Abstract
Introduction
Lenvatinib has been widely used for the treatment of advanced hepatocellular carcinoma (HCC). Some adverse events, including diarrhea, have been reported for lenvatinib. Diarrhea may be associated with the changes in the intestinal microbiome; however, the underlying mechanism has not been elucidated.
Aim
In this study, we aimed to investigate the relationship between the intestinal microbiome and diarrhea caused by lenvatinib via analysis of fecal samples collected before treatment.
Methods
A total of 21 patients with advanced HCC who were treated with lenvatinib were enrolled. Fecal samples were collected from patients. The patients were divided into diarrhea (n = 8) and nondiarrhea groups (n = 12). We compared the characteristics of patients, incidence of adverse events, composition of the intestinal microbiome, and enrichment of functional pathways between both groups using QIIME2 and PICRUSt2.
Results
The median age of the two groups was 73 years. The nondiarrhea group comprised a relatively higher number of male patients than the diarrhea group; however, there were no significant differences in patient characteristics between both groups. The proportion of the microbiome was similar, and alpha and beta diversities were not significantly different between both groups. The relative abundance of order Bacteroidales, including Parabacteroides and Prevotella, was higher in the diarrhea group than in the nondiarrhea group. PICRUSt2 analysis showed some metabolic pathways, including butanoate (butyrate) metabolism, were enriched in the nondiarrhea group when compared with those in the diarrhea group.
Conclusion
Differences in the intestinal microbiomes and their functions may influence the incidence of diarrhea during lenvatinib treatment.
Keywords: Lenvatinib, Microbiome, Diarrhea, Hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide, despite the advances in the treatment of infection with hepatitis B virus (HBV) and hepatitis C virus (HCV) [1]. Lenvatinib is widely used for HCC because its effect on overall survival is superior to that of sorafenib [2, 3]. Both lenvatinib and sorafenib are classified as tyrosine kinase inhibitors (TKIs). Adverse events, such as fatigue, anorexia, hand-foot syndrome (HFS), diarrhea, hypertension, hypothyroidism, and urine protein, associated with the use of TKIs have been reported [3, 4, 5]. In certain cases, such adverse events render the continuity of the treatment difficult. It is important to minimize adverse events and maintain liver function for subsequent treatments, such as regorafenib and cabozantinib [6, 7]. It is also necessary to predict the risk factors associated with adverse events, which remain unknown.
Diarrhea is one of the most important adverse events associated with TKIs; however, the underlying mechanism remains unclear. General anticancer drugs can induce diarrhea due to mucosal damage. However, diarrhea caused by TKIs can be attributed to other mechanisms associated with ErbB signaling, chloride secretion, and the intestinal microbiome [8, 9, 10].
In recent years, many reports have been published examining the relationship between chemotherapy and the intestinal microbiome with respect to efficacy and adverse events [11]. Viaud et al. [12] reported a reduction in the efficacy of cyclophosphamide in germ-free or antibiotic-treated mice. Chemotherapy administered in association with bone marrow transplantation reduces the diversity of the microbiome [13]. Intestinal mucositis and cardiomyopathy induced by doxorubicin are related to changes in oral and intestinal flora [14].
We have previously reported the differences in the microbiome associated with HFS and diarrhea in patients treated with sorafenib [15]. We found that some metabolic pathways, such as glycerophospholipid metabolism, may be associated with diarrhea during treatment with sorafenib.
In this study, we investigated the composition of intestinal bacteria before lenvatinib administration and analyzed the relationship between intestinal bacteria and incidence of diarrhea during lenvatinib treatment. To the best of our knowledge, no reports have been published examining the relationship between lenvatinib-induced diarrhea and the intestinal microbiome.
Patients and Methods
Study Design
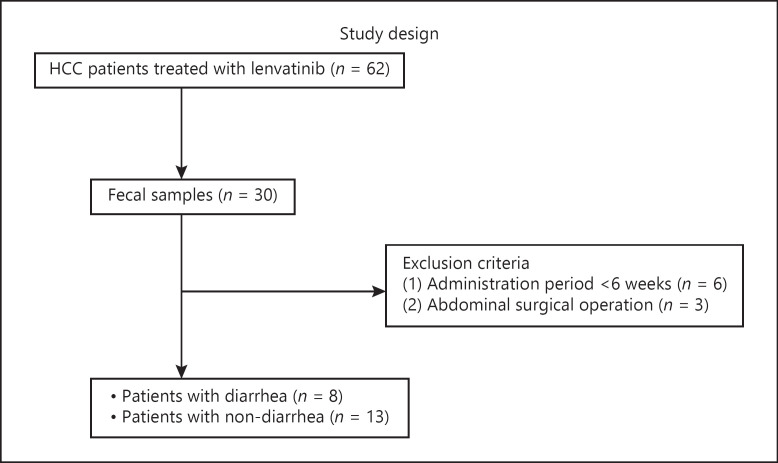
We conducted a single-center, retrospective cohort study. Between March 2018 and December 2020, 62 patients were treated by lenvatinib. We explained the study to all these patients, and then, 30 of these patients agreed to participate in this study. Fecal samples were collected before initiating lenvatinib treatment. Patients who reported diarrhea as an adverse event within the first 6 weeks of treatment were included in the diarrhea group. The remaining patients were assigned to the nondiarrhea group. The study design is illustrated in Figure 1. We analyzed and compared the fecal microbiomes of both groups.
