Key Points
Question
To what extent are shared genetic determinants in the comorbidities and associations between gastrointestinal tract diseases and psychiatry disorders involved in the gut-brain axis?
Findings
In this genome-wide pleiotropic association study using genome-wide association summary statistics from publicly available data sources, pervasive genetic correlations and genetic overlaps between gastrointestinal tract diseases and psychiatric disorders were found. The pleiotropic genetic determinants between them were extensively distributed across the genome.
Meaning
These findings not only support the shared genetic basis underlying the gut-brain axis but also have important implications for intervention and treatment targets of these 2 types of diseases simultaneously.
This genome-wide pleiotropic association study sequentially investigates the pleiotropic associations from genetic and biological pathways to disentangle the underlying shared genetic etiology between 4 gastrointestinal tract diseases and 6 psychiatric disorders.
Abstract
Importance
Comorbidities and genetic correlations between gastrointestinal tract diseases and psychiatric disorders have been widely reported, with the gut-brain axis (GBA) hypothesized as a potential biological basis. However, the degree to which the shared genetic determinants are involved in these associations underlying the GBA is unclear.
Objective
To investigate the shared genetic etiology between gastrointestinal tract diseases and psychiatric disorders and to identify shared genomic loci, genes, and pathways.
Design, Setting, and Participants
This genome-wide pleiotropic association study using genome-wide association summary statistics from publicly available data sources was performed with various statistical genetic approaches to sequentially investigate the pleiotropic associations from genome-wide single-nucleotide variation (SNV; formerly single-nucleotide polymorphism [SNP]), and gene levels and biological pathways to disentangle the underlying shared genetic etiology between 4 gastrointestinal tract diseases (inflammatory bowel disease, irritable bowel syndrome, peptic ulcer disease, and gastroesophageal reflux disease) and 6 psychiatric disorders (schizophrenia, bipolar disorder, major depressive disorder, attention-deficit/hyperactivity disorder, posttraumatic stress disorder, and anorexia nervosa). Data were collected from March 10, 2021, to August 25, 2021, and analysis was performed from January 8 through May 30, 2022.
Main Outcomes and Measures
The primary outcomes consisted of a list of genetic loci, genes, and pathways shared between gastrointestinal tract diseases and psychiatric disorders.
Results
Extensive genetic correlations and genetic overlaps were found among 22 of 24 trait pairs. Pleiotropic analysis under a composite null hypothesis identified 2910 significant potential pleiotropic SNVs in 19 trait pairs, with 83 pleiotropic loci and 24 colocalized loci detected. Gene-based analysis found 158 unique candidate pleiotropic genes, which were highly enriched in certain GBA-related phenotypes and tissues, whereas pathway enrichment analysis further highlighted biological pathways primarily involving cell adhesion, synaptic structure and function, and immune cell differentiation. Several identified pleiotropic loci also shared causal variants with gut microbiomes. Mendelian randomization analysis further illustrated vertical pleiotropy across 8 pairwise traits. Notably, many pleiotropic loci were identified for multiple pairwise traits, such as 1q32.1 (INAVA), 19q13.33 (FUT2), 11q23.2 (NCAM1), and 1p32.3 (LRP8).
Conclusions and Relevance
These findings suggest that the pleiotropic genetic determinants between gastrointestinal tract diseases and psychiatric disorders are extensively distributed across the genome. These findings not only support the shared genetic basis underlying the GBA but also have important implications for intervention and treatment targets of these diseases simultaneously.
Introduction
The comorbidities and associations between gastrointestinal tract diseases and psychiatric disorders have been widely reported,1,2,3 which was likely to be regulated by the gut-brain axis (GBA). The GBA is characterized by bidirectional interactions between the gastrointestinal tract and the central nervous system (CNS) and would link intestinal dysfunction and inflammation with brain function and psychiatric disorders.4,5 Various biological mechanisms were involved in the GBA, such as inflammatory immune responses, the autonomic nervous system, and enteric nervous system, where the role of the composition of gut microbiota and related metabolites has been particularly highlighted.4,5,6,7 Underlying the conceptual framework of the GBA, the shared genetic etiology might be involved in the associations between gastrointestinal tract diseases and psychiatric disorders.
Genome-wide association studies (GWAS) have identified multiple genetic variants (ie, single-nucleotide variations [SNVs]; formerly single-nucleotide polymorphisms [SNPs]) associated with gastrointestinal tract diseases and psychiatric disorders.8,9,10 Genetic correlations have been suggested between these 2 types of diseases using linkage disequilibrium (LD) score regression (LDSC).10,11,12,13,14 However, it remains unclear whether the overall genetic correlation would be attributed to a few loci or across the genome.15 Indeed, there would be genetic overlap even without any genetic correlation. Although previous studies have investigated genetic overlap,16 shared susceptibility genes,17,18 and causal relationships19,20,21 between these 2 types of diseases, they mainly focused on inflammatory bowel disease (IBD) and psychiatric disorders with limited sample sizes. Recently, 2 studies10,22 have conducted GWAS of specific gastrointestinal tract diseases as well as systematic post-GWAS analyses and pointed out the necessity to explore the shared genetic risk across traits to improve the understanding of the disordered brain-gut interactions. Therefore, it is of great importance to further seek out the specific genomic variants or loci accounting for genome-wide genetic correlation and to deeply probe into the shared genetic etiology between these 2 types of diseases. Shared genetic etiology also indicates the potential pleiotropy, which often acts as genetic confounding of the associations between trait pairs.23,24,25 Cross-trait analysis has been proposed to investigate the pleiotropic genetic variants or loci among multiple traits by leveraging the correlation of GWAS signals,23,26,27,28,29 where the pleiotropic loci could serve as intervention targets with the potential to simultaneously prevent or treat these diseases.
In this genome-wide pleiotropic association study, using large-scale GWAS summary data, we performed a genome-wide pairwise trait pleiotropic analysis between 4 gastrointestinal tract diseases (IBD, irritable bowel syndrome [IBS], peptic ulcer disease [PUD], and gastroesophageal reflux disease [GERD]) and 6 psychiatric disorders (schizophrenia, bipolar disorder [BIP], major depressive disorder [MDD], attention-deficit/hyperactivity disorder [ADHD], posttraumatic stress disorder [PTSD], and anorexia nervosa [AN]) through various statistical genetic approaches to sequentially investigate the pleiotropic associations from genome-wide, SNV, and gene levels and biological pathways to disentangle the underlying shared genetic etiology. Of note, under the framework of pleiotropic analysis, we first performed the SNV-level analysis to detect pleiotropic variants and loci, followed by pairwise colocalization analysis to determine colocalized loci and gene-level analysis to identify candidate pleiotropic genes, based on which we further performed parallel phenotype and tissue-specific enrichment analysis to characterize the phenotype and tissue specificity as well as additional gene-level analysis to identify the tissue-specific and cell type-specific pleiotropic genes. We also highlighted the role of gut microbiomes in interpreting the shared genetic etiology, followed by mendelian randomization analysis to evaluate pairwise causal associations and partly characterize different types of pleiotropy (vertical pleiotropy or horizontal pleiotropy).
Methods
GWAS Data Sets
We sought GWAS summary statistics from publicly available data sources with European ancestry, owing to the limited availability of well-powered GWAS with other ancestries, and selected GWAS with sample sizes larger than 50 000 to ensure statistical power. GWAS for GERD, IBD, and PUD were obtained from the same gastrointestinal tract GWAS based on 456 327 individuals from UK Biobank (UKB).10 GWAS for IBS were obtained from a larger meta-analysis with 486 601 individuals (53 400 cases and 433 201 controls).22 GWAS for the 6 psychiatric disorders were from the Psychiatric Genomics Consortium, including schizophrenia,30 BIP,31 MDD,32 ADHD,33 PTSD,34 and AN.35 In addition, GWAS for early age-related macular degeneration (AMD)36 and cataract37 were obtained to serve as a common set of negative controls for both gastrointestinal tract diseases and psychiatric disorders, given these 2 disorders are relatively limited to the pathological lesions of intraocular contents, and previous studies also showed no significant genetic correlations between AMD and MDD38 as well as among cataract, GERD, and psychiatric symptoms.39 All GWAS were approved by relevant ethic committees, and written informed consent was obtained from all participants, with details provided in the eTable 1 in Supplement 1. Data were collected from March 10, 2021, to August 25, 2021, and analyzed from January 8 through May 30, 2022. This genome-wide pleiotropic association study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline.
Statistical Analysis
All analyses were performed after excluding SNVs in the major histocompatibility complex region (chromosome 6: 25-35 megabase [Mb]) due to its complex LD structure and restricted to biallelic SNVs with minor allele frequency larger than 0.01. Details of these methods are provided in Figure 1 and eMethods in Supplement 1.
