; Li-Beisson et al., 2013). The acyl chains from phosphatidylcholine (PC) may also become available for TG synthesis through transfer of an acyl from PC to DAG, which is catalyzed by phospholipid:diacylglycerol acyltransferase (PDAT). Once TG assembly is achieved in the ER, TGs are accumulated between the two layers of phospholipids, resulting in the formation of structures called oil bodies or lipid droplets (LD). These spherical organelles comprise a matrix of TGs surrounded by a phospholipid monolayer where the aliphatic chains are oriented to the TG lumen and the phosphate groups toward the cytosol (Yatsu and Jacks, 1972; Chapman and Ohlrogge, 2012).
Identification of sources of carbon precursors and reductants for FAS using isolated plastids
Besides ATP which can be produced by oxidative phosphorylation and can be translocated from one compartment to the other, the carbon precursor (acetyl-CoA) and reductant do not cross biological membranes, and have therefore to be synthesized in the plastids for FAS and/or the cytosol for FA elongation. As described above ( Figure 1 ), several biochemical steps may lead to the production of acetyl-CoA and NAD(P)H, and their relative importance varies from one species to another. Determining the nature of carbon precursors and reducing power for FAS is crucial to understand and enhance oil synthesis in developing seeds, and also to identify potential bottlenecks.
Early experiments were performed by incubating isolated plastids with radiolabeled substrates to investigate which carbon precursors were stimulating the rate of de novo FAS, and which pathway were producing reductant. Feeding isolated plastids from Brassica napus with 14C-labeled substrates showed that pyruvate, glucose 6-phosphate (G6P), dihydroxyacetone phosphate (DHAP), malate, acetate, and phosphoenolpyruvate (PEP) were utilized as precursors for FAS (Kang and Rawsthorne, 1994; Kubis et al., 2004). Particularly, the utilization of G6P through the OPPP also provided reductant for FAS (Eastmond and Rawsthorne, 2000; Pleite et al., 2005). However, incubation of B. napus isolated plastids with 14C-PEP revealed that it did not only provide carbons, but also supplied additional ATP for FAS (Kubis et al., 2004). In contrast to B. napus, feeding sunflower isolated plastids with 14C-G6P indicated that there was no incorporation of labeling in FAs (Pleite et al., 2005). However, pyruvate utilization in combination with G6P dramatically increased FAS. Due to an insufficient NADPH pool, pyruvate feeding alone was not sufficient to enhance the rate of FAS. Moreover, malate has been demonstrated to be one of the most efficient precursors for FAS. Indeed, in isolated plastids from castor and sunflower embryos, supply of malate stimulated the rates of FAS (Smith et al., 1992; Pleite et al., 2005). In comparison to pyruvate and acetate, malate significantly increased the rate of FAS due to generation of additional NADPH via pNADP-ME (Pleite et al., 2005). However, supply of G6P with malate did not significantly alter the rate of FAS in isolated plastids.
Although experiments conducted with isolated plastids were key to identify the carbon precursors and reductant, these results did not completely reflect what happens in vivo, in the entire developing embryo. Initial experiments in vivo were carried out by incubating embryos with radioisotopes. Labeling studies performed with [U-14C4]-malate in sunflowers embryos in culture showed that malate could provide a source of carbon for the FAS (Alonso et al., 2007), corroborating the previous results on isolated plastids mentioned above (Pleite et al., 2005). However, malate is not a physiological substrate provided from the sunflower plant to the developing embryos. Using culture conditions that mimic the feeding and the development of the sunflower embryos in planta, isotopic steady state labeling experiments demonstrated that the flow of malate towards FAS was minor; the major source of carbon (91-95%) contributing to FAS was from triose phosphates (Alonso et al., 2007). Due to discrepancies between results on isolated plastids and whole embryos in culture, MFA has been the method of choice to study the metabolic pathways involved in FAS under conditions that are physiologically relevant.
Metabolic flux analysis to study fatty acid synthesis in developing embryos
Accessing C partitioning in vivo using 13C-MFA
The goal of MFA is to quantify all the in vivo metabolic fluxes in a given organ or cell, here developing embryos, which results in a metabolic flux map. The determination of intermediary carbon fluxes requires 13C-labeling. Therefore, establishing culture conditions that mimic the development of embryos in planta is decisive to build carbon flux maps. For instance, it has been found for multiple species of the Brassicaceae family that developing embryos readily grow in liquid cultures (Schwender and Ohlrogge, 2002; Lonien and Schwender, 2009; Chen and Shachar-Hill, 2012; Tsogtbaatar et al., 2020). Substrates, furnished by the mother plant, are unloaded in the endosperm liquid and taken up by the embryo. Therefore, the best way to design a liquid growth medium that mimics the in planta liquid environment is to analyze the constituents of the endosperm or the vascular tissue (Alonso et al., 2007; Alonso et al., 2010; Cocuron et al., 2019; Tsogtbaatar et al., 2020). Substrate composition, total osmotic pressure of the medium, and light intensity are important factors influencing plant tissue development (Goffman et al., 2005; Allen et al., 2007; Alonso et al., 2010), and hence have to be optimized. To meet requirements for 13C-MFA, it is important to maintain homeostasis and metabolic steady state in culture embryos. Therefore, photoperiod is not commonly applied to embryos in culture for 13C-MFA studies. Ideal culture conditions for plant embryos are validated when the dry weight gain and the biomass composition of embryos grown in culture are not significantly different from the ones grown in planta (Alonso et al., 2007; Alonso et al., 2010; Alonso et al., 2011; Cocuron et al., 2019; Tsogtbaatar et al., 2020). Parallel labeling experiments, using different 13C-substrates, are usually conducted to have a better coverage of the metabolic network (Schwender et al., 2006; Alonso et al., 2007; Alonso et al., 2010; Antoniewicz, 2015; Crown et al., 2016; Antoniewicz, 2018; Acket et al., 2019; Tsogtbaatar et al., 2020). Labeled embryos are harvested after they have reached an isotopic steady-state, meaning that the labeling in intermediary metabolites and products have reached a constant pattern. The resultant labeling in a range of metabolites is then determined using nuclear magnetic resonance and/or mass spectrometry (MS) (Ratcliffe and Shachar-Hill, 2006; Dieuaide-Noubhani and Alonso, 2014). Several sensitive gas chromatography-mass spectrometry (GC-MS) and liquid chromatography tandem mass spectrometry (LC-MS/MS) methods have been recently developed to follow the labeling directly in key metabolic intermediaries such as sugars, phosphorylated compounds, free amino acids, and organic acids (Alonso et al., 2010; Koubaa et al., 2013; Cocuron and Alonso, 2014; Cocuron et al., 2017; Acket et al., 2019; Cocuron et al., 2020). Compartmentalization in plant cells is usually considered by following different labeling patterns of a few metabolites and hydrolyzed macromolecules whose biosynthesis occur in one compartment (Allen et al., 2007; Allen et al., 2012; Dieuaide-Noubhani and Alonso, 2014; Cocuron et al., 2020). The complete labeling information is entered into a mathematical model describing the metabolic network. This mathematical model includes equations expressing metabolic and isotopic steady states. Finally, the fluxes through the network that correspond to the observed label distribution are calculated using mathematical algorithms that can be computed using available software (Wiechert and De Graaf, 1997; Wiechert and De Graaf, 1997; Möllney et al., 1999; Wiechert et al., 1999; Wiechert et al., 2001). 13C-based MFA has been successfully applied to plant systems to characterize the in vivo carbon fluxes in important metabolic pathways during FAS in developing embryos (Goffman et al., 2005; Alonso et al., 2007; Allen et al., 2009; Lonien and Schwender, 2009; Alonso et al., 2010; Cocuron et al., 2019; Carey et al., 2020; Tsogtbaatar et al., 2020).
Determination of carbon conversion efficiency in developing embryos
Determining the efficiency of the developing embryo in converting carbon sources into oil and other biomass components (e.g. proteins, and carbohydrates) known as carbon conversion efficiency (CCE), is important to evaluate the potential for improving seed oil quantity (Goffman et al., 2005; Alonso et al., 2007; Allen et al., 2009; Lonien and Schwender, 2009; Chen and Shachar-Hill, 2012; Cocuron et al., 2019; Carey et al., 2020; Tsogtbaatar et al., 2020). CCE results from the sum of all catabolic and anabolic metabolic processes, which varies in developing embryos from different oilseeds (Goffman et al., 2005; Allen et al., 2009; Lonien and Schwender, 2009; Carey et al., 2020; Tsogtbaatar et al., 2020). To assess the CCE, embryos are grown for several days culture media and conditions that mimic their development in plant, as explained in the above section. There are two options to determine the percentage of carbon uptaken that was stored into biomass components. The first option uses 14C-labeled substrates in sealed flasks, and quantifies the radiolabeling released as 14CO2, and incorporated as 14C-oil, 14C-proteins, and 14C-carbohydrates (Goffman et al., 2005; Alonso et al., 2007; Allen et al., 2009; Alonso et al., 2010; Alonso et al., 2011; Carey et al., 2020). The second option is the quantify the substrate depletion from the media and the biomass stored during the incubation period (Cocuron et al., 2019; Tsogtbaatar et al., 2020). Table 1 summarizes the biomass composition and the CCE of different embryos from green and non-green oilseeds that have been studied and published so far. Under physiological conditions, the biomass composition of the developing embryos in culture differs among species. Oil content varied from 18% (w/w) for G. max to 56% for B. napus, proteins from 6% for Z. mays LH59 to 40% for G. max, and carbohydrates from 43% for G. max to 60% for Z. mays LH59. Under physiological conditions, T. arvense embryos were found to be the most efficient in converting carbon substrates into biomass (93%) while C. sativa were the least (32%). It is important to note that these differences in CCE reflect the amount of carbon loss as CO2 during the synthesis of biomass components: the lower the CCE, the higher is the loss of carbon as CO2, which has implications for metabolic engineering. For instance, improving oil content in C. sativa may be achieved by reducing the pathways producing CO2. However, this strategy would not work in T. arvense which is already extremely efficient. Instead, for T. arvense, one would have to redirect carbon from another biomass component to increase FAS.
Table 1.
Biomass composition and carbon conversion efficiency.
| Species | Biomass composition in embryo (% w/w) | CCE (%) | References | ||
|---|---|---|---|---|---|
| Oil | Protein | Carbohydrates | |||
| Zea mays LH59 (low oil line)* | 34 | 6 | 60 | 57-71 | (Alonso et al., 2010) |
| Zea mays ALEX (high oil line)* | 48 | 13 | 39 | 61-64 | (Cocuron et al., 2019) |
| Helianthus annuus* | 38 | 18 | 44 | 50 | (Alonso et al., 2007) |
| Glycine max, 35 µE* | 18 | 39 | 43 | 83 | (Allen et al., 2009) |
| Brassica napus, dark | 45 | 60 | (Goffman et al., 2005) | ||
| Brassica napus, 50 µE* | 56 | 86 | (Goffman et al., 2005) | ||
| Brassica napus, 150 µE | 58 | 95 | (Goffman et al., 2005) | ||
| Thlaspi arvense, 20 µE* | 31 | 28 | 41 | 93 | (Tsogtbaatar et al., 2020) |
| Camelina sativa, dark | 29 | 23 | 48 | 21 | (Carey et al., 2020) |
| Camelina sativa, 10 µE* | 35 | 27 | 38 | 32 | (Carey et al., 2020) |
| Camelina sativa, 50 µE | 34 | 20 | 46 | 42 | (Carey et al., 2020) |
| Arabidopsis thaliana (Col), 50 µE | 41 | 24 | 13 | 80 | (Lonien and Schwender, 2009) |
Comparison of the biomass composition and carbon conversion efficiency (CCE) in embryos from crop and model species grown under cultured conditions. Embryos from species highlighted in green are green embryos that may be photosynthetically active; the * denotes physiological conditions.
Except C. sativa, embryos from green seeds had higher CCEs than those seeds that cannot use light ( Table 1 ). The importance of light was further demonstrated by incubating developing embryos at different intensities. In general, higher light intensity significantly increased the CCE. This finding suggests that light has strong effects on the metabolism of developing green embryos—probably due to the additional production of NADPH and ATP via photosynthesis–resulting in faster growth and storage product accumulation, which improved the CCE (Goffman et al., 2005; Alonso et al., 2007; Allen et al., 2009; Lonien and Schwender, 2009; Chen and Shachar-Hill, 2012; Cocuron et al., 2019; Carey et al., 2020; Tsogtbaatar et al., 2020). Indeed, studies on these developing green embryos showed that all the major photosynthetic complexes (PSII, PSI and their antenna complexes, cytochrome b6f complex, and ATP synthase) are present at a necessary stoichiometric ratio, suggesting a high photochemical activity despite being partially blocked from light by the pod and seed coat (Asokanthan et al., 1997; Ruuska et al., 2004; Puthur et al., 2013; Allorent et al., 2015). The light reactions occurring in these green embryos have been associated with the rapid synthesis of ATP and NADPH needed for energetically-expensive FAS. It has also been reported that embryo photosynthesis contributes to a significant amount of oxygen, which fuels energy-generating biochemical pathways, including mitochondrial respiration (Ruuska et al., 2004; Borisjuk et al., 2005; Rolletschek et al., 2005; Tschiersch et al., 2011; Galili et al., 2014).
Differences in the flow of carbon through central metabolic pathways are responsible for the differences in biomass composition and CCE measured in developing embryos from various oilseed species ( Table 1 ). 13C-MFA has been the method of choice to measure in vivo rates of carbon flow, quantifying the metabolic pathways that are active during FAS.
MFA to identity carbon sources and reductants for FAS in developing embryos
13C-MFA was applied to developing embryos from various oilseed species to determine the sources of carbon and reductant for FAS, and to unravel the occurrence of non-conventional pathways that improved the CCE ( Table 2 ) (Schwender et al., 2004; Schwender et al., 2006; Alonso et al., 2007; Allen et al., 2009; Lonien and Schwender, 2009; Alonso et al., 2010; Hay et al., 2014; Cocuron et al., 2019; Acket et al., 2020; Carey et al., 2020; Tsogtbaatar et al., 2020). Glycolysis is a major source of pyruvate for de novo FAS ( Figure 1 ). 13C-labeling and metabolic flux analysis performed in non-photosynthetic and photosynthetic embryos revealed the contribution of the plastidic NADP-dependent malic enzyme (pNADP-ME) for the production of plastidic pyruvate (pPYR) in Z. mays, H. annuus, G. max, T. arvense, and C. sativa ( Table 2 ). The pNADP-ME provided up to 54% of pPYR in the high oil ALEX maize line (Cocuron et al., 2019). In plastids, pNADP-ME catalyzes the conversion of malate to pPYR with the production of CO2 and NADPH ( Figure 1 ). In addition, pPDH further catalyzes the decarboxylation of pPYR to acetyl-CoA with the generation of CO2 and NADH. Although the overall equimolar conversion of malate into plastidic acetyl-CoA results in the production of valuable reductant necessary for FAS (NADH and NADPH), it leads, in return, to the loss of two carbons as CO2, which may affect the overall CCE. Interestingly, 13C-labeling in developing B. napus embryos demonstrated for the first time that Rubisco was fixing this plastidic 13CO2 to produce phosphoglycerate (pPGA) through an unconventional “Rubisco shunt” ( Table 2 ) (Schwender et al., 2004). The fixation of pCO2 by Rubisco contributes to additional sources of carbon for FAS: it compensates for the decarboxylation steps, and channels more substrates into FAS, improving the CCE (Eastmond et al., 1996; Ruuska et al., 2004; Schwender et al., 2004). Indeed, studies in developing B. napus embryos showed a CCE of 86% under physiological conditions due to CO2 refixation by Rubisco into pPGA ( Tables 1 , 2 ) (Schwender et al., 2004). 13C-MFA demonstrated that up to 64% of pPGA was produced by Rubisco, contributing to the synthesis of acetyl-CoA for de novo FAS, and reducing by 40% the carbon lost as CO2 ( Table 2 ) (Schwender et al., 2004). Similar processes have been described in developing embryos of G. max and T. arvense where Rubisco was found to contribute to 14% and 25% of the pPGA, respectively ( Table 2 ) (Allen et al., 2009; Tsogtbaatar et al., 2020).
Table 2.
