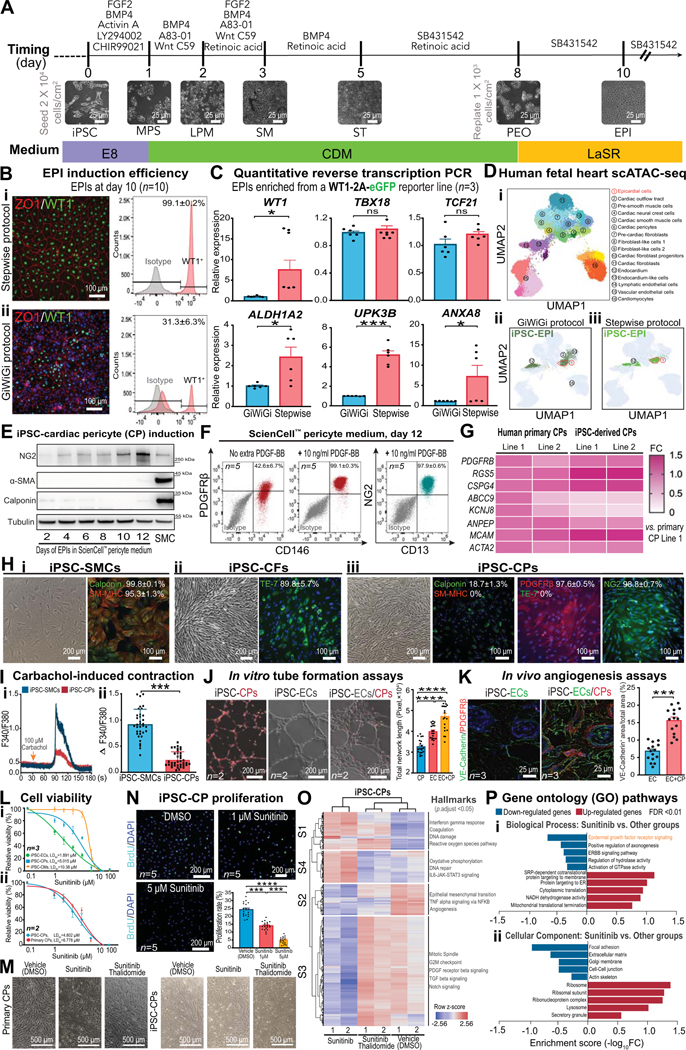
Cardiac pericytes (CPs), a major mural cell type maintaining homeostasis, integrity, and perfusion of the coronary microvasculature, remain the most enigmatic and underappreciated cell population in the heart. Accumulating evidence suggests that CPs play a key role in cardiovascular complications such as coronary vasospasm, no-reflow post myocardial infarction, and cancer drug-induced cardiotoxicity.1 However, the lack of unequivocal cell markers and specific tools for characterization, lineage tracing, and conditional targeting of CPs has precluded a comprehensive understanding of their pathogenic role in coronary microvascular dysfunction. Here, we report a stepwise approach to generate first-of-its-kind CPs from human induced pluripotent stem cells (iPSCs), which were shown to transcriptionally and functionally resemble their primary counterparts.
To derive pure iPSC-CPs, we first generated epicardial cells (EPIs), the predominant progenitor cells giving rise to CPs, in a stepwise fashion (Figure [A]). We found that temporal activation or inhibition of key morphogens at different differentiation stages enabled the generation of pure mesodermal progenitors and EPIs (Figure [Bi]).2 In contrast, using an extant protocol that only manipulated Wnt signaling throughout the entire differentiation process (referred to as GiWiGi protocol),3 we observed a large variation of EPI induction efficiency (ranging from 3%−87%) even among iterative differentiations (Figure [Bii]). Moreover, our stepwise protocol generated more mature EPIs (ALDH1A2, UPK3B, and ANXA8) than did the GiWiGi protocol (Figure [C]). Finally, we performed single-cell ATAC sequencing (scATAC-seq) on stepwise and GiWiGi EPIs and projected them onto human fetal heart scATAC-seq data based on their chromatin landscape similarities.4 Our data showed that >30% of GiWiGi EPIs acquired a fibroblast fate (Figure[Dii]), precluding the generation of pure CPs for cell type-specific studies. In contrast, stepwise EPIs overlapped with primary EPIs and endocardial cells, two progenitors that can give rise to CPs (Figure [Diii]).5
Figure. Phenotypic characterization and functional assessment of stepwise differentiated iPSC-cardiac pericytes.