Fig. 1.
Study design. HCC, hepatocellular carcinoma.
All patients provided written informed consent before enrolment in accordance with the Declaration of Helsinki. This study was approved by the ethics of committee of Nagoya University Hospital (approval number: 2015-0420).
Patient Selection and Data Collection
Of the 30 patients, those who had been administered for less than 6 weeks (6 patients) and those who had previously undergone major intestinal surgery (3 patients) were excluded. Finally, data of 21 patients and their fecal samples were analyzed. After initiating lenvatinib treatment, patients visited our hospital once every 2 weeks for blood tests and were examined for adverse events. Adverse events, including diarrhea, were defined using the Common Toxicity Criteria for Adverse Events (CTCAE; version 4.0). Patients who experienced diarrhea were administered a reduced dose of lenvatinib, antidiarrheal medication, and bowel control medication, depending on the severity of diarrhea. A computed tomography scan was performed every 2–3 months to determine the efficacy. Patients in the diarrhea group reported diarrhea with a CTCAE grade of higher than or equal to 1 within 6 weeks of initiating lenvatinib.
Upon initiating lenvatinib treatment, data related to age, sex, body mass index, etiology of liver disease, blood, liver function, HCC stage, adverse events (diarrhea, HFS, hypertension, hypothyroidism, and urine protein), response to treatment, prior TKI use, diabetes mellitus (DM), use of proton pump inhibitors (PPIs), and consumption of pre/probiotics were collected from the electronic medical record system. HCC stage was classified using the Barcelona Clinic Liver Cancer Classification (BCLC) [16], and the liver function was classified using the Child-Pugh score (CPS) [17]. The relative dose intensity (RDI) was calculated for 6 weeks from the beginning of lenvatinib treatment. Response to lenvatinib was classified as complete response, partial response (PR), stable disease (SD), or progressive disease (PD) according to Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 [18] at 6 weeks from the initiation of lenvatinib treatment. Based on the American Diabetes Association criteria [19], DM was diagnosed according to the following criteria: (1) blood glucose ≥200 mg/dL at any time, (2) fasting blood glucose ≥126 mg/dL, or (3) consumption of anti-diabetes medication. The starting dose of lenvatinib was based on a previous study (patients weighing more than 60 kg were administered 12 mg/day and those weighing less than 60 kg were administered 8 mg/day) [3].
Sample Collection and DNA Isolation
Fecal samples were collected from the hospital before initiating lenvatinib treatment. Samples were immediately stored at −80°C. DNA was isolated using the DNeasy PowerSoil Kit (Qiagen, Hilden, Germany) and stored at −80°C. The following universal PCR primers were used to amplify the V3–4 regions along with the Kapa Hifi Hotstart Ready Mix (Kapa Biosystems, Boston, MA, USA): forward:5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′ and reverse:5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC3′. The sequences presented in bold in the primers correspond to the Illumina overhang adapter. PCR conditions were set as follows: 95°C for 3 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and 72°C for 5 min. PCR amplicons were purified using AMPure XP magnetic purification beads (Beckman Coulter, Brea, CA, USA). Individual samples were barcoded with a second PCR performed under the following conditions: 95°C for 3 min, 8 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and 72°C for 5 min. We pooled PCR products to construct a sequencing library. The products were sequenced with an Illumina MiSeq sequencer to generate paired-end reads using the MiSeq Reagent Kit v3 with 2 × 300 reads and 600 cycles (Illumina, San Diego, CA, USA).
16S rRNA Gene Sequencing
The 16S rRNA sequencing data were processed using QIIME2 (version 2019.11) [20]. After demultiplexing, paired-end reads for 21 microbiome samples derived from patients were imported into QIIME2. Sequence quality control and feature table construction were completed using the Divisive Amplicon Denoising Algorithm 2 QIIME 2 plugin. After the denoising step, a pre-trained naive Bayes classifier was used to explore the taxonomic distribution of the samples. This classifier was trained using SILVA database version 132 [21]. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2) was used to predict the functional abundances of bacterial communities [22].