Comparison of the use of non-conventional pathways in developing embryos.
| Species | Contribution of pNADP-ME to pPYR | Rubisco contribution to pPGA | Reversibility of IDH | Contribution to NADPH for FAS | References |
|---|---|---|---|---|---|
| Zea mays | 30-54% | 0% | No | 74-76% OPPP 30-55% pNADP-ME |
(Alonso et al., 2010; Cocuron et al., 2019) |
| Helianthus annuus | 7% | 0% | No | 212% OPPP 6% pNADP-ME |
(Alonso et al., 2007) |
| Glycine max | <20% | 14% | Yes | <24% OPPP <29% pNADP-ME |
(Allen et al., 2009) |
| Brassica napus | <1% | 36-64% | Yes | 25% OPPP <1% pNADP-ME |
(Schwender et al., 2004; Schwender et al., 2006; Hay et al., 2014) |
| Thlaspi arvense | 20% | 25% | Yes | n.d. | (Tsogtbaatar et al., 2020) |
| Camelina sativa | 9% | 0% | Yes | 6,079% OPPP 15% pNADP-ME |
(Carey et al., 2020) |
| Linum usitatissinum | <1% | 0% | Yes | 186% OPPP <1% pNADP-ME |
(Acket et al., 2019) |
Published data from 13C-labeling and metabolic flux analysis obtained from developing embryos cultured under physiological conditions were used to determine the contribution of the plastidic NADP-dependent malic enzyme (pNADP-ME) to the production of plastidic pyruvate (pPYR); the contribution of Rubisco to plastidic phosphoglycerate (pPGA); the reversibility of the isocitrate dehydrogenase (IDH); and the contribution of the oxidative pentose-phosphate pathway (OPPP) and pNADP-ME to the production of NADPH necessary for fatty acid synthesis (FAS). Embryos from species highlighted in green are green embryos that may be photosynthetically active.
In addition to the role of Rubisco, 13C-labeling and MFA also revealed the non-canonical function of isocitrate dehydrogenase (IDH) in photosynthetic embryos ( Table 2 ). This reaction, assumed to be thermodynamically irreversible, was reported to catalyze the carboxylation of α-ketoglutarate into isocitrate in vivo in developing B. napus embryos (Schwender et al., 2006). It is important to note that the CO2 fixation by IDH may also improve the CCE. This phenomenon was explained by a high demand in citrate for FA elongation in B. napus ( Figure 1 ), and a high concentration of CO2 (40 mM) available in developing oilseeds (Goffman et al., 2004). Similarly, reversibility of IDH was measured in developing green embryos from G. max, even when the concentration of CO2 was lower (Allen et al., 2009), T. arvense (Tsogtbaatar et al., 2020), C. sativa (Schwender et al., 2006), and L. usitatissinum (Acket et al., 2019), but absent in heterotrophic embryos ( Table 2 ). Interestingly, developing embryos that have an active Rubisco and reversible IDH were found to be the more efficient in converting their substrates into biomass ( Tables 1 , 2 ).
Besides carbon, FAS requires reductant that must be synthesized in the plastid. For each mole of acetyl-CoA produced, the plastidic pyruvate dehydrogenase complex generated one mole of NADH, which can be directly used for FAS ( Figure 1 ). The plastidic production of NADPH necessary for de novo FAS may be ensured by the OPPP and/or pNADP-ME, and light reactions in the case of photosynthetic embryos ( Figure 1 ). Knowing that the OPPP and the pNADP-ME generate CO2, their operation may affect negatively the CCE ( Figure 1 ). 13C-labeling and MFA were used to measure the relative contribution of the OPPP and pNADP-ME to the production of NADPH in developing embryos from different species ( Table 2 ). In general, the OPPP supported a higher production of NADPH than the pNADP-ME. For G. max and B. napus, these two combined pathways did not provide sufficient NADPH to support FAS in developing embryos. In those species, the remainder of NADPH may be supplied by the light reactions of photosynthesis and/or catabolism (Goffman et al., 2005; Schwender et al., 2006; Allen et al., 2009). For Z. mays, the production of NADPH by the OPPP and pNAPD-ME was just enough to support FAS in developing embryos ( Table 2 ). More specifically, the pNAPD-ME was found to work at maximal capacity in vivo; it was identified as the limiting step in the provision of pPYR and NADPH for FAS in maize (Alonso et al., 2010; Cocuron et al., 2019). The embryos from the other species, H. annuus, C. sativa, and L. usitatissinum, were producing NADPH via the OPPP and pNADP-ME in excess of the requirements for FAS ( Table 2 ), concomitantly generating CO2, which resulted in the lowest CCE for these species ( Table 1 ).
Overall, the aforementioned studies have demonstrated that MFA is a valuable tool to quantify the carbon partitioning in vivo, and identify the sources of carbon skeletons and reductants necessary for FAS in the developing green and non-green embryos. Information gathered from MFA, combined with transcriptomics and metabolomics, will give insights on the genes that control oil synthesis and create novel approaches for the genetic engineering of oilseed crops. The following sections review some of the target genes and the common genetic engineering strategies for enhancing oil content and quality in seeds.
Metabolic engineering strategies to improve seed oil content and composition
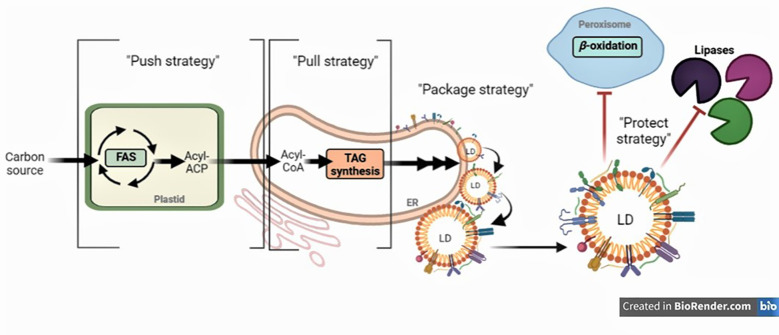
To improve the yield and FA composition in oilseed crops, it is critical to understand how assimilates are partitioned in favor of storage lipids. It is also of great importance to elucidate the mechanisms of TG biosynthesis in different oilseeds to further optimize their FA composition since it is a key determinant of oilseed nutritional value and industrial applications. Several strategies have been employed to genetically manipulate FA composition and plant lipid metabolism in order to increase the FA content oilseeds. To this end, the “push, pull, package, and protect” strategy has been implemented at various degrees; it consists of manipulating the expression of genes to boost the synthesis of FAs (“push”), or increase TG assembly reactions (“pull”), or improve the storage of FAs into lipid droplets (LDs) (“package”), or prevent the degradation of stored lipids (“protect”), or any combination of these ( Figure 2 ) (Vanhercke et al., 2019). The production of novel/unusual FAs presents additional challenges because they may disrupt membrane stability. Therefore, seeds must incorporate and store novel/unusual FAs into TG. Tables 3 – 7 review the genes that were up- or down-regulated in several species, and when available, their effect in total seed oil content and FA composition.
Figure 2.
Improving oil quality in seeds by an integrated metabolic engineering approach. Broadly, strategies aim to (1) increase the carbon flux into de novo fatty acid synthesis (FAS) (“Push strategy”), (2) ensure efficient assembly of nascent acyl chains into triacylglycerol (TG) (“Pull strategy”), (3) facilitate lipid droplet (LD) biogenesis and maximize droplet stability by proper coating (“Package strategy”), and (4) minimize TG turnover (“Protect strategy”).
Table 3.
List of genes encoding TFs manipulated to enhance oil yield and/or change the FA composition in seeds.
| Gene | Gene source | Target species | Strategy | Effect in seed oil content | Effect in fatty Acid composition | References |
|---|---|---|---|---|---|---|
| ABI3 | A. thaliana | A. thaliana | Down-regulation | ↘ seed oil content by up to 30% | ↘ C20:1 | (Yang et al., 2022) |
| Down-regulation | ↘ seed oil content by up to 42% | ↘ C16:0, C16:1, C18:0, C18:1, C18:2, C18:3, C20:0, C20:1, C22:0, C22:1 | (Roscoe et al., 2015) | |||
| Dof1 | G. hirsutum | G. hirsutum | Over-expression | ↗ seed oil content by 16% | (Su et al., 2017) | |
| DREBL | G. max | A. thaliana | Over-expression | ↗ seed oil content by 10% | ↗ C18:1, C18:2, C18:3, C20:1 | (Zhang et al., 2016) |
| FUS3 | B. napus | B. napus | Down-regulation | ↘ seed oil content by up to 12% | ↗ C18:2 | (Elahi et al, 2015) |
| A. thaliana | A. thaliana | Down-regulation | ↘ seed oil content by up to 67% | ↘ C16:0, C16:1, C18:0, C18:1, C18:2, C18:3, C20:0, C20:1, C22:0, C22:1 | (Roscoe et al., 2015) | |
| GLABRA2 (GL2) | A. thaliana | A. thaliana | Down-regulation | ↗ seed oil content by 7% | (Shi et al., 2012) | |
| GRF2 | B. napus | A. thaliana | Over-expression | ↗ seed oil content by 48% | (Liu et al., 2012) | |
| LEC1 | Z. mays | Z. mays | Over-expression | ↗ seed oil content by 48% | (Shen et al., 2010) | |
| B. napus | B. napus | Over-expression | ↗ seed oil content by 2% | ↗ C16:0, C18:0, C18:2, C18:3; ↘ C18:1 | (Tan et al., 2011) | |
| Z. mays | A. thaliana | Over-expression | ↗ seed oil by 20% | (Zhu et al., 2018) | ||
| C. sativa | Over-expression | ↗ seed oil by more than 26% | (Zhu et al., 2018) | |||
| B. napus | B. napus | Over-expression | ↘ seed oil content by up to 16% | ↗ C18:1, C18:3; ↘ C16:0 | (Elahi et al., 2016) | |
| Down-regulation | ↘ seed oil content by up to 12% | no significant difference | (Elahi et al., 2016) | |||
| A. thaliana | A. hypogea | Over-expression | ↗ seed oil by up to 16% | ↗ C18:1, C18:2, C18:3, C20:1; ↘ C16:0, C18:0 | (Tang et al., 2018) | |
| LEC2 | G. max | A. thaliana | Over-expression | ↗ seed oil content by 34% | ↗ C18:0, C18:1; ↘ C16:0, C18:2, C20:1 | (Manan et al., 2017) |
| L1L | B. napus | B. napus | Over-expression | ↗ seed oil content by 16% | ↗ C16:0, C18:0, C18:2, C18:3; ↘ C18:1 | (Tan et al., 2011) |
| MED15 | A. thaliana | A. thaliana | Over-expression | ↗ seed oil content by up to 15% | ↘ C20:1 | (Kim et al., 2016) |
| Down-regulation | ↘ seed oil content by up to 30% | ↘ C18:2; ↗ C18:3, C20:0 | (Kim et al., 2016) | |||
| MIF1 | A. thaliana | A. thaliana | Over-expression | ↗ seed oil content by 13% | ↗ C18:1, C20:0, C20:1; ↘ C18:3, C20:2, C20:3 | (Cheng et al., 2021) |
| Down-regulation | no significant difference | no significant difference | (Cheng et al., 2021) | |||
| MYB1 | J. curcas | A. thaliana | Over-expression | ↗ seed oil content by up to 28% | ↗ C18:3, C20:1; ↘ C18:1 | (Khan et al., 2019) |
| MYB115 or MYB118 | A. thaliana | A. thaliana | Down-regulation | ↘ Total omega-7 fatty acids | (Troncoso-Ponce et al., 2016) | |
| MYB89 | A. thaliana | A. thaliana | Down-regulation | ↗ seed oil content by up to 30% | ↗ C16:0, C18:0, C18:1, C18:2; C18:3; C20:0; C20:1, C22:1 | (Li et al., 2017) |
| MYB96 | C. sativa | C. sativa | Over-expression | ↗ seed oil content by up to 21% | (Kim et al., 2019) | |
| Down-regulation | ↘ seed oil content by up to 15% | ↘ C16:0, C18:0, C18:2, C18:3, C20:1, C20:2 | (Lee et al., 2018) | |||
| WRI1 | A. thaliana | L. campestre | Over-expression | ↗ seed oil by up to 16% in T3 seeds | ↘ C18:1, C18:2, C20:1, C22:1 | (Ivarson et al., 2017) |
| B. napus | A. thaliana | Over-expression | ↗ seed oil by 10–40% | (Liu et al., 2010) | ||
| A. thaliana | C. sativa | Over-expression | ↗ seed oil by 8-31% | ↗ C18:2, C18:3; ↘ C18:1, C20:1 | (An and Suh, 2015) | |
| Z. mays | Z. mays | Over-expression | ↗ seed oil by 9% | ↗ C16:0, C18:0, C18:1, C18:3; ↘ C9:0, C10:0 | (Pouvreau et al., 2011) | |
| G. max | G. max | Over-expression | ↗ seed oil content by up to 13% | ↗ C18:1, C18:2 | (Chen et al., 2018) | |
| C. nucifera | A. thaliana | Over-expression | ↗ seed oil content by up to 20% | ↗ C16:0, C18:0, C18:1; ↘ C20:1 | (Sun et al., 2017) | |
| C. nucifera | O. sativa | Over-expression | ↗ seed oil content by up to 7% | ↗ C16:0, C18:3; ↘ C18:1 | (Sun et al., 2017) | |
| Z. mays | Z. mays | Over-expression | ↗ seed oil by 46% | no significant difference | (Shen et al., 2010) | |
| H. annuus | A. thaliana | Over-expression | ↗ seed oil by 40% | (Lim et al., 2022) | ||
| A. thaliana | A. thaliana | Down-regulation | ↘ seed oil content by up to 33% | ↗ C18:3, C22:1; ↘ C18:1, C18:2, C20:1 | (Baud et al., 2007) | |
| WRKY6 | A. thaliana | A. thaliana | Down-regulation | ↗ seed oil content by 25-27% | ↗ C18:3, C20:0; ↘ C18:2 | (Song et al., 2020) |
Table 7.
List of “protect” genes used to enhance oil yield and/or change the FA composition in seeds.
| Gene | Gene source | Target species | Strategy | Effect in total seed oil content | Effect in fatty Acid composition | References | |
|---|---|---|---|---|---|---|---|
| SFAR | B. napus | B. napus | Down-regulation | ↗ seed oil by 14-28% | ↗ C18:3, C20:0, C20:1, C22:0, C22:1; ↘ C16:0, C16:1 | (Karunarathna et al., 2020) | |
| GDSL1 | A. thaliana | B. napus | Overexpression | ↘ seed oil by 13% | ↗ C18:2, C18:3; ↘ C18:1, C20:0, C20:1 | (Ding et al., 2019) | |
| B. napus | B. napus | Overexpression | ↘ seed oil by 13% | ↗ C18:2, C18:3; ↘ C18:1, C20:0, C20:1 | (Ding et al., 2019) | ||
| B. napus | B. napus | Down-regulation | ↗ seed oil by 12% | ↗ C18:1; ↘ C18:2, C18:3 | (Ding et al., 2019) | ||
| NCP6 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil by 6-8% | ↗ C18:3, C20:0, C20:1; ↘ C16:0 and C18:0 | (Cai et al., 2020) | |
| C. sativa | C. sativa | Overexpression | ↗ seed oil by 6.3% | ↗ C18:3; ↘ C16:0 and C18:0 | (Cai et al., 2020) | ||
| pPLAIIIδ | C. sativa | C. sativa | Overexpression | ↗ seed oil by 14% | ↗ C20:1; ↘ C18:1 | (Li et al., 2015a) | |
| PLIP1 | A. thaliana | A. thaliana | Overexpression | ↘ seed oil by 25% | ↗ C18:3; ↘ C18:1, C18:2 | (Wang et al., 2017) | |
| Down-regulation | ↗ seed oil by 45% | ↗ C18:1, C18:3, C22:1; ↘ C20:0 | (Wang et al., 2017) | ||||
| PUX10 | A. thaliana | A. thaliana | Down-regulation | no significant difference | ↘ C18:2; ↗ C18:3 | (Deruyffelaere et al., 2018) | |
| Down-regulation | no significant difference | no significant difference | (Kretzschmar et al., 2018) | ||||
| SDP1 | J. curcas | A. thaliana | Down-regulation | ↗ seed oil by 13% | ↗ C20:1; ↘ C18:1 | (Kim et al., 2014) | |
| G. max | G. max | Down-regulation | ↗ seed oil by 30% | ↗ C18:1; ↘ C18:2 | (Kanai et al., 2019) | ||
| B. napus | B. napus | Down-regulation | ↗ seed oil by 8% | no significant difference | (Kelly et al., 2013) | ||
Push strategy
This strategy aims to enhance the flux of carbon through FAS to produce additional acyl chains in the plastids for subsequent assembly into TGs in the ER ( Figure 2 ). The “push” strategy is considered to be one of the most extensively used to manipulate oil content. Commonly, over-expression (or even down-regulation) of transcription factors (TFs) offers a great advantage of controlling a number of reactions simultaneously in the oil biosynthetic pathway ( Table 3 ). The best examples of such TFs are LEAFY COTYLEDON1 (LEC1), LEC1-LIKE (L1L), LEC2, and WRINKLED1 (WRI1) (Focks and Benning, 1998; Lotan et al., 1998; Stone et al., 2001; Kwong et al., 2003; Cernac and Benning, 2004; Mu et al., 2008). Other TFs, shown to regulate the expression of genes involved in FAS, have been tested, including dehydration-responsive element-binding (DREB) (Zhang et al., 2016), Dof-type transcription factor 1 (Dof1) (Su et al., 2017), FUSCA3 (FUS3) (Elahi et al, 2015), growth-regulating factor 2-like (GRF2) (Liu et al., 2012), GLABRA2 (GL2) (Shi et al., 2012), and MYB interaction factor 1 (MIF1) (Cheng et al., 2021). Among all the TFs, GRF2, LEC1, WRI1 were shown to be the most efficient in increasing seed oil content (40% to 48% higher compared to wild-type) ( Table 3 ). In parallel, specific enzymes in the first committed steps of de novo FAS have also been targeted, such as acetyl-CoA carboxylase (ACCase) (Dong et al., 2002; Cui et al., 2017), and malonyl CoA-ACP malonyltransferase (MCAMT) (Jung et al., 2019) ( Table 4 ). Finally, several studies demonstrated that overexpression of membrane-intrinsic proteins that mediate the export of FAs from plastids, such as FA export 1 (FAX1) (Li et al., 2015b; Tian et al., 2018; Li et al., 2020b; Cai et al., 2021), FAX2 and FAX4 (Li et al., 2015b; Li et al., 2016a; Li et al., 2020b), BILE ACID : SODIUM SYMPORTER FAMILY PROTEIN 2 (BASS2) (Lee et al., 2017), and fatty acyl-ACP thioesterases (FAT) (Bonaventure et al., 2003; Sinha et al., 2007; Parveez et al., 2010; Belide et al., 2012; Nguyen et al., 2015a; Ozseyhan et al., 2018; Ma et al., 2021; Liu et al., 2022) could also boost lipid accumulation in seeds ( Table 4 ). Using the “push” strategy, the highest TG levels recorded in seed so far were achieved by over-expressing ACCase in maize (+65%) (Dong et al., 2002) ( Table 4 ).