Figure 1. Study Workflow.
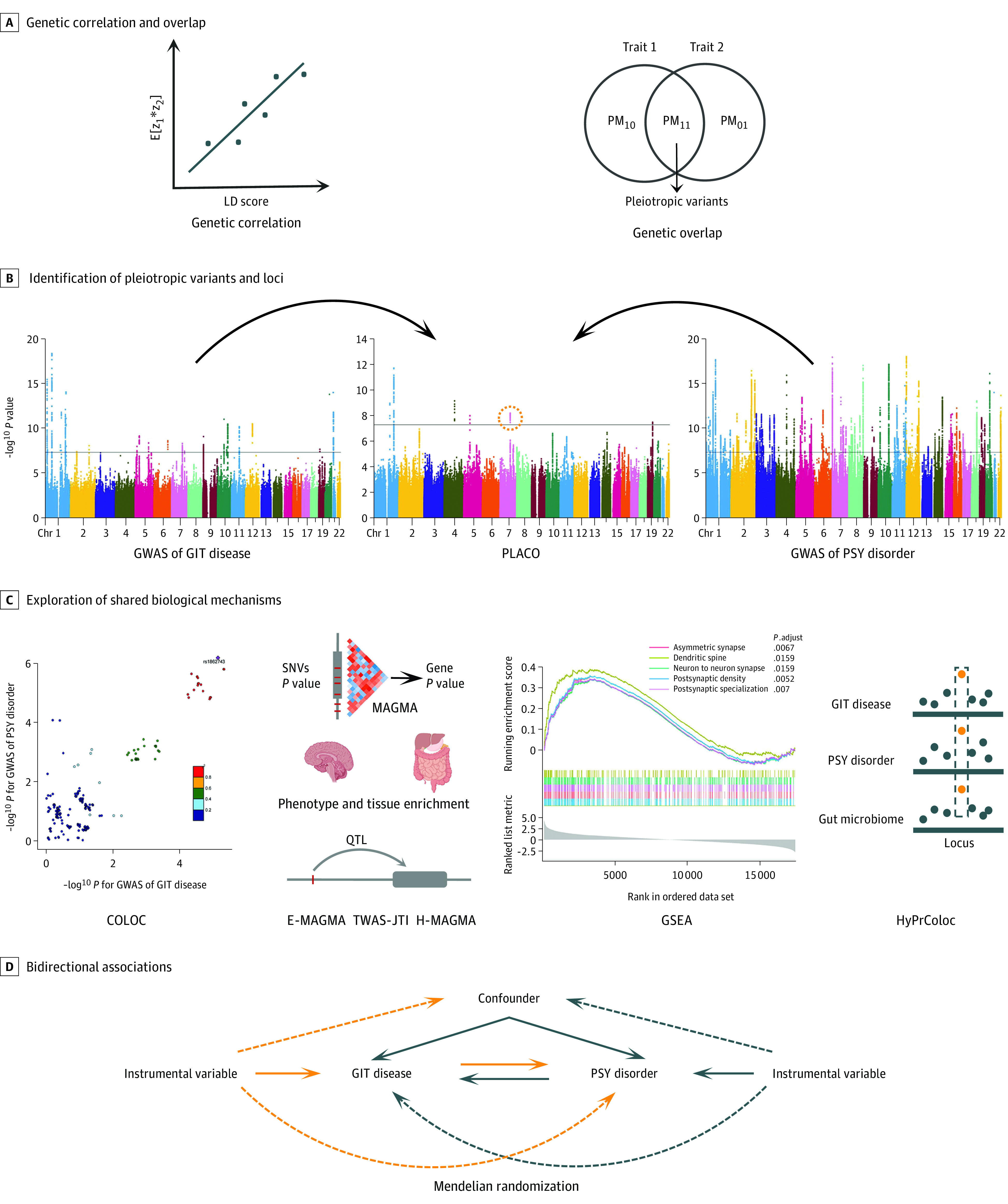
We conducted the comprehensive pleiotropic analysis between 4 gastrointestinal tract (GIT) diseases and 6 psychiatric (PSY) disorders from different perspectives. COLOC indicates colocalization; GSEA, gene set enrichment analysis; GWAS genome-wide association study; E-MAGMA, expression quantitative trait loci–informed multi-marker analysis of GenoMic annotation (MAGMA); H-MAGMA, Hi-C–coupled MAGMA; HyPrColoc, hypothesis prioritization for multi-trait colocalization; LD, linkage disequilibrium; PLACO, pleiotropic analysis under composite null hypothesis; SNV, single-nucleotide variation; and TWAS-JTI, transcriptome-wide association study analysis using joint-tissue imputation.
Both LDSC40 and high-definition likelihood41 were used to assess genome-wide genetic correlations for 24 pairwise traits. The intercept from LDSC could also indicate potential sample overlap between 2 GWAS. Given that genetic correlation only reflects the overall correlation across the genome, we further applied genetic analysis incorporating pleiotropy and annotation (GPA)29 to explore the overall genetic overlap between traits. The Bonferroni-corrected significant threshold was set at P < 2.08 × 10−3 (.05/24). In addition, negative control analysis was performed through LDSC between AMD and cataract and the 4 gastrointestinal tract diseases as well as the 6 psychiatric disorders, with Bonferroni-corrected significant threshold set at P < 2.5 × 10−3 (.05/20).
For the union set of pairwise traits with significant genetic correlation or genetic overlap, we used pleiotropic analysis under composite null hypothesis (PLACO) to identify potential pleiotropic SNVs.26 Single-nucleotide variations with P < 5 × 10−8 for PLACO were considered significant pleiotropic variants. Functional mapping and annotation of genetic associations (FUMA)42 was applied to characterize potential pleiotropic loci, based on which a bayesian colocalization analysis43 was performed to further identify shared causal variants in each pleiotropic locus. We declared a colocalized locus with posterior probability of H4 (PP.H4) larger than 0.7.
Based on PLACO results, we further explored the shared biological mechanisms of these pleiotropic loci. We performed gene-level multimarker analysis of GenoMic annotation (MAGMA)44 analysis on the genes located in or overlapped with the pleiotropic loci based on both PLACO outputs and single-trait GWAS to identify candidate pleiotropic genes, with the significance declared at the locus-specific Bonferroni-corrected 2-sided P < .05 for MAGMA analysis on PLACO results and 2-sided P < .05 for MAGMA analyses based on original single-trait GWAS. Further, we performed phenotype enrichment analysis based on the Mouse Genome Informatics platform45 to characterize the phenotype specificity of these pleiotropic genes against that of nonpleotropic genes by examining the differences in the proportions of genes associated with certain phenotypes in the pleiotropic gene group against that in the nonpleotropic gene group using the Fisher exact test. Then, we performed tissue-specific enrichment analyses to illustrate the tissue specificity of these pleiotropic genes using the deTS tissue-specific enrichment method46 based on 2 different reference panels, Genotype-Tissue Expression project (GTEx)47 and the Encyclopedia of DNA Elements project (ENCODE).48 We declared the significance with a nominal threshold (2-sided P < .05) for these 2 parallel enrichment analyses. In addition, we used E-MAGMA (expression quantitative trait loci [eQTL]–informed MAGMA)49 and transcriptome-wide association study analysis using joint-tissue imputation50 to further investigate the tissue-specific genes, parallelized with H-MAGMA (Hi-C–coupled MAGMA)51 to indicate the cell-type specificity, in which we declared the significance with locus-specific Bonferroni correction. Gene set enrichment analysis was also performed to identify potential biological pathways using the clusterProfiler package.52 Significantly enriched pathways were declared with normalized enrichment score greater than 2 and adjusted P < .05. Multitrait colocalization analysis with HyPrColoc (hypothesis prioritization for multi-trait colocalization)53 incorporating host-microbiome GWAS was performed to highlight the critical role of gut microbiome.