Statistical Analyses
Continuous variables were expressed as medians (interquartile range) and analyzed using the Mann-Whitney U test. Categorical variables were analyzed using the χ2 test or Fisher's exact test. Statistical significance was set at p < 0.05. All statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria). Microbiome comparisons and diversity were visualized and statistically analyzed using the online microbiome data analysis platform (MicrobiomeAnalyst) (https://www.microbiomeanalyst.ca/MicrobiomeAnalyst). Alpha diversity was calculated with the Shannon index using a t test. Beta diversity was estimated using the unweighted UniFrac distance and visualized using the principal coordinate analysis method. The significance of the principal coordinate analysis plot was analyzed using permutational multivariate analysis of variance, which uses distance metrics to confirm the strength and statistical significance of sample groupings. Differential taxonomy between the two groups was compared using linear discriminant analysis effect size (LEfSe) with linear discriminant analysis score >2 and p value <0.05 (http://huttenhower.sph.harvard.edu/galaxy/). Predictive functional genes were evaluated using PICRUSt2 based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We then used Statistical Analysis of Metagenomic Profiles (STAMP) [23] to analyze the identified pathways associated with diarrhea.
Results
Patient Background
Data related to the patient background are shown in Table 1. The median age of the patients was 73. Fourteen patients were males, and hepatitis C was the most common etiological factor. All patients demonstrated CPSs of 5 or 6, BCLC stage B or C, and the median RDI, calculated for 6 weeks of treatment, of 76.1%. Seven patients were treated for DM, 8 patients used PPIs, and 1 patient used probiotics. Eight patients experienced diarrhea (5 patients showed grade 1, 2 patients showed grade 2, and 1 patient showed grade 3 based on CTCAE; 4.0). The presence of other adverse events, such as HFS, hypertension, hypothyroidism, and proteinuria, is also shown in Table 1.
Table 1.
Patient background
| No. | Age | Gender | BMI | Etiology | CPS | BCLC stage | Prior TKI use | RDI, % | Response | DM | PPI | Pre/probiotics | Diarrhea | HFS | Hypertension | Hypothyroidism | Urine protein |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 85 | Female | 17.83 | HCV | 5 | B | No | 57.1 | SD | No | PPI | No | Diarrhea | No | No | No | Urine protein |
| 2 | 77 | Male | 22.72 | HCV | 5 | C | No | 80.9 | SD | No | No | No | No | No | Hypertension | No | Urine protein |
| 3 | 67 | Female | 20.74 | HCV | 6 | B | No | 64.2 | SD | No | PPI | No | Diarrhea | No | No | Hypothyroidism | No |
| 4 | 67 | Male | 18.75 | NBNC | 5 | B | No | 76.1 | PR | DM | No | No | No | HFS | Hypertension | Hypothyroidism | Urine protein |
| 5 | 79 | Female | 20.61 | HBV | 5 | C | No | 41.6 | SD | No | No | No | No | HFS | Hypertension | No | Urine protein |
| 6 | 84 | Female | 19.42 | HCV | 6 | B | No | 86.9 | SD | No | PPI | No | No | No | Hypertension | Hypothyroidism | No |
| 7 | 85 | Male | 30.75 | NBNC | 6 | C | No | 42.8 | SD | DM | No | No | No | No | Hypertension | No | Urine protein |
| 8 | 71 | Female | 25.45 | HCV | 6 | B | No | 95.2 | SD | No | PPI | No | Diarrhea | No | Hypertension | No | No |
| 9 | 73 | Male | 22.66 | NBNC | 6 | C | No | 42.8 | SD | DM | No | No | No | No | Hypertension | No | No |
| 10 | 66 | Male | 24.21 | HCV | 6 | B | No | 67.4 | SD | No | No | No | No | HFS | Hypertension | Hypothyroidism | Urine protein |
| 11 | 78 | Male | 29.73 | NBNC | 5 | B | No | 61.9 | SD | No | PPI | No | No | No | Hypertension | No | Urine protein |
| 12 | 59 | Male | 26.99 | HCV | 6 | B | No | 42.8 | PR | DM | PPI | No | No | No | No | No | No |
| 13 | 69 | Male | 26.92 | HCV | 5 | B | No | 86.5 | PR | No | No | No | No | No | Hypertension | Hypothyroidism | Urine protein |
| 14 | 74 | Female | 33.67 | NBNC | 6 | B | No | 43.6 | SD | DM | No | No | Diarrhea | HFS | Hypertension | Hypothyroidism | Urine protein |
| 15 | 83 | Male | 22.89 | NBNC | 5 | B | No | 100 | SD | DM | No | No | Diarrhea | HFS | Hypertension | Hypothyroidism | Urine protein |
| 16 | 86 | Female | 27.49 | HCV | 6 | B | Sorafenib | 100 | SD | No | PPI | No | Diarrhea | HFS | Hypertension | No | No |
| 17 | 89 | Male | 26.56 | HCV | 5 | B | No | 100 | SD | No | No | No | No | No | Hypertension | No | Urine protein |
| 18 | 60 | Male | 30.47 | HBV | 6 | C | No | 81.7 | SD | No | No | No | Diarrhea | HFS | Hypertension | Hypothyroidism | No |
| 19 | 70 | Male | 20.14 | HCV | 5 | B | No | 100 | PR | No | No | No | No | HFS | Hypertension | Hypothyroidism | Urine protein |
| 20 | 70 | Male | 26.61 | HCV | 6 | C | Sorafenib | 100 | PR | DM | PPI | Probiotics | Diarrhea | HFS | Hypertension | No | Urine protein |
| 21 | 73 | Male | 27.00 | NBNC | 6 | B | No | 47.6 | SD | No | No | No | No | No | No | No | Urine protein |
BMI, body mass index; CPS, Child-Pugh score; BCLC stage, Barcelona Clinic Liver Cancer stage; TKI, tyrosine kinase inhibitor; RDI, relative dose intensity; RECIST, Response Evaluation Criteria in Solid Tumors; DM, diabetes mellitus; PPI, proton pump inhibitor; EIFS, hand-foot syndrome; HCV, hepatitis C virus; SD, stable disease; NBNC, non-B non-C; PR, partial response; HBV, hepatitis B virus. Response to lenvatinib was evaluated using RECIST.