Table 4.
List of “push” genes manipulated to enhance oil yield and/or change the FA composition in seeds.
| Gene | Gene source | Target species | Strategy | Effect in seed oil content | Effect in fatty Acid composition | References |
|---|---|---|---|---|---|---|
| α-carboxyltransferase | P. sativum | A. thaliana | Overexpression | ↗ seed oil content by up to 14% | (Wang et al., 2022) | |
| C. sativa | Overexpression | ↗ seed oil content by up to 14% | ↗ C18:1, C18:3; ↘ C16:0, C18:2 | (Wang et al., 2022) | ||
| ACCase | G. hirsutum | G. hirsutum | Overexpression | ↗ seed oil content by 17-22% | (Cui et al., 2017) | |
| S. italica | Z. mays | Overexpression | ↗ seed oil content by 54-65% | (Dong et al., 2002) | ||
| AAD | B. napus | B. napus | Down-regulation | ↘ seed oil content by up to 30% | ↘ C18:1, C20:1 | (Bryant et al., 2016) |
|
AAD +
FAE1_hairpin + Fat5 + FatB_hairpin + KASII_hairpin |
synthetic A. thaliana C. elegans C. sativa C. sativa |
C. sativa | Overexpression | no significant difference | ↗ Total omega-7 fatty acids | (Nguyen et al., 2015) |
| G. max | Overexpression | no significant difference | ↗ Total omega-7 fatty acids | (Nguyen et al., 2015) | ||
| AAD2 or AAD3 | A. thaliana | A. thaliana | Overexpression | ↗ C16:1 (omega-7), C18:1 (omega-7), C20:1 (omega-7) | (Ettaki et al., 2018) | |
| Down-regulation | ↘ C16:1 (omega-7), C18:1 (omega-7), C20:1 (omega-7) | (Bryant et al., 2016) | ||||
| BASS2 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by 10-37% | no significant difference | (Lee et al., 2017) |
| B. napus | B. napus | Overexpression | ↗ seed oil content by 12% | ↗ C18:0, C18:1; ↘ C16:0, C18:2 | (Tang et al., 2022a) | |
| Down-regulation | ↘ seed oil content by up to 11% | ↗ C16:0, C18:0, C18:2; ↘ C18:1, C20:1 | (Tang et al., 2022a) | |||
| FATA | J. curcas | A. thaliana | Overexpression | ↗ seed oil content by up to 9% | ↗ C16:0, C18:0, C18:1, C18:2, C18:3, C20:0, C22:0, C22:1 | (Liu et al., 2022) |
| FATB | A. thaliana | A. thaliana | Down-regulation | ↗ C16:1, C18:2; ↘C16:0, C18:0, C18:1, C18:3 | (Bonaventure et al., 2003) | |
| C. sativa | C. sativa | Down-regulation | ↗ C18:1, C18:3, C20:1, C22:1; ↘C16:0, C18:0, C20:0, C22:0 | (Ozseyhan et al., 2018) | ||
| E. guineensis | A. thaliana | Overexpression | ↗ C16:0, C18:2, C18:3, C20:0; ↘ C18:0, C18:1 | (Parveez et al., 2010) | ||
| Down-regulation | ↗ C18:2, C20:0; ↘ C18:0, C18:1, C18:3 | (Parveez et al., 2010) | ||||
| A. thaliana | A. thaliana | Down-regulation | ↗ C18:1, C18:2, C18:3; ↘ C16:0, C16:1, C18:0, C18:1, C20:0, C20:2, C22:0 | (Belide et al., 2012) | ||
| G. max | G. max | Down-regulation | ↘ seed oil content by up to 10% | ↗ C18:1; ↘ C16:0, C180, C18:2, C18:3 | (Ma et al., 2021) | |
| J. curcas | A. thaliana | Overexpression | no significant difference | ↗ C16:0, C18:0, C18:1, C18:2, C18:3, C20:0, C22:0; ↘ C22:1 | (Liu et al., 2022) | |
| D. butyracea | B. juncea | Overexpression | ↗ seed oil content by up to 5% | ↗ C16:0, C18:1, C18:2; ↘ C22:1 | (Sinha et al., 2007) | |
| FATB1 + LPAT2 | C. viscosissima + C. viscosissima | T. arvense | Overexpression | ↗C8:0, C10:0, C12:0, C14:0, C16:0, C18:0; ↘ C18:1, C18:2, C18:3, C20:0, C20:1, C22:1, C22:2, C24:1 | (Esfahanian et al., 2021) | |
| FATB1 + LPAT2 + DGAT1 | C. viscosissima + C. viscosissima + C. avigera | T. arvense | Overexpression | ↗ C8:0, C10:0, C12:0, C14:0, C16:0, C18:0; ↘ C18:1, C18:2, C18:3, C20:0, C20:1, C22:1, C22:2, C24:1 | (Esfahanian et al., 2021) | |
| FATB + LPAT | U. californica + C. nucifera | T. arvense | Overexpression | ↗ C8:0, C10:0, C12:0, C14:0, C16:0, C18:0; ↘ C18:1, C18:2, C18:3, C20:0, C20:1, C22:1, C22:2, C24:1 | (Esfahanian et al., 2021) | |
| FATB2 + FatB2 | C. avigera + C. hookeriana | T. arvense | Overexpression | ↗ C8:0, C10:0, C12:0, C14:0, C16:0, C18:0; ↘ C18:1, C18:2, C18:3, C20:0, C20:1, C22:1, C22:2, C24:1 | (Esfahanian et al., 2021) | |
| FAX1 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by up to 34% | (Tian et al., 2018) | |
| A. thaliana | C. sativa | Overexpression | ↗ seed oil content by 4% | ↗ C18:0, C18:1, C18:2; ↘ C18:3, C20:1 | (Cai et al., 2021) | |
| B. napus | A. thaliana | Overexpression | ↗ seed oil content by 17% | ↗ C18:1; ↘ C16:0, C20:0, C20:1 | (Xiao et al., 2021) | |
| FAX2 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by 30% | ↗ C18:2, C18:3 | (Li et al., 2020) |
| Overexpression | ↗ seed oil content by up to 21% | (Tian et al., 2019) | ||||
| Down-regulation | ↘ seed oil content by up to 18% | (Tian et al., 2019) | ||||
| Down-regulation | ↘ C18:2 | (Li et al., 2020) | ||||
| FAX4 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by 30% | ↗ C18:2, C18:3 | (Li et al., 2020) |
| Down-regulation | ↘ C20:1 | (Li et al., 2020) | ||||
| FAX2 + FAX4 | A. thaliana | A. thaliana | Down-regulation | ↘ seed oil content by up to 28% | ↘ C18:3, C20:0 | (Li et al., 2020) |
| GPDH | G. max | G. max | Overexpression | ↗ seed oil content by 24% | ↗ C18:1, C18:2, C18:3 | (Zhao et al., 2021) |
| LACS2 | B. napus | B. napus | Overexpression | ↗ seed oil content by 8% | ↗ C18:2, C20:0, C20:1, C22:0;↘ C18:3, C20:1 | (Ding et al., 2020) |
| Down-regulation | ↘ seed oil content by 20% | no significant difference | (Ding et al., 2020) | |||
| LACS4 + LACS9 | A. thaliana | A. thaliana | Down-regulation | ↘ seed oil content by 5% | ↗ C18:3; ↘ C18:2 | (Jessen et al., 2015) |
| MCAMT | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by 15-20% | ↗ C18:3, C20:1; ↘ C18:2, C22:1 | (Jung et al., 2019) |
| NTT1 | B. napus | B. napus | Overexpression | ↗ seed oil content by 5.5% | (Hong et al., 2022b | |
| NTT2 | B. napus | B. napus | Overexpression | ↗ seed oil content by 8% | ↗ C18:1, C18:3; ↘ C18:2 | (Xia et al., 2022) |
| Down-regulation | no significant difference | ↗ C18:2; ↘ C18:1, C18:3 | (Xia et al., 2022) | |||
| PPT1 | B. napus | B. napus | Overexpression | ↗ seed oil content by 3.3% | no significant difference | (Tang et al., 2022b) |
| Down-regulation | ↘ seed oil content by 9% | no significant difference | (Tang et al., 2022b) | |||
| SAD | X. sorbifolia | A. thaliana | Overexpression | ↘ seed oil content by 8% | ↗ C18:0, C20:0; ↘ C18:1, C18:2, C18:3, C20:1 | (Zhao et al., 2015) |
| X. sorbifolia | Down-regulation | ↗ C16:0, C18:0; ↘ C18:1, C18:2, C18:3 | (Zhao et al., 2015) | |||
| P. ostii | A. thaliana | Overexpression | no significant difference | ↗ C18:1, C18:3, C20:0, C20:1; ↘ C18:0 | (Li et al., 2020a) | |
| Z. mays | Z. mays | Overexpression | ↗ seed oil content by up to 5% | ↘ C18:0 | (Du et al., 2016) | |
| Down-regulation | ↗ seed oil content by up to 10% | ↗ C18:0; ↘ C18:1 | (Du et al., 2016) | |||
| A. thaliana | Overexpression | ↘ seed oil content by up to 5% | ↘ C18:0 | (Du et al., 2016) | ||
| Down-regulation | ↗ seed oil content by up to 23% | ↗ C18:0; ↘ C18:1 | (Du et al., 2016) | |||
| R. communis | H. annuus | Overexpression | ↘ C18:0 | (Rousselin et al., 2002) | ||
| SAD + KASII_hairpin | C. sativa | C. sativa | Overexpression | ↗ C16:1 (omega-7), C18:1 (omega-7), C20:1 (omega-7); ↘ C18:0, C18:1, C18:2, C20:0, C20:1, C20:2, C22:0, C22:1 | (Rodríguez-Rodríguez et al., 2021) |
Pull strategy
The pull strategy aims to ensure efficient assembly of FAs generated in the plastid into TG at the ER ( Figure 2 ). Most of the metabolic engineering studies that attempted to optimize this step in seeds have focused on the over-expression of diacylglycerol acyltransferase (DGAT) (He et al., 2004; Kroon et al., 2006; Lung and Weselake, 2006; Shockey et al., 2006; Yu et al., 2006; Burgal et al., 2008; Misra et al., 2013; Wang et al., 2014; Liu et al., 2015; Savadi et al., 2015; Zhou et al., 2020; Chen et al., 2022) and phospholipid: diacylglycerol acyltransferase 1 (PDAT1) (Zhang et al., 2010; van Erp et al., 2011; Guan et al., 2015; Zhou et al., 2020) which commit acyl chains to storage TGs. Manipulation of other genes encoding for “pull” proteins, such as glycerol-3-phosphate dehydrogenases (GPDH), glycerol-3-phosphate acyltransferase (GPAT), and lysophosphatidic acid acyltransferase (LPAT), have also been reported to increase TG levels in seeds (Liu et al., 2015) ( Table 5 ). Using the “pull” strategy, the combined over-expression of DGAT, GPAT, and LPAT was shown to be the most efficient to increase total seed oil content (+25%) (Shockey et al., 2019) ( Table 5 ).
Table 5.
List of “pull” genes manipulated to enhance oil yield and/or change FA composition in seeds.