A, A schematic showing stage-specific inhibition and activation of morphogens to generate pure epicardial cells (EPI) from human induced pluripotent stem cells (iPSCs). Representative bright-field images for each stage of cell differentiation are demonstrated. MPS, mid-primitive streak; LPM, lateral plate mesoderm; SM, splanchnic mesoderm; ST, septum transversum; PEO, pre-epicardial organ. B, Immunofluorescent images (ZO1 and WT1), flow cytometry (WT1) graphs (n=10 iPSC lines, 5M/5F), and quantitative data showing EPI induction efficiency by stepwise (i) and GiWiGi (ii) protocols. C, Quantitative reverse transcription PCR (RT-qPCR) results showing expression levels of canonical (WT1, TBX18, and TCF21) and mature (ALDH1A2, UPK3B, and ANXA8) markers of EPIs derived by both protocols. D, Single-cell ATAC sequencing (scATAC-seq) of EPIs generated by both GiWiGi (ii) and stepwise (iii) protocols are projected to that of human fetal heart cell clusters (i). Dotted frames indicate the EPI cluster in the human fetal heart scATAC-seq UMAP. E, Immunoblots showing time-dependent changes in cardiac pericytes (CP) and smooth muscle cell (SMC) markers during differentiation. iPSC-SMCs were used as a control. F, Flow cytometry and quantitative data (n=5, 2M/3F) showing iPSC-CP yields with and without exogenous platelet-derived growth factor (PDGF)-BB. G, A heatmap showing transcriptomic similarities of pericyte markers between primary and iPSC-CPs by RT-qPCR. H, Bright field and immunofluorescent images of iPSC-derived SMCs, cardiac fibroblasts (CFs), and endothelial cells (ECs). SM-MHC, smooth muscle-myosin heavy chain; NG2, neural/glial antigen 2. I, Calcium imaging using Fura-2 AM to quantitatively compare carbachol-induced intracellular calcium increases in iPSC-SMCs and iPSC-CPs. J, Tubular networks formed by iPSC-CPs, iPSC-ECs, and in combination in vitro (n=2, 1M/1F). iPSC-CPs were fluorescently labeled with Calcein Red-Orange AM. K, Vasculatures formed by iPSC-ECs alone (1.2×106 cells/Matrigel plug) or in combination with iPSC-CPs (1.0×106 iPSC-ECs and 0.2×106 iPSC-CPs/Matrigel plug) after being implanted in male immunodeficiency NOD SCID mice (n=3 per group) for 7 days. Antibodies targeting iPSC-ECs and iPSC-CPs are human-specific. L, Dose-response curves for (i) iPSC-CMs, iPSC-CPs, and iPSC-ECs (n=3, 1M/2F per group) and (ii) primary and iPSC-CPs (n=2, 1M/1F per group) after 72 hr of sunitinib (0 μM-100 μM) treatment using a PrestoBlue viability assay. M, Bright-filed images showing the cell morphology of primary and iPSC-CPs after 72 hr of vehicle, sunitinib (5 μM), and sunitinib (5 μM)/thalidomide (1 μM) treatment. N, BrdU incorporation assays showing suppressed iPSC-CP proliferation by sunitinib (n=5, 2M/3F per group) in a dose-dependent manner. O, A heatmap showing gene clustering patterns of iPSC-CPs treated with vehicle, sunitinib (5 μM), and sunitinib (5 μM)/thalidomide (1 μM) for 72 hr. Key changes of hallmarks in each cluster are highlighted. P, Gene ontology pathway analysis of iPSC-CPs treated with sunitinib versus other conditions. GraphPad Prism 9 was used for statistical analysis. All data are presented as mean ± sem. Statistical significance was assessed using a non-parametric statistical procedure (Wilcoxon signed-rank test for 2 groups and Kruskal-Wallis test for >2 groups). *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. ns indicates not significant.
Next, we differentiated stepwise EPIs in a commercial pericyte medium and observed that it took at least 12 days to generate iPSC-CPs (Figure [E]) with low efficiency (Figure [F]). Since pericyte-endothelial cell (EC) crosstalk and platelet-derived growth factor receptor (PDGFR) signaling are critical for pericyte development, we successfully generated pure iPSC-CPs (PDGFRβ+/CD146+/NG2+/CD13+) with exogenous PDGF-BB, a ligand predominantly secreted by ECs (Figure [F]). Next, we confirmed comparable expression levels of cell markers between primary and iPSC-CPs using quantitative reverse transcription PCR (Figure [G]). Finally, we showed that iPSC-CPs are negative for smooth muscle cell (SMC) markers and not reactive to an anti-fibroblast antibody TE-7 (Figure [H]), further confirming their cell type specificity.