Last, we conducted bidirectional mendelian randomization analysis for 24 pairwise traits between gastrointestinal tract diseases and psychiatric disorders to investigate the potential causal trait pairs (ie, vertical pleiotropy), along with negative control analysis using AMD and cataract. We used the inverse-variance weighted method54 as main analysis and in total 6 alternative mendelian randomization methods with different model assumptions as additional analyses for further validation. For main analysis, we chose a false discovery rate approach for multiple testing correction with the significance threshold being false discovery rate–adjusted P < .05, given that the commonly used Bonferroni correction is often too stringent for multiple nonindependent tests. The nominally significant threshold (P < .05) was used for alternative mendelian randomization methods. The Latent Heritable Confounder Mendelian Randomization (LHC-MR) method, which could account for sample overlap, was used for pairwise traits with potential sample overlap to further validate the mendelian randomization results.55
Genomic Loci Characterization and Functional Annotation
For significant pleiotropic SNVs from PLACO, we applied FUMA to identify independent variants, characterize genomic risk loci, and annotate the functions of variants using LD information from the 1000 Genome Project phase 3 reference panel of European population.42 We characterized independent SNVs with r2 less than 0.6000 and lead SNVs with r2 less than 0.1000 within 1 Mb. Genomic risk loci were defined by merging genomic regions if the physical distance between lead SNVs was less than 250 kilobase.42
Functional annotations, including ANNOVAR software tool categories (Bioinformatics), combined annotation-dependent depletion (CADD) scores, and RegulomeDB scores, were also provided by FUMA. Single-nucleotide variations with a CADD score larger than 12.37 were considered a potentially deleterious variant.42 Single-nucleotide variations with P < 5 × 10−8 in each single-trait GWAS were also annotated by FUMA for comparison.
Results
Genetic Correlations and Genetic Overlaps Between Gastrointestinal Tract Diseases and Psychiatric Disorders
Using genome-wide association summary statistics from publicly available data sources, pervasive significant genome-wide genetic correlations and genetic overlaps were found across 24 pairwise traits, among which 14 were identified from LDSC and 21 were identified from GPA (Table 1). Notably, 8 trait pairs were identified with nonsignificant genetic correlation but with significant genetic overlap, producing a final union set of 22 pairwise traits for subsequent analysis. In addition, the results from the LDSC were highly consistent with those from high-definition likelihood (eTable 2 and eFigure 1 in Supplement 1) and suggested significant sample overlap for 5 trait pairs (Table 1). For negative control analysis, no significant genetic correlations were detected as expected (eTable 3 in Supplement 1).
Table 1. Genetic Correlation and Genetic Overlap Estimations Between 24 Pairwise Traitsa.
| Trait pair | Genetic correlation | Genetic overlap | |||||
|---|---|---|---|---|---|---|---|
| Genetic correlation (SE) | P value for LDSC | Intercept (SE) | P value for intercept | PM 11 | PARb | P value for GPA | |
| IBD-MDDc,d | 0.1706 (0.0407) | 2.82 × 10−5 | 0.0003 (0.0055) | 9.57 × 10−1 | 0.0116 | 0.0473 | 1.41 × 10−3 |
| IBD-PTSD | 0.1742 (0.0377) | 7.39 × 10−2 | 0.0049 (0.0051) | 3.37 × 10−1 | 0.0038 | 0.0189 | 2.27 × 10−1 |
| IBD-SCZd | 0.0359 (0.0403) | 3.73 × 10−1 | 0.0004 (0.0062) | 9.49 × 10−1 | 0.0167 | 0.0605 | 6.07 × 10−15 |
| IBD-ADHD | –0.0045 (0.0599) | 9.40 × 10−1 | 0.0121 (0.0057) | 3.38 × 10−2 | 0.0121 | 0.0512 | 5.16 × 10−2 |
| IBD-BIPd | 0.0221 (0.0478) | 6.45 × 10−1 | 0.0064 (0.0063) | 3.10 × 10−1 | 0.0194 | 0.0828 | 3.80 × 10−28 |
| IBD-ANd | –0.0296 (0.0630) | 6.38 × 10−1 | 0.0019 (0.0063) | 7.63 × 10−1 | 0.0206 | 0.0851 | 8.85 × 10−11 |
| IBS-MDDc,d,e | 0.5705 (0.0256) | 1.14 × 10−109 | 0.0795 (0.0066) | 2.05 × 10−33 | 0.1571 | 0.7791 | <1 × 10−300 |
| IBS-PTSDc,d,e | 0.4720 (0.0795) | 2.87 × 10−9 | 0.0275 (0.0049) | 2.00 × 10−8 | 0.0422 | 0.1504 | 1.27 × 10−5 |
| IBS-SCZc,d | 0.1711 (0.0285) | 1.85 × 10−9 | –0.0025 (0.006) | 6.77 × 10−1 | 0.1246 | 0.4130 | <1 × 10−300 |
| IBS-ADHDc,d | 0.2060 (0.0405) | 3.73 × 10−7 | 0.0138 (0.0059) | 1.93 × 10−2 | 0.1155 | 0.3895 | 1.67 × 10−87 |
| IBS-BIPc,d,e | 0.1269 (0.0305) | 3.12 × 10−5 | 0.0219 (0.0058) | 1.59 × 10−4 | 0.1183 | 0.4579 | 3.54 × 10−268 |
| IBS-ANc,d | 0.1536 (0.0413) | 2.00 × 10−4 | 0.0003 (0.0064) | 9.63 × 10−1 | 0.1207 | 0.4059 | 3.98 × 10−86 |
| PUD-MDDc,d,e | 0.4438 (0.0437) | 3.31 × 10−24 | 0.0333 (0.0059) | 1.66 × 10−8 | 0.0987 | 0.4011 | 1.04 × 10−102 |
| PUD-PTSDc | 0.5448 (0.1120) | 1.15 × 10−6 | 0.0074 (0.0046) | 1.08 × 10−1 | 0.0274 | 0.1052 | 3.65 × 10−2 |
| PUD-SCZc,d | 0.1306 (0.0402) | 1.20 × 10−3 | –0.0007 (0.006) | 9.07 × 10−1 | 0.0832 | 0.2713 | 7.89 × 10−55 |
| PUD-ADHDc,d | 0.4771 (0.0579) | 1.63 × 10−16 | 0.0072 (0.0058) | 2.14 × 10−1 | 0.0886 | 0.3409 | 1.27 × 10−37 |
| PUD-BIPd | 0.0691 (0.0437) | 1.14 × 10−1 | 0.0023 (0.0054) | 6.70 × 10−1 | 0.0679 | 0.2491 | 1.31 × 10−34 |
| PUD-ANd | 0.0366 (0.0539) | 4.97 × 10−1 | 0.0014 (0.0056) | 8.03 × 10−1 | 0.0448 | 0.1441 | 8.03 × 10−4 |
| GERD–MDDc,d,e | 0.4596 (0.0267) | 3.50 × 10−66 | 0.0692 (0.0053) | 5.83 × 10−39 | 0.1762 | 0.6824 | <1 × 10−300 |
| GERD-PTSDc,d | 0.4193 (0.0690) | 1.24 × 10−9 | 0.0156 (0.0051) | 2.22 × 10−3 | 0.0538 | 0.1567 | 3.43 × 10−4 |
| GERD-SCZd | 0.0313 (0.0273) | 2.51 × 10−1 | 0.0107 (0.0062) | 8.44 × 10−2 | 0.1299 | 0.3596 | 1.07 × 10−189 |
| GERD-ADHDc,d | 0.4919 (0.0371) | 3.32 × 10−40 | –0.0003 (0.0063) | 9.62 × 10−1 | 0.1543 | 0.4844 | 6.81 × 10−132 |
| GERD-BIPd | 0.0332 (0.0308) | 2.81 × 10−01 | 0.0048 (0.0059) | 4.16 × 10−1 | 0.1279 | 0.4050 | 1.27 × 10−174 |
| GERD-ANd | 0.0179 (0.0386) | 6.43 × 10−1 | 0.0038 (0.0063) | 5.46 × 10−1 | 0.1265 | 0.3597 | 1.24 × 10−61 |
Abbreviations: ADHD, attention-deficit/hyperactivity disorder; AN, anorexia nervosa; BIP, bipolar disorder; GERD, gastroesophageal reflux disease; GPA, genetic analysis incorporating pleiotropy and annotation method; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome; LDSC, linkage disequilibrium score regression; MDD, major depressive disorder; PAR, pleiotropy association ratio; PM 11, proportion of genetic variants associated with both traits; PTSD, posttraumatic stress disorder; PUD, peptic ulcer disease; SCZ, schizophrenia.
Genetic correlation and genetic overlap were estimated by LDSC and GPA methods, respectively. Bonferroni-corrected significance threshold was set at P < 2.83 × 10−3 (.05/24), producing a final union set of 22 pairwise traits with significant genetic correlation or genetic overlap for subsequent analysis.
We introduced PAR as PM 11/(PM 10 + PM 01 + PM 11) to represent the proportion of pleiotropic single-nucleotide variations (SNVs) associated with both traits against the proportion of SNVs associated with at least 1 trait.
Pairwise trait with significant genetic correlation.
Pairwise trait with significant genetic overlap.
Genome-wide association study summary data with potentially significant sample overlap.