| Gene | Gene source | Target species | Strategy | Effect in total seed oil content | Effect in fatty Acid composition | References |
|---|---|---|---|---|---|---|
| ABCA9 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by 24% | no significant difference | (Kim et al., 2013) |
| Down-regulation | ↘ seed oil content by 16% | ↗ C18:2↘ C18:3 | (Kim et al., 2013) | |||
| C. sativa | Overexpression | ↗ seed oil content by 22% | (Cai et al., 2021) | |||
| ACP-desaturase | D. unguis-cati | A. thaliana | Overexpression | ↗ C16:0, C18:2, C18:3, C20:1, C22:1; ↘ C16:1, C18:0, C18:1, C20:0 | (Bondaruk et al., 2007) | |
| B. napus | Overexpression | ↗ C18:1; ↘ C16:0, C16:1, C18:0, C18:3 | (Bondaruk et al., 2007) | |||
| AAD | B. napus | B. napus | Down-regulation | ↘ seed oil content by up to 30% | ↘ C18:1, C20:1 | (Bryant et al., 2016a) |
| ACBP | O. sativa | O. sativa | Overexpression | ↗ seed oil content by 10% | ↗ C14:0, C16:0, C18:0, C18:1, C18:2, C18:3, C20:0, C22:0; ↘ C18:1 | (Guo et al., 2019) |
| Down-regulation | ↘ seed oil content by 20% | ↗ C16:1, C18:1, C20:1; ↘ C14:0 | (Guo et al., 2019) | |||
| DGAT | R. communis | FAH transgenic A. thaliana | Overexpression | ↗ C18:1-OH, C18:2-OH | (Burgal et al., 2008) | |
| DGAT + WRI1 + down-regulated SDP1 | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by 16% | no significant difference | (van Erp et al., 2014) |
| DGAT + WRI1 | A. thaliana | G. max | Over-expression | no significant difference | ↗ C16:0, C18:0, C18:1, C18:3 | (Arias et al., 2022) |
| FAD | C. abyssinica | A. thaliana | Down-regulation | ↗ C18:1; ↘ C18:2, C20:1 | (Li et al., 2016b) | |
| FAD+FAE | C. abyssinica | A. thaliana | Down-regulation | ↗ C18:1, C18:2; ↘ C18:3, C20:1 | (Li et al., 2016b) | |
| C. abyssinica | Down-regulation | ↗ C16:0, C18:0, C18:1, C20:1; ↘ C18:2, C18:3, C20:2, C22:0, C22:1, C24:0, C24:1 | (Li et al., 2016b) | |||
| FAD2 | A. hypogaea | A. hypogaea | Down-regulation | ↗ C18:1; ↘ C18:2 | (Yin et al., 2007) | |
| G. max | G. max | Down-regulation | ↗ C18:1; ↘ C16:0, C18:2, C18:3 | (Haun et al., 2014) | ||
| B. napus | B. napus | Down-regulation | ↘ seed oil content by up to 40% | (Chapman et al., 2008) | ||
| T. arvense | T. arvense | Down-regulation | ↗ C18:1, C20:1, C22:1; ↘ C16:0, C16:1, C18:2, C18:3 | (Jarvis et al., 2021) | ||
| FAD3 | P. suffruticosa | A. thaliana | Overexpression | ↗ C16:0, C18:0, C18:3; ↘ C18:1, C18:2 | (Yin et al., 2018) | |
| A. thaliana | A. thaliana | Down-regulation | ↗ C18:2; ↘ C16:0, C18:1, C18:3 | (Yin et al., 2018) | ||
| G. max | O. sativa | Overexpression | ↗ C18:3 | (Anai et al., 2003) | ||
| FAD6 | P. irregulare | B. juncea | Overexpression | ↗ C18:3 (gamma); ↘ C18:0; C18:1, C18:2, C18:3 | (Hong et al., 2002) | |
| FAD6 + FAD15 | B. officinalis/A. thaliana | G. max | Overexpression | ↗ C18:3 (alpha and gamma), C18:4 | (Eckert et al., 2006) | |
| FADX | P. granatum | A. thaliana | Overexpression | ↗ C18:1, punic acid; ↘ C16:0, C18:0, C18:2, C18:3 | (Mietkiewska et al., 2014) | |
| FADX +FAD2 | P. granatum | A. thaliana | Overexpression | ↗ punic acid; ↘ C16:0, C18:0, C18:2, C18:3 | (Mietkiewska et al., 2014) | |
| FAE | C. abyssinica | A. thaliana | Down-regulation | ↗ C18:1; ↘ C18:2, C20:1 | (Li et al., 2016b) | |
| B. juncea | B. juncea | Down-regulation | ↗ seed oil content by up to 11% | ↗ C16:0, C18:0, C18:1, C18:2; ↘ C18:3, C22:1 | (Sinha et al., 2007) | |
| T. majus | A. thaliana | Overexpression | ↗ C22:0, C22:1, C24:0; ↘ C18:0, C20:0, C20:1 | (Mietkiewska et al., 2004) | ||
| FAE1 | E. hyemalis | B. carinata | Overexpression | ↗ C16:0, C18:0, C20:0, C20:1, C20:2, C20:3, C24:1; ↘ C18:1, C18:2, C18:3 | (Meesapyodsuk et al., 2021) | |
| A. thaliana | B. napus | Overexpression | ↗ seed oil content by up to 11% | ↗ C22:1 | (Katavic et al., 2000) | |
| C. abyssinica | A. thaliana | Overexpression | ↗ C22:0, C22:1, C24:0, C24:1; ↘ C18:0, C18:1, C20:0, C20:1 | (Mietkiewska et al., 2007) | ||
| B. carinata | Overexpression | ↗ C18:1, C22:0, C22:1, C24:0; ↘ C16:0, C18:0, C18:2, C18:3, C20:0, C20:1 | (Mietkiewska et al., 2007) | |||
| T. arvense | T. arvense | Down-regulation | ↗ C16:0, C16:1, C18:0, C18:1, C18:2, C18:3; ↘ C20:1, C22:1, C24:1 | (Jarvis et al., 2021) | ||
| FAE1 + FAD2 | T. arvense | T. arvense | Down-regulation | ↗ C18:0, C18:1; ↘ C16:0, C16:1, C18:2, C18:3, C20:1, C22:1, C24:1 | (Jarvis et al., 2021) | |
| FAE1 + ROD1 | T. arvense | T. arvense | Down-regulation | ↗ C16:0, C18:0, C18:1, C18:3; ↘ C16:1, C18:2, C20:1, C22:1, C24:1 | (Jarvis et al., 2021) | |
| FAH | R. communis | A. thaliana | Overexpression | ↘ seed oil content by up to 50% | ↗ HFA | (Bates et al., 2014) |
| A. thaliana | Overexpression | ↗ HFA | (Smith et al., 2003) | |||
| FAH + (DGAT or PDAT) | R. communis | A. thaliana | Overexpression | ↘ seed oil content by up to 15% | ↗ HFA | (Bates et al., 2014) |
| FAH + FAE | P. fendleri/R. communis | C. sativa | Overexpression | ↗ C18:1-OH, C18:2-OH, C20:1-OH, C20:2-OH | (Snapp et al., 2014) | |
| KCS | C. graeca | A. thaliana | Overexpression | ↗ C22:1, C24:1, C26:1; ↘ C20:1 | (Taylor et al., 2009) | |
| B. carinata | Overexpression | ↗ C22:1, C24:1; ↘ C18:1, C18:2, C18:3 | (Taylor et al., 2009) | |||
| KCS+KCR+HCD+ECR | L. annua, A. thaliana | C. sativa | Overexpression | ↗ C16:0, C18:3, C22:0, C22:1, C24:0, C24:1; ↘ C18:0, C18:1, C18:2, C20:0, C20:1 | (Huai et al., 2015) | |
| LPAAT2 + FAE1 | E. hyemalis | B. carinata | Overexpression | ↗ seed oil content by up to 2% | ↗ C16:0, C18:0, C20:2, C22:1, C22:2, C22:3, C24:1, C24:2, C24:3; ↘ C18:1, C20:1, C20:2 | (Meesapyodsuk et al., 2021) |
| LPAT+GPAT+DGAT | R. communis | FAH transgenic A. thaliana | Overexpression | ↗ seed oil content by up to 25% | ↗ HFA | (Shockey et al., 2019) |
| LPCAT | C. abyssinica | C. abyssinica | Down-regulation | ↗ C18:1, C20:1; ↘ C22:1 | (Guan et al., 2015) | |
| PDAT | C. abyssinica | C. abyssinica | Down-regulation | ↗ C18:1; ↘ C18:2 | (Guan et al., 2015) | |
| R. communis | FAH transgenic A. thaliana | Overexpression | ↗ C18:1-OH, C18:2-OH | (van Erp et al., 2011) | ||
| PDCT | C. abyssinica | C. abyssinica | Down-regulation | ↗ C18:1; ↘ C18:2 | (Guan et al., 2015) | |
| PDCT+LPCAT | C. abyssinica | C. abyssinica | Down-regulation | ↗ C18:1, C20:1; ↘ C18:2, C22:1 | (Guan et al., 2015) | |
| ROD1 | T. arvense | T. arvense | Down-regulation | ↗ C16:0, C16:1, C18:1, C18:3, C20:1; ↘ C18:0, C24:1 | (Jarvis et al., 2021) | |
| Δ6 and Δ5 desaturase + FAE | M. alpina (fungus) | G. max | Overexpression | ↗ C16:0, C18:0, C18:3 (gamma), C20:2, C20:3, C20:4; ↘ C18:1, C18:2, C18:3 (alpha) | (Chen et al., 2006) | |
| Δ6 desaturase | G. max | Down-regulation | (Chen et al., 2006) | |||
| Δ9 elongase | I. galbana | A. thaliana | Overexpression | ↗ C20:2, C20:3, C20:4, C20:5; ↘ C18:0, C18:1, C18:2, C18:3 | (Petrie et al., 2012) | |
| Δ8 and Δ5 desaturases | P. salina | |||||
| Δ9 elongase | I. galbana | B. napus | Overexpression | ↗ C16:0, C20:2, C20:3, C20:4, C20:5; ↘ C18:0, C18:1, C18:2, C18:3, C20:0 | (Petrie et al., 2012) | |
| Δ8 and Δ5 desaturases | P. salina | |||||
| Δ12-oleate hydroxylase | P. fendleri | A. thaliana | Overexpression | ↗ C18:1-OH, C18:2-OH, C20:1-OH, C20:2-OH | (Smith et al., 2000) |
Package strategy
As TGs accumulate within the ER phospholipid bilayer, the outer layer starts to expand into the cytosol, forming nascent LDs that bud off to form LDs ( Figure 2 ). The “package” strategy aims to facilitate LD biogenesis and maximize droplet stability by proper coating. Proteomic analyses of seed LDs indicated that they contain 20–30 coat proteins which were found to be involved in LD formation and/or stability (Jolivet et al., 2004; Jolivet et al., 2009). These coat proteins might also help blocking the accessibility of the TG within the LD core to lipases allowing more LD formation. Recent evidence shows that seed-specific overexpression or down-regulation of some genes encoding these proteins increased LD size and/or number. Such proteins include lipid droplet-associated protein (LDAP) (Gidda et al., 2016), LDAP-interacting protein (LDIP) (Pyc et al., 2017), oleosin (Lu et al., 2006, Lu et al., 2018; Siloto et al., 2006; Shimada et al., 2008; Hu et al., 2009; Liu et al., 2013; Zhang et al., 2019), and seipins (Lunn et al., 2018) ( Table 6 ). Interestingly, several studies also used mammalian “package” genes, such as fat-specific protein 27 (FSP27) and fat storage-inducing transmembrane protein 2 (FIT2) from mouse in Arabidopsis. These resulted not only in improvement of seed oil content but also stimulation of LD clustering and fusion (Cai et al., 2017; Price et al., 2020) ( Table 6 ). Among all the mentioned “package” genes, over-expression of seipin in Arabidopsis gave the highest increase in seed oil content (+62%), which enhanced hydroxy-fatty acid (HFA) levels too (Lunn et al., 2018) ( Table 6 ).
Table 6.
List of “package” genes manipulated to enhance oil yield and/or change the FA composition in seeds.
| Gene | Gene source | Target species | Strategy | Effect in total seed oil content | Effect in fatty Acid composition | References |
|---|---|---|---|---|---|---|
| FIT2 | Mouse | A. thaliana | Overexpression | ↗ seed oil content by up to 13% | ↗ C16:0, C18:1; ↘ C20:2 | (Cai et al., 2017) |
| FSP27 | Mouse | A. thaliana | Overexpression | ↗ seed oil content by up to 9% | (Price et al., 2020) | |
| LDAP | A. thaliana | A. thaliana | Overexpression | no significant difference | ↗ C16:0, C18:3; ↘ C18:1, C18:2, C20:1 | (Gidda et al., 2016) |
| Down-regulation | no significant difference | no significant difference | (Gidda et al., 2016) | |||
| LDIP | A. thaliana | A. thaliana | Down-regulation | ↗ seed oil content by up to 22% | ↗ C18:1, C18:2, C18:3, C20:1 | (Pyc et al., 2017) |
| Oleosin | C. tinctorius | A. thaliana | Overexpression | ↗ seed oil content by up to 30% | (Lu et al., 2018) | |
| G. max | O. sativa | Overexpression | ↗ seed oil content by up to 46% | no significant difference | (Liu et al., 2013) | |
| G. max | Overexpression | ↗ seed oil content by up to 10.6% | ↗ C18:2 | (Zhang et al., 2019) | ||
| Seipin | A. thaliana | A. thaliana | Overexpression | ↗ seed oil content by up to 62% | ↗ HFA | (Lunn et al., 2018) |
| Overexpression | ↗ seed oil content by up to 10% | (Cai et al., 2015) |
Protect strategy
Coat proteins can be hydrolyzed by endogenous proteases and TGs can be degraded into free FAs by various lipases to produce acetyl‐CoA via β‐oxidation particularly during seed germination and seedling growth (Pracharoenwattana and Smith, 2008; Li-Beisson et al., 2013; Borek et al., 2015) ( Figure 2 ). Blocking the breakdowns of coat proteins and TGs (also known as “protect” strategy) is therefore an attractive strategy to increase oil content in seeds. The initial step in lipase-induced TG mobilization is the ubiquitination of the respective coat proteins, particularly oleosin and caleosin, and then subsequent digestion by the proteasome (Sorokin et al., 2009; Hsiao and Tzen, 2011; Deruyffelaere et al., 2015), which was extensively reviewed (Sorokin et al., 2009; Shao et al., 2019; Guzha et al., 2023). Important insights into the mechanism regulating the turnover of oleosins in plants has been recently published (Deruyffelaere et al., 2018; Kretzschmar et al., 2018). Two key components, PUX10 (a member of the plant ubiquitin regulatory X (UBX)-domain containing protein family) and CDC48A (the AAA ATPase, Cell Division Cycle 48) were identified. PUX10 localizes to LDs, binds to the ubiquitinated oleosins, and interacts with ubiquitin and CDC48A, respectively. As an adaptor, PUX10 recruits CDC48A to ubiquitinated oleosins, leading to dislocation of oleosins from LDs via the segregase activity of CDC48A (Deruyffelaere et al., 2018; Kretzschmar et al., 2018). In Arabidopsis pux10 mutant seeds, PUX10 deficiency impaired the degradation of ubiquitinated oleosins from LDs. However, this did not change the total FA content in seeds of mutant genotypes in comparison to wild-type (Deruyffelaere et al., 2018; Kretzschmar et al., 2018) ( Table 7 ). On the other hand, suppression of TG degradation to enhance FA content was achieved by down-regulating the genes that code for TG breakdown enzymes. Such genes are seed fatty acid reducer (SFAR) (Karunarathna et al., 2020), Gly-Asp-Ser-Leu (GDSL)-motif lipases (Ding et al., 2019), plastid lipase1 (PLIP1), and Sugar-dependent1 (SDP1) (Kelly et al., 2013; Kim et al., 2014; Kanai et al., 2019) ( Table 7 ). Interestingly, recent studies reported that the over-expression of several genes with major role in TG degradation have a positive effect on seed oil content, which may seem counterintuitive. Such genes include patatin‐related phospholipase (pPLAIIIδ) (Li et al., 2013; Li et al., 2015a), and nonspecific phospholipase C6 (NPC6) (Cai et al., 2020) ( Table 7 ). An intriguing question arising from these studies is how the over-expressed proteins increased lipid accumulation in seeds. One hypothesis is that phospholipase-mediated phospholipid turnover facilitates the movement of FAs from the plastid to the ER. Phosphatidylcholine (PC) is the most abundant class of phospholipids in plants and plays pivotal roles in TG production (Wang, 2005; Chapman and Ohlrogge, 2012; Karki et al., 2019). When hydrolyzed, PC can serve as a substrate for FA desaturation and also provides free FAs or DAG for TG synthesis (Mhaske et al., 2005; Lu et al., 2009; Zhang et al., 2010; Pokotylo et al., 2013). Therefore, over-expression of these phospholipases could enhance the acyl flux into ER and increase the overall levels of TGs in seeds. Among the genes tested as part of the “protect” strategy, it was shown that the down-regulation of PLIP1 in Arabidopsis gave the highest increase in seed oil content (+45%) (Wang et al., 2017) ( Table 7 ).
Modifying FA composition
Metabolic engineering strategies have been used to produce novel high-value FAs or to improve their synthesis in seeds of established crops and other model species ( Tables 3 - 7 ). For instance, over-expression and down-regulation of desaturases, such as acyl-acyl carrier protein desaturase (AAD), fatty acid desaturases (FADs), and stearoyl-acyl carrier protein desaturase (SAD), and FAT, and/or down-regulation of fatty acid elongase (FAE) genes were attempted to divert the carbon flux towards the synthesis of beneficial FAs required for human health and nutrition, such as gamma-linolenic acid (C18:3), alpha-linolenic acid (C18:3), docosadienoic acid (C22:2), docosatrienoic acid (C22:3) (Hong et al., 2002; Anai et al., 2003; Eckert et al., 2006; Huai et al., 2015; Yin et al., 2018; Meesapyodsuk et al., 2021), and oils enriched in omega-7 monounsaturated FAs such as palmitoleic acid (C16:1) and its elongation products vaccenic acid (C18:1) and paullinic acid (C20:1) (Nguyen et al., 2015; Ettaki et al., 2018; Rodríguez-Rodríguez et al., 2021) ( Tables 4 - 5 ). Also, several oilseed plants produce industrially-relevant FAs, such as erucic acid (C22:1), lesquerolic acid (C20:1-OH), and nervonic acid (C24:1), providing renewable alternatives to petrochemicals for the manufacture of lubricants, coatings, or polymers. However, most plants producing these FAs usually have undesirable traits and are not economically viable crops. It is hence important to improve the production of these valuable FAs in other established crops via metabolic engineering technology. One example is the improvement of the production of erucic and nervonic acids. VLCFAs, such as erucic acid (C22:1) and nervonic acid (C24:1), are important in plastic, cosmetic, nylon, and lubricant industries (Mastebroek and Marvin, 2000). Nervonic acid has only been found in the seed oils of a few known plants, and among these, only Lunaria annua has been considered as a niche crop for future development. However, this plant is a biennial with highly variable seed yields (800–2,000 kg/ha) and has a major problem of seed shattering, making it an uneconomical source of nervonic acid (Mastebroek and Marvin, 2000). Consequently, there is a high demanded to improve the production of this valuable FA in other oilseed crops via metabolic engineering technology (Taylor et al., 2009). Over-expression of 3-ketoacyl-CoA synthase (KCS) and FAE genes were shown to increase the production of erucic and nervonic acids in Brassica napus, Camelina sativa, and Arabidopsis thaliana (Katavic et al., 2000; Mietkiewska et al., 2004; Taylor et al., 2009; Huai et al., 2015) ( Table 5 ). Another example is the production of HFAs in a toxin free oilseed crop to replace castor oil as a renewable source for numerous industrial applications. This was achieved through over-expression of fatty acid hydroxylase and elongase genes, and also acyltransferase genes from species producing HFAs, such as castor bean (Ricinus communis) and lesquerella (Physaria fendleri), in the seeds of the model species Arabidopsis and in the industrial oilseed crop Camelina (Smith et al., 2003; van Erp et al., 2011; Bates et al., 2014; Snapp et al., 2014; Shockey et al., 2019) ( Table 5 ).