A comparison of patient characteristics between diarrhea and nondiarrhea groups is shown in Table 2. A relatively higher number of male patients was observed in the nondiarrhea group, and the RDI was relatively higher in the diarrhea group. Two patients in the diarrhea group had a history of TKI use (both used sorafenib). There were no significant differences between both groups with respect to patient characteristics, and no patients discontinued treatment due to diarrhea.
Table 2.
Comparison of patient characteristics between diarrhea and nondiarrhea groups
| Diarrhea (n = 8) | Nondiarrhea (n = 13) | p value | |
|---|---|---|---|
| Age, years† | 73 (69–84) | 73 (69–79) | 0.913 |
| Gender (male/female) | 3/5 | 11/2 | 0.056 |
| BMI, kg/m2† | 26.0 (22.4–28.2) | 24.2 (20.6–27.0) | 0.515 |
| Etiology (HBV/HCV/NBNC/alcohol) | 1/5/2/0 | 1/7/5/0 | 0.830 |
| CPS (5/6) | 2/6 | 7/6 | 0.367 |
| BCLC stage (B/C) | 6/2 | 9/4 | 1.000 |
| Prior TKI use (yes/no) | 2/6 | 0/13 | 0.133 |
| RDI for 6 weeks, %† | 88.5 (62.4–100) | 67.4 (42.8–86.5) | 0.165 |
| mRECIST (SD/PR) | 7/1 | 9/4 | 0.606 |
| DM (yes/no) | 3/5 | 4/9 | 1.000 |
| PPI (yes/no) | 5/3 | 3/10 | 0.164 |
| Pre/probiotics (yes/no) | 1/7 | 0/13 | 0.381 |
| HFS (yes/no) | 5/3 | 4/9 | 0.203 |
| Hypertension (yes/no) | 6/2 | 11/2 | 0.618 |
| Hypothyroidism (yes/no) | 4/4 | 5/8 | 0.673 |
| Urine protein (yes/no) | 4/4 | 10/3 | 0.346 |
BMI, body mass index; HBV, hepatitis B virus; HCV, hepatitis C virus; NBNC, non-B non-C; BCLC stage, Barcelona Clinical Liver Cancer stage; TKI, tyrosine kinase inhibitor; RDI, relative dose intensity; mRECIST, modified Response Evaluation Criteria in Solid Tumors; SD, stable disease; PR, partial response; PPI, proton pump inhibitor; HFS, hand-foot syndrome.
Values are expressed as median (interquartile range).
Microbiome Profiling and Comparison of Alpha and Beta Diversities
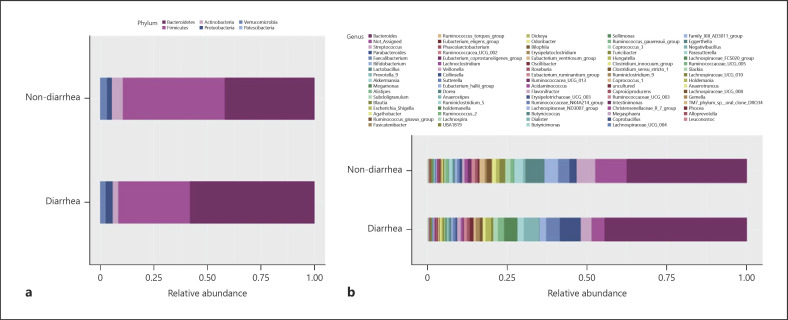
Figure 2a shows relative abundance at the phylum level. Bacteroidetes (diarrhea group vs. nondiarrhea group: 58.3% vs. 41.9% p = 0.011), Firmicutes (33.3% vs. 47.5%), Actinobacteria (2.8% vs. 5.5%), Proteobacteria (3.3% vs. 2.3%), and Verrucomicrobia (2.2% vs. 2.8%) were the major bacterial communities with a relative abundance of higher than 1%. There was no significant difference (p > 0.05) in them except for Bacteroidetes.
Fig. 2.
Relative abundances at phylum (a) and genus levels in diarrhea and nondiarrhea groups (b).