Next, we examined the relative cell contraction capacities of iPSC-CPs versus iPSC-SMCs by measuring intracellular calcium transients after carbachol treatment. While both cell types showed a stereotypical contraction pattern to carbachol, iPSC-CPs had a much smaller calcium amplitude, indicating weaker contraction due to lower intracellular calcium increase and lower expression levels of contractile proteins (Figure [I]). We also tested the pro-angiogenic potentials of iPSC-CPs. We observed that iPSC-CPs not only spontaneously formed a tubular network in vitro, but also improved tube formation in vitro and neovessel maturation in vivo when they were mixed with iPSC-ECs (Figure [J and K]).
A previous study reported that CPs were the primary targets of sunitinib-induced coronary microvascular abnormalities and cardiac dysfunction in mice.1 To validate whether our iPSC-CPs can recapitulate this phenotype, we treated iPSC-CPs, iPSC-ECs, and iPSC-cardiomyocytes (CMs) with a wide dose range of sunitinib. In line with the in vivo findings,1 we observed that iPSC-CPs were more sensitive than iPSC-CMs to sunitinib-induced cell death (Figure [Li]). In contrast, primary and iPSC-CPs showed a similar cytotoxicity profile to sunitinib treatment (Figure [Lii and M]). Sunitinib significantly suppressed iPSC-CP proliferation in a dose-dependent manner (Figure [N]). Notably, thalidomide, a drug that can rescue mice from sunitinib-induced CP loss and cardiac dysfunction,1 profoundly rescued CP death (Figure [M]) and restored genes associated with cell cycle, pericyte function, and DNA damage to a baseline level (Figure [O and P]). Because the precise mechanisms by which sunitinib-induced CP cytotoxicity remain elusive, our iPSC model, if combined with high throughput screening tools, may help identify novel therapeutics to prevent cardiotoxicity induced by sunitinib or other agents that primarily inhibit PDGFR signaling.
In summary, our human iPSC-CPs represent a novel model system essential to understanding coronary microvascular dysfunction. The high resemblance between iPSC-CPs and their in vivo counterparts will help researchers understand genetic or environmental factor-induced coronary microvasculature malformation, spur the development of more effective pro-angiogenic cell therapies for patients experiencing myocardial infarction, and spark novel approaches for drug toxicity evaluation and discovery.
Human heart tissue samples, animals, and iPSCs used in the study were approved by the Stanford Institutional Review Board (#42056), Administrative Panel on Laboratory Animal Care (#34262), and Stem Cell Research Oversight Committee (#563), respectively. All scATAC-seq data can be accessed at GSE181346 and RNA-seq data at GSE210652. Raw data can be made available upon request from the corresponding author.
Acknowledgement
We thank Blake Wu for proof-reading of the manuscript and Dr. Lingfeng Luo for technical support in OCT sample preparation.
Sources of Funding
This study was supported by research grants from Tobacco-Related Disease Research Program (TRDRP) 30FT0852 (M.S.); American Heart Association (AHA) Career Development Award 19CDA34760019 (C.L.); American Heart Association-Allen Initiative 19PABHI34580007 (J.C.W); and the National Institutes of Health (NIH) R01 HL126527, R01 HL130020, R01 HL146690, R01 HL141851, and P01 HL141084 (J.C.W).
J.C.W. is a cofounder of Greenstone Biosciences but has no competing interests, as the work presented was performed independently.
Footnotes
Disclosures
The other authors declare no competing interests.
References
- 1.Chintalgattu V, Rees ML, Culver JC, Goel A, Jiffar T, Zhang J, Dunner K Jr., Pati S, Bankson JA, Pasqualini R, et al. Coronary microvascular pericytes are the cellular target of sunitinib malate-induced cardiotoxicity. Sci Transl Med. 2013;5:187ra169. doi: 10.1126/scitranslmed.3005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen M, Liu C, Wu JC. Generation of embryonic origin-specific vascular smooth muscle cells from human induced pluripotent stem cells. Methods Mol Biol (Clifton, NJ). 2022;2429:233–246. doi: 10.1007/978-1-0716-1979-7_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao X, Lian X, Hacker TA, Schmuck EG, Qian T, Bhute VJ, Han T, Shi M, Drowley L, Plowright A, et al. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat Biomed Eng. 2016;1. doi: 10.1038/s41551-016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameen M, Sundaram L, Shen M, Banerjee A, Kundu S, Nair S, Shcherbina A, Gu M, Wilson KD, Varadarajan A, Vadgama N, Balsubramani A, Wu JC, Engreitz J, Farh K, Karakikes I, Wang KC, Quertermous T, Greenleaf W, Kundaje A. Integrative single cell analysis of human cardiogenesis. Cell. doi: 10.1016/j.cell.2022.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q, Zhang H, Liu Y, Adams S, Eilken H, Stehling M, Corada M, Dejana E, Zhou B, Adams RH. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat Commu. 2016;7:12422. doi: 10.1038/ncomms12422 [DOI] [PMC free article] [PubMed] [Google Scholar]