Shared Loci Between Gastrointestinal Tract Diseases and Psychiatric Disorders
A total of 2910 SNVs (2284 unique) were identified as potential pleiotropic variants by PLACO in 19 trait pairs (eFigure 2 in Supplement 1). FUMA identified 83 independent genomic risk loci as pleiotropic loci, involving 54 unique chromosomal regions (Figure 2 and eTables 4 and 5 in Supplement 1). Some pleiotropic regions were identified in multiple pairwise traits, such as 11q23.2 (mapped gene: NCAM1 [OMIM 116930]) and 1q32.1 (mapped gene: INAVA [OMIM 618051]), suggesting the extensive pleiotropic effects of these loci. The top SNVs in 83 FUMA-annotated pleiotropic loci showed mixed directions of allelic associations (eTable 6 in Supplement 1). Overall, 53 of 83 top SNVs (64%) showed concordant associations with a certain pair of traits, indicating these variants could simultaneously decrease or increase the risk of gastrointestinal tract diseases and psychiatric disorders. The remaining 30 of 83 top SNVs (36%) showed discordant associations, suggesting the possibly distinct biological mechanisms.
Figure 2. The Overall Landscape of the Pleiotropic Associations Across 4 Gastrointestinal Tract Diseases and 6 Psychiatric Disorders.
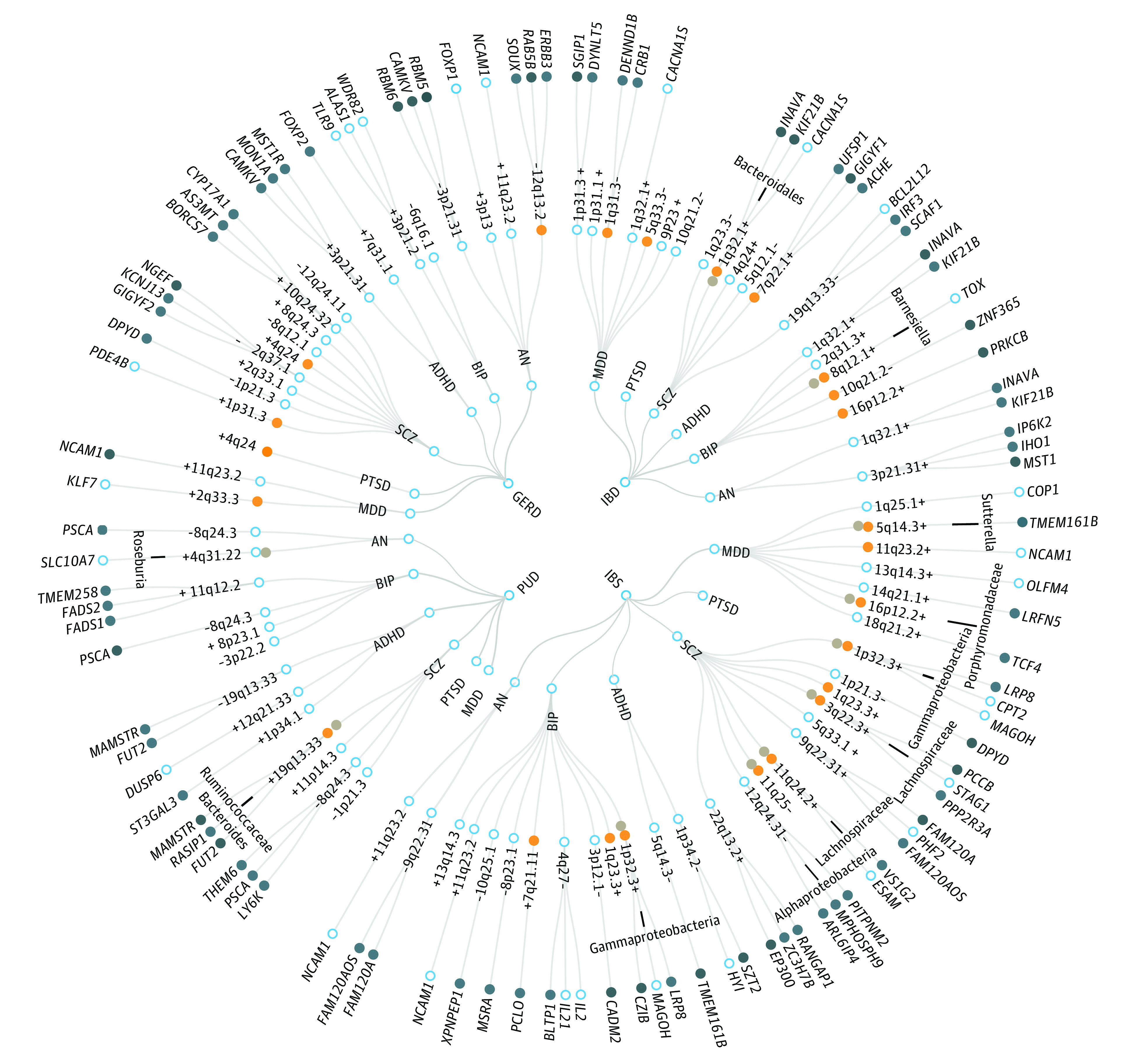
A circular dendrogram included 4 gastrointestinal tract diseases (inner circle, including gastroesophageal reflux disease [GERD], inflammatory bowel disease [IBD], irritable bowel syndrome [IBS], and peptic ulcers disease [PUD]) and 6 psychiatric disorders (second circle; attention-deficit/hyperactivity disorder [ADHD], anorexia nervosa [AN], bipolar disorder [BIP], major depressive disorder [MDD], posttraumatic stress disorder [PTSD], and schizophrenia [SCZ]), resulting in 24 trait pairs. A total of 83 pleiotropic loci were identified across 19 trait pairs (third circle; no pleiotropic loci were identified for IBS-PTSD, PUD-MDD, and PUD-PTSD), in which 53 of 83 top single nucleotide variations (SNVs) showed concordant associations (marked by a plus sign) with a certain pair of traits while the remaining top SNVs showed discordant associations (marked by a minus sign); 24 pleiotropic loci were colocalized for pairwise traits (orange dots, posterior probability of H4 [PP.H4]>0.7); and 11 were colocalized for certain gastrointestinal tract diseases, psychiatric disorders, and gut microorganisms (gray dots, PP>0.7, with the specific microorganism annotated on the lines). A total of 196 significant pleiotropic genes (158 unique) were further identified by multimarker analysis of GenoMic annotation (MAGMA). For the trait pairs with more than 3 pleiotropic genes, we only showed the top 3 pleiotropic genes according to the prioritization of candidate pleiotropic genes, which were shown with the statistical prioritization attenuation manifested by clockwise direction (fourth circle), the genes with the tissue specificity in at least one of gastrointestinal tract or brain tissues and in other tissues identified by E-MAGMA were also highlighted (dark blue dots). Detailed information for 83 pleiotropic loci (eTable 6 in Supplement 1), colocalization results (Table 2), multitrait colocalization results (eTable 17 Supplement 1), significant pleiotropic genes with annotations of tissue specificity (eTable 15 in Supplement 1) was also provided.
ANNOVAR category annotation illustrated that 34 of 83 index SNVs (41%) were intronic variants and 27 of 83 (33%) were intergenic variants. Only 7 of 83 index SNVs (8%) (6 unique) were exonic variants, including 5 messenger RNA (mRNA) exonic variants and 2 noncoding RNA exonic variants (eTable 5 in Supplement 1). Of note, 5 mRNA exonic variants (4 unique) were located in 3 loci and mapped to 3 genes. Specifically, the index SNV rs20551 at 22q13.2 locus (PLACO P = 5.07 × 10−11 for IBS-schizophrenia) was a significant eQTL for EP300 (OMIM 602700) encoding p300 protein, which plays an important role in cell proliferation and differentiation. The index SNVs rs681343 (PLACO P = 1.36 × 10−12 for PUD-schizophrenia) and rs601338 (PLACO P = 5.07 × 10−11 for PUD-ADHD) at 22q13.2 were in high LD (r2 = 0.996) and had almost identical eQTL regulation information (eTable 7 in Supplement 1), regulating the expression of FUT2 (OMIM 182100). The index SNV rs13107325 at 4q24 locus (PLACO P = 1.78 × 10−14 for GERD-schizophrenia; PLACO P = 4.19 × 10−8 for GERD-PTSD) was a significant eQTL for SLC39A8 (OMIM 608732), which encodes ZIP8 metal cation transporter. In addition, 7 index SNVs (6 unique) were identified with CADD scores larger than 12.37, in which 2 mRNA exonic variants had higher CADD scores, including rs601338 (CADD score: 52; mapped gene: FUT2 [OMIM 182100]) and rs13107325 (CADD score: 34; mapped gene: SLC39A8).
