Abstract
Although the information of cDNAs is indispensable for analyzing gene function, most of the cDNA sequences stored in current databases are imperfect in the sense that they lack the precise information of 5′ end termini. To overcome this difficulty, we have developed the oligo-capping method to obtain full-length cDNAs, the information of which has been partly deposited in public databases. In this study, we further constructed human cDNA libraries enriched in clones containing the cap structure to systematically explore the 5′ end structure of expressed genes. Of approximately 217 402 5′ end sequences obtained, 111 382 have been matched to cDNA sequences of known genes (7889 genes) and are presented in our new database, DataBase of Transcriptional Start Sites (DBTSS; http://elmo.ims.u-tokyo.ac.jp/dbtss/). Sequence comparison between our entries and those of a reference sequence database, RefSeq, revealed that 4683 (34%) of RefSeq sequences should be extended towards the 5′ ends. We also mapped each sequence on the human draft genome sequence to identify its transcriptional start site, which provides us with more detailed information on distribution patterns of transcriptional start sites and adjacent regulatory regions.
INTRODUCTION
Even after the human genome was sequenced as a rough draft (1,2), the need for its cDNA analyses is still unchanged to locate the position of genes and to examine their expression status. Since conventional methods for determining exact transcriptional start sites (TSS), such as 5′ RACE (3,4) or primer extension (5), are laborious, the 5′ end positions of most cDNA sequences deposited so far in public databases are incomplete. In addition, computational methods to predict the location of promoters are still immature (6). Nevertheless, positional data of TSS is a rich source of biological information. First, precise determination of TSS is indispensable to identifying the promoter region, which is located just proximal to TSS in many cases. Secondly, when multiple TSS are observed, it enables us to examine the dynamic nature of the transcriptional initiation events. Loose specification of TSS may reflect slippery interaction between the promoter and the transcription machinery, whereas tight specification may reflect the rigid interaction (7). In any case, it would be intriguing to find correlations between distribution patterns of TSS and their upstream sequences. Finally, since TSS marks the 5′ end limit of the cDNA, the amino acid sequence of the exact N-terminus of the encoding protein could be deduced, which is essential to examine the presence of protein sorting signals.
To obtain the full-length cDNAs, we have developed the ‘oligo-capping’ method (8,9). In this paper, we present a novel database containing the result of systematic 5′ end sequencing of human full-length cDNAs [the DataBase of human Transcriptional Start Sites and full-length cDNAs (DBTSS)]. Our database will be useful not only as a full-length version of the RefSeq database (10,11) but also as a resource for analyzing regulatory information, such as differential TSS and promoter uses.
CONSTRUCTION OF FULL-LENGTH cDNA LIBRARIES
Here we briefly explain the oligo-capping method (reviewed in 8,9). With this method, the cap structure of mRNA is replaced with the synthetic oligonucleotide enzymatically in the following three steps: bacterial alkaline phosphatase hydrolyses the phosphate of truncated mRNA 5′ ends whose cap structures have been broken down. Tobacco acid pyrophosphatase removes the cap structure, leaving the phosphate at the 5′ end. T4 RNA ligase, which requires a phosphate at the 5′ end as its substrate, selectively ligates the 5′ oligo to the 5′ end that originally had the cap structure. Each mRNA product of the ‘oligo-capping’ contains the sequence tags at both ends, which are poly(A) at the 3′ end and the cap-replaced oligo at the 5′ end. With the oligo-capped mRNA as a starting material, a new system is developed to selectively clone the cDNAs that contain both of the sequence tags at the respective ends. Thus, cDNA libraries were constructed in which the content of ‘full-length’ cDNA is significantly enriched (‘full-length-enriched’ cDNA library). We obtained 217 402 sequences derived from 132 libraries. More information of the used libraries is listed in the database. We will continue to produce 5′ end sequences by the oligo-capping method and will update DBTSS accordingly.