Combining multiple strategies
To further increase the TG levels in seed oil, several studies have combined genes from different strategies. It is important to note that this combinatorial strategy has been mostly used to increase TG content in non-seed organs, such as leaves and roots (Slocombe et al., 2009; Fan et al., 2013; Kelly et al., 2013; Vanhercke et al., 2013; Winichayakul et al., 2013), and were extensively reviewed (Cahoon et al., 2007; Taylor et al., 2011; Xu et al., 2018; Vanhercke et al., 2019; Singh et al., 2021). However, it was shown that seed-specific overexpression of AtWRI1 (“push”) and AtDGAT1 (“pull”) combined with suppression of the triacylglycerol lipase SUGAR- DEPENDENT1 (AtSDP1) (“protect”) resulted in a higher seed oil content than manipulation of each gene individually (van Erp et al., 2014a) ( Table 5 ). Similarly, seed specific over-expression of Seipin (“package”) in transgenic Arabidopsis expressing R. communis fatty acid hydroxylase (FAH) (“pull”) not only increased the total oil content but also increased the HFA composition in seeds (Lunn et al., 2018) ( Table 6 ). However, this combinatorial strategy is not guaranteed to succeed: several attempts did not increase TG in seeds, such as the over-expression of WRI1 (“push”) and DGAT1(“pull”) from Arabidopsis in soybean, suggesting that some species put in place counteracting processes to keep oil content stable (Arias et al., 2022) ( Table 5 ). On the other hand, combining multiple strategies has also been successful to modify FA composition in seeds ( Tables 4 - 5 ). For instance, co-expression of Cuphea viscosissima FATB (“Push”) and LPAT (“pull”) with Cuphea avigera DGAT (“pull”) increased the accumulation of TGs rich in medium-chain FAs (C6-C14) in pennycress for industrial, jet fuel and improved biodiesel applications (Esfahanian et al., 2021) ( Table 4 ). Another example is the co-expression of Δ6 and Δ5 desaturases (“pull”) and FAE (“pull”) from a fungal species (Mortierella alpinia) in soybean, which caused the seed to accumulate novel FAs that are not naturally produced in soybean: γ-linolenic acid (GLA), eicosa-8, 11-dienoic acid (EDA), dihomo-γ-linolenic acid (DGLA), and arachidonic acid (C20:4), which are important for human health (Chen et al., 2006) ( Table 5 ).
Perspectives and challenges
The demands in seed oils for food and feed is rapidly increasing with growing population, urbanization, and industrialization. Enhancing oil production and improving FA composition in oilseed crops through metabolic engineering is a promising venue to meet these demands; the challenge is in identifying the right target(s). 13C-MFA has played a pivotal role in advancing our understanding of plant FAS at the systems level. The flux maps of primary metabolism that have emerged from MFA provide information on regulatory steps and pathways to be assessed within the context of the whole network, leading to the identification of candidate genes to be engineered for oil improvement. Current advances in mass spectrometry imaging techniques coupled with 13C-isotopic pulse labeling would also allow to assess spatial and temporal resolution of metabolic fluxes (Romsdahl et al., 2021). Recent discoveries of a number of genes involved in the “push, pull, package, and protect” steps of oil synthesis have enabled successful engineering of oil content and composition in different crops. Effective strategies combined the overexpression of TFs that upregulate FAS and genes involved in TG assembly, combined with the downregulation of TG catabolic enzymes. The challenge is that a given strategy may work in a species but not in others: as demonstrated by 13C-MFA studies, developing embryos from different species use different pathways, sometimes even non-conventional reactions, for FAS. Finally, the comprehensive understanding of multi-”omics” technologies and advanced genome-editing capabilities offer the possibility of rapid assembly and introduction of multiple candidate genes for further improvements in seed oil quality. Popular genome-editing tools, such as CRISPR/Cas9, could be used to remove or minimize metabolic competition while directing metabolic flux toward TG biosynthesis, or to edit specific amino acids in oil biosynthesis enzymes to improve/modify enzymatic activities (Park and Kim, 2022).
Author contributions
AA conceived the idea. JS and UY drafted the manuscript and designed the figures. AA reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We acknowledge Drs. Christopher Johnston and Cintia Arias at the UNT BioDiscovery Institute and Department of Biological Sciences for their helpful comments to improve the review. We also acknowledge the BioRender tool that we used to build the Figures 1 and 2 .
Funding Statement
This review was funded by the Agriculture and Food Research Initiative competitive grant # 2021-67013-33777 from the USDA National Institute of Food and Agriculture, the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program grant no. DE-SC0020325, and the United Soybean Board project no. 2332-203-0102 to AA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Acket S., Degournay A., Rossez Y., Mottelet S., Villon P., Troncoso-Ponce A., et al. (2019). 13c-metabolic flux analysis in developing flax (Linum usitatissinum l.) embryos to understand storage lipid biosynthesis. Metabolites 10, 14. doi: 10.3390/metabo10010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D. K., Laclair R. W., Ohlrogge J. B., Shachar-Hill Y. (2012). Isotope labelling of rubisco subunits provides in vivo information on subcellular biosynthesis and exchange of amino acids between compartments. Plant Cell Environ. 35, 1232–1244. doi: 10.1111/j.1365-3040.2012.02485.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen D. K., Ohlrogge J. B., Shachar-Hill Y. (2009). The role of light in soybean seed filling metabolism. Plant J. 58, 220–234. doi: 10.1111/j.1365-313X.2008.03771.x [DOI] [PubMed] [Google Scholar]
- Allen D. K., Shachar-Hill Y., Ohlrogge J. B. (2007). Compartment-specific labeling information in 13C metabolic flux analysis of plants. Phytochemistry 68, 2197–2210. doi: 10.1016/j.phytochem.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Allorent G., Osorio S., Ly Vu J., Falconet D., Jouhet J., Kuntz M., et al. (2015). Adjustments of embryonic photosynthetic activity modulate seed fitness in arabidopsis thaliana. New Phytol. 205, 707–719. doi: 10.1111/nph.13044 [DOI] [PubMed] [Google Scholar]
- Alonso A. P., Dale V. L., Shachar-Hill Y. (2010). Understanding fatty acid synthesis in developing maize embryos using metabolic flux analysis. Metab. Eng. 12, 488–497. doi: 10.1016/j.ymben.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Alonso A. P., Goffman F. D., Ohlrogge J. B., Shachar-Hill Y. (2007). Carbon conversion efficiency and central metabolic fluxes in developing sunflower (Helianthus annuus l.) embryos. Plant J. 52, 296–308. doi: 10.1111/j.1365-313X.2007.03235.x [DOI] [PubMed] [Google Scholar]
- Alonso A. P., Val D. L., Shachar-Hill Y. (2011). Central metabolic fluxes in the endosperm of developing maize seeds and their implications for metabolic engineering. Metab. Eng. 13, 96–107. doi: 10.1016/j.ymben.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Anai T., Koga M., Tanaka H., Kinoshita T., Rahman S. M., Takagi Y. (2003). Improvement of rice (Oryza sativa l.) seed oil quality through introduction of a soybean microsomal omega-3 fatty acid desaturase gene. Plant Cell Rep. 21, 988–992. doi: 10.1007/s00299-003-0609-6 [DOI] [PubMed] [Google Scholar]
- An D., Suh M. C. (2015). Overexpression of arabidopsis WRI1 enhanced seed mass and storage oil content in camelina sativa. Plant Biotechnol. Rep. 9, 137–148. doi: 10.1007/s11816-015-0351-x [DOI] [Google Scholar]
- Antoniewicz M. R. (2015). Parallel labeling experiments for pathway elucidation and 13C metabolic flux analysis. Curr. Opin. Biotechnol. 36, 91–97. doi: 10.1016/j.copbio.2015.08.014 [DOI] [PubMed] [Google Scholar]
- Antoniewicz M. R. (2018). A guide to 13C metabolic flux analysis for the cancer biologist. Exp. Mol. Med. 50, 1–13. doi: 10.1038/s12276-018-0060-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C. L., Quach T., Huynh T., Nguyen H., Moretti A., Shi Y., et al. (2022). Expression of AtWRI1 and AtDGAT1 during soybean embryo development influences oil and carbohydrate metabolism. Plant Biotechnol. J. 20, 1327–1345. doi: 10.1111/pbi.13810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asokanthan P. S., Johnson R. W., Griffith M., Krol M. (1997). The photosynthetic potential of canola embryos. Physiol. Plant 101, 353–360. doi: 10.1111/j.1399-3054.1997.tb01008.x [DOI] [Google Scholar]
- Atadashi I. M., Aroua M. K., Abdul Aziz A. R., Sulaiman N. M. N. (2012). Production of biodiesel using high free fatty acid feedstocks. Renewable Sustain. Energy Rev. 16, 3275–3285. doi: 10.1016/j.rser.2012.02.063 [DOI] [Google Scholar]
- Bates P. D., Johnson S. R., Cao X., Li J., Nam J. W., Jaworski J. G., et al. (2014). Fatty acid synthesis is inhibited by inefficient utilization of unusual fatty acids for glycerolipid assembly. Proc. Natl. Acad. Sci. U.S.A. 111, 1204–1209. doi: 10.1073/pnas.1318511111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S., Mendoza M. S., To A., Harscoët E., Lepiniec L., Dubreucq B. (2007). WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in arabidopsis. Plant J. 50, 825–838. doi: 10.1111/j.1365-313X.2007.03092.x [DOI] [PubMed] [Google Scholar]
- Belide S., Petrie J. R., Shrestha P., Singh S. P. (2012). Modification of seed oil composition in arabidopsis by artificial microRNA-mediated gene silencing. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G., Salas J. J., Pollard M. R., Ohlrogge J. B. (2003). Disruption of the FATB gene in arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 15, 1020–1033. doi: 10.1105/TPC.008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondaruk M., Johnson S., Degafu A., Boora P., Bilodeau P., Morris J., et al. (2007). Expression of a cDNA encoding palmitoyl-acyl carrier protein desaturase from cat’s claw (Doxantha unguis-cati l.) in arabidopsis thaliana and brassica napus leads to accumulation of unusual unsaturated fatty acids and increased stearic acid content in the. Plant Breed. 126, 186–194. doi: 10.1111/j.1439-0523.2007.01316.x [DOI] [Google Scholar]
- Borek S., Ratajczak W., Ratajczak L. (2015). Regulation of storage lipid metabolism in developing and germinating lupin (Lupinus spp.) seeds. Acta Physiol. Plant 37, 1–11. doi: 10.1007/S11738-015-1871-2/FIGURES/4 [DOI] [Google Scholar]
- Borisjuk L., Nguyen T. H., Neuberger T., Rutten T., Tschiersch H., Claus B., et al. (2005). Gradients of lipid storage, photosynthesis and plastid differentiation in developing soybean seeds. New Phytol. 167, 761–776. doi: 10.1111/J.1469-8137.2005.01474.X [DOI] [PubMed] [Google Scholar]
- Bryant F. M., Munoz-Azcarate O., Kelly A. A., Beaudoin F., Kurup S., Eastmond P. J. (2016). ACYL-ACYL CARRIER PROTEIN DESATURASE2 and 3 are responsible for making omega-7 fatty acids in thearabidopsis aleurone. Plant Physiol. 172, 154–162. doi: 10.1104/pp.16.00836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgal J., Shockey J., Lu C., Dyer J., Larson T., Graham I., et al. (2008). Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol. J. 6, 819–831. doi: 10.1111/j.1467-7652.2008.00361.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon E. B., Shockey J. M., Dietrich C. R., Gidda S. K., Mullen R. T., Dyer J. M. (2007). Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr. Opin. Plant Biol. 10, 236–244. doi: 10.1016/j.pbi.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Cai G., Fan C., Liu S., Yang Q., Liu D., Wu J., et al. (2020). Nonspecific phospholipase C6 increases seed oil production in oilseed brassicaceae plants. New Phytol. 226, 1055–1073. doi: 10.1111/nph.16473 [DOI] [PubMed] [Google Scholar]
- Cai Y., Goodman J. M., Pyc M., Mullen R. T., Dyer J. M., Chapmana K. D. (2015). Arabidopsis SEIPIN proteins modulate triacylglycerol accumulation and influence lipid droplet proliferation. Plant Cell 27, 2616–2636. doi: 10.1105/tpc.15.00588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., McClinchie E., Price A., Nguyen T. N., Gidda S. K., Watt S. C., et al. (2017). Mouse fat storage-inducing transmembrane protein 2 (FIT2) promotes lipid droplet accumulation in plants. Plant Biotechnol. J. 15, 824–836. doi: 10.1111/pbi.12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai G., Wang G., Kim S. C., Li J., Zhou Y., Wang X. (2021). Increased expression of fatty acid and ABC transporters enhances seed oil production in camelina. Biotechnol. Biofuels 14, 49. doi: 10.1186/s13068-021-01899-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Huang A. H. C. (1986). Diacylglycerol acyltransferase in maturing oil seeds of maize and other species. Plant Physiol. 82, 813–820. doi: 10.1104/pp.82.3.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.-Z., Huang A. H. C. (1987). Acyl coenzyme a preference of diacylglycerol acyltransferase from the maturing seeds of cuphea , maize, rapeseed, and canola. Plant Physiol. 84, 762–765. doi: 10.1104/pp.84.3.762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey L. M., Clark T. J., Deshpande R. R., Cocuron J. C., Rustad E. K., Shachar-Hill Y. (2020). High flux through the oxidative pentose phosphate pathway lowers efficiency in developing camelina seeds. Plant Physiol. 182, 493–506. doi: 10.1104/pp.19.00740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A., Benning C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in arabidopsis. Plant J. 40, 575–585. doi: 10.1111/J.1365-313X.2004.02235.X [DOI] [PubMed] [Google Scholar]
- Chapman K. D., Neogi P. B., Hake K. D., Stawska A. A., Speed T. R., Cotter M. Q., et al. (2008). Reduced oil accumulation in cottonseeds transformed with a brassica nonfunctional allele of a delta-12 fatty acid desaturase (FAD2). Crop Sci. 48, 1470–1481. doi: 10.2135/cropsci2007.11.0618 [DOI] [Google Scholar]
- Chapman K. D., Ohlrogge J. B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287, 2288–2294. doi: 10.1074/jbc.R111.290072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Zhao P., Ren Y., Zou J., Sun M. X. (2021). AtMIF1 increases seed oil content by attenuating GL2 inhibition. New Phytol. 229, 2152–2162. doi: 10.1111/nph.17016 [DOI] [PubMed] [Google Scholar]
- Chen G., Harwood. J. L., Lemieux M. J., Stone S. J., Weselakea R. J. (2022). Acyl-CoA:diacylglycerol acyltransferase: Properties, physiological roles, metabolic engineering and intentional control. Prog. Lipid Res. 88, 101181. doi: 10.1016/j.plipres.2022.101181 [DOI] [PubMed] [Google Scholar]
- Chen R., Matsui K., Ogawa M., Oe M., Ochiai M., Kawashima H., et al. (2006). Expression of Δ6, Δ5 desaturase and GLELO elongase genes from mortierella alpina for production of arachidonic acid in soybean [Glycine max (L.) Merrill] seeds. Plant Sci. 170, 399–406. doi: 10.1016/j.plantsci.2005.09.006 [DOI] [Google Scholar]
- Chen X., Shachar-Hill Y. (2012). Insights into metabolic efficiency from flux analysis. J. Exp. Bot. 63, 2343–2351. doi: 10.1093/jxb/ers057 [DOI] [PubMed] [Google Scholar]
- Chen L., Zheng Y., Dong Z., Meng F., Sun X., Fan X., et al. (2018). Soybean (Glycine max) WRINKLED1 transcription factor, GmWRI1a, positively regulates seed oil accumulation. Mol. Genet. Genomics 293, 401–415. doi: 10.1007/s00438-017-1393-2 [DOI] [PubMed] [Google Scholar]
- Cocuron J. C., Alonso A. P. (2014). Liquid chromatography tandem mass spectrometry for measuring 13C-labeling in intermediates of the glycolysis and pentose phosphate pathway. Methods Mol. Biol. 1090, 131–142. doi: 10.1007/978-1-62703-688-7_9 [DOI] [PubMed] [Google Scholar]
- Cocuron J. C., Koubaa M., Kimmelfield R., Ross Z., Alonso A. P. (2019). A combined metabolomics and fluxomics analysis identifies steps limiting oil synthesis in maize embryos. Plant Physiol. 181, 961–975. doi: 10.1104/PP.19.00920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J.-C., Ross Z., Alonso A. P. (2020). Liquid chromatography tandem mass spectrometry quantification of 13C-labeling in sugars. Metabolites 10, 30. doi: 10.3390/metabo10010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J. C., Tsogtbaatar E., Alonso A. P. (2017). High-throughput quantification of the levels and labeling abundance of free amino acids by liquid chromatography tandem mass spectrometry. J. Chromatogr A 1490, 148–155. doi: 10.1016/j.chroma.2017.02.028 [DOI] [PubMed] [Google Scholar]