Further colocalization analysis identified 24 of 83 potential pleiotropic loci (29%) with PP.H4 larger than 0.7, in which 22 top SNVs of corresponding loci were identified as candidate shared causal variants (Table 2, Figure 2, and eFigure 3 in Supplement 1). Interestingly, the 19q13.33 locus, which was identified as pleiotropic loci for 2 pairs of traits, was only colocalized between PUD and schizophrenia (PP.H4 = 0.8927) rather than PUD and ADHD (PP.H4 = 0.3552), with the same potential shared causal variant rs681343 identified (mapped gene: FUT2). In addition, 7 pleiotropic loci were identified with PP.H3 larger than 0.7000, indicating there might be different causal variants in these loci (eTable 8 in Supplement 1).
Table 2. 24 Colocalized Loci Identified by Colocalization Analysis Performed on 83 Pleiotropic Loci.
| Trait pair | Top SNV | Locus boundarya | Region | Nearest gene | PP.H3 | PP.H4 | Best causal | SNV.PP.H4 |
|---|---|---|---|---|---|---|---|---|
| IBD-MDD | rs12118513 | 1:197342380–197781198 | 1q31.3 | DENND1B | 0.0395 | 0.7372 | rs12118513b | 0.2022 |
| IBD-MDD | rs60689680 | 5:158827769–158856513 | 5q33.3 | AC008703.1 | 0.0614 | 0.7034 | rs60689680b | 0.1184 |
| IBD-SCZ | rs905634 | 1:200874229–201027055 | 1q32.1 | INAVA | 0.0813 | 0.9139 | rs905634b | 0.4835 |
| IBD-SCZ | rs492430 | 7:100219167–100523241 | 7q22.1 | EPO | 0.0801 | 0.9116 | rs492430b | 0.1452 |
| IBD-BIP | rs56073120 | 8:59800835–59925249 | 8q12.1 | TOX | 0.0435 | 0.9044 | rs56073120b | 0.0632 |
| IBD-BIP | rs7090073 | 10:64387108–64441247 | 10q21.2 | ZNF365 | 0.0257 | 0.9719 | rs7090073b | 0.1992 |
| IBD-BIP | rs196001 | 16:23892887–23962504 | 16p12.2 | PRKCB | 0.0656 | 0.8724 | rs196001b | 0.0811 |
| IBS-MDD | rs3099439 | 5:87514778–87822672 | 5q14.3 | TMEM161B | 0.0536 | 0.9428 | rs3099439b | 0.2131 |
| IBS-MDD | rs4937872 | 11:112826867–112912811 | 11q23.2 | RP11-629G13.1 | 0.0845 | 0.9030 | rs4937872b | 0.1163 |
| IBS-MDD | rs1862743 | 16:60665658–60743834 | 16p12.2 | GNPATP | 0.0108 | 0.9578 | rs1862743b | 0.4157 |
| IBS-SCZ | rs12031155 | 1:53658317–53752134 | 1p32.3 | LRP8 | 0.0518 | 0.9368 | rs12031155b | 0.1577 |
| IBS-SCZ | rs7542202 | 1:163616199–163766672 | 1q23.3 | RP4-640E24.1 | 0.0550 | 0.9299 | rs7542202b | 0.0691 |
| IBS-SCZ | rs1280622 | 3:135807609–136673157 | 3q22.3 | RP11-731C17.1 | 0.1145 | 0.8386 | rs7432375 | 0.0965 |
| IBS-SCZ | rs11604175 | 11:124619407–124624854 | 11q24.2 | VSIG2 | 0.0011 | 0.7717 | rs11604175b | 0.5730 |
| IBS-SCZ | rs12277680 | 11:134576216–134595774 | 11q25 | RP11-469N6.2 | 0.0106 | 0.7331 | rs12277680b | 0.4057 |
| IBS-BIP | rs5177 | 1:53658317–53752134 | 1p32.3 | LRP8 | 0.0553 | 0.8665 | rs5177b | 0.1081 |
| IBS-BIP | rs2345964 | 1:163582980–163768927 | 1q23.3 | RP4-640E24.1 | 0.0254 | 0.9684 | rs2345964b | 0.1043 |
| IBS-BIP | rs13239217 | 7:82387493–82583609 | 7q21.11 | PCLO | 0.0939 | 0.8493 | rs13239217b | 0.1919 |
| PUD-SCZ | rs681343 | 19:49103447–49254955 | 19q13.33 | FUT2 | 0.0951 | 0.8927 | rs681343b | 0.2834 |
| GERD-MDD | rs1263674 | 2:208017033–208088987 | 2q33.3 | AC007879.1:AC007879.2 | 0.0567 | 0.8596 | rs62188630 | 0.0638 |
| GERD-PTSD | rs13107325 | 4:102938709–103438709 | 4q24 | SLC39A8 | 0.0111 | 0.7904 | rs13107325b | 0.7793 |
| GERD-SCZ | rs1892346 | 1:66304167–66333877 | 1p31.3 | PDE4B | 0.0210 | 0.8141 | rs1892346b | 0.0854 |
| GERD-SCZ | rs13107325 | 4:102702364–103387161 | 4q24 | SLC39A8 | 0.0020 | 0.9966 | rs13107325b | 0.8466 |
| GERD-AN | rs1873914 | 12:56368708–56478658 | 12q13.2 | RAB5B | 0.0484 | 0.8134 | rs1873914b | 0.1042 |
Abbreviations: ADHD, attention-deficit/hyperactivity disorder; AN, anorexia nervosa; Best causal, the candidate causal single-nucleotide variation (SNV); BIP, bipolar disorder; GERD, gastroesophageal reflux disease; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome; MDD, major depressive disorder; PP.H3, posterior probability of H3; PP.H4, posterior probability of H4; PTSD, posttraumatic stress disorder; PUD, peptic ulcer disease; SCZ, schizophrenia; SNV.PP.H4, posterior probability of the best causal variant.
Locus boundary of each pleiotropic genomic risk locus was denoted as “chromosome: start-end” defined by FUMA for the corresponding trait pair.
The top SNV in this locus was also identified as a candidate causal SNV.
Prioritization of Candidate Pleiotropic Genes and Characterization of Phenotype and Tissue Specificity
MAGMA analysis based on 295 potential pleiotropic genes that located in or overlapped with 83 pleiotropic loci identified 196 significant pleiotropic genes (158 unique), in which 38 genes were detected in 2 or more trait pairs (eTable 9 in Supplement 1). For example, NCAM1 was identified as a significant pleiotropic gene in 5 pairs of traits, followed by INAVA, CACNA1S (OMIM 114208), and KIF21B (OMIM 608322) in 4 pairs of traits and BANK1 (OMIM 610292), CAMKV (OMIM 614993), DPYD (OMIM 612779), MON1A (OMIM 611464), MST1R (OMIM 600168), PSCA (OMIM 602470), RBM5 (OMIM 606884), RBM6 (OMIM 606886), and SLC39A8 in 3 pairs of traits.