Figure 2b shows the proportion of relative abundance at the genus level. Bacteroides, Streptococcus, Parabacteroides, Faecalibacterium, Bifidobacterium, Lactobacillus, Prevotella, Akkermansia, and Megamonas were the major genera across the groups. Alpha and beta diversities did not differ significantly between both groups (online suppl. Fig. 1; for all online suppl. material, see www.karger.com/doi/10.1159/000524298). Thus, although there was no difference in diversity, Bacteroides and Parabacteroides belonging to the phylum Bacteroidetes alone accounted for more than 50% of the total microbiome in the diarrhea group, which may reduce the relative abundance of each of the other bacteria.
Comparison of Relative Abundance of Bacterial Communities between Diarrhea and Nondiarrhea Groups with LEfSe
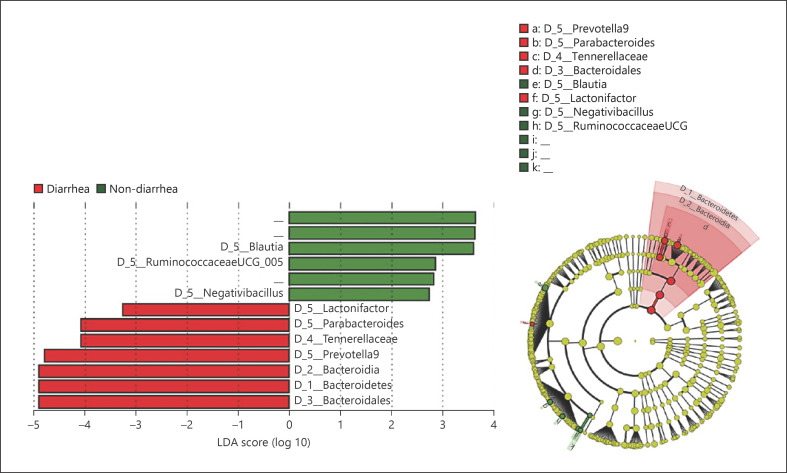
To identify bacterial genera associated with diarrhea, we used LEfSe to compare bacterial abundance (Fig. 3). Red bars and green bars in Figure 3 indicate bacterial communities that were increased significantly in diarrhea and nondiarrhea groups, respectively. One family (Tannerellaceae) and three genera (Lactonifactor, Parabacteroides, and Prevotella) were enriched in the diarrhea group, while three genera (Blautia, RuminococcaceaeUCG_005, and Negativibacillus) were more abundant in the nondiarrhea group. Bacterial communities belonging to the order Bacteroidales were enriched in the diarrhea group. As shown in the circular diagram, the diarrhea group had significantly higher levels of several bacterial communities belonging to the order Bacteroidales than the nondiarrhea group. In contrast, the nondiarrhea group showed increased levels of bacterial communities belonging to Clostridiales, although no clear clusters were observed. This might indicate that specific bacterial communities belonging to the Bacteroidales increased in the diarrhea group, while bacterial communities belonging to the Clostridiales decreased in general rather than specific bacterial communities.
Fig. 3.
Comparison of relative abundance of bacterial communities between groups performed with LEfSe. The graph shows a comparison of the relative abundance of bacterial communities between both groups performed with LEfSe. Red bars show bacterial communities with a higher relative abundance in the diarrhea group, and green bars represent bacterial communities that were more abundant in the nondiarrhea group. The genera Parabacteroides and Prevotella9 and the family Tannerellaceae are members of the order Bacteroidales. LEfSe, linear discriminant analysis effect size.
Functional Differences Based on Marker Gene Sequences Analyzed Using PICRUSt2
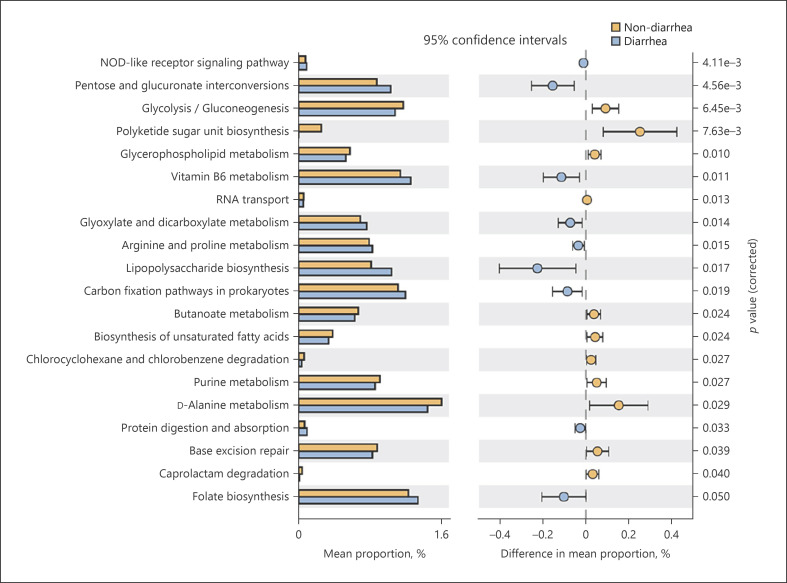
Finally, we compared the differences in KEGG pathways in the microbiome between the two groups using PICRUSt2 (Fig. 4). Twenty pathways of KEGG were significantly different at level 3. Among these pathways, 11 were predominantly enriched in the nondiarrhea group, and nine were enriched in the diarrhea group. Most of these pathways belonged to the metabolic class. Seven of the 11 pathways enriched in the nondiarrhea group were classified as metabolic in the KEGG. The pathways are listed as follows: “glycolysis/gluconeogenesis” and “purine metabolism” (central carbohydrate metabolism), “glycerophospholipid metabolism” (lipid metabolism), “butanoate (butyrate) metabolism” and “D-alanine metabolism” (amino acid metabolism), biosynthesis of unsaturated fatty acids (fatty acid metabolism), and caprolactam degradation (xenobiotics biodegradation and metabolism). Six of the nine pathways enriched in the diarrhea group were also classified as metabolic pathways. The pathways are listed as follows: “pentose and glucuronate interconversions” and “glyoxylate and dicarboxylate metabolism” (carbohydrate metabolism), “vitamin B6 metabolism” and “folate biosynthesis” (cofactor and vitamin metabolism), “lipopolysaccharide (LPS) biosynthesis” (glycan metabolism), and “carbon fixation pathways in prokaryotes” (energy metabolism). The results suggest that the metabolic functions of the two groups of bacteria were different. Different metabolic functions indicate that the metabolites synthesized by bacteria and intestinal environments may be different.