- Crown S. B., Kelleher J. K., Rouf R., Muoio D. M., Antoniewicz M. R. (2016). Comprehensive metabolic modeling of multiple13C-isotopomer data sets to study metabolism in perfused working hearts. Am. J. Physiol. Heart Circ. Physiol. 311, H881–H891. doi: 10.1152/ajpheart.00428.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Liu Z., Zhao Y., Wang Y., Huang Y., Li L., et al. (2017). Overexpression of heteromeric GhACCase subunits enhanced oil accumulation in upland cotton. Plant Mol. Biol. Rep. 35, 287–297. doi: 10.1007/s11105-016-1022-y [DOI] [Google Scholar]
- Deruyffelaere C., Bouchez I., Morin H., Guillot A., Miquel M., Froissard M., et al. (2015). Ubiquitin-mediated proteasomal degradation of oleosins is involved in oil body mobilization during post-germinative seedling growth in arabidopsis. Plant Cell Physiol. 56, 1374–1387. doi: 10.1093/pcp/pcv056 [DOI] [PubMed] [Google Scholar]
- Deruyffelaere C., Purkrtova Z., Bouchez I., Collet B., Cacas J. L., Chardot T., et al. (2018). PUX10 IS a CDC48A adaptor protein that regulates the extraction of ubiquitinated oleosins from seed lipid droplets in arabidopsis. Plant Cell 30, 2116–2136. doi: 10.1105/tpc.18.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieuaide-Noubhani M., Alonso A. P. (2014). Application of metabolic flux analysis to plants. Methods Mol. Biol. 1090, 1–17. doi: 10.1007/978-1-62703-688-7_1 [DOI] [PubMed] [Google Scholar]
- Ding L. N., Guo X. J., Li M., Fu Z. L., Yan S. Z., Zhu K. M., et al. (2019). Improving seed germination and oil contents by regulating the GDSL transcriptional level in brassica napus. Plant Cell Rep. 38, 243–253. doi: 10.1007/s00299-018-2365-7 [DOI] [PubMed] [Google Scholar]
- Ding L. N., Gu S. L., Zhu F. G., Ma Z. Y., Li J., Li M., et al. (2020). Long-chain acyl-CoA synthetase 2 is involved in seed oil production in brassica napus. BMC Plant Biol. 20, 1–14. doi: 10.1186/s12870-020-2240-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Zhao H., He J., Huai J., Lin H., Zheng J., et al. (2002). Overexpression of a foxtail millet acetyl-CoA carboxylase gene in maize increases sethoxydim resistance and oil conten. Afr J. Biotechnol. 10, 3986–3995. doi: 10.5897/AJB11.053 [DOI] [Google Scholar]
- Du H., Huang M., Hu J., Li J. (2016). Modification of the fatty acid composition in arabidopsis and maize seeds using a stearoyl-acyl carrier protein desaturase-1 (ZmSAD1) gene. BMC Plant Biol. 16, 1–10. doi: 10.1186/s12870-016-0827-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett T. P., Benning C., Ohlrogge J. (2008). Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. doi: 10.1111/j.1365-313X.2008.03442.x [DOI] [PubMed] [Google Scholar]
- Eastmond P., Koláčná L., Rawsthorne S. (1996). Photosynthesis by developing embryos of oilseed rape (Brassica napus l.). J. Exp. Bot. 47, 1763–1769. doi: 10.1093/JXB/47.11.1763 [DOI] [Google Scholar]
- Eastmond P. J., Rawsthorne S. (2000). Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol. 112, 767–774. doi: 10.1104/pp.122.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert H., LaVallee B., Schweiger B. J., Kinney A. J., Cahoon E. B., Clemente T. (2006). Co-Expression of the borage Δ6 desaturase and the arabidopsis Δ15 desaturase results in high accumulation of stearidonic acid in the seeds of transgenic soybean. Planta 224, 1050–1057. doi: 10.1007/s00425-006-0291-3 [DOI] [PubMed] [Google Scholar]
- Elahi N., Duncan R. W., Stasolla C. (2015). Decreased seed oil production in FUSCA3 brassica napus mutant plants. Plant Physiol. Biochem. 96, 222–230. doi: 10.1016/j.plaphy.2015.08.002 [DOI] [PubMed] [Google Scholar]
- Elahi N., Duncan R. W., Stasolla C. (2016). Modification of oil and glucosinolate content in canola seeds with altered expression of brassica napus LEAFY COTYLEDON1. Plant Physiol. Biochem. 100, 52–63. doi: 10.1016/j.plaphy.2015.12.022 [DOI] [PubMed] [Google Scholar]
- Esfahanian M., Nazarenus T. J., Freund M. M., McIntosh G., Phippen W. B., Phippen M. E., et al. (2021). Generating pennycress (Thlaspi arvense) seed triacylglycerols and acetyl-triacylglycerols containing medium-chain fatty acids. Front. Energy Res. 9. doi: 10.3389/fenrg.2021.620118 [DOI] [Google Scholar]
- Ettaki H., Troncoso-Ponce M. A., To A., Barthole G., Lepiniec L., Baud S. (2018). Overexpression of MYB115, AAD2, or AAD3 in arabidopsis thaliana seeds yields contrasting omega-7 contents. PloS One 13, 1–19. doi: 10.1371/journal.pone.0192156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Yan C., Zhang X., Xu C. (2013). Dual role for Phospholipid:Diacylglycerol acyltransferase: Enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in arabidopsis leaves. Plant Cell 25, 3506–3518. doi: 10.1105/TPC.113.117358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N., Benning C. (1998). wrinkled1: A novel, low-Seed-Oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118, 91. doi: 10.1104/PP.118.1.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili G., Avin-Wittenberg T., Angelovici R., Fernie A. R. (2014). The role of photosynthesis and amino acid metabolism in the energy status during seed development. Front. Plant Sci. 0. doi: 10.3389/FPLS.2014.00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda S. K., Park S., Pyc M., Yurchenko O., Cai Y., Wu P., et al. (2016). Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 170, 2052–2071. doi: 10.1104/pp.15.01977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman F. D., Alonso A. P., Schwender J., Shachar-Hill Y., Ohlrogge J. B. (2005). Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol. 138, 2269–2279. doi: 10.1104/pp.105.063628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman F. D., Ruckle M., Ohlrogge J., Shachar-Hill Y. (2004). Carbon dioxide concentrations are very high in developing oilseeds. Plant Physiol. Biochem. 42, 703–708. doi: 10.1016/J.PLAPHY.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Guan R., Li X., Hofvander P., Zhou X. R., Wang D., Stymne S., et al. (2015). RNAi targeting putative genes in phosphatidylcholine turnover results in significant change in fatty acid composition in crambe abyssinica seed oil. Lipids 50, 407–416. doi: 10.1007/s11745-015-4004-1 [DOI] [PubMed] [Google Scholar]
- Guo Z. H., Haslam R. P., Michaelson L., Yeung E. C., Lung S. C., Napier J. A., et al. (2019). The overexpression of rice ACYL-CoA-BINDING PROTEIN2 increases grain size and bran oil content in transgenic rice. Plant J. 100, 1132–1147. doi: 10.1111/tpj.14503 [DOI] [PubMed] [Google Scholar]
- Guzha A., Whitehead P., Ischebeck T., Chapman K. D. (2023). Lipid droplets: Packing hydrophobic molecules within the aqueous cytoplasm. 1–29. Annu. Rev. Plant Biol. 74, 1–29. doi: 10.1146/annurev-arplant-070122-021752 [DOI] [PubMed] [Google Scholar]
- Hasan M. M., Rahman M. M. (2017). Performance and emission characteristics of biodiesel–diesel blend and environmental and economic impacts of biodiesel production: A review. Renewable Sustain. Energy Rev. 74, 938–948. doi: 10.1016/j.rser.2017.03.045 [DOI] [Google Scholar]
- Haun W., Coffman A., Clasen B. M., Demorest Z. L., Lowy A., Ray E., et al. (2014). Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 12, 934–940. doi: 10.1111/pbi.12201 [DOI] [PubMed] [Google Scholar]
- Hay J. O., Shi H., Heinzel N., Hebbelmann I., Rolletschek H., Schwender J. (2014). Integration of a constraint-based metabolic model of brassica napus developing seeds with 13C-metabolic flux analysis. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Turner C., Chen G. Q., Lin J. T., McKeon T. A. (2004). Cloning and characterization of a cDNA encoding diacylglycerol acyltransferase from castor bean. Lipids 39, 311–318. doi: 10.1007/S11745-004-1234-2 [DOI] [PubMed] [Google Scholar]
- Hong H., Datla N., Reed D. W., Covello P. S., MacKenzie S. L., Qiu X. (2002). High-level production of γ-linolenic acid in brassica juncea using a Δ6 desaturase from pythium irregulare. Plant Physiol. 129, 354–362. doi: 10.1104/pp.001495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Xia H., Li X., Fan R., Li Q., Ouyang Z., et al. (2022). Brassica napus BnaNTT1 modulates ATP homeostasis in plastids to sustain metabolism and growth. Cell Rep. 40, 111060. doi: 10.1016/j.celrep.2022.111060 [DOI] [PubMed] [Google Scholar]
- Hsiao E. S. L., Tzen J. T. C. (2011). Ubiquitination of oleosin-h and caleosin in sesame oil bodies after seed germination. Plant Physiol. Biochem. 49, 77–81. doi: 10.1016/j.plaphy.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Huai D., Zhang Y., Zhang C., Cahoon E. B., Zhou Y. (2015). Combinatorial effects of fatty acid elongase enzymes on nervonic acid production in camelina sativa. PloS One 10, e0131755. doi: 10.1371/journal.pone.0131755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Wang X., Zhan G., Liu G., Hua W., Wang H. (2009). Unusually large oilbodies are highly correlated with lower oil content in brassica napus. Plant Cell Rep. 28, 541–549. doi: 10.1007/S00299-008-0654-2 [DOI] [PubMed] [Google Scholar]
- Ivarson E., Leiva-Eriksson N., Ahlman A., Kanagarajan S., Bülow L., Zhu L. H. (2017). Effects of overexpression of WRI1 and hemoglobin genes on the seed oil content of lepidium campestre. Front. Plant Sci. 7, 2032. doi: 10.3389/fpls.2016.02032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis B. A., Romsdahl T. B., McGinn M. G., Nazarenus T. J., Cahoon E. B., Chapman K. D., et al. (2021). CRISPR/Cas9-induced fad2 and rod1 mutations stacked with fae1 confer high oleic acid seed oil in pennycress (Thlaspi arvense l.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.652319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J., Cahoon E. B. (2003). Industrial oils from transgenic plants. Curr. Opin. Plant Biol. 6, 178–184. doi: 10.1016/S1369-5266(03)00013-X [DOI] [PubMed] [Google Scholar]
- Jessen D., Roth C., Wiermer M., Fulda M. (2015). Two activities of long-chain acyl-coenzyme a synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in arabidopsis. Plant Physiol. 167, 351–366. doi: 10.1104/pp.114.250365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. L., Luethy M. H., Miernyk J. A., Randall D. D. (1997). Cloning and molecular analyses of the arabidopsis thaliana plastid pyruvate dehydrogenase subunits. Biochim. Biophys. Acta Bioenerg 1321, 200–206. doi: 10.1016/S0005-2728(97)00059-5 [DOI] [PubMed] [Google Scholar]
- Jolivet P., Boulard C., Bellamy A., Larré C., Barre M., Rogniaux H., et al. (2009). Protein composition of oil bodies from mature brassica napus seeds. Proteomics 9, 3268–3284. doi: 10.1002/PMIC.200800449 [DOI] [PubMed] [Google Scholar]
- Jolivet P., Roux E., D’Andrea S., Davanture M., Negroni L., Zivy M., et al. (2004). Protein composition of oil bodies in arabidopsis thaliana ecotype WS. Plant Physiol. Biochem. 42, 501–509. doi: 10.1016/J.PLAPHY.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Jung S. H., Kim R. J., Kim K. J., Lee D. H., Suh M. C. (2019). Plastidial and mitochondrial malonyl CoA-ACP malonyltransferase is essential for cell division and its overexpression increases storage oil content. Plant Cell Physiol. 60, 1239–1249. doi: 10.1093/pcp/pcz032 [DOI] [PubMed] [Google Scholar]
- Kanai M., Yamada T., Hayashi M., Mano S., Nishimura M. (2019). Soybean (Glycine max l.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-45331-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F., Rawsthorne S. (1994). Starch and fatty acid synthesis in plastids from developing embryos of oilseed rape (Brassica napus l.). Plant J. 6, 795–805. doi: 10.1046/j.1365-313X.1994.6060795.x [DOI] [Google Scholar]
- Karki N., Johnson B. S., Bates P. D. (2019). Metabolically distinct pools of phosphatidylcholine are involved in trafficking of fatty acids out of and into the chloroplast for membrane production. Plant Cell 31, 2768–2788. doi: 10.1105/tpc.19.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunarathna N. L., Wang H., Harloff H. J., Jiang L., Jung C. (2020). Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 18, 2251–2266. doi: 10.1111/pbi.13381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katavic V., Friesen W., Barton D. L., Gossen K. K., Giblin E. M., Luciw T., et al. (2000). “Utility of the arabidopsis FAEI and yeast SLCI-I genes for improvements in erucic acid and oil content in rapeseed,” in Biochemical society transactions (Portland Press; ), 935–937. doi: 10.1042/bst0280935 [DOI] [PubMed] [Google Scholar]
- Kelly A. A., Shaw E., Powers S. J., Kurup S., Eastmond P. J. (2013). Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus l.). Plant Biotechnol. J. 11, 355–361. doi: 10.1111/pbi.12021 [DOI] [PubMed] [Google Scholar]
- Khan K., Kumar V., Niranjan A., Shanware A., Sane V. A. (2019). JcMYB1, a jatropha R2R3MYB transcription factor gene, modulates lipid biosynthesis in transgenic plants. Plant Cell Physiol. 60, 462–475. doi: 10.1093/pcp/pcy223 [DOI] [PubMed] [Google Scholar]
- Kim M. J., Jang I. C., Chua N. H. (2016). The mediator complex MED15 subunit mediates activation of downstream lipid-related genes by the WRINKLED1 transcription factor. Plant Physiol. 171, 1951–1964. doi: 10.1104/pp.16.00664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R. J., Kim H. U., Suh M. C. (2019). Development of camelina enhanced with drought stress resistance and seed oil production by co-overexpression of MYB96A and DGAT1C. Ind. Crops Prod 138, 111475. doi: 10.1016/j.indcrop.2019.111475 [DOI] [Google Scholar]
- Kim T. Y., Sohn S. B., Kim Y., Kim W. J., Lee S. Y. (2012). Recent advances in reconstruction and applications of genome-scale metabolic models. Curr. Opin. Biotechnol. 23, 617–623. doi: 10.1016/j.copbio.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Kim S., Yamaoka Y., Ono H., Kim H., Shim D., Maeshima M., et al. (2013). AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 110, 773–778. doi: 10.1073/pnas.1214159110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Yang S. W., Mao H. Z., Veena S. P., Yin J. L., Chua N. H. (2014). Gene silencing of sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in jatropha curcas. Biotechnol. Biofuels 7, 1–16. doi: 10.1186/1754-6834-7-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. P., Badger M. R., Furbank R. T. (1998). CO2 refixation characteristics of developing canola seeds and silique wall. Aust. J. Plant Physiol. 25, 377–386. doi: 10.1071/PP97157 [DOI] [Google Scholar]
- Koubaa M., Cocuron J. C., Thomasset B., Alonso A. P. (2013). Highlighting the tricarboxylic acid cycle: Liquid and gas chromatography-mass spectrometry analyses of 13C-labeled organic acids. Anal. Biochem. 436, 151–159. doi: 10.1016/j.ab.2013.01.027 [DOI] [PubMed] [Google Scholar]
- Kretzschmar F. K., Mengel L. A., Müller A. O., Schmitt K., Blersch K. F., Valerius O., et al. (2018). PUX10 is a lipid droplet-localized scaffold protein that interacts with CELL DIVISION CYCLE48 and is involved in the degradation of lipid droplet proteins. Plant Cell 30, 2137–2160. doi: 10.1105/tpc.18.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon J. T. M., Wei W., Simon W. J., Slabas A. R. (2006). Identification and functional expression of a type 2 acyl-CoA:diacylglycerol acyltransferase (DGAT2) in developing castor bean seeds which has high homology to the major triglyceride biosynthetic enzyme of fungi and animals. Phytochemistry 67, 2541–2549. doi: 10.1016/J.PHYTOCHEM.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Kruger N. J., Masakapalli S. K., Ratcliffe R. G. (2012). Strategies for investigating the plant metabolic network with steady-state metabolic flux analysis: Lessons from an arabidopsis cell culture and other systems. J. Exp. Bot. 63, 2309–2323. doi: 10.1093/jxb/err382 [DOI] [PubMed] [Google Scholar]