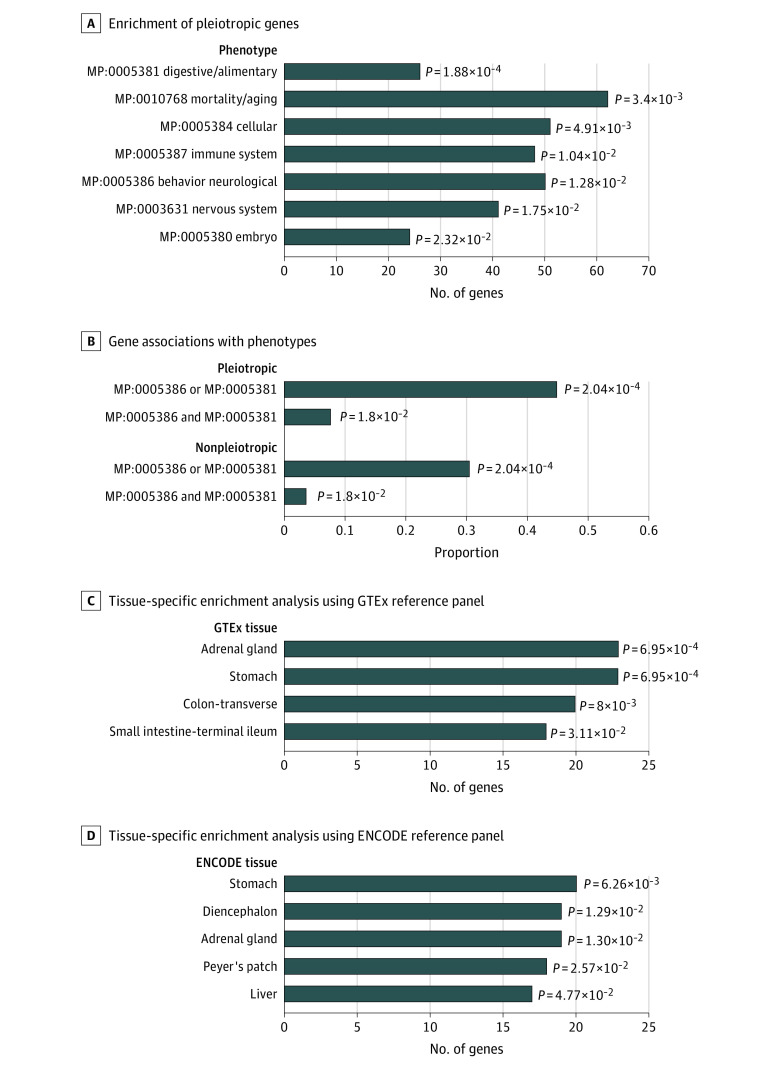
Further parallel phenotype and tissue-specific enrichment analysis suggested both higher phenotype and tissue specificity (Figure 3). Specifically, we obtained in total 19 326 genes with available phenotype annotations resorting to Mouse Genome Informatics platform, in which 144 of 158 unique pleiotropic genes were included. These pleiotropic genes were associated with several GBA-related phenotypes with the analyses performed on each phenotype (Figure 3A) and were associated with the 2 phenotypes simultaneously (behavior-neurological phenotype and digestive-alimentary phenotype), contrasting with that of nonpleotropic genes with less association with these phenotypes (7.64% vs 3.61%; P = 1.80 × 10−2) (Figure 3B). Details of these genes were provided in eTable 10 in Supplement 1. Tissue-specific enrichment analysis using deTS based on GTEx and ENCODE reference panels suggested higher tissue specificity (Figure 3) in several gastrointestinal tract and brain tissues, such as stomach, transverse colon, terminal ileum of small intestine, Peyer’s patch, and diencephalon. The hypothalamic-pituitary-adrenal axis is an important component of the GBA and could provide primary biological response to stressful stimuli, with adrenal gland especially highlighted. Details of these tissue-specific genes were also provided (eFigure 4 and eTables 11 and 12 in Supplement 1).
Figure 3. Phenotype and Tissue Specificity for Candidate Pleiotropic Genes.
Phenotype enrichment analysis based on existing mouse/human orthology with phenotype annotations from the Mouse Genome Informatics platform suggested 7 phenotypes in which the significant pleiotropic genes were enriched (A) and both the proportions of the genes associated with at least 1 of 2 gut-brain axis–related phenotypes or associated with both phenotypes (behavior-neurological phenotype and digestive-alimentary phenotype) in the pleiotropic gene group were higher (B). Detailed information of phenotype annotations for these genes was provided in eTable 10 in Supplement 1. Tissue-specific enrichment analysis using the deTS method based on 2 reference panels showed higher tissue specificity, 4 significantly enriched tissues were identified when using the Genotype-Tissue Expression project (GTEx) reference panel (C), and 5 were identified when using the Encyclopedia of DNA Elements project (ENCODE) reference panel (D). Detailed information of tissue-specific genes was provided in eTables 11 and 12 in Supplement 1. We declared the nominally significant threshold (2-sided P < .05) in both analyses.
In addition, E-MAGMA and joint-tissue imputation analysis also confirmed the tissue specificity not only in gastrointestinal tract and brain tissues but also in other GBA-related tissues. In total, 52 pleiotropic genes (22 unique) across 8 pleiotropic loci were significantly detected in multiple GBA-related tissues in at least 2 trait pairs, for example, PSCA (8q24.3) for 3 pairwise traits (PUD-schizophrenia, PUD-BIP, and schizophrenia-AN), LPR8 (1p32.3) for 2 pairwise traits (IBS-schizophrenia and IBS-BIP), and FUT2 (19q13.33) for 2 pairwise traits (PUD-schizophrenia and PUD-ADHD). H-MAGMA analysis further suggested the cell-type specificity of these pleiotropic genes. Details of these gene-level analyses were also provided (eTables 13-15 in Supplement 1).
Synaptic and Immune-Related Mechanisms Shared Between Gastrointestinal Tract Diseases and Psychiatric Disorders
Gene set enrichment analysis identified 127 significantly enriched pathways (normalized enrichment score >2 and adjusted P < .05), including 99 Gene Ontology (GO) terms and 28 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (eTable 16 in Supplement 1). These enriched pathways are mainly involved in cell adhesion, synaptic structure and function, and immune cell differentiation. For example, homophilic cell adhesion via plasma membrane adhesion molecules (GO:0007156), which plays important roles in neural and immune mechanisms, was identified in 5 pairs of traits. A γ-aminobutyric acid (GABA)–modifying synapse (GO:0098982), which typically functions as an inhibitory synapse using GABA as neurotransmitter, was significantly enriched in 3 pairs of traits. Notably, both T helper 1 (TH1) and TH2 cell differentiation (KEGG has04658) and TH17 cell differentiation (KEGG has04659), which involve in immunoinflammatory responses, were highlighted in 2 pairs of traits.
Multitrait Colocalization Analysis to Pinpoint Critical Gut Microbiomes
Multitrait colocalization analysis from HyPrColoc highlighted 15 pleiotropic loci (11 unique) that were colocalized to share a causal variant, supporting the significant role of gut microorganisms (Figure 2 and eTable 17 in Supplement 1). Seven of 11 unique loci were identified to be colocalized among schizophrenia, gastrointestinal tract diseases, and gut microorganisms. For example, 19q13.33 was particularly colocalized between PUD and schizophrenia not only with the prevalence of Ruminococcaceae species but with the abundance of Bacteroides species, where the index SNV rs681343 (mapped gene: FUT2) was also identified as shared causal variant.
Mendelian Randomization and Associations Between Gastrointestinal Tract Diseases and Psychiatric Disorders
Bidirectional mendelian randomization analyses using inverse-variance weighted method showed 11 significant positive associations (eFigure 5 and eTable 18 in Supplement 1). In addition, no significant bidirectional associations were detected in negative control analysis (eTable 19 in Supplement 1). The results from several alternative mendelian randomization methods were largely consistent with that from main analysis (eTables 18 and 19 in Supplement 1). Notably, among 5 pairwise traits with potential sample overlap, LHC-MR further validated the results (eTable 20 in Supplement 1).
Discussion
In this genome-wide pleiotropic association study, we found extensive genome-wide genetic correlations and genetic overlaps between gastrointestinal tract diseases and psychiatric disorders. Further comprehensive analyses highlighted the pleiotropic genetic variants and loci, potential shared causal variants, pleiotropic genes, biological pathways, and a potential genetic basis associated with gut microbiome. All these findings supported the role of GBA in the shared genetic etiology underlying these 2 types of diseases.
Using multiple methods with different model assumptions could provide complementary evidence and allow deep investigation of the underlying pleiotropic associations from different perspectives. The GPA identified substantial genetic overlap between several trait pairs even without any significant genetic correlation. The mixed directions across the genomic risk loci, which indicate the existence of concordant and discordant pleiotropy, also help explain the polygenic overlap despite nonsignificant genetic correlation. PLACO analysis further determines the pleiotropic variants, followed by colocalization analysis to identify potential shared causal variants in each pleiotropic locus. Mendelian randomization analysis could detect causal trait pairs and partially reflect vertical pleiotropy. Of note, we conducted LDSC to examine potential sample overlap and performed pleiotropic analysis using PLACO as well as mendelian randomization analysis using LHC-MR to alleviate sample overlap issue. The LDSC and mendelian randomization analyses were of comparable consistency with previous studies, with detailed comparisons provided (eTables 21-23 in Supplement 1).
Overall, pleiotropic variants between gastrointestinal tract diseases and psychiatric disorders were extensively distributed, with several loci especially highlighted between certain trait pairs, such as 1q32.1 (INAVA), 19q13.33 (FUT2), 11q23.2 (NCAM1), and 1p32.3 (LRP8). Several loci previously identified to be associated with gastrointestinal tract diseases were illustrated to be potential pleiotropic loci shared with psychiatric disorders and vice versa (eDiscussion in Supplement 1). The pleiotropic genes were more likely to be enriched in GBA-related phenotypes, such as behavioral, neurological, and digestive phenotypes, as well as GBA-related tissues, especially gastrointestinal and brain tissues.