Fig. 4.
Comparison of bacterial function predicted by PICRUSt2. The figure demonstrates the pathways that showed significant differences in KEGG pathway analysis between diarrhea and nondiarrhea groups. Orange bars represent the pathways enriched in the nondiarrhea group, and blue bars represent the pathways enriched in the diarrhea group. Eight pathways, including glycerophospholipid metabolism and butanoate metabolism, were enriched in the nondiarrhea group. PICRUSt2, phylogenetic investigation of communities by reconstruction of unobserved states; KEGG, Kyoto Encyclopedia Genes and Genomes.
Discussion
To improve the treatment of unresectable HCC using TKIs, it is important to predict and manage adverse events. Since several options are available among TKIs, selecting a drug with fewer adverse events may improve the quality of life of patients. In this study, we investigated pretreatment fecal microbiome profiles of patients with HCC treated with lenvatinib. We found differences among bacterial communities of the microbiome between diarrhea and nondiarrhea groups. The pathway related to butyrate metabolism may be associated with diarrhea caused by the treatment.
Lenvatinib has been widely used as systemic chemotherapy for unresectable HCC since it showed superior effects compared to those of sorafenib in the REFLECT study [3]. The RDI of lenvatinib is related to its effects [24]. Therefore, increasing the dose and administration time is crucial for better efficacy. Hence, management of the side effects associated with the treatment is crucial.
Diarrhea may be associated with the changes in the intestinal microbiome induced by small-molecule TKIs. Secombe et al. [10] reported that chemotherapy-induced diarrhea is caused by high mucosal toxicity, whereas TKI-induced diarrhea may be caused by changes in the intestinal function attributed to alterations in the intestinal microbiome, in addition to drug-induced mucosal damage. The mechanism may involve drug-induced, direct inhibition-driven dysbiosis and butyrate, and chloride secretion.
In our study, a significant increase in Bacteroidales was observed in the diarrhea group compared to that in the nondiarrhea group. Changes associated with a bacterial strain similar to those reported in our study have been demonstrated in association with diarrhea caused by other TKIs. Pal et al. [9] reported an increase in Bacteroides in patients experiencing diarrhea after TKI treatment for renal cell carcinoma. Hahn et al. [25] showed that progression-free survival was prolonged when antibiotics affecting Bacteroides were used. There may be an association between TKIs and Bacteroides in terms of drug concentration.
We also found some differences in the KEGG pathways between the two groups. One thing that seems to be closely related to diarrhea is butyrate metabolism. Butyrate induces the development of regulatory T cells that maintain immune tolerance. It also maintains a balance between Th17 cells and regulatory T cells [26]. This balance is critical for regulating gut inflammation. In addition, short-chain fatty acids, including butyrate, may be used as antidiarrheals as they stimulate NaCl absorption and inhibit Cl− secretion by decreasing intracellular cAMP production in the intestinal tract [27, 28].
Differences were also observed in the pathways associated with glycerophospholipid metabolism. Li et al. [29] reported that this pathway may be strongly associated with radiation enteritis. They speculated that the glycerophospholipid metabolism plays an important role in maintaining the integrity of the intestinal epithelium. Disruption of the intestinal epithelial barrier is an important factor that affects the progression of radiation enteritis.
We previously reported on the pretreatment microbiome of patients treated with sorafenib [15]. We compared the microbiome of patients divided into two groups with or without diarrhea due to adverse events. This showed that microbiome of patients with diarrhea had significantly fewer genes related to “glycerophospholipid metabolism.” This is consistent with the results of the present study. Despite different patient groups and drugs, both studies detected a significant difference in the aforementioned pathway with a low p value.