SEQUENCE DATA PROCESSING
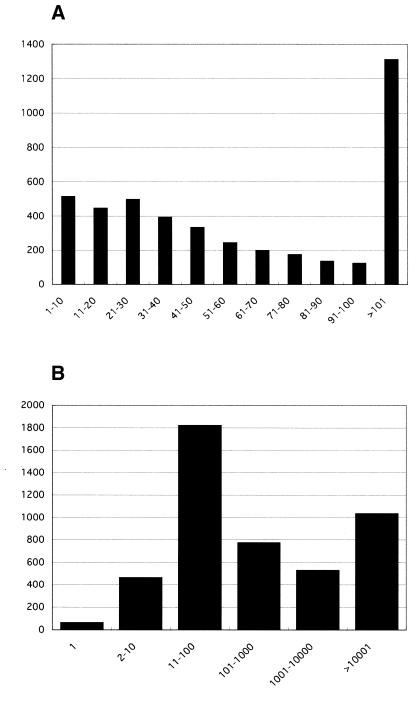
Each sequence produced by the oligo-capping method was first processed to trim its vector site and its low quality parts. Then, they were compared with human reference sequences (RefSeq) using the BLAST program (12). If a homolog was found with >95% identity and less than 10–100 in e-value, it was regarded as identical to the RefSeq sequence. In total, 142 021 sequences were matched to the RefSeq NM (i.e. curated mRNA) human sequences. In order to identify precise TSS information, sequences that have multiple homologs in RefSeq were discarded. Besides, we removed sequences that were not mapped on the human genome working draft sequence (Golden Path: http://genome.ucsc.edu/) database. Using the sim4 program (13), 111 382 sequences were mapped to the human genome sequence. These 111 382 clones were classified into 7889 RefSeq NM entries and are stored in our database. Sequence comparison between these data indicated that 4683 (34%) of the 7889 RefSeq sequences could be extended towards the 5′ ends. Figure 1A shows the length distribution of added sequences. On average, our data extended the RefSeq sequence by 87 bases. The predicted full-length sequences which are the RefSeq sequences elongated with our data are called ‘Ref-Full’ and are obtainable via FTP.
Figure 1.
Histogram of the lengths of elongated sequences. There are 4683 RefSeq genes that are elongated with our data. The vertical axis represents the number of clones while the horizontal axis represents the class of elongated lengths in mRNA level (A) or in genomic level (B).
Figure 1B represents the distribution of the distance between the 5′ ends of Ref-Full and RefSeq on genomic sequence. In many genes, the distance between newly identified TSS as the 5′ end of Ref-Full and the 5′ end of pre-existing RefSeq was so large that our data should be indispensable to identifying the promoter region, which is located adjacent to the TSS.
USAGE OF DBTSS
From our DBTSS web page at http://elmo.ims.u-tokyo.ac.jp/dbtss/, users can retrieve the TSS information of a specified gene in several ways. For example, users can use a gene name search or can directly enter the RefSeq IDs (such as ‘NM_005718’), LocusLink IDs, UniGene IDs or gene symbols. Another way of entry specification is to enter a partial mRNA sequence. Then, entries hit by a BLAST search will be displayed.