Shared genetic determinants also reflect common biological pathways, among which the TH1, TH2, and TH17 cell differentiation pathways were highlighted between IBD and schizophrenia, BIP, and AN. The TH17 cells play important roles in the development of inflammatory responses and autoimmune diseases.56,57,58 Previous studies suggested the significant role of dysfunction of TH17 cell differentiation and the accumulation of TH17-related cytokines in the pathological process of IBD.59,60 In addition, TH17 cells and related cytokines would provoke CNS neuroinflammation and neurotoxicity under pathological status and disrupt the blood-brain barrier and thus alter the permeability of the blood-brain barrier.61,62 Bacterially produced bile acid metabolites have been shown to inhibit TH17 cell differentiation, which may be related to the pathophysiology of IBD.63 Therefore, TH17 cell differentiation and function play a vital role in the regulation of brain-gut-microbiome axis.
Limitations
Our study is not without limitations. First, we were unable to assess the causal effects of gut microbiome on gastrointestinal tract diseases or psychiatric disorders through mendelian randomization analyses since SNVs with associated gut microorganisms were hard to obtain due to the limited sample size of gut microbiome GWAS. Therefore, caution should be used on the interpretation of the role of gut microorganisms. Second, our study was restricted to European ancestry and may not generalize to other ancestries.
Conclusions
Given strong evidence of genetic correlation and genetic overlap between gastrointestinal tract diseases and psychiatric disorders, we found that pleiotropic genetic variants, loci, and genes were extensively distributed across the genome, with higher phenotype and tissue specificity. We highlighted some genetic determinants previously associated with gastrointestinal tract diseases that were shared with psychiatric disorders and vice versa. More importantly, shared biological mechanisms concerning immune response, synaptic structure and function, and potential gut microbiome were identified. Our findings not only support the shared genetic basis underlying GBA but provide novel insight into the intervention and treatment targets of these diseases.
eMethods, Study Analyses
eDiscussion. Study Outcomes
eFigure 1. Fourteen Pairs of Traits With Significant Genetic Correlations Identified by Both HDL and LDSC Methods
eFigure 2. Quantile-Quantile (Q-Q) Plots of PLACO Results for 22 Pairwise Traits
eFigure 3. LocusZoom and LocusCompare Plots of 24 Significantly Colocalized Loci
eFigure 4. Gene Expression Heatmap of 158 Significant Pleiotropic Genes in 25 Tissues
eFigure 5. The Bidirectional Causal Effects Estimated by IVW Method
eTable 1. Details of GWAS Summary Data Sources
eTable 2. Genetic Correlations Between 4 Gastrointestinal Tract Diseases and 6 Psychiatric Disorders Estimated by HDL
eTable 3. Bivariate LDSC Estimates in Negative Control Analysis
eTable 4. Summary of Genome-Wide Significant Pleiotropic SNVs and FUMA-Annotated Pleiotropic Genomic Risk Loci for Each Pair of Traits
eTable 5. 83 Pleiotropic Genomic Loci Identified by FUMA Using PLACO Results
eTable 6. Effect Sizes and P Values of Top SNPs in 83 Pleiotropic Loci From Original GWAS Summary Statistics
eTable 7. The eQTL Regulatory Information of rs601338 and rs681343 on FUT2 Gene in Gastrointestinal Tract and Brain Tissues
eTable 8. The Remaining 59 Loci in Colocalization Analysis
eTable 9. Candidate Pleiotropic Genes Identified by MAGMA
eTable 10. Phenotype Enrichment Results With Existing Phenotype Annotations of the Pleiotropic Genes
eTable 11. Pleiotropic Genes Identified With Tissue Specificity in GTEx Reference Panel
eTable 12. Pleiotropic Genes Identified With Tissue Specificity in ENCODE Reference Panel
eTable 13. Twenty-Five Tissue Types Used for E-MAGMA Analysis
eTable 14. Six Tissue/Cell Types Used for H-MAGMA Analysis
eTable 15. Tissue Specificity and Cell-Type Specificity of the Identified Pleiotropic Genes in E-MAGMA, JTI, and H-MAGMA Analysis
eTable 16. Significantly Enriched GO and KEGG Pathways in GSEA Analysis
eTable 17. Significantly Colocalized Loci Identified by Multitrait Colocalization Using HyPrColoc
eTable 18. Results of Bidirectional Mendelian Randomization Analysis From Main Analysis and Alternative Methods Between 4 Gastrointestinal Diseases and 6 Psychiatric Disorders
eTable 19. Results of Bidirectional Mendelian Randomization Analysis From Main Analysis and Alternative Methods for Negative Control Analysis
eTable 20. Results of Mendelian Randomization Analysis Using LHC-MR Method
eTable 21. Comparisons of Data Sources of Psychiatric Disorders-Related GWAS in Genetic Correlation and Mendelian Randomization Analysis
eTable 22. Summary of Genetic Correlation Results Between 4 Gastrointestinal Tract Diseases and Psychiatric Disorders
eTable 23. Summary of Associations Between Gastrointestinal Tract Diseases and Psychiatric Disorders in Mendelian Randomization Analysis
eReferences.
Data Sharing Statement
References
- 1.Shah E, Rezaie A, Riddle M, Pimentel M. Psychological disorders in gastrointestinal disease: epiphenomenon, cause or consequence? Ann Gastroenterol. 2014;27(3):224-230. [PMC free article] [PubMed] [Google Scholar]
- 2.Person H, Keefer L. Psychological comorbidity in gastrointestinal diseases. Prog Neuropsychopharmacol Biol Psychiatry. 2021;107:110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barberio B, Zamani M, Black CJ, Savarino EV, Ford AC. Prevalence of symptoms of anxiety and depression in patients with inflammatory bowel disease. Lancet Gastroenterol Hepatol. 2021;6(5):359-370. [DOI] [PubMed] [Google Scholar]
- 4.Cryan JF, O’Riordan KJ, Cowan CSM, et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99(4):1877-2013. [DOI] [PubMed] [Google Scholar]
- 5.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125(3):926-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carabotti M, Scirocco A, Maselli MA, Severi C. The gut-brain axis. Ann Gastroenterol. 2015;28(2):203-209. [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Q, Xing C, Long W, Wang HY, Liu Q, Wang RF. Impact of microbiota on central nervous system and neurological diseases. J Neuroinflammation. 2019;16(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan PF. The psychiatric GWAS consortium. Neuron. 2010;68(2):182-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Murray GK, Byrne EM, Sidorenko J, Visscher PM, Wray NR. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat Commun. 2021;12(1):1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tylee DS, Lee YK, Wendt FR, et al. An atlas of genetic correlations and genetically informed associations linking psychiatric and immune-related phenotypes. JAMA Psychiatry. 2022;79(7):667-676. doi: 10.1001/jamapsychiatry.2022.0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tylee DS, Sun J, Hess JL, et al. ; 23 and Me Research Team; Inflammation Working Group of the CHARGE Consortium; METASTROKE Consortium of the International Stroke Genetics Consortium; Netherlands Twin Registry; neuroCHARGE Working Group; Obsessive Compulsive and Tourette Syndrome Working Group of the Psychiatric Genomics Consortium . Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am J Med Genet B Neuropsychiatr Genet. 2018;177(7):641-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pouget JG, Han B, Wu Y, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . Cross-disorder analysis of schizophrenia and 19 immune-mediated diseases identifies shared genetic risk. Hum Mol Genet. 2019;28(20):3498-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan LE, Shen H, Ballon JS, Hardy KV, Noordsy DL, Levinson DF. Genetic correlation profile of schizophrenia mirrors epidemiological results and suggests link between polygenic and rare variant (22q11.2) cases of schizophrenia. Schizophr Bull. 2018;44(6):1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Lu Q, Ye Y, et al. SUPERGNOVA: local genetic correlation analysis reveals heterogeneous etiologic sharing of complex traits. Genome Biol. 2021;22(1):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, Yang C, Gelernter J, Zhao H. Pervasive pleiotropy between psychiatric disorders and immune disorders revealed by integrative analysis of multiple GWAS. Hum Genet. 2015;134(11-12):1195-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uellendahl-Werth F, Maj C, Borisov O, et al. Cross-tissue transcriptome-wide association studies identify susceptibility genes shared between schizophrenia and inflammatory bowel disease. Commun Biol. 2022;5(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu S, He M, Liu Z, Qin Z, Wang Z, Duan L. Shared genetic susceptibilities for irritable bowel syndrome and depressive disorder in Chinese patients uncovered by pooled whole-exome sequencing. J Adv Res. 2020;23:113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J, Xu Z, Noordam R, van Heemst D, Li-Gao R. Depression and inflammatory bowel disease. J Crohns Colitis. 2022;16(4):633-642. [DOI] [PubMed] [Google Scholar]