- Kubis S. E., Pike M. J., Everett C. J., Hill L. M., Rawsthorne S. (2004). The import of phosphoenolpyruvate by plastids from developing embryos of oilseed rape, brassica napus (L.), and its potential as a substrate for fatty acid synthesis. J. Exp. Bot. 55, 1455–1462. doi: 10.1093/jxb/erh157 [DOI] [PubMed] [Google Scholar]
- Kwong R. W., Bui A. Q., Lee H., Kwong L. W., Fischer R. L., Goldberg R. B., et al. (2003). LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15, 5–18. doi: 10.1105/TPC.006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. G., Kim H., Suh M. C., Kim H. U., Seo P. J. (2018). The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in arabidopsis seeds. Plant Cell Physiol. 59, 1432–1442. doi: 10.1093/pcp/pcy073 [DOI] [PubMed] [Google Scholar]
- Lee E. J., Oh M., Hwang J. U., Li-Beisson Y., Nishida I., Lee Y. (2017). Seed-specific overexpression of the pyruvate transporter BASS2 increases oil content in arabidopsis seeds. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. Y., Park J. M., Kim T. Y. (2011). “Application of metabolic flux analysis in metabolic engineering,” in Methods in enzymology (Academic Press Inc; ), 67–93. doi: 10.1016/B978-0-12-385120-8.00004-8 [DOI] [PubMed] [Google Scholar]
- Li M., Bahn S. C., Fan C., Li J., Phan T., Ortiz M., et al. (2013). Patatin-related phospholipase pPLAIIIδ increases seed oil content with long-chain fatty acids in arabidopsis. Plant Physiol. 162, 39–51. doi: 10.1104/PP.113.216994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y., Shorrosh B., Beisson F., Andersson M. X., Arondel V., Bates P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book / Am. Soc. Plant Biologists 11, e0161. doi: 10.1199/TAB.0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libourel I. G. L., Shachar-Hill Y. (2008). Metabolic flux analysis in plants: From intelligent design to rational engineering. Annu. Rev. Plant Biol. 59, 625–650. doi: 10.1146/annurev.arplant.58.032806.103822 [DOI] [PubMed] [Google Scholar]
- Li N., Gügel I. L., Giavalisco P., Zeisler V., Schreiber L., Soll J., et al. (2015. b). FAX1, a novel membrane protein mediating plastid fatty acid export. PloS Biol. 13, e1002053. doi: 10.1371/JOURNAL.PBIO.1002053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Jin C., Duan S., Zhu Y., Qi S., Liu K., et al. (2017). MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 173, 1211–1225. doi: 10.1104/pp.16.01634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li Y., Wang R., Chao L., Xiu Y., Wang H. (2020. a). Characterization of the stearoyl-ACP desaturase gene (PoSAD) from woody oil crop paeonia ostii var. lishizhenii in oleic acid biosynthesis. Phytochemistry 178, 112480. doi: 10.1016/j.phytochem.2020.112480 [DOI] [PubMed] [Google Scholar]
- Li X., Mei D., Liu Q., Fan J., Singh S., Green A., et al. (2016). Down-regulation of crambe fatty acid desaturase and elongase in arabidopsis and crambe resulted in significantly increased oleic acid content in seed oil. Plant Biotechnol. J. 14, 323–331. doi: 10.1111/pbi.12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Meng H., Li S., Zhang Z., Zhao X., Wang S., et al. (2020. b). Two plastid fatty acid exporters contribute to seed oil accumulation in arabidopsis. Plant Physiol. 182, 1910–1919. doi: 10.1104/PP.19.01344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A. R. Q., Kong Q., Singh S. K., Guo L., Yuan L., Ma W. (2022). Sunflower WRINKLED1 plays a key role in transcriptional regulation of oil biosynthesis. Int. J. Mol. Sci. 23, 3054. doi: 10.3390/ijms23063054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Han J., Li Z., Jiang Z., Luo L., Zhang Y., et al. (2022). Heterologous expression of jatropha curcas fatty acyl-ACP thioesterase a (JcFATA) and b (JcFATB) affects fatty acid accumulation and promotes plant growth and development in arabidopsis. Int. J. Mol. Sci. 23, 4209. doi: 10.3390/IJMS23084209/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Hua W., Yang H. L., Zhan G. M., Li R. J., Deng L.B., et al. (2012). The BnGRF2 gene (GRF2-like gene from brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 63, 3727–3740. doi: 10.1093/jxb/ers066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Hua W., Zhan G., Wei F., Wang X., Liu G., et al. (2010). Increasing seed mass and oil content in transgenic arabidopsis by the overexpression of wri1-like gene from brassica napus. Plant Physiol. Biochem. 48, 9–15. doi: 10.1016/j.plaphy.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Liu W. X., Liu H. L., Qu L. Q. (2013). Embryo-specific expression of soybean oleosin altered oil body morphogenesis and increased lipid content in transgenic rice seeds. Theor. Appl. Genet. 126, 2289–2297. doi: 10.1007/s00122-013-2135-4 [DOI] [PubMed] [Google Scholar]
- Liu F., Xia Y., Wu L., Fu D., Hayward A., Luo J., et al. (2015). Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene 557, 163–171. doi: 10.1016/j.gene.2014.12.029 [DOI] [PubMed] [Google Scholar]
- Li M., Wei F., Tawfall A., Tang M., Saettele A., Wang X. (2015. a). Overexpression of patatin-related phospholipase AIIIδ altered plant growth and increased seed oil content in camelina. Plant Biotechnol. J. 13, 766–778. doi: 10.1111/pbi.12304 [DOI] [PubMed] [Google Scholar]
- Li N., Xu C., Li-Beisson Y., Philippar K. (2016. a). Fatty acid and lipid transport in plant cells. Trends Plant Sci. 21, 145–158. doi: 10.1016/j.tplants.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Lonien J., Schwender J. (2009). Analysis of metabolic flux phenotypes for two arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 151, 1617–1634. doi: 10.1104/pp.109.144121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M. A., Matsudaira Yee K., West M. A. L., Lo R., Kwong R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4 [DOI] [PubMed] [Google Scholar]
- Lu Y., Chi M., Li L., Li H., Noman M., Yang Y., et al. (2018). Genome-wide identification, expression profiling, and functional validation of oleosin gene family in carthamus tinctorius l. Front. Plant Sci. 9, 1393. doi: 10.3389/fpls.2018.01393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Fulda M., Wallis J. G., Browse J. (2006). A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic arabidopsis. Plant J. 45, 847–856. doi: 10.1111/J.1365-313X.2005.02636.X [DOI] [PubMed] [Google Scholar]
- Lung S. C., Weselake R. J. (2006). Diacylglycerol acyltransferase: A key mediator of plant triacylglycerol synthesis. Lipids 41, 1073–1088. doi: 10.1007/S11745-006-5057-Y [DOI] [PubMed] [Google Scholar]
- Lunn D., Wallis J. G., Browse J. (2018). Overexpression of Seipin1 increases oil in hydroxy fatty acid-accumulating seeds. Plant Cell Physiol. 59, 205–214. doi: 10.1093/pcp/pcx177 [DOI] [PubMed] [Google Scholar]
- Lu C., Xin Z., Ren Z., Miquel M., Browse J. (2009). An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 18837–18842. doi: 10.1073/PNAS.0908848106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manan S., Ahmad M. Z., Zhang G., Chen B., Haq B. U., Yang J., et al. (2017). Soybean LEC2 regulates subsets of genes involved in controlling the biosynthesis and catabolism of seed storage substances and seed development. Front. Plant Sci. 8, 1604. doi: 10.3389/fpls.2017.01604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C., Nikovics K., To A., Lepiniec L., Baud S. (2014). Transcriptional regulation of fatty acid production in higher plants: Molecular bases and biotechnological outcomes. Eur. J. Lipid Sci. Technol. 116, 1332–1343. doi: 10.1002/ejlt.201400027 [DOI] [Google Scholar]
- Mastebroek H. D., Marvin H. J. P. (2000). Breeding prospects of lunaria annua l. Ind. Crops Prod 11, 139–143. doi: 10.1016/S0926-6690(99)00056-4 [DOI] [Google Scholar]
- Ma J., Sun S., Whelan J., Shou H. (2021). CRISPR/Cas9-mediated knockout of GmFATB1 significantly reduced the amount of saturated fatty acids in soybean seeds. Int. J. Mol. Sci. 22, 3877. doi: 10.3390/IJMS22083877/S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meesapyodsuk D., Chen Y., Ye S., Chapman R. G., Qiu X. (2021). Co-Expressing eranthis hyemalis lysophosphatidic acid acyltransferase 2 and elongase improves two very long chain polyunsaturated fatty acid production in brassica carinata. Metab. Eng. Commun. 12, e00171. doi: 10.1016/j.mec.2021.e00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhaske V., Beldjilali K., Ohlrogge J., Pollard M. (2005). Isolation and characterization of an arabidopsis thaliana knockout line for phospholipid: diacylglycerol transacylase gene (At5g13640). Plant Physiol. Biochem. 43, 413–417. doi: 10.1016/J.PLAPHY.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Mietkiewska E., Brost J. M., Giblin E. M., Barton D. L., Taylor D. C. (2007). Cloning and functional characterization of the fatty acid elongase 1 (FAE1) gene from high erucic crambe abyssinica cv. prophet. Plant Biotechnol. J. 5, 636–645. doi: 10.1111/j.1467-7652.2007.00268.x [DOI] [PubMed] [Google Scholar]
- Mietkiewska E., Giblin E. M., Wang S., Barton D. L., Dirpaul J., Brost J. M., et al. (2004). Seed-specific heterologous expression of a nasturtium FAE gene in arabidopsis results in a dramatic increase in the proportion of erucic acid. Plant Physiol. 136, 2665–2675. doi: 10.1104/pp.104.046839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mietkiewska E., Miles R., Wickramarathna A., Sahibollah A. F., Greer M. S., Chen G., et al. (2014). Combined transgenic expression of punica granatum conjugase (FADX) and FAD2 desaturase in high linoleic acid arabidopsis thaliana mutant leads to increased accumulation of punicic acid. Planta 240, 575–583. doi: 10.1007/s00425-014-2109-z [DOI] [PubMed] [Google Scholar]
- Misra A., Khan K., Niranjan A., Nath P., Sane V. A. (2013). Over-expression of JcDGAT1 from jatropha curcas increases seed oil levels and alters oil quality in transgenic arabidopsis thaliana. Phytochemistry 96, 37–45. doi: 10.1016/j.phytochem.2013.09.020 [DOI] [PubMed] [Google Scholar]
- Möllney M., Wiechert W., Kownatzki D., de Graaf A. A. (1999). Bidirectional reaction steps in metabolic networks: IV. optimal design of isotopomer labeling experiments. Biotechnol. Bioeng 66, 86–103. doi: [DOI] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu Y., Liang Y., Zhang J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in arabidopsis. Plant Physiol. 148, 1042–1054. doi: 10.1104/pp.108.126342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T., Park H., Koster K. L., Cahoon R. E., Nguyen H. T. M., Shanklin J., et al. (2015). Redirection of metabolic flux for high levels of omega-7 monounsaturated fatty acid accumulation in camelina seeds. Plant Biotechnol. J. 13, 38–50. doi: 10.1111/PBI.12233 [DOI] [PubMed] [Google Scholar]
- O’Grady J., Schwender J., Shachar-Hill Y., Morgan J. A. (2012). Metabolic cartography: Experimental quantification of metabolic fluxes from isotopic labelling studies. J. Exp. Bot. 63, 2293–2308. doi: 10.1093/jxb/ers032 [DOI] [PubMed] [Google Scholar]
- Ozseyhan M. E., Li P., Na G. N., Li Z., Wang C., Lu C. (2018). Improved fatty acid profiles in seeds of camelina sativa by artificial microRNA mediated FATB gene suppression. Biochem. Biophys. Res. Commun. 503, 621–624. doi: 10.1016/J.BBRC.2018.06.051 [DOI] [PubMed] [Google Scholar]
- Park M. E., Kim H. U. (2022). Applications and prospects of genome editing in plant fatty acid and triacylglycerol biosynthesis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.969844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveez G. K. A., Othman A., Ramin N., Bohari B. (2010)Functional analysis of oil palm palmitoyl-acp thioesterase (FatB) gene via down-regulation in a model plant: Arabidopsis thaliana (Accessed September 30, 2022).
- Petrie J. R., Shrestha P., Belide S., Mansour M. P., Liu Q., Horne J., et al. (2012). Transgenic production of arachidonic acid in oilseeds. Transgenic Res. 21, 139–147. doi: 10.1007/s11248-011-9517-7 [DOI] [PubMed] [Google Scholar]
- Pleite R., Pike M. J., Garcés R., Martínez-Force E., Rawsthorne S. (2005). The sources of carbon and reducing power for fatty acid synthesis in the heterotrophic plastids of developing sunflower (Helianthus annuus l.) embryos. J. Exp. Bot. 56, 1297–1303. doi: 10.1093/jxb/eri130 [DOI] [PubMed] [Google Scholar]
- Pokotylo I., Pejchar P., Potocký M., Kocourková D., Krčková Z., Ruelland E., et al. (2013). The plant non-specific phospholipase c gene family. novel competitors in lipid signalling. Prog. Lipid Res. 52, 62–79. doi: 10.1016/J.PLIPRES.2012.09.001 [DOI] [PubMed] [Google Scholar]
- Pouvreau B., Baud S., Vernoud V., Morin V., Py C., Gendrot G., et al. (2011). Duplicate maize wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 156, 674–686. doi: 10.1104/pp.111.173641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I., Smith S. M. (2008). When is a peroxisome not a peroxisome? Trends Plant Sci. 13, 522–525. doi: 10.1016/J.TPLANTS.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Price A. M., Doner N. M., Gidda S. K., Jambunathan S., James C. N., Schami A., et al. (2020). Mouse fat-specific protein 27 (FSP27) expressed in plant cells localizes to lipid droplets and promotes lipid droplet accumulation and fusion. Biochimie 169, 41–53. doi: 10.1016/j.biochi.2019.08.002 [DOI] [PubMed] [Google Scholar]
- Puthur J. T., Shackira A. M., Saradhi P. P., Bartels D. (2013). Chloroembryos: A unique photosynthesis system. J. Plant Physiol. 170, 1131–1138. doi: 10.1016/j.jplph.2013.04.011 [DOI] [PubMed] [Google Scholar]
- Pyc M., Cai Y., Gidda S. K., Yurchenko O., Park S., Kretzschmar F. K., et al. (2017). Arabidopsis lipid droplet-associated protein (LDAP) – interacting protein ( LDIP ) influences lipid droplet size and neutral lipid homeostasis in both leaves and seeds. Plant J. 92, 1182–1201. doi: 10.1111/tpj.13754 [DOI] [PubMed] [Google Scholar]
- Ratcliffe R. G., Shachar-Hill Y. (2006). Measuring multiple fluxes through plant metabolic networks. Plant J. 45, 490–511. doi: 10.1111/j.1365-313X.2005.02649.x [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez M. F., Moreno-Pérez A. J., Makni S., Troncoso-Ponce M. A., Acket S., Thomasset B., et al. (2021). Lipid profiling and oil properties of camelina sativa seeds engineered to enhance the production of saturated and omega-7 fatty acids. Ind. Crops Prod 170, 113765. doi: 10.1016/j.indcrop.2021.113765 [DOI] [Google Scholar]
- Rolletschek H., Radchuk R., Klukas C., Schreiber F., Wobus U., Borisjuk L. (2005). Evidence of a key role for photosynthetic oxygen release in oil storage in developing soybean seeds. New Phytol. 167, 777–786. doi: 10.1111/J.1469-8137.2005.01473.X [DOI] [PubMed] [Google Scholar]
- Romsdahl T. B., Kambhampati S., Koley S., Yadav U. P., Alonso A. P., Allen D. K., et al. (2021). Analyzing mass spectrometry imaging data of13c-labeled phospholipids in camelina sativa and thlaspi arvense (Pennycress) embryos. Metabolites. 11, 148. doi: 10.3390/metabo11030148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe T. T., Guilleminot J., Bessoule J. J., Berger F., Devic M. (2015). Complementation of seed maturation phenotypes by ectopic expression of ABSCISIC ACID INSENSITIVE3, FUSCA3 and LEAFY COTYLEDON2 in arabidopsis. Plant Cell Physiol. 56, 1215–1228. doi: 10.1093/pcp/pcv049 [DOI] [PubMed] [Google Scholar]
- Rousselin P., Molinier J., Himber C., Schontz D., Prieto-Dapena P., Jordano J., et al. (2002). Modification of sunflower oil quality by seed-specific expression of a heterologous Δ9-stearoyl-(acyl carrier protein) desaturase gene. Plant Breed. 121, 108–116. doi: 10.1046/j.1439-0523.2002.00682.x [DOI] [Google Scholar]
- Ruuska S. A., Schwender J., Ohlrogge J. B. (2004). The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 136, 2700–2709. doi: 10.1104/pp.104.047977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarth N. B., Mahanwar P. A. (2015). Modified vegetable oil based additives as a future polymeric material–review. Open J. Organic Polymer Materials 05, 1–22. doi: 10.4236/ojopm.2015.51001 [DOI] [Google Scholar]
- Savadi S., Naresh V., Kumar V., Bhat S. R. (2015). Seed-specific overexpression of arabidopsis DGAT1 in Indian mustard (Brassica juncea) increases seed oil content and seed weight. Botany 94, 177–184. doi: 10.1139/cjb-2015-0218 [DOI] [Google Scholar]