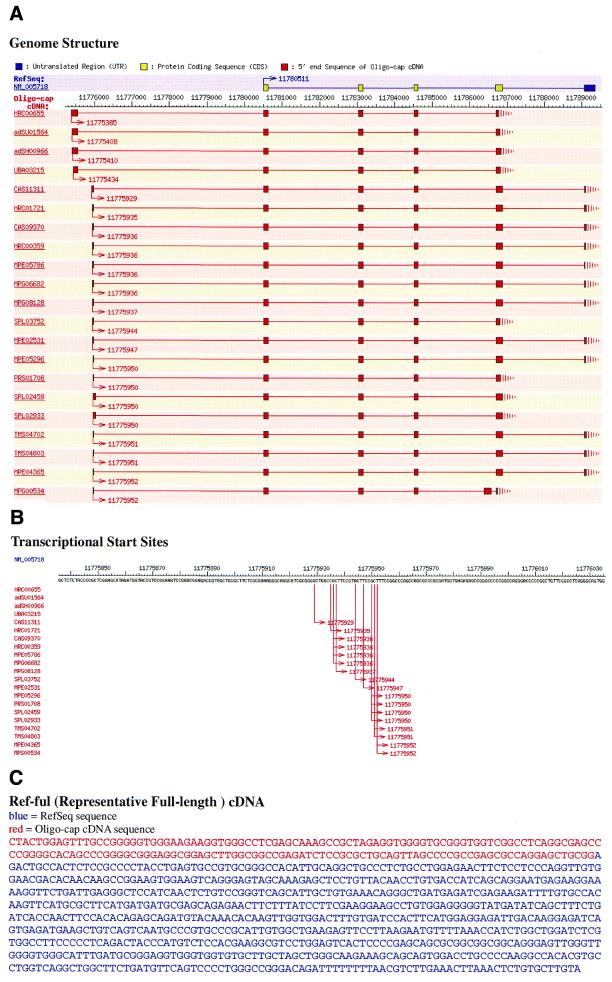
Figure 2 shows an example of NM_005718 (actin related protein 2/3 complex, subunit 4). One of the major contents of the entry is Ref-Full sequence, i.e. representative full-length cDNA sequence (Fig. 2C). While the RefSeq entry NM_005718 starts at position 11 780 511 of chromosome 3, our clone, HRC00655, starts at 11 775 385. Then, we added 175 bases of the 5′ end region from HRC00655 to NM_005718, defining a Ref-Full sequence of NM_005718. Although the 5′ end sequence extended to the RefSeq sequence is short, this part is essential to identify the first exon and adjacent promoter region which are separated from the other exons by a large intron.
Figure 2.
An example of DBTSS web page for NM_005718. (A) Graphical overview of the multiple TSS and exon–intron structures. The top yellow (ORF regions) and blue (5′- and 3′-UTR regions) boxes demonstrate the RefSeq exons, while Red boxes represent our clones. Lines connecting boxes indicate introns and arrows indicate the TSS of each clone. (B) Closer look at the TSS flanking region. (C) Ref-Full sequence of NM_005718.
Another feature of DBTSS is that it enables us to see the exon–intron structures and the distribution patterns of TSS with various clones. For example, Figure 2A shows that there are two types of alternative first exons in this gene. Figure 1B, shows that even in either of the exons, the exact positions of the TSS are highly variable, which is consistent with our recent observations (7). Our database should provide versatile information on the diverse nature of the human gene transcripts.
Acknowledgments
ACKNOWLEDGEMENTS
We thank H. Hata and every member of the HGC-IMSUT sequencing team for their excellent sequencing work. We are also thankful to T. Hasui and J. M. Sugano for helpful discussions, and to Y. Makita for technical support in database construction. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and by special coordination funds for promoting science and technology (SCF), both from the Ministry of Education, Culture, Sports, Science and Technology in Japan.
REFERENCES
- 1.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,H. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Frohman M.A., Dush,M.K. and Martin,G.R. (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl Acad. Sci. USA, 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaefer B.C. (1995) Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem., 227, 255–273. [DOI] [PubMed] [Google Scholar]
- 5.McKnight S.L. and Kingsbury,R. (1982) Transcriptional control signals of a eukaryotic protein-coding gene. Science, 217, 316–324. [DOI] [PubMed] [Google Scholar]
- 6.Ohler U. and Niemann,H. (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet., 17, 56–60. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y., Taira,H., Tsunoda,T., Mizushima-Sugano,J., Sese,J., Hata,H., Ota,T., Isogai,T., Tanaka,T., Morishita,S. et al. (2001) Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep., 2, 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maruyama K. and Sugano,S. (1994) Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene, 138, 171–174. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y., Yoshitomo-Nakagawa,K., Maruyama,K., Suyama,A. and Sugano,S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene, 200, 149–156. [DOI] [PubMed] [Google Scholar]
- 10.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 11.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 17, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]