- 20.Qian L, He X, Gao F, et al. Estimation of the bidirectional relationship between schizophrenia and inflammatory bowel disease using the mendelian randomization approach. Schizophrenia (Heidelb). 2022;8(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadik A, Dardani C, Pagoni P, et al. ; iPSYCH Autism Spectrum Disorder Working Group . Parental inflammatory bowel disease and autism in children. Nat Med. 2022;28(7):1406-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eijsbouts C, Zheng T, Kennedy NA, et al. ; 23andMe Research Team; Bellygenes Initiative . Genome-wide analysis of 53 400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat Genet. 2021;53(11):1543-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14(7):483-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sivakumaran S, Agakov F, Theodoratou E, et al. Abundant pleiotropy in human complex diseases and traits. Am J Hum Genet. 2011;89(5):607-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray D, Chatterjee N. A powerful method for pleiotropic analysis under composite null hypothesis identifies novel shared loci between type 2 diabetes and prostate cancer. PLoS Genet. 2020;16(12):e1009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackinger S, Zeggini E. Statistical methods to detect pleiotropy in human complex traits. Open Biol. 2017;7(11):170125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee CH, Shi H, Pasaniuc B, Eskin E, Han B. PLEIO: a method to map and interpret pleiotropic loci with GWAS summary statistics. Am J Hum Genet. 2021;108(1):36-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10(11):e1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardiñas AF, Holmans P, Pocklington AJ, et al. ; GERAD1 Consortium; CRESTAR Consortium . Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50(3):381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullins N, Forstner AJ, O’Connell KS, et al. ; HUNT All-In Psychiatry . Genome-wide association study of more than 40 000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53(6):817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard DM, Adams MJ, Clarke TK, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demontis D, Walters RK, Martin J, et al. ; ADHD Working Group of the Psychiatric Genomics Consortium (PGC); Early Lifecourse & Genetic Epidemiology (EAGLE) Consortium; 23andMe Research Team . Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51(1):63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nievergelt CM, Maihofer AX, Klengel T, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10(1):4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson HJ, Yilmaz Z, Thornton LM, et al. ; Anorexia Nervosa Genetics Initiative; Eating Disorders Working Group of the Psychiatric Genomics Consortium . Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019;51(8):1207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkler TW, Grassmann F, Brandl C, et al. Genome-wide association meta-analysis for early age-related macular degeneration highlights novel loci and insights for advanced disease. BMC Med Genomics. 2020;13(1):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe K, Stringer S, Frei O, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet. 2019;51(9):1339-1348. [DOI] [PubMed] [Google Scholar]
- 38.Amare AT, Vaez A, Hsu YH, et al. Bivariate genome-wide association analyses of the broad depression phenotype combined with major depressive disorder, bipolar disorder or schizophrenia reveal eight novel genetic loci for depression. Mol Psychiatry. 2020;25(7):1420-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choquet H, Melles RB, Anand D, et al. ; 23andMe Research Team . A large multiethnic GWAS meta-analysis of cataract identifies new risk loci and sex-specific effects. Nat Commun. 2021;12(1):3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ning Z, Pawitan Y, Shen X. High-definition likelihood inference of genetic correlations across human complex traits. Nat Genet. 2020;52(8):859-864. [DOI] [PubMed] [Google Scholar]
- 42.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10(5):e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blake JA, Baldarelli R, Kadin JA, Richardson JE, Smith CL, Bult CJ; Mouse Genome Database Group . Mouse Genome Database (MGD): knowledgebase for mouse-human comparative biology. Nucleic Acids Res. 2021;49(D1):D981-D987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pei G, Dai Y, Zhao Z, Jia P. deTS: tissue-specific enrichment analysis to decode tissue specificity. Bioinformatics. 2019;35(19):3842-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Consortium GT; GTEx Consortium . The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580-585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium EP; ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerring ZF, Mina-Vargas A, Gamazon ER, Derks EM. E-MAGMA: an eQTL-informed method to identify risk genes using genome-wide association study summary statistics. Bioinformatics. 2021;37(16):2245-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou D, Jiang Y, Zhong X, Cox NJ, Liu C, Gamazon ER. A unified framework for joint-tissue transcriptome-wide association and mendelian randomization analysis. Nat Genet. 2020;52(11):1239-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sey NYA, Hu B, Mah W, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat Neurosci. 2020;23(4):583-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foley CN, Staley JR, Breen PG, et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat Commun. 2021;12(1):764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darrous L, Mounier N, Kutalik Z. Simultaneous estimation of bi-directional causal effects and heritable confounding from GWAS summary statistics. Nat Commun. 2021;12(1):7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J, Sundrud MS, Skepner J, Yamagata T. Targeting TH17 cells in autoimmune diseases. Trends Pharmacol Sci. 2014;35(10):493-500. [DOI] [PubMed] [Google Scholar]
- 57.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and TH17 cells. Annu Rev Immunol. 2009;27(1):485-517. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J, Lu Q, Liu Y, et al. TH17 cells in inflammatory bowel disease: cytokines, plasticity, and therapies. J Immunol Res. 2021;2021:8816041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rovedatti L, Kudo T, Biancheri P, et al. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut. 2009;58(12):1629-1636. [DOI] [PubMed] [Google Scholar]
- 60.Fujino S, Andoh A, Bamba S, et al. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52(1):65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kebir H, Kreymborg K, Ifergan I, et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med. 2007;13(10):1173-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huppert J, Closhen D, Croxford A, et al. Cellular mechanisms of IL-17-induced blood-brain barrier disruption. FASEB J. 2010;24(4):1023-1034. [DOI] [PubMed] [Google Scholar]
- 63.Paik D, Yao L, Zhang Y, et al. Human gut bacteria produce TH17-modulating bile acid metabolites. Nature. 2022;603(7903):907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods, Study Analyses
eDiscussion. Study Outcomes
eFigure 1. Fourteen Pairs of Traits With Significant Genetic Correlations Identified by Both HDL and LDSC Methods
eFigure 2. Quantile-Quantile (Q-Q) Plots of PLACO Results for 22 Pairwise Traits
eFigure 3. LocusZoom and LocusCompare Plots of 24 Significantly Colocalized Loci
eFigure 4. Gene Expression Heatmap of 158 Significant Pleiotropic Genes in 25 Tissues
eFigure 5. The Bidirectional Causal Effects Estimated by IVW Method
eTable 1. Details of GWAS Summary Data Sources
eTable 2. Genetic Correlations Between 4 Gastrointestinal Tract Diseases and 6 Psychiatric Disorders Estimated by HDL
eTable 3. Bivariate LDSC Estimates in Negative Control Analysis
eTable 4. Summary of Genome-Wide Significant Pleiotropic SNVs and FUMA-Annotated Pleiotropic Genomic Risk Loci for Each Pair of Traits
eTable 5. 83 Pleiotropic Genomic Loci Identified by FUMA Using PLACO Results
eTable 6. Effect Sizes and P Values of Top SNPs in 83 Pleiotropic Loci From Original GWAS Summary Statistics
eTable 7. The eQTL Regulatory Information of rs601338 and rs681343 on FUT2 Gene in Gastrointestinal Tract and Brain Tissues
eTable 8. The Remaining 59 Loci in Colocalization Analysis
eTable 9. Candidate Pleiotropic Genes Identified by MAGMA
eTable 10. Phenotype Enrichment Results With Existing Phenotype Annotations of the Pleiotropic Genes
eTable 11. Pleiotropic Genes Identified With Tissue Specificity in GTEx Reference Panel
eTable 12. Pleiotropic Genes Identified With Tissue Specificity in ENCODE Reference Panel
eTable 13. Twenty-Five Tissue Types Used for E-MAGMA Analysis
eTable 14. Six Tissue/Cell Types Used for H-MAGMA Analysis
eTable 15. Tissue Specificity and Cell-Type Specificity of the Identified Pleiotropic Genes in E-MAGMA, JTI, and H-MAGMA Analysis
eTable 16. Significantly Enriched GO and KEGG Pathways in GSEA Analysis
eTable 17. Significantly Colocalized Loci Identified by Multitrait Colocalization Using HyPrColoc
eTable 18. Results of Bidirectional Mendelian Randomization Analysis From Main Analysis and Alternative Methods Between 4 Gastrointestinal Diseases and 6 Psychiatric Disorders
eTable 19. Results of Bidirectional Mendelian Randomization Analysis From Main Analysis and Alternative Methods for Negative Control Analysis
eTable 20. Results of Mendelian Randomization Analysis Using LHC-MR Method
eTable 21. Comparisons of Data Sources of Psychiatric Disorders-Related GWAS in Genetic Correlation and Mendelian Randomization Analysis
eTable 22. Summary of Genetic Correlation Results Between 4 Gastrointestinal Tract Diseases and Psychiatric Disorders
eTable 23. Summary of Associations Between Gastrointestinal Tract Diseases and Psychiatric Disorders in Mendelian Randomization Analysis
eReferences.
Data Sharing Statement