When we focused on the metabolic pathways that were predominant in the diarrhea group, the pathway of LPS biosynthesis was increased. LPS is a major component of the cell wall of Gram-negative bacilli and is known to trigger several types of inflammation and infection in macrophages and other cells with CD14 receptors, and it has been suggested that LPS may cause diarrhea by producing various inflammatory cytokines [30, 31]. Although the present patients did not have diarrhea at the time of fecal collection, patients in the diarrhea group may have been more susceptible to inflammatory cytokines.
The presence or absence of diarrhea during TKI administration may be related to differences in intestinal microbiome as described above. However, there is insufficient evidence regarding the efficacy of prebiotics and probiotics in preventing anticancer drug-induced diarrhea [32]. Therefore, it is important to examine methods for preventing diarrhea in patients.
Our study showed that the abundance of Bacteroidales was increased in the diarrhea group. The results also suggest that the differences in metabolic pathways, including butyrate metabolism, may be important in the development of diarrhea during treatment with lenvatinib. The metabolic pathways of intestinal microbiome other than those described above have not yet been elucidated in relation to diarrhea caused by TKIs, and further research is desirable.
Limitation
Because of the small number of cases in this single-center, retrospective study, it was difficult to correct for false positives. Owing to the strong interindividual variability associated with the intestinal microbiome, further studies with a larger patient cohort are needed. In addition, we were not able to measure the blood concentration of lenvatinib, fatty acids, and butyrate concentrations in the stool. Although PICRUSt2 is a useful program, it is less accurate than shotgun sequencing.
Conclusion
In patients with HCC receiving lenvatinib, differences in the intestinal microbiome, including butyrate metabolism, may influence the incidence of diarrhea during treatment.
Statement of Ethics
This study protocol followed the ethical guidelines of the Helsinki Declaration and was approved by the Local Ethics Committee (approval number:2015-0420). Written informed consent was obtained from all patients prior to inclusion in the study.
Conflict of Interest Statement
None of the authors have any conflicts of interest related to the contents of this article.
Funding Sources
This study was not funded.
Author Contributions
Yosuke Inukai, Kenta Yamamoto, Takashi Honda, Takanori Ito, Norihiro Imai, Yoji Ishizu, and Ishigami Masatoshi were involved in study design and sample collection. Yosuke Inukai, Kenta Yamamoto, Takashi Honda, Takanori Ito, Norihiro Imai, Yoji Ishizu, Masanao Nakamura, Hiroki Kawashima, and Masatoshi Ishigami were involved in the data analysis. Yosuke Inukai, Kenta Yamamoto, and Takashi Honda performed statistical analysis and wrote the paper. All authors read and approved the final manuscript.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. Further inquiries can be directed to the corresponding author.
Supplementary Material
Supplementary data
Supplementary data
Funding Statement
This study was not funded.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68((6)):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ikeda K, Kudo M, Kawazoe S, Osaki Y, Ikeda M, Okusaka T, et al. Phase 2 study of lenvatinib in patients with advanced hepatocellular carcinoma. J Gastroenterol. 2017 Apr;52((4)):512–519. doi: 10.1007/s00535-016-1263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018 Mar;391((10126)):1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 4.Hatanaka T, Kakizaki S, Nagashima T, Namikawa M, Tojima H, Shimada Y, et al. Analyses of objective response rate, progression‐free survival, and adverse events in hepatocellular carcinoma patients treated with lenvatinib: a multicenter retrospective study. Hepatol Res. 2020 Mar;50((3)):382–395. doi: 10.1111/hepr.13460. [DOI] [PubMed] [Google Scholar]
- 5.Ogushi K, Chuma M, Uojima H, Hidaka H, Numata K, Kobayashi S, et al. Safety and efficacy of lenvatinib treatment in child-pugh a and b patients with unresectable hepatocellular carcinoma in clinical practice: a multicenter analysis. Clin Exp Gastroenterol. 2020;13:385–396. doi: 10.2147/CEG.S256691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017 Jan;389((10064)):56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 7.Abou-Alfa GK, Meyer T, Cheng AL, El-Khoueiry AB, Rimassa L, Ryoo BY, et al. Cabozantinib in patients with advanced and progressing hepatocellular carcinoma. N Engl J Med. 2018 Jul;379((1)):54–63. doi: 10.1056/NEJMoa1717002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Sebille YZ, Gibson RJ, Wardill HR, Bowen JM. ErbB small molecule tyrosine kinase inhibitor (TKI) induced diarrhoea: chloride secretion as a mechanistic hypothesis. Cancer Treat Rev. 2015 Jul;41((7)):646–652. doi: 10.1016/j.ctrv.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Pal SK, Li SM, Wu X, Qin H, Kortylewski M, Hsu J, et al. Stool bacteriomic profiling in patients with metastatic renal cell carcinoma receiving vascular endothelial growth factor-tyrosine kinase inhibitors. Clin Cancer Res. 2015 Dec;21((23)):5286–5293. doi: 10.1158/1078-0432.CCR-15-0724. [DOI] [PubMed] [Google Scholar]