- Schwender J., Goffman F., Ohlrogge J. B., Shachar-Hill Y. (2004). Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432, 779–782. doi: 10.1038/nature03145 [DOI] [PubMed] [Google Scholar]
- Schwender J., Ohlrogge J. B. (2002). Probing in vivo metabolism by stable isotope labeling of storage lipids and proteins in developing brassica napus embryos. Plant Physiol. 130, 347–361. doi: 10.1104/pp.004275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwender J., Shachar-Hill Y., Ohlrogge J. B. (2006). Mitochondrial metabolism in developing embryos of brassica napus. J. Biol. Chem. 281, 34040–34047. doi: 10.1074/jbc.M606266200 [DOI] [PubMed] [Google Scholar]
- Shachar-Hill Y. (2013). Metabolic network flux analysis for engineering plant systems. Curr. Opin. Biotechnol. 24, 247–255. doi: 10.1016/j.copbio.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Shao Q., Liu X., Su T., Ma C., Wang P. (2019). New insights into the role of seed oil body proteins in metabolism and plant development. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer H. L., Turpin D. H., Dennis D. T. (2004). Characterization of NADP-dependent malic enzyme from developing castor oil seed endosperm. Arch. Biochem. Biophys. 429, 134–144. doi: 10.1016/j.abb.2004.07.001 [DOI] [PubMed] [Google Scholar]
- Shen B., Allen W. B., Zheng P., Li C., Glassman K., Ranch J., et al. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153, 980–987. doi: 10.1104/pp.110.157537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Katavic V., Yu Y., Kunst L., Haughn G. (2012). Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J. 69, 37–46. doi: 10.1111/j.1365-313X.2011.04768.x [DOI] [PubMed] [Google Scholar]
- Shimada T. L., Shimada T., Takahashi H., Fukao Y., Hara-Nishimura I. (2008). A novel role for oleosins in freezing tolerance of oilseeds in arabidopsis thaliana. Plant J. 55, 798–809. doi: 10.1111/J.1365-313X.2008.03553.X [DOI] [PubMed] [Google Scholar]
- Shockey J. M., Gidda S. K., Chapital D. C., Kuan J. C., Dhanoa P. K., Bland J. M., et al. (2006). Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18, 2294–2313. doi: 10.1105/TPC.106.043695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J., Lager I., Stymne S., Kotapati H. K., Sheffield J., Mason C., et al. (2019). Specialized lysophosphatidic acid acyltransferases contribute to unusual fatty acid accumulation in exotic euphorbiaceae seed oils. Planta 249, 1285–1299. doi: 10.1007/S00425-018-03086-Y/FIGURES/8 [DOI] [PubMed] [Google Scholar]
- Siloto R. M. P., Findlay K., Lopez-Villalobos A., Yeung E. C., Nykiforuk C. L., Moloney M. M. (2006). The accumulation of oleosins determines the size of seed oilbodies in arabidopsis. Plant Cell 18, 1961–1974. doi: 10.1105/TPC.106.041269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S. D., Greer M. S., Mietkiewska E., Pan X., Weselake R. J. (2013). “Genetic engineering of lipid biosynthesis in seeds,” in Biotechnology of crucifers (Springer New York; ), 111–149. doi: 10.1007/978-1-4614-7795-2_7 [DOI] [Google Scholar]
- Singh R., Arora A., Singh V. (2021). Biodiesel from oil produced in vegetative tissues of biomass – a review. Bioresour Technol. 326, 124772. doi: 10.1016/j.biortech.2021.124772 [DOI] [PubMed] [Google Scholar]
- Sinha S., Jha J. K., Maiti M. K., Basu A., Mukhopadhyay U. K., Sen S. K. (2007). Metabolic engineering of fatty acid biosynthesis in Indian mustard (Brassica juncea) improves nutritional quality of seed oil. Plant Biotechnol. Rep. 1, 185–197. doi: 10.1007/s11816-007-0032-5 [DOI] [Google Scholar]
- Slocombe S. P., Cornah J., Pinfield-Wells H., Soady K., Zhang Q., Gilday A., et al. (2009). Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnol. J. 7, 694–703. doi: 10.1111/j.1467-7652.2009.00435.x [DOI] [PubMed] [Google Scholar]
- Smith R. G., Gauthier D. A., Dennis D. T., Turpin D. H. (1992). Malate- and pyruvate-dependent fatty acid synthesis in leucoplasts from developing castor endosperm. Plant Physiol. 98, 1233–1238. doi: 10.1104/pp.98.4.1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. A., Moon H., Chowrira G., Kunst L. (2003). Heterologous expression of a fatty acid hydroxylase gene in developing seeds of arabidopsis thaliana. Planta 217, 507–516. doi: 10.1007/s00425-003-1015-6 [DOI] [PubMed] [Google Scholar]
- Smith M., Moon H., Kunst L. (2000). “Production of hydroxy fatty acids in the seeds of arabidopsis thaliana,” in Biochemical society transactions (Portland Press; ), 947–950. doi: 10.1042/bst0280947 [DOI] [PubMed] [Google Scholar]
- Snapp A. R., Kang J., Qi X., Lu C. (2014). A fatty acid condensing enzyme from physaria fendleri increases hydroxy fatty acid accumulation in transgenic oilseeds of camelina sativa. Planta 240, 599–610. doi: 10.1007/s00425-014-2122-2 [DOI] [PubMed] [Google Scholar]
- Song G., Li X., Munir R., Khan A. R., Azhar W., Yasin M. U., et al. (2020). The WRKY6 transcription factor affects seed oil accumulation and alters fatty acid compositions in Arabidopsis thaliana . Physiol. Plant 169, 612–624. doi: 10.1111/ppl.13082 [DOI] [PubMed] [Google Scholar]
- Sorokin A., Kim E. R., Ovchinnikov L. P. (2009). Proteasome system of protein degradation and processing. Biochem. (Moscow) 74, 1411–1442. doi: 10.1134/S000629790913001X [DOI] [PubMed] [Google Scholar]
- Stone S. L., Kwong L. W., Yee K. M., Pelletier J., Lepiniec L., Fischer R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811. doi: 10.1073/PNAS.201413498/ASSET/33629AFE-C5C7-4684-9F25-24BA60D45D9F/ASSETS/GRAPHIC/PQ2014134006.JPEG [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Liang W., Liu Z., Wang Y., Zhao Y., Ijaz B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017 [DOI] [PubMed] [Google Scholar]
- Sun R. H., Ye R., Gao L., Zhang L., Wang R., Mao T., et al. (2017). Characterization and ectopic expression of CoWR1, an AP2/EREBP domain-containing transcription factor from coconut (cocos nucifera l.) endosperm, changes the seeds oil content in transgenic arabidopsis thaliana and rice (oryza sativa l.). Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Guo N., Tang Q., Peng F., Liu Y., Xia H., et al. (2022. a). Pyruvate transporter BnaBASS2 impacts seed oil accumulation in brassica napus. Plant Biotechnol. J. 20, 2406–2417. doi: 10.1111/pbi.13922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Peng F., Tang Q., Liu Y., Xia H., Yao X., et al. (2022. b). BnaPPT1 is essential for chloroplast development and seed oil accumulation in brassica napus. J. Adv. Res. 42, 29–40. doi: 10.1016/j.jare.2022.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Xu P., Ma W., Wang F., Liu Z., Wan S., et al. (2018). Seed-specific expression of AtLEC1 increased oil content and altered fatty acid composition in seeds of peanut (Arachis hypogaea l.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H., Yang X., Zhang F., Zheng X., Qu C., Mu J., et al. (2011). Enhanced seed oil production in canola by conditional expression of brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 156, 1577–1588. doi: 10.1104/pp.111.175000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. C., Francis T., Guo Y., Brost J. M., Katavic V., Mietkiewska E., et al. (2009). Molecular cloning and characterization of a KCS gene from cardamine graeca and its heterologous expression in brassica oilseeds to engineer high nervonic acid oils for potential medical and industrial use. Plant Biotechnol. J. 7, 925–938. doi: 10.1111/j.1467-7652.2009.00454.x [DOI] [PubMed] [Google Scholar]
- Taylor D. C., Smith M. A., Fobert P., Mietkiewska E., Weselake R. J. (2011). “Metabolic engineering of higher plants to produce bio-industrial oils,” in Comprehensive biotechnology, second edition (Elsevier Inc; ). doi: 10.1016/B978-0-08-088504-9.00256-7 [DOI] [Google Scholar]
- Tian Y., Lv X., Xie G., Wang L., Dai T., Qin X., et al. (2019). FAX2 mediates fatty acid export from plastids in developing arabidopsis seeds. Plant Cell Physiol. 60, 2231–2242. doi: 10.1093/pcp/pcz117 [DOI] [PubMed] [Google Scholar]
- Tian Y., Lv X., Xie G., Zhang J., Xu Y., Chen F. (2018). Seed-specific overexpression of AtFAX1 increases seed oil content in arabidopsis. Biochem. Biophys. Res. Commun. 500, 370–375. doi: 10.1016/j.bbrc.2018.04.081 [DOI] [PubMed] [Google Scholar]
- Troncoso-Ponce M. A., Barthole G., Tremblais G., To A., Miquel M., Lepiniec L., et al. (2016). Transcriptional activation of two delta-9 palmitoyl-acp desaturase genes by myb115 and myb118 is critical for biosynthesis of omega-7 monounsaturated fatty acids in the endosperm of arabidopsis seeds. Plant Cell 28, 2666–2682. doi: 10.1105/tpc.16.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch H., Borisjuk L., Rutten T., Rolletschek H. (2011). Gradients of seed photosynthesis and its role for oxygen balancing. BioSystems 103, 302–308. doi: 10.1016/j.biosystems.2010.08.007 [DOI] [PubMed] [Google Scholar]
- Tsogtbaatar E., Cocuron J.-C., Alonso A. P. (2020). Non-conventional pathways enable pennycress (Thlaspi arvense l.) embryos to achieve high efficiency of oil biosynthesis. J. Exp. Bot. 71, 3037–3051. doi: 10.1093/jxb/eraa060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H., Bates P. D., Burgal J., Shockey J., Browse J. (2011). Castor phospholipid:Diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic arabidopsis. Plant Physiol. 155, 683–693. doi: 10.1104/pp.110.167239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp H., Kelly A. A., Menard G., Eastmond P. J. (2014). Multigene engineering of triacylglycerol metabolism boosts seed oil content in arabidopsis. Plant Physiol. 165, 30–36. doi: 10.1104/pp.114.236430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhercke T., Dyer J. M., Mullen R. T., Kilaru A., Rahman M. M., Petrie J. R., et al. (2019). Metabolic engineering for enhanced oil in biomass. Prog. Lipid Res. 74, 103–129. doi: 10.1016/j.plipres.2019.02.002 [DOI] [PubMed] [Google Scholar]
- Vanhercke T., el Tahchy A., Shrestha P., Zhou X. R., Singh S. P., Petrie J. R. (2013). Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 587, 364–369. doi: 10.1016/j.febslet.2012.12.018 [DOI] [PubMed] [Google Scholar]
- Wang X. (2005). Regulatory functions of phospholipase d and phosphatidic acid in plant growth, development, and stress responses. Plant Physiol. 139, 566–573. doi: 10.1104/pp.105.068809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Froehlich J. E., Zienkiewicz A., Hersh H. L., Benning C. (2017). A plastid phosphatidylglycerol lipase contributes to the export of acyl groups from plastids for seed oil biosynthesis. Plant Cell 29, 1678–1696. doi: 10.1105/tpc.17.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Garneau M. G., Poudel A. N., Lamm D., Koo A. J., Bates P. D., et al. (2022). Overexpression of pea α-carboxyltransferase in arabidopsis and camelina increases fatty acid synthesis leading to improved seed oil content. Plant J. 110, 1035–1046. doi: 10.1111/tpj.15721 [DOI] [PubMed] [Google Scholar]
- Wang Z., Huang W., Chang J., Sebastian A., Li Y., Li H., et al. (2014). Overexpression of SiDGAT1, a gene encoding acyl-CoA:diacylglycerol acyltransferase from sesamum indicum l. increases oil content in transgenic arabidopsis and soybean. Plant Cell Tissue Organ Cult 119, 399–410. doi: 10.1007/s11240-014-0543-z [DOI] [Google Scholar]
- Wiechert W., De Graaf A. A. (1997). Bidirectional reaction steps in metabolic networks: I. modeling and simulation of carbon isotope labeling experiments. Biotechnol. Bioeng 55, 101–117. doi: 10.1002/(SICI)1097-0290(19970705)55:1 [DOI] [PubMed] [Google Scholar]
- Wiechert W., Möllney M., Isermann N., Wurzel M., de Graaf A. A. (1999). Bidirectional reaction steps in metabolic networks: III. explicit solution and analysis of isotopomer labeling systems. Sons Inc. Biotechnol. Bioeng 66, 69–85. doi: [DOI] [PubMed] [Google Scholar]
- Wiechert W., Möllney M., Petersen S., de Graaf A. A. (2001). A universal framework for 13C metabolic flux analysis. Metab. Eng. 3, 265–283. doi: 10.1006/mben.2001.0188 [DOI] [PubMed] [Google Scholar]
- Winichayakul S., William Scott R., Roldan M., Bertrand Hatier J. H., Livingston S., Cookson R., et al. (2013). In vivo packaging of triacylglycerols enhances arabidopsis leaf biomass and energy density. Plant Physiol. 162, 626–639. doi: 10.1104/pp.113.216820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Hong Y., Li X., Fan R., Li Q., Ouyang Z., et al. (2022). BnaNTT2 regulates ATP homeostasis in plastid to sustain lipid metabolism and plant growth in brassica napus. Mol. Breed. 42, 1–19. doi: 10.1007/s11032-022-01322-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z., Tang F., Zhang L., Li S., Wang S., Huo Q., et al. (2021). The brassica napus fatty acid exporter FAX1-1 contributes to biological yield, seed oil content, and oil quality. Biotechnol. Biofuels 14, 190. doi: 10.1186/s13068-021-02035-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. Y., Yang H. K., Singh S. P., Sharp P. J., Liu Q. (2018). Genetic manipulation of non-classic oilseed plants for enhancement of their potential as a biofactory for triacylglycerol production. Engineering 4, 523–533. doi: 10.1016/j.eng.2018.07.002 [DOI] [Google Scholar]
- Yang Z., Liu X., Wang K., Li Z., Jia Q., Zhao C., et al. (2022). ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation. J. Exp. Bot. 73, 2077–2092. doi: 10.1093/jxb/erab524 [DOI] [PubMed] [Google Scholar]
- Yatsu L. Y., Jacks T. J. (1972). Spherosome membranes. Plant Physiol. 49, 937–943. doi: 10.1104/pp.49.6.937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin D., Deng S., Zhan K., Cui D. (2007). High-oleic peanut oils produced by HpRNA-mediated gene silencing of oleate desaturase. Plant Mol. Biol. Rep. 25, 154–163. doi: 10.1007/s11105-007-0017-0 [DOI] [Google Scholar]
- Yin D. D., Xu W. Z., Shu Q. Y., Li S. S., Wu Q., Feng C. Y., et al. (2018). Fatty acid desaturase 3 (PsFAD3) from paeonia suffruticosa reveals high α-linolenic acid accumulation. Plant Sci. 274, 212–222. doi: 10.1016/j.plantsci.2018.05.027 [DOI] [PubMed] [Google Scholar]
- Yu K., McCracken C. T., Li R., Hildebrand D. F. (2006). Diacylglycerol acyltransferases from vernonia and stokesia prefer substrates with vernolic acid. Lipids 41, 557–566. doi: 10.1007/S11745-006-5005-X [DOI] [PubMed] [Google Scholar]
- Zhang M., Fan J., Taylor D. C., Ohlrogge J. B. (2010). DGAT1 and PDAT1 acyltransferases have overlapping functions in arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21, 3885–3901. doi: 10.1105/TPC.109.071795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Q., Lu X., Zhao F. Y., Li Q. T., Niu S. L., Wei W., et al. (2016). Soybean GmDREBL increases lipid content in seeds of transgenic arabidopsis. Sci. Rep. 6, 1–13. doi: 10.1038/srep34307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zhang H., Hu Z., Chu S., Yu K., Lv L., et al. (2019). Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PloS Genet. 15, 1008267. doi: 10.1371/journal.pgen.1008267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Cao P., Cui Y., Liu D., Li J., Zhao Y., et al. (2021). Enhanced production of seed oil with improved fatty acid composition by overexpressing NAD+-dependent glycerol-3-phosphate dehydrogenase in soybean. J. Integr. Plant Biol. 63, 1036–1053. doi: 10.1111/jipb.13094 [DOI] [PubMed] [Google Scholar]
- Zhao N., Zhang Y., Li Q., Li R., Xia X., Qin X., et al. (2015). Identification and expression of a stearoyl-ACP desaturase gene responsible for oleic acid accumulation in xanthoceras sorbifolia seeds. Plant Physiol. Biochem. 87, 9–16. doi: 10.1016/j.plaphy.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Zhou B., Fei W., Yang S., Yang F., Qu G., Tang W., et al. (2020). Alteration of the fatty acid composition of brassica napus l. via overexpression of phospholipid: Diacylglycerol acyltransferase 1 from sapium sebiferum (L.) roxb. Plant Sci. 298, 110562. doi: 10.1016/j.plantsci.2020.110562 [DOI] [PubMed] [Google Scholar]
- Zhu Y., Xie L., Chen G. Q., Lee M. Y., Loque D., Scheller H. V. (2018). A transgene design for enhancing oil content in arabidopsis and camelina seeds. Biotechnol. Biofuels 11, 46. doi: 10.1186/s13068-018-1049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]