- 10.Secombe KR, Van Sebille YZA, Mayo BJ, Coller JK, Gibson RJ, Bowen JM. Diarrhea induced by small molecule tyrosine kinase inhibitors compared with chemotherapy: potential role of the microbiome. Integr Cancer Ther. 2020;19:1534735420928493. doi: 10.1177/1534735420928493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342((6161)):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342((6161)):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montassier E, Batard E, Massart S, Gastinne T, Carton T, Caillon J, et al. 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol. 2014 Jan;67((3)):690–699. doi: 10.1007/s00248-013-0355-4. [DOI] [PubMed] [Google Scholar]
- 14.Rigby RJ, Carr J, Orgel K, King SL, Lund PK, Dekaney CM. Intestinal bacteria are necessary for doxorubicin-induced intestinal damage but not for doxorubicin-induced apoptosis. Gut Microbes. 2016 Sep;7((5)):414–423. doi: 10.1080/19490976.2016.1215806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto K, Kuzuya T, Honda T, Ito T, Ishizu Y, Nakamura M, et al. Relationship between adverse events and microbiomes in advanced hepatocellular carcinoma patients treated with sorafenib. Anticancer Res. 2020;40((2)):665–676. doi: 10.21873/anticanres.13996. [DOI] [PubMed] [Google Scholar]
- 16.Galle PR, Forner A, Llovet JM, Mazzaferro V, Piscaglia F, Raoul JL, et al. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018 Jul;69((1)):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60((8)):646–649. doi: 10.1002/bjs.1800600817. [DOI] [PubMed] [Google Scholar]
- 18.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009 Jan;45((2)):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 19.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2014 Jan;37 Suppl 1:S81–90. doi: 10.2337/dc14-S081. [DOI] [PubMed] [Google Scholar]
- 20.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019 Aug;37((8)):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013 Jan;41((Database issue)):D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol. 2020 Jun;38((6)):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014 Nov;30((21)):3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirino S, Tsuchiya K, Kurosaki M, Kaneko S, Inada K, Yamashita K, et al. Relative dose intensity over the first four weeks of lenvatinib therapy is a factor of favorable response and overall survival in patients with unresectable hepatocellular carcinoma. PLoS One. 2020 Apr;15((4)):e0231828. doi: 10.1371/journal.pone.0231828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn AW, Froerer C, VanAlstine S, Rathi N, Bailey EB, Stenehjem DD, et al. Targeting bacteroides in stool microbiome and response to treatment with first-line VEGF tyrosine kinase inhibitors in metastatic renal-cell carcinoma. Clin Genitourin Cancer. 2018 Oct;16((5)):365–368. doi: 10.1016/j.clgc.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012 Apr;336((6080)):489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binder HJ. Role of colonic short-chain fatty acid transport in diarrhea. Annu Rev Physiol. 2010 Mar;72((1)):297–313. doi: 10.1146/annurev-physiol-021909-135817. [DOI] [PubMed] [Google Scholar]
- 28.Krishnan S, Ramakrishna BS, Binder HJ. Stimulation of sodium chloride absorption from secreting rat colon by short-chain fatty acids. Dig Dis Sci. 1999;44((9)):1924–1930. doi: 10.1023/a:1018871412748. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Yan H, Zhang Y, Li Q, Yu L, Li Q, et al. Alterations of the gut microbiome composition and lipid metabolic profile in radiation enteritis. Front Cell Infect Microbiol. 2020 Oct;10:541178. doi: 10.3389/fcimb.2020.541178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chorawala MR, Chauhan S, Patel R, Shah G. Cell wall contents of probiotics (Lactobacillus species) protect against lipopolysaccharide (LPS)-induced murine colitis by limiting immuno-inflammation and oxidative stress. Probiotics Antimicrob Proteins. 2021 Aug;13((4)):1005–1017. doi: 10.1007/s12602-020-09738-4. [DOI] [PubMed] [Google Scholar]
- 31.Liang YC, Liu HJ, Chen SH, Chen CC, Chou LS, Tsai LH. Effect of lipopolysaccharide on diarrhea and gastrointestinal transit in mice: roles of nitric oxide and prostaglandin E2. World J Gastroenterol. 2005 Jan;11((3)):357–361. doi: 10.3748/wjg.v11.i3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wardill HR, Van Sebille YZA, Ciorba MA, Bowen JM. Prophylactic probiotics for cancer therapy-induced diarrhoea: a meta-analysis. Curr Opin Support Palliat Care. 2018 Jun;12((2)):187–197. doi: 10.1097/SPC.0000000000000338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Data Availability Statement
All data generated or analyzed during this study are included in this article and its online supplementary material. Further inquiries can be directed to the corresponding author.